Summary
Liver progenitor cells (LPCs) have a remarkable contribution to the hepatocytes and ductal cells when normal hepatocyte proliferation is severely impaired. As a biomarker for LPCs, Sry-box 9 (Sox9) plays critical roles in liver homeostasis and repair in response to injury. However, the regulation mechanism of Sox9 in liver physiological and pathological state remains unknown. In this study, we found that miR-126 positively regulated the expression of Sox9, the proliferation and differentiation of SOX9+ LPCs by suppressing the translation of homeobox b6 (Hoxb6). As a transcription factor, HOXB6 directly binds to the promoter of Sox9 to inhibit Sox9 expression, resulting in the destruction of the properties of SOX9+ LPCs in CCl4-induced liver injury. These findings revealed the role of miR-126 in regulating SOX9+ LPCs fate by targeting Hoxb6 in liver injury repair. Our findings suggest the potential role of miR-126 as a nucleic acid therapy drug target for liver failure.
KEYWORDS: MiR-126, liver stem cells, injury repair, proliferation, differentiation
Graphical Abstract
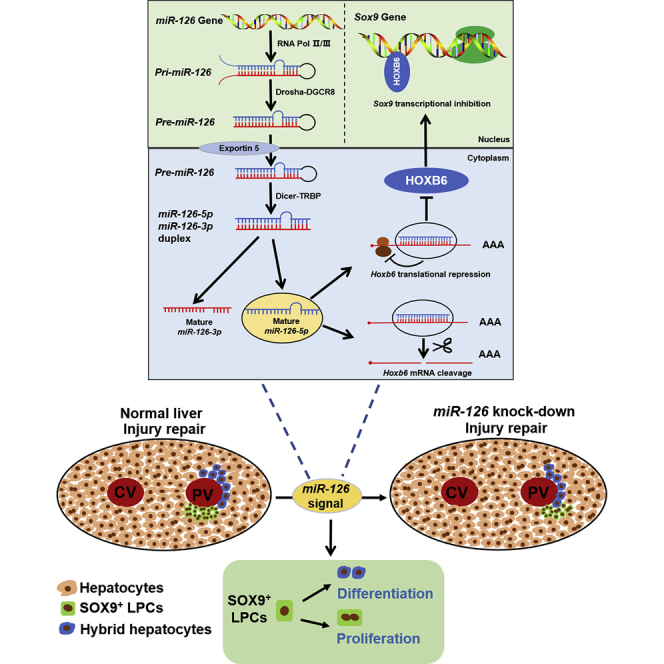
Highlights
-
•
miR-126 promotes Sox9 expression and maintains SOX9+ LPCs in adult mouse livers
-
•
HOXB6 suppresses properties of SOX9+ LPCs in chronic liver injury model
-
•
HOXB6 negatively regulates Sox9 trans-activity
-
•
miR-126 regulates properties of SOX9+ LPCs by targeting Hoxb6
LPCs have a remarkable contribution to the physiological maintenance of liver homeostasis. However, the regulation mechanism of LPCs in liver physiology and pathology remains unknown. In this article, Zhang and his colleagues revealed the role of miR-126 in regulating SOX9+ LPCs fate by targeting Hoxb6 in liver injury repair. These findings suggest the potential role of miR-126 in developing nucleic acid therapy drugs for liver failure.
Introduction
MicroRNAs (miRNAs), a class of endogenous, small non-coding RNAs composed of ∼22 nucleotides, bind to partial complementary sequences in the 3ʹ untranslated region (UTR) of their target transcripts and recruit RNA-induced silencing complex to inhibit the translation of these transcripts, they are involved in the regulation of cellular functions (Bartel, 2018; Mori et al., 2019). A recent study suggests that miRNAs, which are abundant in liver, can modulate a wide range of hepatocellular functions (Su et al., 2018). Our previous research showed that hepatic miR-657 enhanced nuclear factor κB activity to promote hepatocellular carcinoma cell growth and transformation (Zhang et al., 2013). Moreover, miR-122 has been identified as a biomarker of acute liver failure in mice and humans (John et al., 2014; Luna et al., 2017). The newly reported miR-221-3p reduced secretion of CCR2 through post-transcription regulation of Gnai2, thus mitigating liver fibrosis (Tsay et al., 2019). The above-mentioned examples indicated that miRNAs play a critical role in liver physiological and pathological regulation. However, the specific roles of miRNAs in liver regeneration and repair, especially in regulating hepatic stem cell properties remain to be examined.
Our data showed that miR-126 is encoded in the intron of Egfl7 (Yan et al., 2020). Previous research reported that miR-126 was highly expressed in normal hematopoietic stem cells (HSCs) and hematopoietic progenitor cells and restrained cell-cycle progression during hematopoiesis (Lechman et al., 2012). Recently, miR-126 has been reported to regulate the self-renewal of leukemia stem cells in chronic myelogenous leukemia (Zhang et al., 2018). Moreover, our previous research has shown that miR-126 was involved in regulating liver aging. To be more specific, the knockdown of miR-126 in bone marrow stromal cells (BMSCs) accelerated cell aging and inhibited hepatic repair functions of BMSCs (Yan et al., 2019). These results indicated that miR-126 might be involved in hepatic aging through liver stem cells (LSCs) or liver progenitor cells (LPCs). Although the roles of miR-126 in stem cell function regulation and hepatic repair have been intensively studied, it remains largely unknown whether miR-126 contributes to liver regeneration by regulating the LPC properties.
A few markers, including Sox9, Axin2, Cd44, and Lgr5 were reported to be used to identify LSCs or LPCs (Huch et al., 2013; Wang et al., 2015). SOX9, a member of the sry-related high-mobility group box transcription factors, is closely related to cell proliferation and differentiation, and it regulates the stem cell homeostasis and differentiation (Ko et al., 2019; Mori-Akiyama et al., 2007). SOX9 is expressed throughout the biliary and pancreatic ductal epithelia (Alison and Lin, 2011). A previous study used Sox9-IRES-CreERT2 lineage tracing approach and found that the SOX9+ biliary compartment contributed to most new hepatocytes even during normal liver homeostasis. In addition, SOX9+ LPCs contributed to the formation of hepatocytes after liver injury (Furuyama et al., 2011; Tarlow et al., 2014). SOX9 was also weakly expressed in a population of periportal (PP) hepatocytes (named hybrid hepatocytes [HybHPs]) located in the portal triads of livers. HybHPs underwent extensive proliferation and replenished liver mass after chronic hepatocyte-depleting injury (Font-Burgada et al., 2015). The above-mentioned studies have demonstrated that SOX9 plays a critical role in liver regeneration and repair. However, the regulatory mechanisms of Sox9 involved in liver regeneration and repair remain unclear.
In this study, we examined the effect of miR-126 as regulatory factors on the properties of SOX9+ LPCs. Moreover, we revealed the regulatory mechanism by which HOXB6 was involved in miR-126-mediated liver injury repair. Our data demonstrated that the interaction between miR-126-5p and Hoxb6 mRNA induced post-transcriptional silencing, leading to the stable expression of Sox9, thereby maintaining the stem cell properties of SOX9+ LPCs during the liver repair.
