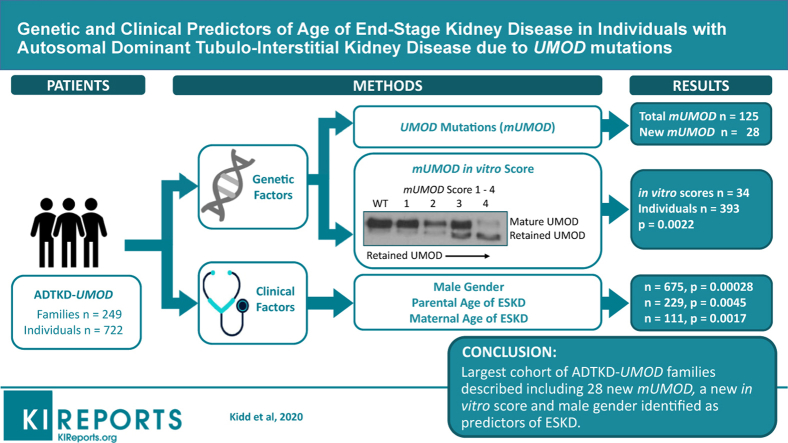
Abstract
Introduction
Autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD) is a rare condition associated with high variability in the age of end-stage kidney disease (ESKD). The minor allele of rs4293393, located in the promoter of the UMOD gene, is present in 19% of the population and downregulates uromodulin production by approximately 50% and might affect the age of ESKD. The goal of this study was to better understand the genetic and clinical characteristics of ADTKD-UMOD and to perform a Mendelian randomization study to determine if the minor allele of rs4293393 was associated with better kidney survival.
Methods
An international group of collaborators collected clinical and genetic data on 722 affected individuals from 249 families with 125 mutations, including 28 new mutations. The median age of ESKD was 47 years. Men were at a much higher risk of progression to ESKD (hazard ratio 1.78, P < 0.001).
Results
The allele frequency of the minor rs4293393 allele was only 11.6% versus the 19% expected (P < 0.01), resulting in Hardy-Weinberg disequilibrium and precluding a Mendelian randomization experiment. An in vitro score reflecting the severity of the trafficking defect of uromodulin mutants was found to be a promising predictor of the age of ESKD.
Conclusion
We report the clinical characteristics associated with 125 UMOD mutations. Male gender and a new in vitro score predict age of ESKD.
Keywords: autosomal dominant uromodulin kidney disease, genotype, phenotype, rs4293393, uromodulin
Graphical abstract
The cardinal manifestations of ADTKD-UMOD include autosomal dominant inheritance, precocious gout in some individuals, and slowly progressive chronic kidney disease.1 Progression to ESKD is variable, occurring between ages 20 and 70 years.2, 3, 4, 5 The reasons for this variation are unknown. Identification of the causes would lead to a better understanding of the pathogenesis of ADTKD-UMOD, identify individuals at risk of progression for clinical trials, and provide information about prognosis for patients.
In ADTKD-UMOD, retention of mutant uromodulin (mUMOD) protein in the endoplasmic reticulum (ER) of tubular epithelial cells in the thick ascending limb leads to ER stress, tubular cell death, and chronic kidney disease.4,6, 7, 8, 9, 10, 11 Uromodulin has a high cysteine content, resulting in a slow transit through the ER as disulfide bonds form.12 Approximately two-thirds of the mutations causing ADTKD-UMOD (mUMOD) involve cysteine residues, and no mutations have been found resulting in truncation or loss of transcription.4 Umod knockout mice also do not develop the ADTKD phenotype.13 These findings implicate mUMOD as the principal pathophysiologic cause of ADTKD-UMOD. Based on these hypotheses, one could theorize the amount of mUMOD expressed and the type of mutation may contribute to the pathophysiology and age of ESKD onset.
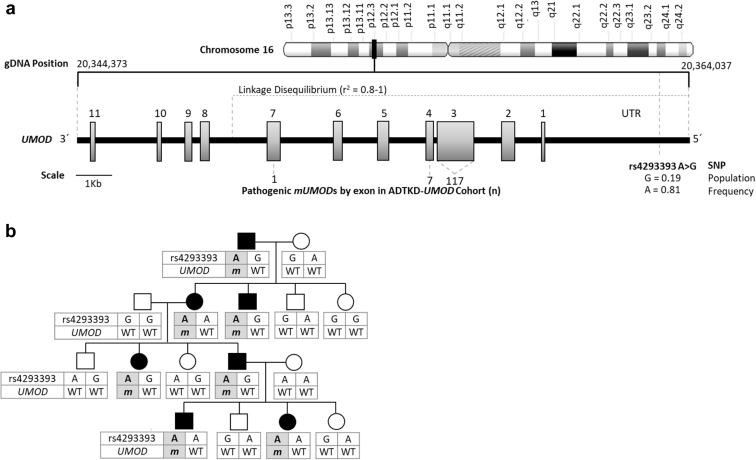
There is a genetic variant (known as single nucleotide polymorphism [SNP] rs4293393) present in the UMOD promoter (Figure 1a). In 19% of Europeans, this SNP has a guanosine residue and results in an approximately 50% reduction in uromodulin expression as compared with the remaining 81% of the European population, which has an adenosine residue at this site.14 As rs4293393 resides in the promoter of the UMOD gene, it is virtually always inherited together with the UMOD gene on the same allele. Indeed, UMOD promoter variants are within a region of complete linkage disequilibrium that spans exons 1 to 5. Because more than 95% of UMOD mutations are within exons 3 and 4, this implies that for virtually all pedigrees the variants in the UMOD promoter and the causal UMOD mutation cosegregate. In a given family with ADTKD-UMOD, all affected individuals who inherit the mUMOD gene will inherit the same rs4293393 allele adjacent to mUMOD (Figure 1b). The rs4293393 variant that is present on the wild-type allele will be inherited from the unaffected parent and will not be the same for all affected family members (Figure 1b).
Figure 1.