Results
MiR-126 Promotes SOX9 Expression and Induces SOX9+ LPCs
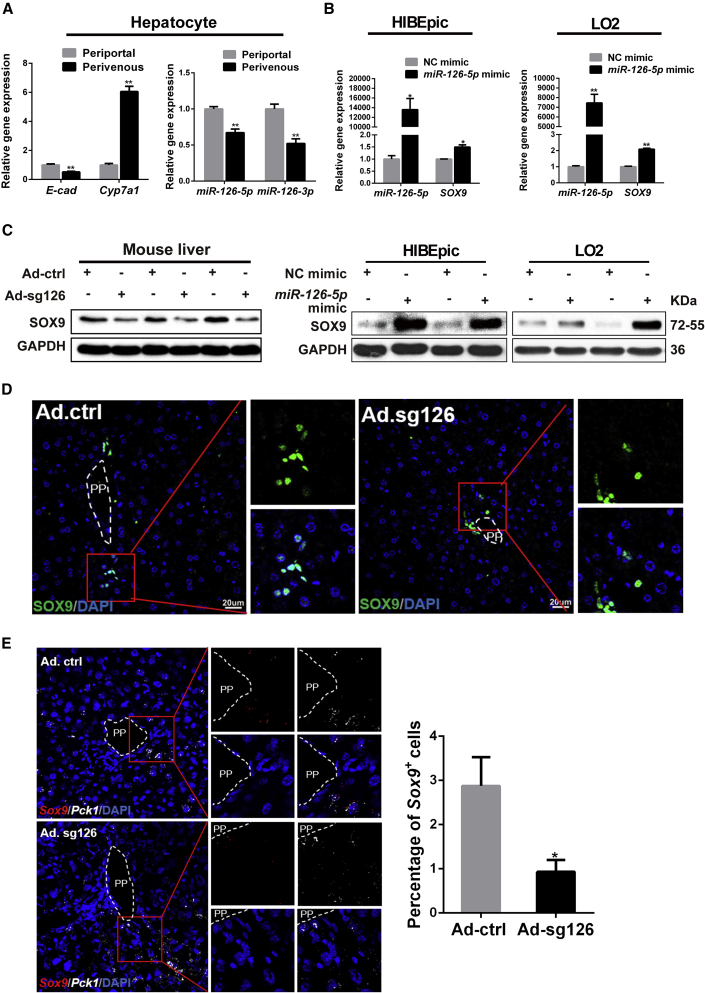
Recent investigation has revealed that miRNAs are involved in liver regeneration and might serve as the therapeutic approach to liver fibrosis (John et al., 2014; Tsay et al., 2019). Our previous research has shown that miR-126 contributes to hepatic anti-aging and damage repair (Yan et al., 2019). To reveal the effect of miR-126 on anti-aging, we first detected the expression of stem cell-associated genes in C3H10 cells from which the miR-126 has been deleted. The result showed that there were no significant changes in the expression levels of Axin2, Cd44, and Lgr5, but Sox9 abundance was significantly reduced, compared with that of control C3H10 cells (Figure S1A). Then, we measured the expression of the previously reported hepatic PP (E-cad) and perivenous (PV) (Cyp7a1) zonation genes in the purified hepatocytes separated from mouse livers (Han et al., 2019; Pu et al., 2016; Rocha et al., 2015). Expression levels of miR-126-5p and miR-126-3p genes were significantly higher in the PP hepatocytes than in the PV hepatocytes (Figures 1A and S1B). Previous studies reported that there were much more SOX9-expressing hepatocytes detected in the PP area than in the PV area (Font-Burgada et al., 2015; Halpern et al., 2017). Based on these findings, we hypothesized that there might be a correlation between these two genes.
Figure 1.
miR-126 Regulates Liver Progenitor Cell Marker Sox9 Expression and SOX9+ Hepatocyte-like Cell Number
(A) Periportal (PP) hepatocytes and perivenous (PV) hepatocytes in 8-week-old mouse were isolated by using the digitonin/collagenase perfusion technique. Quantification of mature miR-126-5p and miR-126-3p, PP marker (E-cad), and PV marker (Cyp7a1) expressions in primary cultured PP hepatocytes and PV hepatocytes.
(B) HIBEpic and LO2 cells were transfected with miR-126-5p mimics or negative control (NC) mimics for 48 h. Quantification of miR-126-5p and SOX9 expressions in NC group and miR-126-5p group. The protein level of SOX9 was detected by western blotting.
(C) Expression of SOX9 was measured at the protein level in Ad.ctrl and Ad.sg126 mouse livers.
(D) Immunofluorescence was performed on the sections from paraffin-embedded tissue samples of Ad.ctrl mouse livers and Ad.sg126 mouse livers. Immunofluorescence of SOX9 was performed. DAPI (blue) shows nuclei. Scale bar represents 20 μm.
(E) Representative images from RNAscope assays for Sox9 mRNA levels and quantification of Sox9+ cells in livers of Ad.ctrl and Ad.sg126 mice. Red presents Sox9. White (Pck1) marks hepatocytes in the PP zone. Scale bar represents 20 μm.
Data are expressed as means ± SD, n = 3 independent experiments containing three replicates (A and B), n = 6 mice per group containing three replicates (C–E). Significant difference is presented at the levels of ∗p < 0.05 and ∗∗p < 0.01 by two-tailed Student's t test. See also Figure S1.
To confirm the hypothesis, miR-126-5p was overexpressed by transfecting the miR-126-5p mimics into HIBEpic and LO2 cells, and we found that SOX9 expression was dramatically increased at the mRNA and protein levels (Figures 1B and S1B). We also inhibited miR-126 expression by infecting BMSCs with sg126-expressing lentivirus (sg126). The result showed that the expression of Sox9 in BMSCs was significantly decreased after infection with sg126-expressing lentivirus (Figure S1C). To further investigate the effects of miR-126 on SOX9 expression in the liver, we disrupted miR-126 by delivering sg126-expressing adenovirus (Ad.sg126) into mouse livers, and we found that the SOX9 level in the Ad.sg126 group was much lower than that in the control adenovirus (Ad.ctrl) group by western blot (Figure 1C). Interestingly, a drastic decrease in the number of SOX9+ hepatocyte-like cells was observed around the PP area in miR-126-deleted mouse liver (Figures 1D and 1E).
Hoxb6 Is a Target of miR-126-5p
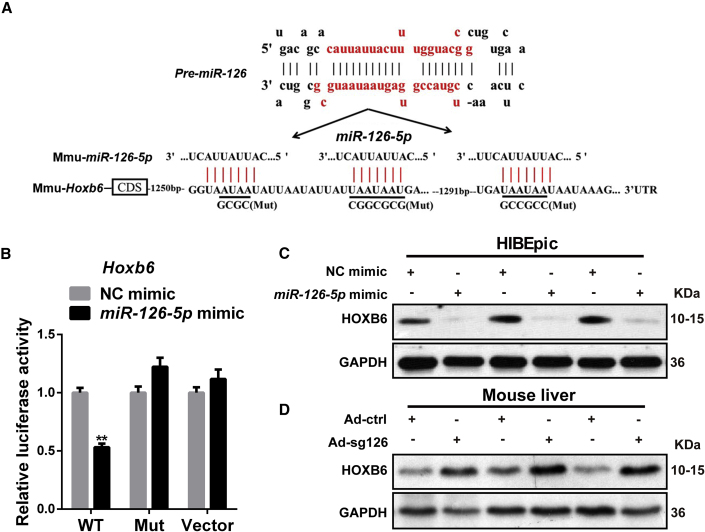
To identify putative targets of miR-126-5p contributing to the increase in SOX9+ LPCs in liver, we searched the candidate target genes by TargetScan and miRbase. Among various potential targets, we focused on Hoxb6 gene since SOX9 was continuously expressed in Hoxb6 mutant pancreata (Larsen et al., 2015), and Hoxb6 regulates the generation, proliferation, or survival of erythroid progenitor cells in fetal livers, and disruption of the Hoxb6 resulted in increased numbers of early erythrocyte progenitors (Kappen, 2000). So, we hypothesized that miR-126 induces SOX9+ progenitors through Hoxb6. The 3′ UTR of the Hoxb6 gene contains binding sites for miR-126-5p (Figure 2A). To determine whether Hoxb6 is a direct target of miR-126-5p, we constructed luciferase reporter vector in which the Hoxb6 3′ UTR is placed behind the luciferase gene. We found that miR-126-5p inhibited luciferase activity, whereas no inhibition was observed when the miR-126-5p target site was mutated (Figures 2A, 2B, and S2A). Consistently, miRNA mimics-mediated overexpression of miR-126-5p suppressed the expression of HOXB6 in HIBEpic cells and HepG2 cells (Figures 2C and S2B). However, antagomir-mediated inhibition of miR-126-5p promoted the expression of HOXB6 (Figure S2B). Furthermore, lentivirus-mediated deletion of miR-126 resulted in the upregulation of HOXB6 protein level in BMSCs and C3H10 cells (Figure S2C).
Figure 2.
Hoxb6 Is a Target of miR-126-5p
(A) The 3′ UTR of Hoxb6 gene contains binding sites for miR-126-5p according to TargetScan (http://www.targetscan.org/). Black underlined sequences indicate the point mutations used to generate mouse Hoxb6 3′ UTR Mut constructs.
(B) Relative luciferase activity assays of luciferase reporter plasmid with mouse WT or Mut Hoxb6 3′ UTR, and that of vector were performed after co-transfection with miR-126-5p mimics or NC mimics in HeLa cells. Hoxb6 3′ UTR Mut seed complementary sites were shown in (A).
(C) HOXB6 protein levels were measured by western blotting after transient transfection with miR-126-5p mimics or NC into HIBEpic cells.
(D) Expression of HOXB6 was measured at the protein level in Ad.ctrl and Ad.sg126 mouse livers.
Data are expressed as means ± SD, n = 3 independent experiments containing three replicates. Significant difference is presented at the level of ∗∗p < 0.01 by two-tailed Student's t test. See also Figure S2.
Consistent with the in vitro data, western blot analysis of the liver samples showed that deletion of miR-126 increased hepatic HOXB6 levels (Figure 2D). Taken together, these results demonstrated that miR-126-5p directly targeted Hoxb6.
HOXB6 Negatively Regulates SOX9 Expression In Vitro and In Vivo
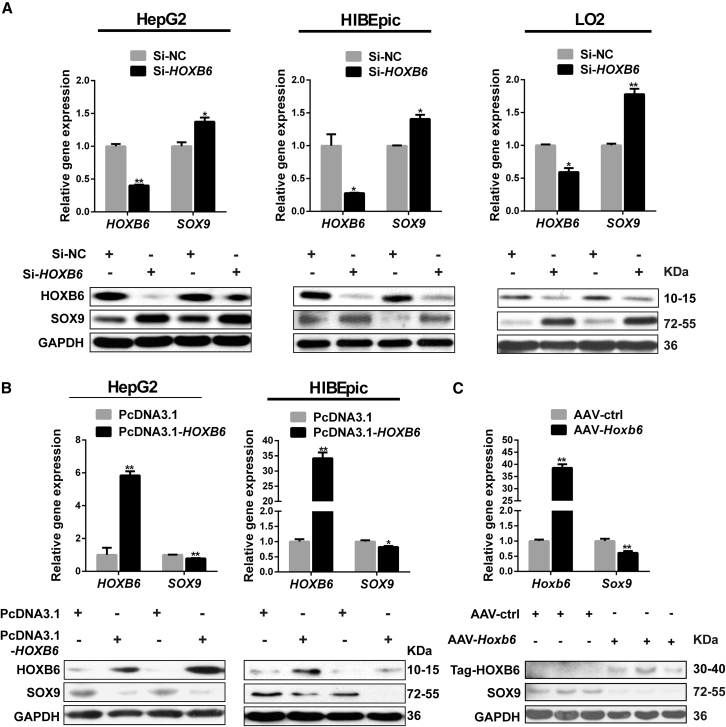
Based on the results described above, we explored whether HOXB6 regulated the expression of SOX9. HOXB6 siRNA (Si-HOXB6) or PcDNA3.1-HOXB6 was transfected into HepG2, HIBEpic, and LO2 cells to reduce or enhance HOXB6 function, respectively. Expression level of SOX9 was significantly increased in Si-HOXB6-transfected HepG2, HIBEpic, and LO2 cells (Figure 3A), whereas SOX9 was significantly reduced in PcDNA3.1-HOXB6-transfected HepG2 and HIBEpic cells (Figure 3B), indicating that HOXB6 suppressed SOX9 expression in vitro.
Figure 3.
HOXB6 Negatively Regulates SOX9 Expression In Vitro and In Vivo
(A) HepG2 cells, HIBEpic cells, and LO2 cells were transfected with the HOXB6 siRNA (Si-HOXB6) or NC. At 48 h after transfection, western blotting and qRT-PCR analysis of HOXB6 and SOX9 expression in the NC group and the Si-HOXB6 group were performed.
(B) Western blotting and qRT-PCR analysis were conducted to detect HOXB6 and SOX9 expression in HepG2 and HIBEpic cells after HOXB6 exogenous overexpression.
(C) Western blotting and qRT-PCR analysis of HOXB6 and SOX9 expression in AAV-ctrl and AAV-Hoxb6 mice livers were performed.
Data are expressed as means ± SD, n = 3 independent experiments containing three replicates (A and B), n = 6 mice per group containing three replicates (C). ∗p < 0.05 and ∗∗p < 0.01 by two-tailed Student's t test.
To confirm this observation, HOXB6 was overexpressed by tail vein injection of the adeno-associated virus (AAV)-Hoxb6 or AAV control into mouse livers. We found that overexpression of HOXB6 significantly reduced Sox9 mRNA abundance compared with the control group (Figure 3C). In line with the mRNA levels, SOX9 was even more profoundly downregulated at the protein level in the AAV-Hoxb6 group (Figure 3C). Taken together, our data indicated that HOXB6 negatively regulated the expression of the LPC gene Sox9.
HOXB6 Negatively Regulates SOX9 trans-Activity
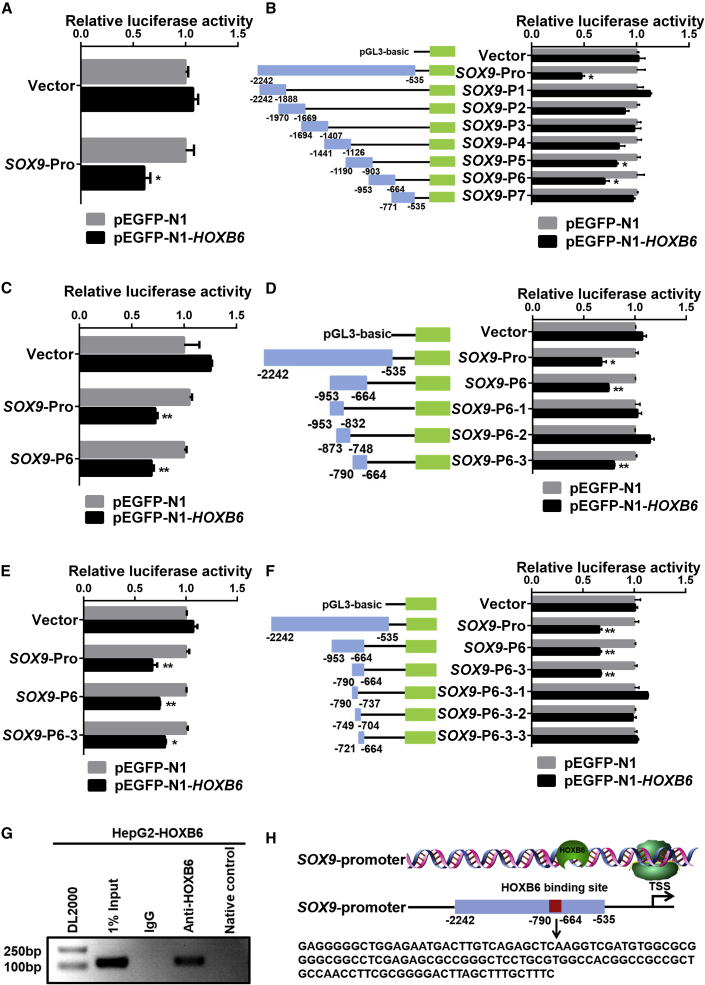
HOXB6 and other HOX genes have been thought to function as transcription factors, and they bind to DNA targets containing a TAAT sequence (Porcelli et al., 2019). Based on the description above, we hypothesized that HOXB6 could regulate SOX9 transcription. We next attempted to identify the binding sites of HOXB6 by serially truncating regions within the SOX9 promoter. At ∼3 kb immediately upstream of the SOX9 transcriptional start site, we identified six potential TAAT-HOXB6 binding sites in the SOX9 promoter region from −2,242 to −535 bp (Figure S3A). Next, we constructed a series of truncated promoter fragments aimed at removing these sites in a stepwise manner. As shown in Figures 4A and S3B, human or mouse SOX9 promoter (SOX9-Pro), a full-length construct, exhibited a dramatic decrease in transcriptional activity by HOXB6 overexpression.
Figure 4.
HOXB6 Negatively Regulates SOX9 trans-Activity
(A) Relative luciferase activity analysis of the SOX9 promoter-reporter constructs. pGL3-basic plasmid with a 1,700-base-pair fragment of the human SOX9 promoter (position −2,242 to −535 relative to the transcription start site, SOX9-Pro) was transfected into HEK293T cells with the phRL-TK and pEGFP-N1 or pEGFP-N1-HOXB6 eukaryotic expression plasmids.
(B) Relative luciferase activity assays were performed in HEK293T cells by transiently co-transfecting SOX9-Pro (−2,242 to −535), SOX9-P1 (−2,242 to −1,888), SOX9-P2 (−1,970 to −1,669), SOX9-P3 (−1,694 to −1,407), SOX9-P4 (−1,441 to −1,126), SOX9-P5 (−1,190 to −903), SOX9-P6 (−953 to −664), SOX9-P7 (−771 to −535), and vector with expression plasmids of pEGFP-N1-HOXB6 or pEGFP-N1.
(C) Relative luciferase activity assays were performed in HEK293T cells by transiently co-transfecting SOX9-Pro (−2,242 to −535), SOX9-P6 (−953 to −664), and vector in a combination of expression plasmids of pEGFP-N1 or pEGFP-N1-HOXB6.
(D) SOX9-P6 (−953 to −664) as well as various truncated fragments were inserted into pGL3-basic plasmid with luciferase reporter genes, including SOX9-P6-1 (−953 to 832), SOX9-P6-2 (−873 to −748), and SOX9-P6-3 (−790 to −664). Relative luciferase activity assays were performed in HEK293T cells by transiently transfecting the above plasmids with pEGFP-N1 or pEGFP-N1-HOXB6.