Genetic map of rs4293393 and UMOD with a representative autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD) pedigree demonstrating rs4293393-UMOD haplotype inheritance. (a) Genetic map of rs4293393 and UMOD, showing the linkage disequilibrium of rs4293393 with the UMOD mutations (mUMODs) found in this cohort study; 124 of 125 mUMODs occurred in exons 3 and 4 and were in a region of complete linkage disequilibrium with rs4293393. (b) Representative 4-generation ADTKD-UMOD pedigree. Genetically affected individuals are represented by black symbols, and the rs4293393-mUMOD haplotype is shaded gray. The rs4293393 allele (in this case “A”) is in phase with the mUMOD (designated m). In all genetically affected family members (due to linkage disequilibrium of rs4293393 and the UMOD gene [A]), the rs4293393-mUMOD haplotype is inherited together. In contrast, the rs4293393–wild-type UMOD (designated WT) haplotype inherited from the unaffected parent varies in subsequent generations based on the rs4293393-WT UMOD haplotypes of the unaffected parent. gDNA, genomic DNA; SNP, single nucleotide polymorphism; UTR, untranslated region.
The primary aims of this investigation were to better characterize the genetic and clinical findings of ADKTD-UMOD in a large population of affected families and to perform a Mendelian randomization study of individuals affected with ADTKD-UMOD. In a Mendelian randomization study, one studies the effects of genetic variants randomly distributed in a population on an outcome (e.g., kidney failure). We hypothesized that the presence of the minor rs4293393 variant in the promoter of the mUMOD allele would lead to a decreased expression of mUMOD. Thus, families with the minor rs4293393 variant in the mUMOD promoter should have decreased expression of mUMOD, which might ameliorate mutant protein deposition, preserve the tubulo-interstitium, and slow progression of chronic kidney disease and development of ESKD. This decreased production of mUMOD would be similar to the administration of a medication from birth onward (with 100% compliance) that lowers mUMOD production by approximately 50%. Our goal was to determine if the presence of the rs4293393 minor SNP variant with the mUMOD allele results in a later age of onset of ESKD.
Another factor that could affect the age of ESKD onset is the nature of the mUMOD mutation and its effect on the transit of uromodulin and mUMOD through the ER and on apoptosis. Some mutations may have a more deleterious effect on uromodulin trafficking and consequently ER function.7,15 To this end, we quantified the mUMOD trafficking defect for 35 selected mutations (Supplementary Table S1) through an in vitro score and investigated whether this score correlated with the age of onset of ESKD.
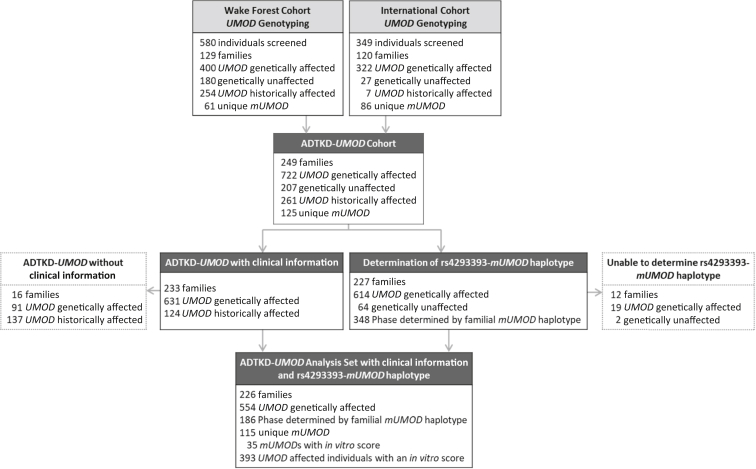
The dataset included genetic information and age of ESKD from 12 international ADTKD research teams (international cohort), as well as the Wake Forest ADTKD registry (WF cohort), which included additional clinical information (Figure 2). The data from the WF cohort and from the international cohort were combined to analyze genetic factors and gender, and the WF cohort data were then further analyzed to explore other factors that could affect the age of ESKD, including body mass index, smoking status, presence of gout, age of gout onset, and the mean age of ESKD for family members.
Figure 2.
Flow diagram showing contribution of the Wake Forest and International cohorts to the development of the autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD) registry. A total of 929 individuals underwent genetic testing and 722 were documented to have a UMOD mutation (UMOD genetically affected), and 633 underwent further genetic testing for rs4293393. Historically affected individuals were from families genetically diagnosed with ADTKD-UMOD but in whom a DNA sample was unavailable. These individuals suffered from at least chronic kidney disease stage 3 and clinical findings were consistent with ADTKD-UMOD. Clinical information was available in most individuals from the Wake Forest cohort. In vitro scores were developed for 35 UMOD mutations to further understand disease progression.
Methods
The study was approved by the Wake Forest School of Medicine Institutional Review Board, all institutional review boards of participating centers, and was carried out in accordance with the Declaration of Helsinki.
Recruitment
(i) Most participants were from the WF cohort16 (Figure 2). Families were either referred by their physician or self-referred by a family member. A genetic diagnosis was made in the index case and then in as many family members as possible. A family tree was constructed that included the age of onset of ESKD in both living and deceased family members. A questionnaire containing demographic and clinical information was completed by as many affected family members as possible. (ii) Data obtained from affected individuals and families from the international cohort included the UMOD mutation, rs4293393 genotype when available, gender, and current kidney function or age of ESKD onset.
Genetic Evaluation
An index case in each family underwent mutational analysis and was found to have a UMOD mutation as described in the Supplementary Methods. As many family members as possible then underwent genetic testing for the at-risk UMOD mutation. Individuals found to be genetically affected underwent rs4293393 genotyping (Supplementary Methods).
In 209 of 240 families, genetic linkage was used to identify which of the rs4293393 alleles was present in the promoter of the mUMOD allele. In 18 families in whom DNA was available, the phase of the rs4293393 allele and mUMOD was established via cloning, genotyping, and sequencing of long-range polymerase chain reaction products encompassing the promoter and UMOD genomic sequence (Supplementary Methods). If the genotype and phase of the rs4293393 variant were established in at least 1 affected family member, it was assumed that all affected family members had the same rs4293393 variant, given the very high linkage disequilibrium. All available family members underwent rs4293393 variant testing to determine if results were consistent.
DNA could not be obtained on some family members, most often because they were deceased. Individuals were considered historically affected if they met the following criteria: (i) a DNA sample for genetic diagnosis could not be obtained; (ii) there was a clinical history of at least chronic kidney disease stage 3 (estimated glomerular filtration rate <60 ml/min per 1.73 m2) that was consistent with ADTKD-UMOD; and (iii) the familial inheritance pattern was consistent with the individual being genetically affected.
In Vitro Score Determination
See Supplementary Methods for a full description. Thirty-five UMOD mutations were selected based on cohort prevalence and affected families having the youngest and oldest mean ages of ESKD onset. MDCK and/or HEK293 cell lines were transfected with expression vectors for each selected mUMOD and cell lysates analyzed by Western blots to evaluate uromodulin trafficking defects. UMOD mutation scoring was performed by quantifying the ratio between low- and high-molecular weight uromodulin glycoforms in 3 independent experiments.