(E) Relative luciferase activity assays were performed in HEK293T cells by transiently transfecting SOX9-Pro, SOX9-P6, SOX9-P6-3, and vector in a combination of expression plasmids of pEGFP-N1 or pEGFP-N1-HOXB6.
(F) SOX9-P6-3 (−790 to −664) as well as various truncated fragments were inserted into the pGL3-basic plasmid with luciferase reporter gene, including SOX9-P6-3-1, SOX9-P6-3-2, and SOX9-P6-3-3.
(G) Chromatin immunoprecipitation-PCR analysis. Chromatin was prepared and immunoprecipitated with specific antibodies against HOXB6 or IgG. The input DNA and DNA isolated from the precipitated chromatin were amplified by PCR using primers spanning the HOXB6 binding site, and the obtained PCR product was separated on a 1.5% agarose gel. Lanes: 1, marker; 2, input; 3, IgG; 4, HOXB6 antibody; 5, negative control.
(H) Sequence of the SOX9 promoter region. The listed nucleotide is the HOXB6 binding site.
Data are expressed as means ± SD, n = 3 independent experiments containing three replicates. Significant difference is presented at the levels of ∗p < 0.05 and ∗∗p < 0.01 by two-tailed Student's t test. See also Figure S3.
To determine the binding site in which the transcriptional activity of SOX9 promoter was inhibited by HOXB6, seven ∼300-bp SOX9 promoter-truncated fragments were constructed using SOX9-Pro as a template. These seven constructs were named SOX9-P1, SOX9-P2, SOX9-P3, SOX9-P4, SOX9-P5, SOX9-P6, and SOX9-P7 (Figure 4B). As shown in Figure 4B, luciferase activity of SOX9-P5 and SOX9-P6 were observed to be inhibited by HOXB6. But SOX9-P6 demonstrated a dramatically decreased transcriptional activity by HOXB6. Thus, these results revealed that the SOX9-P6 (−953 to −664 bp) region was responsible for SOX9 transcriptional activity inhibition by HOXB6, and HOXB6 had the same effect on SOX9-P6 and SOX9-Pro (Figure 4C). Next, we constructed a series of SOX9 promoter-truncated fragments with SOX9-P6 as a template. We named the three constructs SOX9-P6-1 (−953 to −832), SOX9-P6-2 (−873 to −748), and SOX9-P6-3 (−790 to −664) (Figure 4D). We detected the reduction in luciferase activity of the constructs containing only SOX9-P6-3 (−790 to −664) regions after overexpression of HOXB6 (Figures 4D and 4E). However, no reduction in luciferase activity was observed from three truncated fragments containing SOX9-P6-3-1 (−790 to −737), SOX9-P6-3-2 (−749 to −704), or SOX9-P6-3-3 (−721 to −664) promoter regions after overexpression of HOXB6 (Figure 4F).
To confirm the binding sites of HOXB6 to the SOX9 promoter, we performed a chromatin immunoprecipitation (ChIP) assay. The crosslinked extracts were immunoprecipitated with antibodies against HOXB6 or control anti-IgG antibody. The crosslinked DNA was analyzed by using PCR with the primers designed to amplify the HOXB6-responsive region containing the SOX9-P6-3 (−790 to −664). HOXB6 was determined to be associated with the SOX9 promoter region containing the SOX9-P6-3 fragments (Figure 4G). Taken together, our results suggested that SOX9 was directly negatively regulated by HOXB6 with the binding sequences of SOX9 promoter by HOXB6 shown in Figure 4H.
AAV-Mediated Overexpression of HOXB6 Suppresses Proliferation and Differentiation of SOX9+ LPCs to Aggravate Damage in a CCl4 Chronic Liver Injury Model
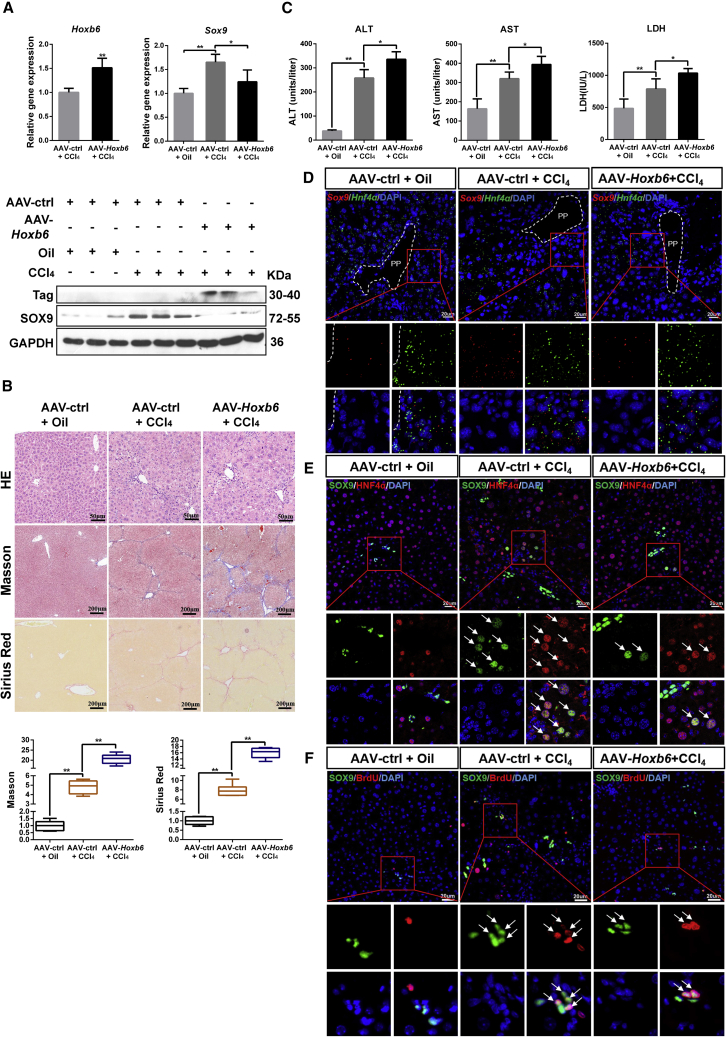
Based on the above-mentioned regulation mechanism in vitro, an in vivo experiment was performed to reveal the mechanism by which HOXB6 regulates SOX9 in CCl4-induced liver injury model. To elucidate the regulation mechanism in liver, we first demonstrated the expression of HOXB6 and SOX9 in primary hepatocyte (Figure S4). Compared with control group, CCl4 treatment group exhibited a higher level of SOX9 expression (Figure 5A). After tail vein injection of 1 × 1010 particles of AAV-mediated overexpression of HOXB6 into the CCl4-treated mice, the expression of SOX9 was strongly suppressed at mRNA and protein levels (Figure 5A). Hematoxylin and eosin (H&E) staining results indicated that CCl4 treatment induced hepatotoxicity (Figure 5B). In addition, Masson and Sirius red staining results showed that CCl4 treatment promoted liver fibrosis (Figure 5B). At the same time, the levels of aspartate aminotransferase (ALT), alanine aminotransferase (AST), and lactic dehydrogenase (LDH) were observed significantly higher in the CCl4 group than in the control group (Figure 5C). Serological and histological analysis revealed that the treatment with AAV-Hoxb6 resulted in a significantly increased liver damage and fibrosis as well as a significant increase in ALT, AST, and LDH levels (Figures 5B and 5C). Our results indicated that AAV-mediated overexpression of HOXB6 was able to obviously aggravate liver injury caused by CCl4.
Figure 5.
AAV-Mediated Overexpression of HOXB6 Suppresses the Proliferation and Differentiation of SOX9+ LPCs to Aggravate Damage in a CCl4 Chronic Liver Injury Model
AAV-ctrl and AAV-Hoxb6 were administered to 8-week-old C57 mice via tail vein injection. After 2 weeks, mice were treated with CCl4 (diluted in oil with 1:4) twice per week for 4 weeks, then livers and blood were collected.
(A) Hepatic expression levels of SOX9 and HOXB6 were measured in AAV-ctrl, AAV-HOXB6 mice after CCl4 injury by qRT-PCR and western blot.
(B) Representative H&E, Masson, and Sirius red staining in AAV-ctrl mice liver and AAV-Hoxb6 mice liver after chronic hepatic injury by CCl4 treatment. Fibrosis was quantified by morphometric measurement of Masson and Sirius red.
(C) Serum ALT, AST, and LDH levels of AAV-ctrl mice and AAV-HOXB6 mice after chronic hepatic injury by CCl4 treatment.
(D) Representative images from RNAscope assays of Hnf4α and Sox9 in livers of AAV-ctrl and AAV-Hoxb6 mice after CCl4 injury. Red presents Sox9. Blue (DAPI) shows nuclei. Green (Hnf4α) marks hepatocytes. Scale bar represents 20 μm.
(E) SOX9 and HNF4α double staining in periportal areas in the livers of AAV-ctrl and AAV-Hoxb6 mice after CCl4 injury. Blue (DAPI) shows nuclei. Red (HNF4α) marks hepatocytes. Green presents SOX9. Scale bar represents 20 μm.
(F) SOX9 and BrdU double staining in periportal areas in livers of AAV-ctrl and AAV-Hoxb6 mice after CCl4 injury. Blue (DAPI) shows nuclei. Red (BrdU) marks the proliferating cells. Green presents SOX9. Scale bar represents 20 μm.
Data are expressed as means ± SD, n = 6 mice per group containing three replicates. Significant difference is presented at the levels of ∗p < 0.05 and ∗∗p < 0.01 by two-tailed Student's t test. See also Figure S4.
To investigate the effect of HOXB6 on liver regeneration and injury repair through regulation of SOX9, we analyzed the behavior of SOX9+ cells during the regeneration period. Since SOX9+/HNF4α+ HybHPs was regenerated in the injured liver without developing into cancer (Font-Burgada et al., 2015), we focused on the Hoxb6 effects on the HybHPs. Hnf4α and Sox9 were co-stained to assess Sox9+ cell differentiation using an RNAscope assay. We found that the number of Sox9+/Hnf4α+ hepatocytes around the PP area in the CCl4 treatment group was larger than that in the control group. After AAV-Hoxb6 treatment, CCl4-injured mice presented a smaller number of Sox9+/Hnf4α+ hepatocytes than the AAV control group (Figure 5D). The above results were also consistent with immunofluorescence results that HOXB6 decreased the number of SOX9+ hepatocytes (Figure 5E).
To investigate whether HOXB6 regulated SOX9+ LPC proliferation, we performed proliferation analysis in liver sections. Immunofluorescent double staining of the proliferation marker bromodeoxyuridine (BrdU) and SOX9 results showed an obvious increased proliferation of SOX9+ LPCs in the livers of the CCl4 injury group compared with that in control livers. After treatment with AAV-Hoxb6, a significant decrease in proliferation of SOX9+ LPCs was seen in the livers of the CCl4 injury group (Figure 5F). Collectively, these data demonstrated that overexpression of HOXB6 suppressed SOX9+ stem cell proliferation and propagation.
Taken together, our data indicated the downregulation of the LPC marker Sox9 and the resultant decreased proliferation and differentiation of SOX9+ LPCs after HOXB6 exogenous expression in CCl4 chronic liver injury.
Adenovirus-Mediated Deletion of miR-126 Suppresses Proliferation and Differentiation of SOX9+ LPCs to Aggravate Damage in CCl4 a Chronic Liver Injury Model by Targeting Hoxb6
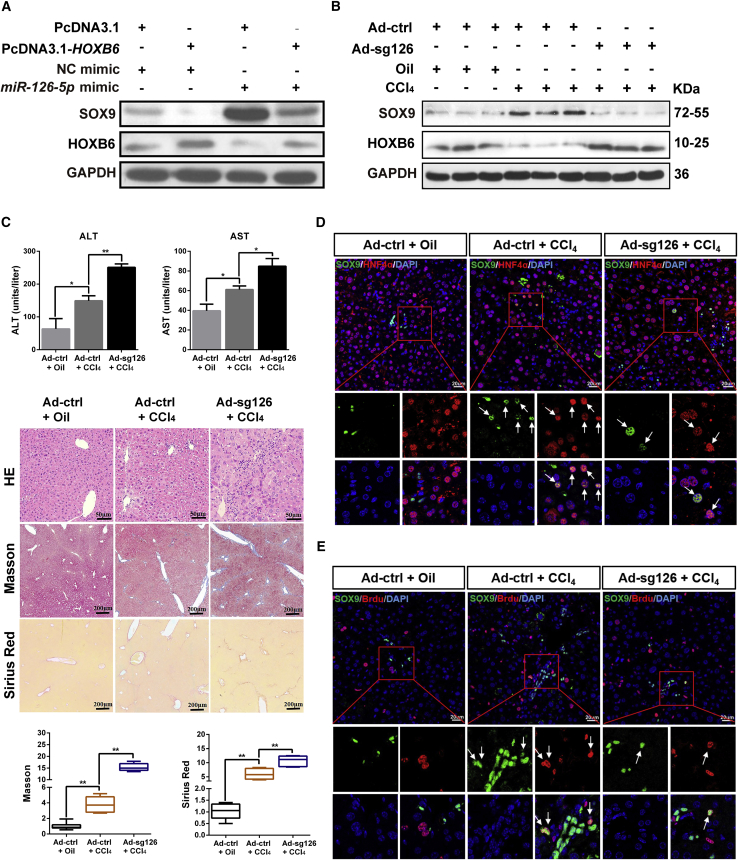
To further examine whether miR-126 regulated SOX9 by targeting HOXB6, HepG2 cells were transfected with negative control mimics, miR-126-5p mimics, PcDNA3.1, or PcDNA3.1-HOXB6, respectively. The qRT-PCR and western blot results revealed that HOXB6 overexpression abrogated the miR-126-5p mimics-induced promotion of SOX9 levels, suggesting that miR-126-5p regulated SOX9 by targeting HOXB6 (Figure 6A). The above results were consistent with in the cell line C3H10, from which the miR-126 has been deleted (Figure S5A).
Figure 6.
Adenovirus-Mediated Deletion of miR-126 Suppresses the Proliferation and Differentiation of SOX9+ LPCs to Aggravate Liver Damage through Hoxb6 in a CCl4 Chronic Injury Model
(A) PcDNA3.1 or PcDNA3.1-HOXB6 was co-transfected with miR-126-5p mimics or NC into HepG2 cells, respectively. HOXB6 and SOX9 levels were measured by western blotting.
(B) Hepatic expression levels of SOX9 and HOXB6 were measured by western blot in CCl4-injured mice following CRISPR/Cas9-mediated miR-126 gene disruption.
(C) Hepatotoxicity and ALT/AST enzymes activities change in CCl4-injured mice following miR-126 gene disruption. Representative H&E, Masson, and Sirius red staining in Ad-ctrl- and Ad-sg126-treated mice liver after chronic hepatic injury by CCl4. Fibrosis was quantified by morphometric measurement of Masson and Sirius red.
(D) SOX9 and HNF4α double staining in periportal areas in livers of Ad-ctrl- or Ad-sg126-treated mice after CCl4 injury. Blue (DAPI) shows nuclei. Red (HNF4α) marks hepatocytes. Green presents SOX9. Scale bar represents 20 μm.
(E) SOX9 and BrdU double staining in periportal areas in livers of Ad-ctrl- or Ad-sg126-treated mice after CCl4 injury. Blue (DAPI) shows nuclei. Red (BrdU) marks the proliferating cells. Green presents SOX9. Scale bar represents 20 μm.
Data are expressed as means ± SD, n = 6 mice per group containing three replicates. Significant difference is presented at the levels of ∗p < 0.05 and ∗∗p < 0.01 by two-tailed Student's t test. See also Figure S5.
We further studied Sox9 regulation by miR-126 in vivo in a CCl4-induced chronic liver injury model. Mice were repeatedly administered a low dose of CCl4 to induce chronic liver injury. After tail vein injection of 1 × 1010 particles of Ad.ctrl or Ad.sg126 into the CCl4-treated mice, the expression of miR-126 in liver was reduced (Figures S5B and S5C). To investigate whether miR-126 contributed to liver injury repair, we collected the livers and blood for further analysis. Compared with the control group, the CCl4 treatment group exhibited a higher expression level of SOX9 and a lower level of HOXB6 (Figure 6B). After deletion of miR-126 in the CCl4-treated mice, the drastically decreased expression of SOX9 and the increased expression of HOXB6 were observed (Figure 6B). We found that CCl4 treatment induced hepatotoxicity. Serological and histological analysis revealed that, after treatment with Ad.sg126, a significant increase in the severity of liver damage and fibrosis and in ALT and AST levels in the CCl4 injury group was observed (Figure 6C). In addition, the deletion of miR-126 promoted a CCl4-induced increase in liver/body weight ratio (Figure S5D). These results indicated that deletion of miR-126 could obviously aggravate liver injury by targeting Hoxb6.