Statistical Analysis
Descriptive statistics are shown as counts and proportions for categorical variables and mean ± SD for continuous variables. For each variable, comparisons between WF and international cohorts were made using χ2 and Fisher’s exact tests for categorical variables and Wilcoxon rank-sum tests for continuous variables.
Testing for Hardy-Weinberg equilibrium was performed by adopting bootstrapping resampling methods with 1000 repeated sampling on the cohort subsets to estimate variance for the minor allele frequency.
A pedigree structure was built using Sequential Oligogenetic Linkage Analysis Routines (SOLAR) software (http://solar-eclipse-genetics.org) based on the reported family trees and used to create a kinship matrix with the R package “kinship2” (https://cran.r-project.org/web/packages/kinship2/kinship2.pdf). Survival analysis was then performed with the outcome being age of ESKD (defined as starting dialysis, receiving a kidney transplant, or dying from kidney failure without receiving dialysis). Individuals were censored if they had not yet received dialysis or if they died before developing ESKD. Cox mixed effects models were built with the R package “coxme” (https://cran.r-project.org/web/packages/coxme/coxme.pdf) to incorporate the kinship matrix as the correlation structure and adjust for familial relationships. Univariate models were created for all parameters. Significant predictors from the univariate models were used develop a best-fit multivariate model. Because of the absence of some data for individual variables (for example, parental age of ESKD), variables were included in modeling only if 100 events had occurred to protect the robustness of the data. This modeling adjusted for variable family sizes. The multivariate model was created in a forward stepwise manner with entry criteria of a P value <0.05. The model that resulted in the highest C-statistic was considered the most effective model.
Results
In the WF cohort, 580 individuals from families with ADTKD-UMOD underwent genotyping, with 180 of 580 (31%) individuals being genetically unaffected and 400 of 580 (69%) individuals being genetically affected (Figure 2). There were 61 unique mutations in 129 families. An additional 254 individuals were considered historically affected, meaning that individuals were known to have ADTKD-UMOD as demonstrated by clinical findings and inheritance but did not undergo genotyping because they were deceased or otherwise unavailable to provide a DNA sample. The remaining data concern only individuals who were genetically affected or historically affected. Information regarding smoking history, body mass index, gout, and parental age of ESKD were available only in the WF cohort (Table 1).
Table 1.
Characteristics of individuals with ADTKD-UMOD who were genetically or historically affected, by cohort
| Characteristic | WF cohort |
International cohort | P value | ||
|---|---|---|---|---|---|
| ADTKD-UMOD genotyped | ADTKD-UMOD historic | All | |||
| ADTKD-UMOD genotyped | n = 400 | 400 (61%) | 322 (98%) | ||
| ADTKD-UMOD historic | n = 254 | 254 (39%) | 7 (2%) | ||
| Number of individuals who reached ESKD, n (%) | 159 (40) 4 (1) unknown |
144 (57) 77 (30) unknown |
303 (53) 81 (12) unknown |
123 (37) 79 (24) unknown |
0.33a |
| Age of ESKD | 46.37 | 51.17 | 48.65 | 48.88 | 0.88 |
| Male gender, n (%) | 180 (48) | 145 (57) | 325 (50) | 162 (49) 2 (1) unknown |
0.20b |
| Race, n (%) | <0.0001b | ||||
| White | 380 (95) | 247 (97) | 627 (96) | 271 (82) | |
| Black | 3 (1) | 0 | 3 (0.5) | 0 | |
| Hispanic | 0 | 0 | 0 | 0 | |
| Asian/Pacific Islander | 11 (3) | 5 (2) | 16 (2) | 0 | |
| From India | 1 (0.3) | 1 (0.4) | 2 (0.3) | 0 | |
| Other | 0 | 0 | 0 | 0 | |
| Unreported | 5 (1) | 1 (0.4) | 6 (1) | 58 (18) | |
| Ethnicity, n (%) | <0.0001b | ||||
| Hispanic or Latino | 6 (2) | 0 | 6 (1) | 24 (7) | |
| Not Hispanic or Latino | 374 (94) | 239 (94) | 613 (94) | 251 (76) | |
| Other | 8 (2) | 5 (2) | 13 (2) | 0 | |
| Unreported | 12 (3) | 10 (4) | 22 (3) | 54 (16) | |
| Smoking, n (%) | <0.0001a,c | ||||
| Never | 228 (57) | 8 (3) | 236 (36) | ||
| Current | 23 (6) | 1 (0.4) | 24 (4) | ||