To investigate whether miR-126 affected liver regeneration and injury repair through Sox9, we analyzed the behavior of SOX9+ stem cells during the regeneration period after CCl4-induced liver injury. We found that the number of SOX9+/HNF4α+ hepatocytes around the PP area in the CCl4 administration group was larger than in the control group. After Ad.sg126 treatment, CCl4-injured mice presented a smaller number of SOX9+/HNF4α+ hepatocytes (Figure 6D). These results suggested that the deletion of miR-126 suppressed the differentiation of SOX9+ LPCs into hepatocytes. Furthermore, immunofluorescent double staining of BrdU and SOX9 showed an obvious increase in SOX9+ LPC proliferation in the livers of the CCl4 injury group compared with control livers. After treatment with Ad.sg126, an obvious decrease in SOX9+ LPC proliferation in the livers of CCl4 injury group was observed (Figure 6E). The above results indicated that the deletion of miR-126 suppressed SOX9+ LPC proliferation. Taken together, our data suggested that upregulation of the LPC marker Sox9 and the resultant increased proliferation and differentiation of SOX9+ LPCs possibly explained why miR-126 might contribute to liver regeneration and repair.
Discussion
The liver plays a pivotal role in the metabolism and detoxification of xenobiotics, which increases its possibility to be subjected to toxic damage, resulting in rapid loss of hepatic function, and high risk of losing regenerative and repair capacity (Font-Burgada et al., 2015). One previous study has shown that miR-302b and miR-20a repress hepatic functions by regulating transforming growth factor β (TGF-β) (Wei et al., 2013). Other previous studies have reported that miRNAs functioned by stimulating hepatocytes or by repressing cholangiocyte gene expression in mature hepatocytes (Gailhouste et al., 2013; Laudadio et al., 2012; Rogler et al., 2009). Our study found that miR-126 was expressed in hepatocytes and biliary epithelial cells, and it was necessary for maintaining SOX9+ LSCs properties. Thus, the molecular mechanisms of miR-126 regulation in LSCs are important for understanding liver repair.
LSCs or LPSs play important roles in the generation of hepatocytes and cholangiocytes (Suzuki et al., 2000). During persistent and severe liver damage, part of hepatocytes undergo dedifferentiation into LPCs for liver regeneration (Itoh and Miyajima, 2014). Although it has been well reported that the properties of LPCs were regulated by a number of intracellular and extracellular signaling pathways (Kitade et al., 2013; Parent and Beretta, 2008), the relationships among these signaling pathways and the contribution of miR-126 in LPCs are largely unknown. To identify possible miR-126 target genes involved in the regulation of LPCs properties, we performed a computational screen for genes with complementary sites of miR-126 in their 3′ UTR by using open-access software. Here, we identified Hoxb6 to be a miR-126 target gene involved in regulating Sox9 levels.
The Hox family of homeobox genes, as major transcriptional regulators in the body, encodes DNA binding proteins that play a crucial role in early body morphogenesis and hematopoietic development, and Hox family exhibits important effects on stem cell renewal, lineage commitment, and differentiation (Amsellem et al., 2003; Krumlauf, 1994; Larsen et al., 2015; Thorsteinsdottir et al., 1997). Hoxb6, as a member of the Hox family, was found to regulate HSC self-renewal, and its overexpression in mice resulted in HSC and myeloid progenitor cell expansion (Bhatlekar et al., 2018; Fischbach et al., 2005). However, HOXB6 functions in liver and the related molecular signals and mechanisms have not been investigated. The cell types that could respond to HOXB6 activation also remain unknown. The present study demonstrated that HOXB6 inhibited SOX9+ LPC proliferation and differentiation by reducing SOX9 levels in liver injury repair. Our results are also consistent with previous research findings that SOX9+ LPCs were a preexisting group around the PP area with the high regenerative and proliferative capability (Font-Burgada et al., 2015; Li et al., 2016), and the ability to differentiate into hepatocytes in livers (Ko et al., 2019). These studies support our results that upregulation of SOX9 promoted SOX9+ LPC proliferation and differentiation during liver fibrosis.
SOX9 has been reported to be regulated in a variety of cytokines and signaling pathways in different tissues. The deletion of BMP type I receptor gene in chondro-osteo progenitor cells led to chondrodysplasia and reduction in SOX9 (Yoon et al., 2005). In addition, SOX9 was induced by TGF-β in the kidney fibroblast, and it acted as an important downstream mediator of TGF-β signaling to promote renal fibrosis (Li et al., 2018). However, the regulation mechanism of Sox9 transcription in the progression of liver injury repair remained unknown. Considering the principal role of HOXB6 in the HSC self-renewal and differentiation (Bhatlekar et al., 2018), and the fact that the HOXB6 protein repressed globin transcript levels in a DNA binding-dependent manner, thus repressing the erythroid phenotype in human leukemic cells (Shen et al., 2004), we investigated the effects of HOXB6 on SOX9 expression and properties of SOX9+ LPCs.
Although HOX homeodomain proteins were thought to function as transcription factors, most full-length HOX proteins, including HOXB6, bound only weakly to DNA targets containing a TAAT sequence, and the transcription of target genes was weakly activated or repressed (Catron et al., 1993; Graba et al., 1997; Sánchez-Higueras and Rastogi, 2019). In addition, in one previous study, transient reporter gene analysis results revealed that HOXB6 and other HOX proteins did not change the activity of luciferase reporter vectors containing synthetic TAAT multimers or putative gene regulatory regions (Shen et al., 2001). Similarly, in this study, our observations provided evidence that a binding region of HOXB6 was in the upstream of SOX9 promoter, and that HOXB6 was responsible for the downregulation of SOX9 in hepatocytes and cholangiocytes. However, we found that HOXB6 did not change the luciferase activity in transient reporter gene assays using putative SOX9 regulatory regions containing a TAAT sequence, but luciferase activity was found to be changed in SOX9 promoter regions containing no TAAT sequences (SOX9-P5 and SOX9-P6). This result indicated that HOXB6 might exert transcriptional repression by binding to other specific sequences in the downstream target gene promoter region, which requires further study in the future. These findings indicated that HOXB6 affected the proliferation and differentiation of SOX9+ LPC by transcriptionally regulating the expression of SOX9. To our knowledge, this is the first attempt to report a LPC regulatory event during liver regeneration and repair.
Furthermore, the overall contribution of miR-126 to the whole repair process, especially to liver injury repair, remains elusive. In this study, we found that, during liver injury repair, the expression of HOXB6 increases with deletion of miR-126, which leads to the inhibition of proliferation and differentiation in SOX9+ LPCs. In addition, we overexpressed miR-126 or/and HOXB6 in liver cancer cell lines, respectively, and we found that overexpression of HOXB6 abrogated miR-126-induced increases in stem cell-related gene SOX9 expression. These results suggest that miR-126-5p can restrict the inhibition effect of HOXB6 on SOX9+ LPCs properties, thus contributing to hepatic repair and regeneration.
The miR-126 has broad biological and physiological implications. Thus, it is vital to fully understand the biological and physiological properties and functions of miR-126. In conclusion, we have identified miR-126 as a regulator of stem cell-related gene Sox9 expression and SOX9+ LPCs properties in liver repair by targeting Hoxb6. Our principal findings may have a clinical implication for treating liver diseases. In future studies, the molecular mechanisms of regulating the expression and function of miR-126 in hepatocytes and cancer cells require further clarification and analysis. Knowledge of the molecular properties of liver injury repair and carcinogenesis will contribute to the development of anticarcinogenic agents.
Experimental Procedures
Detailed methods are provided in the Supplemental Information.
Mice and Injury Regimens
Adult C57BL/6J male mice were given standard rodent chow and water ad libitum under a standard 12-h light/dark cycle. For CCl4 injury experiments, CCl4 was injected into mice intraperitoneally at the dose of 2 mL/kg body weight, twice per week for 4 weeks (Tu et al., 2015). BrdU (Sigma-Aldrich, St. Louis, MO) was injected at the dose of 50 mg/kg 2 h before sacrifice (Zhang et al., 2015). Tissue and serum were collected at the end of the experiments. All procedures followed the Huazhong Agricultural University Guidelines for the Care and Use of Laboratory Animals.
Cell Culture and Transient Transfection
The cell lines used in this study included HepG2, HIBEpic, LO2, BMSC, HeLa, and C3H10 cells. All cells were seeded into 6- or 24-well plates, and grown in high glucose DMEM (HyClone, Logan, UT) supplied with 10% (v/v) fetal bovine serum (Gibco BRL, Grand Island, NY) and 1% (v/v) penicillin-streptomycin. The following day, cells were transfected with plasmid, mimics, or siRNA. Transient transfection was performed using lipofectamine 2000 or lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). The primer sequences are listed in Table S1.