| Former | 69 (17) | 5 (2) | 74 (11) | ||
| Uncertain | 80 (20) | 240 (94) | 320 (49) | ||
| Weight (kg) | 0.24c | ||||
| Male and female | 76.9 ± 19.9 (n = 287) | 83.0 ± 22.2 (n = 20) | 77.3 ± 20.2 (n = 307) | ||
| Male | 86.3 ± 19.2 (n = 127) | 87.9 ± 18.4 (n = 11) | 86.4 ± 19.1 (n = 138) | ||
| Female | 69.4 ± 17.1 (n = 160) | 77.0 ± 25.9 (n = 9) | 69.8 ± 17.6 (n = 169) | ||
| Height (cm) | 0.56c | ||||
| Male and Female | 169.5 ± 12.0 (n = 287) | 170.9 ± 10.0 (n = 20) | 169.6 ± 11.8 (n = 307) | ||
| Male | 177.5 ± 11.8 (n = 127) | 177.1 ± 6.3 (n = 11) | 177.4 ± 11.5 (n = 138) | ||
| Female | 163.2 ± 7.5 (n = 160) | 163.4 ± 8.4 (n = 9) | 163.2 ± 7.5 (n = 169) | ||
| BMI (kg/m2) | 26.5 ± 5.8 (n = 286) | 28.15 ± 6.4 (n = 20) | 26.6 ± 5.8 (n = 306) | 0.27c | |
| Gout, n (%) | <0.0001a,c | ||||
| Yes | |||||
| Male and female | 202 (50) | 43 (17) | 245 (37) | ||
| Male | 106 (59) | 31 (21) | 137 (42) | ||
| Female | 96 (44) | 12(11) | 108 (33) | ||
| No | |||||
| Male and female | 189 (47) | 13 (5) | 202 (31) | ||
| Male | 72 (40) | 6(4) | 78 (24) | ||
| Female | 117 (53) | 7(6) | 124 (38) | ||
| Uncertain | |||||
| Male and female | 9 (2) | 198 (78) | 207 (32) | ||
| Male | 2 (1) | 108 (7) | 110 (34) | ||
| Female | 7 (3) | 90 (83) | 97 (29) | ||
| Age of gout onset (y) | 0.15c | ||||
| Male and female | 30.5 ± 11.5 (n = 197) | 27.2 ± 9.5 (n = 20) | 30.2 ± 11.4 (n = 217) | ||
| Male | 29.1 ± 9.9 (n = 105) | 28.2 ± 10.4 (n = 13) | 29.0 ± 9.9 (n = 118) | ||
| Female | 32.2 ± 12.9 (n = 92) | 25.4 ± 8.0 (n = 7) | 31.7 ± 12.7 (n = 99) | ||
| Mutation type, n (%) | |||||
| p.H177-R185del (18%) | 108 (27) | 65 (26) | 173 (26) | 0 | |
| p.V93-G97delinsAASC (8%) | 42 (10) | 30 (12) | 72 (11) | 2 (1) | |
| p.R178P (5%) | 21 (5) | 27 (11) | 48 (7) | 0 | |
| p.C106F (5%) | 26 (6) | 21 (8) | 47 (7) | 0 | |
| p.C148Y (3%) | 12 (3) | 11 (4) | 23 (4) | 2 (1) | |
| p.G88D (2%) | 0 | 0 | 0 | 23 (7) | |
| p.P236L (2%) | 10 (2) | 7 (3) | 17 (3) | 3 (1) | |
| p.C135Y (2%) | 14 (4) | 3 (1) | 17 (3) | 0 | |
| p.S91del (2%) | 0 | 1 (0.4) | 1 (0.2) | 16 (5) | |
| p.P236R (1%) | 3 (1) | 1 (0.4) | 4 (1) | 5 (2) | |
| Other mutation (54%) | 164 (41) | 88 (35) | 252 (39) | 278 (84) | |
ADTKD-UMOD, autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations; BMI, body mass index; ESKD, end-stage kidney disease; WF, Wake Forest.
Chi-squared test.
Fisher’s exact test.
WF genotyped versus historic.
For the international cohort, 349 individuals were screened from 120 affected families. There were 322 genetically affected, 27 genetically unaffected, and 7 historically affected. Table 1 compares characteristics of the 2 cohorts.
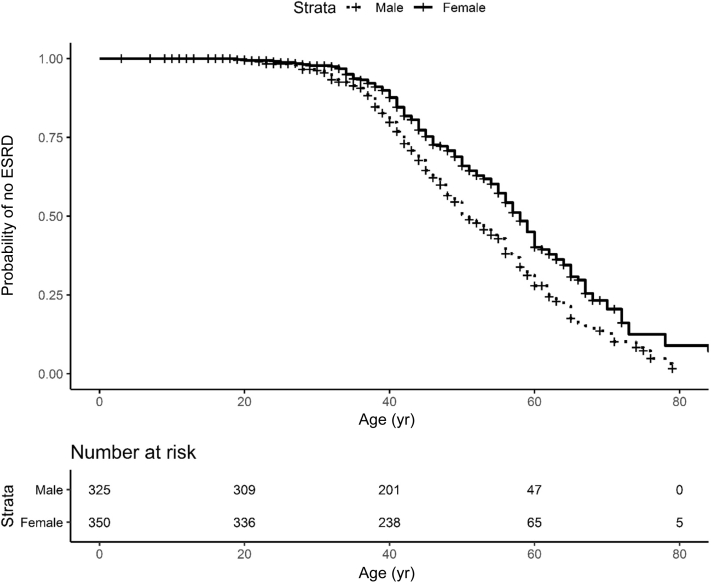
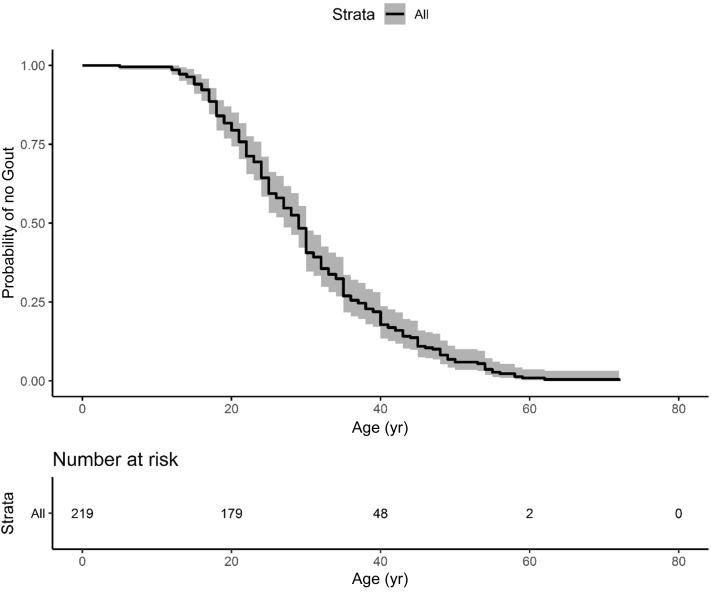
There was a total of 722 individuals from 249 families affected with ADTKD-UMOD, with 125 unique mUMOD mutations, including 28 mutations not previously described. Of the mutations, 117 (94%) were in exon 3, 7 (6%) in exon 4, and 1 in exon 7; 47% of mutations resulted in the loss of a cysteine residue, 6% in the gain of a cysteine residue, and 6% resulted in a gain in hydrophobicity. Characteristics of individuals affected with the most common UMOD mutations are listed in Table 2 and Supplementary Table S1 for all mutations. This table is updated regularly at http://j.mp/2q7Fi8f. The median age of ESKD for the entire cohort was 47 years (range 18–87) and mean age 48.7 ± 12.7 years. Gender had a marked association with earlier age of ESKD (Figure 3), with an odds ratio of 1.78 (P = 0.00028). ESKD was uncommon before age 30, with approximately 50% of the male cohort reaching ESKD between 30 and 50, and 50% of the female cohort reaching ESKD between 30 and 60. In the WF cohort, 55% (245 of 447) of patients with ADTKD-UMOD developed gout. Figure 4 shows a survival curve with event defined as onset of gout for individuals with gout in whom an age of gout onset was known. The median age of gout onset was 28 years, with gout most commonly developing between ages 15 and 40. Of 180 men with information available, 106 (59%) developed gout at a mean age of 29.1 ± 9.9 years. Of 220 women with information available, 96 (44%) developed gout at a mean age of 32.2 ± 12.9 years.
Table 2.
Most common UMOD mutations (mUMOD) with in vitro score and age of ESKD
| mUMOD | In vitro score | Families (n) | Individuals (n) | Median age ESKD | Range of ESKD | Families linked to major variant | Families linked to minor variant | Families with unknown linkage |
|---|---|---|---|---|---|---|---|---|
| p.H177_R185del | 3 | 25 | 173 | 46 | 20–87 | 25 | 0 | 0 |
| p.V93_G97delins AASC |
2 | 11 | 74 | 48 | 27–75 | 10 | 0 | 1 |
| p.R178P | 4 | 9 | 48 | 53 | 39–79 | 8 | 0 | 1 |
| p.C106F | 1 | 11 | 47 | 55 | 35–72 | 11 | 0 | 0 |
| p.C148Y | 4 | 4 | 25 | 44 | 25–66 | 4 | 0 | 0 |
| p.G88D | 1 | 8 | 23 | 65.5 | 55–76 | 7 | 0 | 1 |
| p.P236L | 3 | 4 | 20 | 45 | 43–67 | 4 | 0 | 0 |
| p.C135Y | 4 | 2 | 17 | 41 | 37–47 | 2 | 0 | 0 |
| p.S91del | 2 | 5 | 17 | 50 | 37–66 | 5 | 0 | 0 |
| p.P236R | 4 | 5 | 9 | 40 | 24–50 | 4 | 0 | 1 |
ESKD, end-stage kidney disease.
Range of ESKD is earliest and latest ages of ESKD.
Figure 3.
End-stage kidney disease (ESKD) survival according to gender in autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD). This analysis included 675 individuals with ADTKD-UMOD with known gender and clinical information. An event was defined as starting dialysis, receiving a transplant, or dying of kidney failure. Censoring occurred for death before ESKD or if the individual had not reached ESKD by the end of the study period. ESKD rarely occurred before age 30, with most patients requiring dialysis by age 70. Male gender was associated with an increased risk of reaching ESKD at an earlier age (hazard ratio 0.562, 0.00028). ESRD, end-stage renal disease.
Figure 4.
Survival curve of gout onset in individuals with autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD). This analysis included only 219 individuals who developed gout. An event was defined as age to onset of gout. Gout rarely occurred before age 15, with the vast majority of affected individuals developing gout between the ages of 20 and 40.
Genetic Analysis
Genetic analysis was performed on both the WF and international cohorts, including 929 individuals from 249 families with 125 distinct UMOD mutations. Supplementary Table S1 shows a complete list of the number of individuals with each mutation, the median age and range of ESKD, and the in vitro score. A family was defined as a group of individuals from which 1 individual was referred, and in whom there were no family members known related to other referred families at the time of referral. The most common mUMOD was p.H177_R185del, with 173 individuals and 25 families from the WF cohort. The second most common mUMOD was p.V93_G97delinsAASC with 74 individuals and 11 families. This mutation originated in England.17 The statistical analysis performed (see Methods) adjusted for the large number of family members.
Genotyping of rs4293393 was performed in 633 of 722 genetically affected individuals and in 64 of 207 genetically unaffected individuals (Figure 2). With the aid of long-range sequencing in 18 families, it was possible to determine whether the minor or major allele was in phase with mUMOD in 614 individuals and 227 of 240 families. In 614 of 614 cases, rs4293393 testing results were consistent with inheritance of the same rs4293393-mUMOD haplotype throughout each family. In 348 individuals (92 genetically affected and 256 historically affected) who were not rs4293393 genotyped, the allele in phase with mUMOD was assigned based on familial results.
Distribution of the rs4293393 SNP Variant in Families With ADTKD-UMOD
When performing a Mendelian randomization study, one must first determine that the genetic variant under study is randomly distributed in the population being investigated. Approximately 19% of the European population has the minor rs4293393 allele (G) (defined as the minor allele frequency [MAF]).14 In patients with ADTKD-UMOD, for the rs4293393 allele inherited from the unaffected parent, the MAF was 0.17, which is similar to reference populations (Table 3). We then sought to determine if the rs4293393 minor variant linked to the mUMOD allele was distributed in the same proportion as the general population. As some of the families under study might be distantly related, we assumed that all individuals with the same rs4293393-mUMOD haplotype were one family. Using this approach, there were 123 families with the major rs4293393 allele linked to mUMOD and 24 families with the minor rs4293393 allele linked to mUMOD. For 9 mUMODs, there were families with the minor allele-mUMOD haplotype and also families with the major allele-mUMOD haplotype. For the rs4293393 variant linked to -mUMOD, the MAF was only 0.12 versus the MAF of 0.19 to 0.20 in the TOPMED, gnomAD, and 1000 genome populations, which statistically deviated from expected proportions (P = 0.0037, Table 3). In other words, the SNP minor allele, associated with lower uromodulin production, is underrepresented when associated with mUMOD mutation, whereas this is not the case when it is associated with a wild-type UMOD allele. Given that the rs4293393 variant was not randomly distributed in this population, a Mendelian randomization experiment could not be performed.
Table 3.
Comparison of minor allele frequencies of the rs42993393 SNP in phase and out of phase with the mutated UMOD (mUMOD)
| Testing | Population | n | Observed MAF (minor, major) | Comparison database | MAF in comparison group | P value |
|---|---|---|---|---|---|---|
| rs4293993 in phase with mUMOD | 1 per haplotype present in cohort | 129a | 0.1163 (G, A) | TOPMED | 0.18639 | 0.0147 |
| gnomAD | 0.1924 | 0.0082 | ||||
| 1000 genomes | 0.20 | 0.0037 | ||||
| rs4293993 out of phase with mUMOD | UMOD genotyped, rs4293393 genotyped | 554 | 0.1645 (G, A) | TOPMED | 0.18639 | 0.2693 |
| gnomAD | 0.1924 | 0.1629 | ||||
| 1000 Genomes | 0.20 | 0.07958 |
MAF, minor allele frequency.
Conservative testing was used, in which it was assumed that all individuals with the same rs4293393-mUMOD haplotype were related. The MAF of the test population was compared with the MAF found in TOPMED, gnomAD, and 1000 genomes registries.18 All comparisons showed that the MAF deviates from expected population frequencies. The MAF was then determined in available samples for the UMOD allele that was inherited from the unaffected parents. This allele was found to have similar allele frequencies as the control populations.
There were 129 unique rs4293393/mUMOD haplotypes.