Immunofluorescent Analysis
Liver tissues were immobilized with 4% paraformaldehyde (PFA), dehydrated, embedded in paraffin, sectioned at 5 μm, and processed for immunofluorescent staining. SOX9 antibody (AB5535, Millipore, USA) and HNF4α antibody (Ab41898, Abcam, Cambridge, England) were used to incubate sections at 4°C overnight. Slides were washed with PBS and incubated with corresponding secondary antibodies for 1 h, followed by PBS washes. The incubated slides were added with DAPI for nuclear staining and mounting. Images were acquired with a laser scanning confocal microscope (LSM710, Carl Zeiss Microscopy), were analyzed by Zen software with fixed parameters.
Liver Histology and Immunohistochemical Staining
Liver tissues were immobilized with 4% PFA, dehydrated, embedded in paraffin, sectioned at 5 μm, and processed for H&E, Masson trichrome, and Sirius red staining.
In Situ mRNA Hybridization
In situ detection of Sox9 and Hnf4α RNA transcripts was carried out on OCT-embedded tissue sections using the RNAscope Multiplex Fluorescent Reagent Kit v.2 (Advanced Cell Diagnostics). RNAscope probes for Sox9, Hnf4α, standard negative probe for 4-hydroxy-tetrahydrodipicolinate reductase, and positive probe for peptidylprolyl isomerase B were used for in situ detection. The probe information for RNAscope assay is listed in Table S2.
RNA Isolation and qRT-PCR
The RNAiso Plus (Takara, Japan) was used to isolate total RNA, including low-molecular-weight RNA from frozen samples and cell lines, according to the manufacturer's protocol. Then the first-strand cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). Real-time PCR was performed using the MonAmp SYBR Green qPCR Mix (Low ROX). The relative levels were calculated using the comparative-Ct method (2−ΔΔCt method). The primer sequences are listed in Table S3.
Western Blots
For whole-cell protein extraction, liver tissues were prepared in lysis buffer (Beyotime, Jiangsu, China) according to the manufacturer's instructions. Protein lysates were separated by SDS-PAGE. Next, the gel was transferred to polyvinylidene difluoride membranes (Millipore). After being blocked with 5% skimmed milk, the membranes were incubated overnight with the anti-SOX9 (AB5535, Millipore), anti-HOXB6 (sc-166950X, Santa Cruz Biotechnology), anti-GAPDH (60004-I-Ig, Proteintech, Chicago), or Tag-3∗Flag-antibody (66008-3-Ig, Proteintech) at 4°C. Then, the membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies at room temperature for 1.5 h. Finally, the membranes were visualized with enhanced chemiluminescence (Bio-Rad, USA).
Biochemical Evaluation
Plasma was collected from blood after centrifugation (at 3,000 rpm) for 10 min at 4°C. Plasma ALT, AST, and LDH were determined to evaluate liver injury using a Multiskan MK3 microplate reader (Thermo Electron Corporation, USA) and commercial kits (Nanjing Jiancheng Bioengineering Institute, China), according to the manufacturer's instructions.
Lentivirus, Adenovirus, and AAV Plasmid Construction and Production
Two pairs of CRISPR guide RNAs (gRNAs) targeting pre-miR-126 gene were initially screened in NIH3T3cells. The gRNA targeting sgRNA3 displayed ∼50% mutagenesis at the on-target site in pre-miR-126, as determined by PCR and a T7EN1 cleavage assay (Yan et al., 2019). After targeting efficiency was confirmed, the sgRNAs were constructed into lentivirus and adenovirus plasmids. To completely overexpress HOXB6 in vivo, the Hoxb6-CDS sequence was inserted into the multiple cloning site in the pHBAAV-CMV-Hoxb6-3flag-T2A-ZsGreen plasmid. The two recombinant AAV plasmids were transfected into HEK293 cells with pAAV-RC and pHelper by using lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). AAV-Hoxb6 and AAV-ctrl (Hanbio, Shanghai, China) were packaged in AAV-293 cells.
Intravenous Virus Injection for Liver Transduction
To intravenously inject adenovirus or AAVs, mice were restrained in a rodent restrainer, their tails were dilated using a heat lamp or warm water and sterilized by 70% ethanol, and 100 μL of concentrated AAV (1 × 1010 particles per mouse) was injected into the tail vein of each mouse.
ChIP
ChIP assays were performed using the ChIP Assay kit (Beyotime) according to the user manual. In brief, HepG2 cells were transfected with pEGFP-N1-HOXB6 for 24 h, and then incubated with formaldehyde at a final concentration of 1% (v/v) for 10 min at 37°C to crosslink the nuclear proteins to DNA. Subsequently, cells were sonicated and then immunoprecipitated with the antibody against HOXB6 (sc-166950, Santa Cruz Biotechnology), taking IgG as a negative control, or without anti-HOXB6 and IgG in the reaction as mock control. The captured chromatin was eluted and un-crosslinked, and the DNA was recovered. The ChIP-isolated DNA was subjected to PCR amplification using the primer pair spanning the SOX9-P6-3 in the SOX9 promoter region. The primer sequences are listed in Table S4.
Statistical Methods
Data are expressed as the means ± SD and were analyzed using Prism 6 (GraphPad). Statistical details of the experiments can be found in the Results and figure legends. Student's two-tailed t test (unpaired) was used to determine statistical significance differences between groups. Statistical significance was presented at the level of ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data and Code Availability
The sequences of all primers are included in the paper.
Authors Contributions
Y.Y. and Lisheng Zhang conceived and designed the study. Y.Y. provided the experimental data. Y.Y. and R.W. performed the cell and animal experiments. X.H., S.W., Liang Zhang, and C.H. provided assistance in molecular assays. Y.Y. and R.W. discussed and drafted the manuscript. Lisheng Zhang organized the data and wrote the manuscript.
Acknowledgments
This work was supported by National Key R&D plan nos. 2017YFA0103202 and 2017YFA0103200, the Fundamental Research Funds for the Central Universities (2662017PY106, 2662016PY087, and 2662019YJ008), and HZAU Startup funds to L.Z.
Published: August 6, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.07.005.
Supplemental Information
References
- Alison M.R., Lin W.R. Hepatocyte turnover and regeneration: virtually a virtuoso performance. Hepatology. 2011;53:1393–1396. doi: 10.1002/hep.24252. [DOI] [PubMed] [Google Scholar]
- Amsellem S., Pflumio F., Bardinet D., Izac B., Charneau P., Romeo P.H., Dubart-Kupperschmitt A., Fichelson S. Ex vivo expansion of human hematopoietic stem cells by direct delivery of the HOXB4 homeoprotein. Nat. Med. 2003;9:1423–1427. doi: 10.1038/nm953. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatlekar S., Fields J.Z., Boman B.M. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron K.M., Iler N., Abate C. Nucleotides flanking a conserved TAAT core dictate the DNA binding specificity of three murine homeodomain proteins. Mol. Cell. Biol. 1993;13:2354–2365. doi: 10.1128/mcb.13.4.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach N.A., Rozenfeld S., Shen W., Fong S., Chrobak D., Ginzinger D., Kogan S.C., Radhakrishnan A., Le Beau M.M., Largman C. HOXB6 overexpression in murine bone marrow immortalizes a myelomonocytic precursor in vitro and causes hematopoietic stem cell expansion and acute myeloid leukemia in vivo. Blood. 2005;105:1456–1466. doi: 10.1182/blood-2004-04-1583. [DOI] [PubMed] [Google Scholar]
- Font-Burgada J., Shalapour S., Ramaswamy S., Hsueh B., Rossell D., Umemura A., Taniguchi K., Nakagawa H., Valasek M.A., Ye L. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., Hosokawa S., Elbahrawy A., Soeda T., Koizumi M. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat. Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Gailhouste L., Gomez-Santos L., Hagiwara K., Hatada I., Kitagawa N., Kawaharada K., Thirion M., Kosaka N., Takahashi R.U., Shibata T. miR-148a plays a pivotal role in the liver by promoting the hepatospecific phenotype and suppressing the invasiveness of transformed cells. Hepatology. 2013;58:1153–1165. doi: 10.1002/hep.26422. [DOI] [PubMed] [Google Scholar]