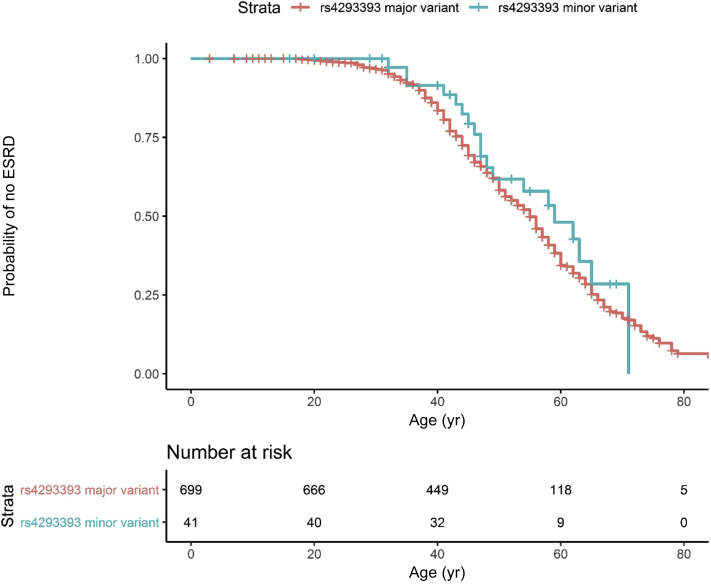
ESKD Survival by rs4293393 Allele
Figure 5 shows the ESKD survival curves for individuals according to the rs4293393-mUMOD haplotype. There were 699 (94.5%) genotyped and historic individuals with the major allele (A) in phase with mUMOD and 41 (5.5%) genotyped and historic individuals with the minor rs4293393 allele (G) in phase with mUMOD. A univariate model (Table 4) was underpowered because of the presence of only 41 individuals with the minor allele phased with mUMOD.
Figure 5.
Survival curves according to the rs4293393 allele in phase with mutant UMOD (mUMOD). This analysis included 668 individuals with autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD) with clinical information available in addition to determined rs4293393-mUMOD haplotype. An event was defined as end-stage kidney disease (ESKD) by starting dialysis, receiving a transplant, or dying of kidney failure. Censoring occurred for death before ESKD or if the individual had not reached ESKD by the end of the study period. There were only 41 individuals with the minor allele (G) in phase with mUMOD, resulting in insufficient power to detect a difference in survival. ESRD, end-stage renal disease.
Table 4.
Univariate models for individuals with ADTKD-UMOD
| Parameter | Observations (n) | Events | Reference category | Hazard ratio | C-statistic | P value |
|---|---|---|---|---|---|---|
| Gender | 675 | 342 | Male | 1.78 | 0.556 | 0.00028 |
| rs4293393 minor variant in phase with mUMOD | 668 | 337 | A (major) | 0.6885 | 0.507 | 0.36 |
| rs4293393 minor variant in phase with wtUMOD | 494 | 221 | A (major) | 0.6152 | 0.529 | 0.064 |
| rs4293393 variant (at least 1 G allele present) | 499 | 224 | 2 A alleles present | 0.6943 | 0.528 | 0.13 |
| mUMOD type | 675 | 342 | p.(H177_R185del) | 0.561 | ||
| Cysteine gain | 0.9234 | 0.88 | ||||
| Cysteine loss | 0.9995 | 1.0 | ||||
| Deletion/insertion | 0.6369 | 0.3 | ||||
| Hydrophobic amino acid gain | 0.3197 | 0.066 | ||||
| Proline gain | 0.5321 | 0.13 | ||||
| Other | 0.6155 | 0.15 | ||||
| In vitro score | 393 | 198 | 1.5457 | 0.591 | 0.0022 | |
| Cysteine-rich domains | 675 | 342 | All other domains | 0.8247 | 0.521 | 0.38 |
ADTKD-UMOD, autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations; mUMOD, mutant uromodulin; wtUMOD, wild-type uromodulin.
Data combine Wake Forest and International cohorts.
Scoring of UMOD Mutations
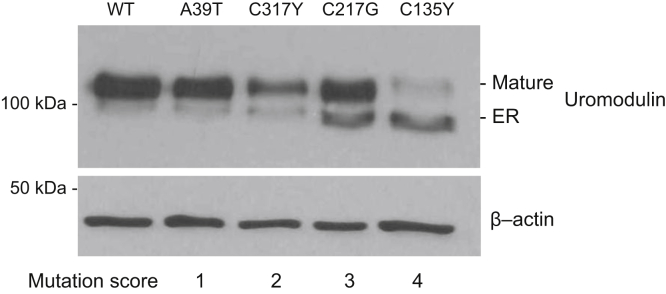
The trafficking of 35 mUMOD isoforms was characterized in vitro (Figure 6, Table 2, Supplementary Table S1, and Supplementary Figures S1 and S2). MDCK and/or HEK293 cell lines were transfected with expression vectors coding for different mUMODs. As previously described,8 Western blot revealed 2 bands: a lower molecular weight glycoform corresponding to the uromodulin precursor in the ER that carries Endo H-sensitive N-glycans, and a higher molecular weight glycoform that carries post-Golgi, Endo H-resistant type of glycans. The higher molecular weight form corresponds to fully glycosylated, mature protein that proceeded along the secretory pathway into post-Golgi compartments (trans-Golgi network, secretory vesicles, plasma membrane). UMOD mutation scoring was performed by quantifying the ratio between low- and high-molecular weight uromodulin glycoforms in cell lysates, as a measure of trafficking defect. We normalized these values to the ratio obtained for the well-characterized, paradigm mutation C150S that consistently shows strong ER retention. Mutations were then subdivided into 4 distinct subgroups based on the generated ratio before statistical correlation with age of ESKD (Figure 6).
Figure 6.
Representative Western blot results from mutant UMOD (mUMOD) in vitro experiments (Supplementary Methods). Western blot analysis of HEK cells stably expressing wild-type (WT) or mutant uromodulin isoforms. An in vitro score reflecting the severity of trafficking defect was assigned to each mutation. The scoring was performed by quantifying the ratio between endoplasmic reticulum (ER)–retained and mature uromodulin glycoforms in cell lysates.
The different cellular phenotypes of mUMOD forms and their severity were reproducible between independent experiments (Supplementary Figure S1) and conserved when expressed in different cell lines (e.g., HEK293 vs. MDCK) (Figure 6 and Supplementary Figure S3), regardless of the expression system (transient or stable transfection) (Supplementary Figure S4).
UMOD Mutation Type and ESKD Survival
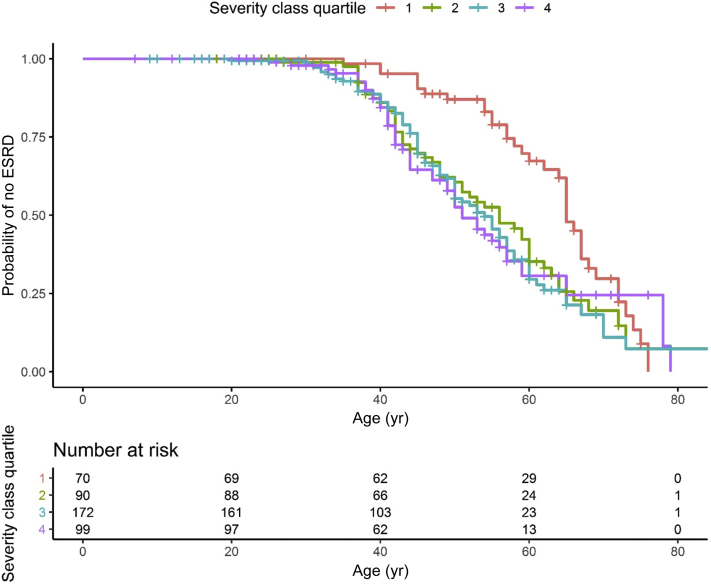
In univariate models, we found no difference in survival for the different mutation types but did show an association with the in vitro score (Table 5 and ESKD survival curve in Figure 7). For individuals with mUMOD mutations that less hindered uromodulin export to the cell surface (Group 1), renal survival was significantly improved.
Table 5.
Mean age of end-stage kidney disease in individuals with ADTKD-UMOD according to in vitro score
| In vitro score | Observations (n) | Age (mean ± SD) |
|---|---|---|
| 1 | 42 | 59.4 ± 11.0 |
| 2 | 56 | 50.4 ± 11.0 |
| 3 | 90 | 48.5 ± 12.1 |
| 4 | 59 | 47.2 ± 11.7 |
| Not available | 179 | 46.3 ± 13.1 |
ADTKD-UMOD, autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations.
Data include only individuals who reached end-stage kidney disease.
Figure 7.
End-stage kidney disease (ESKD) survival in individuals with autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations (ADTKD-UMOD) according in vitro score. This analysis included 393 individuals with 1 of the 35 UMOD mutations receiving an in vitro score. An event was defined as ESKD by starting dialysis, receiving a transplant, or dying of kidney failure. Censoring occurred for death before ESKD or if the individual had not reached ESKD by the end of the study period. ESRD, end-stage renal disease.
Multivariate Model
A multivariate model was then created based on the genetic factors present in both cohorts. In the multivariate model, the presence of gender and in vitro score provided the highest correlation (Table 6). We did not find that the type of mutation was predictive of age of ESKD (data not shown). There were no significant interaction terms.
Table 6.
Best-fit multivariate model for individuals with ADTKD-UMOD using gender and in vitro score
| Parameter | Observations (n) | Events | Reference category | Hazard ratio | C-statistic | P value |
|---|---|---|---|---|---|---|
| Gender | 393 | 198 | Male | 0.53 | 0.61 | 0.002 |
| In vitro score | 1.5 | 0.00085 |
ADTKD-UMOD, autosomal dominant tubulo-interstitial kidney disease due to UMOD mutations.
Analysis of Genetic and Clinical Factors for the WF Cohort
Analysis of clinical and genetic factors was performed on the WF cohort (Table 7). The presence of gout was not associated with age of ESKD; however, a younger age of gout for individuals who developed gout was highly correlated with a younger age of ESKD (P < 0.0001). In univariate analysis (Table 7), maternal age was highly associated with survival (P = 0.0017), whereas the paternal age was not (P = 0.23). When looking at subgroups, daughter and maternal age of ESKD were most highly correlated, followed by son and maternal age of ESKD (Supplementary Figures S1 and S2).
Table 7.
Univariate models for clinical characteristics for the WF cohort
| Parameter | Observations (n) | Events | Reference category | Hazard ratio | C-statistic | P value |
|---|---|---|---|---|---|---|
| Weight | 301 | 130 | 0.9967 | 0.545 | 0.23 | |
| BMI | 300 | 130 | 0.9617 | 0.564 | 0.069 | |
| Smoking (active or former) | 337 | 197 | Nonsmoker | 0.7185 | 0.533 | 0.17 |
| Gout (y/n) | 417 | 178 | No | 1.2549 | 0.52 | 0.29 |
| Age of gout onset | 209 | 110 | 0.9452 | 0.666 | 0.0000053 | |
| Parental age ESKD | 229 | 101 | 0.9701 | 0.629 | 0.0045 | |
| Family mean age ESKD | 429 | 217 | 0.9706 | 0.619 | 0.009 | |
| Mother’s age ESKD | 111 | 53 | 0.9593 | 0.667 | 0.0017 | |
| Father’s age ESKD | 114 | 47 | 0.9816 | 0.559 | 0.23 |
BMI, body mass index; ESKD, end-stage kidney disease; WF, Wake Forest.
Multivariate clinical and genetic/clinical models were then created using the WF cohort (Table 8). The combination of gender and the family mean age of ESKD were found to be the best predictors of survival. There were no significant interaction terms.
Table 8.
Multivariate model for the Wake Forest cohort
| Parameter | Observations (n) | Events | Reference category | Hazard ratio | C-statistic | P value |
|---|---|---|---|---|---|---|
| Gender | 429 | 217 | Male | 0.6380 | 0.634 | 0.0054 |
| Family mean age ESKD | 0.9591 | <0.0001 |
ESKD, end-stage kidney disease.
Discussion
This is the largest study that has been performed in individuals with ADTKD-UMOD and is the result of a large multinational collaboration that included 13 different research groups. The age of ESKD onset in individuals with ADTKD-UMOD ranges from 20 to >70, and the goal of this study was to identify factors associated with the highly variable age of ESKD onset. The principal findings of the study included the following: (i) the rs4293393 SNP in the mutant UMOD promoter was not randomly distributed in families with ADTKD-UMOD, with the minor allele (G) postulated to produce decreased mUMOD significantly underrepresented. (ii) An in vitro score that measured the effect of the specific mUMOD mutation on uromodulin trafficking was significantly correlated with kidney survival. (iii) Women had significantly better renal survival than men (hazard ratio 1.78, P < 0.001). (iv) The maternal age of ESKD was significantly associated with the child’s age of ESKD, particularly for women; however, there remained significant variation. (v) A younger age of gout was associated with a younger age of ESKD. (vi) Twenty-eight new mutations were described.