- Graba Y., Aragnol D., Pradel J. Drosophila Hox complex downstream targets and the function of homeotic genes. BioEssays. 1997;19:379–388. doi: 10.1002/bies.950190505. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Shenhav R., Matcovitch-Natan O., Toth B., Lemze D., Golan M., Massasa E.E., Baydatch S., Landen S., Moor A.E. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang Y., Pu W., Huang X., Qiu L., Li Y., Yu W., Zhao H., Liu X., He L. Lineage tracing reveals the bipotency of SOX9+ hepatocytes during liver regeneration. Stem Cell Reports. 2019;12:624–638. doi: 10.1016/j.stemcr.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M., Dorrell C., Boj S.F., van Es J.H., Li V.S., van de Wetering M., Sato T., Hamer K., Sasaki N., Finegold M.J. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Miyajima A. Liver regeneration by stem/progenitor cells. Hepatology. 2014;59:1617–1626. doi: 10.1002/hep.26753. [DOI] [PubMed] [Google Scholar]
- John K., Hadem J., Krech T., Wahl K., Manns M.P., Dooley S., Batkai S., Thum T., Schulze-Osthoff K., Bantel H. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology. 2014;60:1346–1355. doi: 10.1002/hep.27250. [DOI] [PubMed] [Google Scholar]
- Kappen C. Disruption of the homeobox gene Hoxb-6 in mice results in increased numbers of early erythrocyte progenitors. Am. J. Hematol. 2000;65:111–118. doi: 10.1002/1096-8652(200010)65:2<111::aid-ajh4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Kitade M., Factor V.M., Andersen J.B., Tomokuni A., Kaji K., Akita H., Holczbauer A., Seo D., Marquardt J.U., Conner E.A. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes Dev. 2013;27:1706–1717. doi: 10.1101/gad.214601.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S., Russell J.O., Tian J., Gao C., Kobayashi M., Feng R., Yuan X., Shao C., Ding H., Poddar M. Hdac1 regulates differentiation of bipotent liver progenitor cells during regeneration via Sox9b and Cdk8. Gastroenterology. 2019;156:187–202.e14. doi: 10.1053/j.gastro.2018.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Larsen B.M., Hrycaj S.M., Newman M., Li Y., Wellik D.M. Mesenchymal Hox6 function is required for mouse pancreatic endocrine cell differentiation. Development. 2015;142:3859–3868. doi: 10.1242/dev.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudadio I., Manfroid I., Achouri Y., Schmidt D., Wilson M.D., Cordi S., Thorrez L., Knoops L., Jacquemin P., Schuit F. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology. 2012;142:119–129. doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Lechman E.R., Gentner B., van Galen P., Giustacchini A., Saini M., Boccalatte F.E., Hiramatsu H., Restuccia U., Bachi A., Voisin V. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Li W., Hui L. Hybrid hepatocyte: a newly identified player for regeneration in hepatic injuries. Hepatology. 2016;64:2244–2246. doi: 10.1002/hep.28837. [DOI] [PubMed] [Google Scholar]
- Li H., Cai H., Deng J., Tu X., Sun Y., Huang Z., Ding Z., Dong L., Chen J., Zang Y. TGF-beta-mediated upregulation of Sox9 in fibroblast promotes renal fibrosis. Biochim. Biophys. Acta. 2018;1864:520–532. doi: 10.1016/j.bbadis.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Luna J.M., Barajas J.M., Teng K.Y., Sun H.L., Moore M.J., Rice C.M., Darnell R.B., Ghoshal K. Argonaute CLIP defines a deregulated miR-122-bound transcriptome that correlates with patient survival in human liver cancer. Mol. Cell. 2017;67:400–410.e7. doi: 10.1016/j.molcel.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y., van den Born M., van Es J.H., Hamilton S.R., Adams H.P., Zhang J., Clevers H., de Crombrugghe B. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Mori M.A., Ludwig R.G., Garcia-Martin R., Brandão B.B., Kahn C.R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 2019;30:656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent R., Beretta L. Translational control plays a prominent role in the hepatocytic differentiation of HepaRG liver progenitor cells. Genome Biol. 2008;9:R19. doi: 10.1186/gb-2008-9-1-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcelli D., Fischer B., Russell S., White R. Chromatin accessibility plays a key role in selective targeting of Hox proteins. Genome Biol. 2019;20:115. doi: 10.1186/s13059-019-1721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W., Zhang H., Huang X., Tian X., He L., Wang Y., Zhang L., Liu Q., Li Y., Li Y. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat. Commun. 2016;7:13369. doi: 10.1038/ncomms13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha A.S., Vidal V., Mertz M., Kendall T.J., Charlet A., Okamoto H., Schedl A. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- Rogler C.E., Levoci L., Ader T., Massimi A., Tchaikovskaya T., Norel R., Rogler L.E. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- Sánchez-Higueras C., Rastogi C. In vivo Hox binding specificity revealed by systematic changes to a single cis regulatory module. Nat. Commun. 2019;10:3597. doi: 10.1038/s41467-019-11416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W., Chrobak D., Krishnan K., Lawrence H.J., Largman C. HOXB6 protein is bound to CREB-binding protein and represses globin expression in a DNA binding-dependent, PBX interaction-independent process. J. Biol. Chem. 2004;279:39895–39904. doi: 10.1074/jbc.M404132200. [DOI] [PubMed] [Google Scholar]
- Shen W.F., Krishnan K., Lawrence H.J., Largman C. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell Biol. 2001;21:7509–7522. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Kumar V., Sud N., Mahato R.I. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv. Drug Deliv. Rev. 2018;129:54–63. doi: 10.1016/j.addr.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Zheng Y., Kondo R., Kusakabe M., Takada Y., Fukao K., Nakauchi H., Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- Tarlow B.D., Finegold M.J., Grompe M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology. 2014;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Sauvageau G., Hough M.R., Dragowska W., Lansdorp P.M., Lawrence H.J., Largman C., Humphries R.K. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay H.C., Yuan Q., Balakrishnan A., Kaiser M., Mobus S., Kozdrowska E., Farid M., Tegtmeyer P.K., Borst K., Vondran F.W.R. Hepatocyte-specific suppression of microRNA-221-3p mitigates liver fibrosis. J. Hepatol. 2019;70:722–734. doi: 10.1016/j.jhep.2018.12.016. [DOI] [PubMed] [Google Scholar]
- Tu X., Zheng X., Li H., Cao Z., Chang H., Luan S., Zhu J., Chen J., Zang Y., Zhang J. MicroRNA-30 protects against carbon tetrachloride-induced liver fibrosis by attenuating transforming growth factor beta signaling in hepatic stellate cells. Toxicol. Sci. 2015;146:157–169. doi: 10.1093/toxsci/kfv081. [DOI] [PubMed] [Google Scholar]
- Wang B., Zhao L., Fish M., Logan C.Y., Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Hou J., Alder O., Ye X., Lee S., Cullum R., Chu A., Zhao Y., Warner S.M., Knight D.A. Genome-wide microRNA and messenger RNA profiling in rodent liver development implicates mir302b and mir20a in repressing transforming growth factor-beta signaling. Hepatology. 2013;57:2491–2501. doi: 10.1002/hep.26252. [DOI] [PubMed] [Google Scholar]
- Yan Y., Qin D., Hu B., Zhang C., Liu S., Wu D., Huang W., Huang X., Wang L., Chen X. Deletion of miR-126a promotes hepatic aging and inflammation in a mouse model of cholestasis. Mol. Ther. Nucleic Acids. 2019;16:494–504. doi: 10.1016/j.omtn.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Wang S., Wang R., Jiang P., Chen Y., Zhang L., Hou C., Zhang L. Transcriptional regulation of microRNA-126a by farnesoid X receptor in vitro and in vivo. Biotechnol. Lett. 2020;27:2020. doi: 10.1007/s10529-020-02864-7. [DOI] [PubMed] [Google Scholar]
- Yoon B.S., Ovchinnikov D.A., Yoshii I., Mishina Y., Behringer R.R., Lyons K.M. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc. Natl. Acad. Sci. U S A. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Nguyen L.X.T., Li L., Zhao D., Kumar B., Wu H., Lin A., Pellicano F., Hopcroft L. Bone marrow niche trafficking of miR-126 controls the self-renewal of leukemia stem cells in chronic myelogenous leukemia. Nat. Med. 2018;24:450–462. doi: 10.1038/nm.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang L., Liu X., Chen W., Chang L., Chen L., Loera S., Chu P., Huang W.C., Liu Y.R. MicroRNA-657 promotes tumorigenesis in hepatocellular carcinoma by targeting transducin-like enhancer protein 1 through nuclear factor kappa B pathways. Hepatology. 2013;57:1919–1930. doi: 10.1002/hep.26162. [DOI] [PubMed] [Google Scholar]
- Zhang R.R., Zheng Y.W., Li B., Tsuchida T., Ueno Y., Nie Y.Z., Taniguchi H. Human hepatic stem cells transplanted into a fulminant hepatic failure Alb-TRECK/SCID mouse model exhibit liver reconstitution and drug metabolism capabilities. Stem Cell Res. Ther. 2015;6:49. doi: 10.1186/s13287-015-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences of all primers are included in the paper.