A major finding of the study was that the MAF of the rs4293393 minor variant linked to mUMOD was only 11% in affected families as opposed to the 18% to 20% found in the general population (P < 0.001) and as opposed to the observed MAF of the rs4293393 allele in phase with wild-type UMOD allele (17%). We postulate that the decreased allele frequency is due to decreased mUMOD expression and hence a milder form of ADTKD-UMOD associated with later age of ESKD onset. Families with a later age of ESKD onset would be less likely to be identified with ADTKD-UMOD and less likely to be referred for evaluation and potential entry into a disease registry. Because of the rs4293393 MAF of 19% in the general population, we anticipated the need to collect a large number of affected individuals for the study. The further decrease in allele frequency resulted in a marked decrease in the number of individuals with the minor allele and hence a loss of statistical power for the study. As the rs4293393 minor SNP variant was not randomly distributed in the ADTKD-UMOD population, we were not able to perform a Mendelian randomization study. We did not find a difference in survival between families with major versus minor rs4293393-mUMOD haplotypes, which may have been the result of lack of statistical power, the fact that there was no difference, or the fact that families with milder disease due to minor allele rs4293393-mUMOD haplotypes were undetected.
We found that mutation class (e.g., loss or gain of polarity or a cysteine residue) was not correlated with the age of ESKD. This absence of correlation may have been because of the conservative statistical analysis, which adjusted for the large number of individuals in different families. An in vitro score reflecting the severity of the trafficking defect caused by specific mUMOD mutations was found to be a promising predictor of the age of ESKD. In addition, this finding points to enhanced transit of mUMOD through the ER as a potential therapeutic approach. Supplementary Table S1 provides information on all in vitro scores, as well as the median age of ESKD for each mutation. Based on the in vitro score, we would predict that the type of mutation, but not the class of amino acid substitution (e.g., cysteine vs. noncysteine substitution) does have an effect on ESKD survival. Other investigators have also studied the interaction between genotype and phenotype in ADTKD-UMOD. Bollee and colleagues3 reported on 109 patients from 45 families with 37 distinct UMOD mutations and a median age of ESKD of 54. These authors found a high intrafamilial variability in the age of ESKD, with only a modest, nonsignificant effect of the type of mutation on survival. In a review of the literature, Moskowitz and colleagues4 identified 202 patients from 74 families with 59 different UMOD mutations and a median age of ESKD of 56. Onset of ESKD was significantly earlier with mutations in the epidermal growth factor domains 2 and 3 (range 45–52 years) compared with the cysteine-rich domains (range 60–65 years) using a shared frailty model. The in vitro score is novel and should be considered a research test at this time. Further development and validation are required to assess its relevance as a clinical test that could be useful in predicting the age of ESKD for individuals with de novo mutations or from smaller families in which the age of ESKD is not well characterized.
Male gender was a significant predictor of worse renal outcomes, with a hazard ratio of 1.78, P < 0.001. This was similar to the finding of Moskowitz et al.4 of an increased risk of ESKD in men (hazard ratio 2.09, P = 0.04) in their cohort.
As part of this study, we were able to produce a catalog of UMOD mutations, including the median age of ESKD onset and in vitro score, which is included in Supplementary Table S1 and available in an updated form online at http://j.mp/2q7Fi8f. We believe that this information will be helpful to clinicians working with families with ADTKD-UMOD.
A primary weakness of this article was the lack of power to better detect statistical differences among groups, a major obstacle in the study of rare diseases. Another weakness was the retrospective nature of the study and limited clinical data from the international cohort. For instance, information on gout was missing from many of the historically affected individuals, and this may have affected our ability to identify significant findings. Formation of a registry of individuals with ADTKD who contribute genetic samples, as well as clinical information, will be helpful to overcome this obstacle in the future. In addition, we were not able to explain fully the interfamilial and intrafamilial variation in the age of ESKD onset.
In summary, we studied genetic and clinical factors associated with the age of ESKD onset. An in vitro score of mUMOD transit was a predictor of the age of onset of ESKD, as was the presence of gout, age of gout onset, and parental age of ESKD. The rs4293393 UMOD minor allele, associated with decreased uromodulin production, was underrepresented in families with ADTKD-UMOD.
Disclosure
All the authors declared no competing interests.
Acknowledgments
We thank all participating patients and families, and the referring physicians. We acknowledge Sebastiano Regina (San Luigi Gonzaga University Hospital) and Alessandra Cuccurullo (University of Turin) for genotyping, Alessandra Pelle (University of Turin) for genetic counseling, and Elena Pasqualetto (San Raffaele Scientific Institute) for in vitro studies.
This study was funded by National Institutes of Health (NIH)–National Institute of Diabetes and Digestive and Kidney Diseases R21 DK106584. Wake Forest also thanks the Black-Brogan Foundation for support. YMC was supported by NIH grants R01 DK105056A1, R03DK106451, and K08DK089015; The Assistant Secretary of Defense for Health Affairs endorsed by the Department of Defense, Award Number W81XWH-19–1–0320. PV, MŽ, and SK were supported by grant NV17–29786A from the Ministry of Health of the Czech Republic and by institutional programs of Charles University in Prague (UNCE/MED/007 and PROGRES-Q26/LF1); they thank The National Center for Medical Genomics (LM2015091) for help in genotyping. EO is supported by the Fonds National de la Recherche Luxembourg (6903109). OD is supported by the European Reference Network for Rare Kidney Diseases (ERKNet), project ID No. 739532; the National Centre for Competence in Research Kidney CH program; and the Swiss National Science Foundation 310030–189044. LR was supported by the Italian Society of Nephrology (SIN) under the “Adotta un progetto di ricerca” program, Telethon-Italy (GGP14263); the Ministry of Health of Italy (grant RF-2010–2319394 and RF-2016–02362623), Soli Deo Gloria.
Footnotes
Table S1. Clinical characteristics according to UMOD mutation.
Figure S1. Subgroup comparison of parental age of ESKD versus child’s age of ESKD.
Figure S2. Histogram of difference in years between daughter’s age of ESKD versus mother’s age of ESKD.
Figure S3. Western blot analysis of the indicated uromodulin mutant isoforms stably expressed in MDCK cells.
Figure S4. Western blot analysis of the indicated uromodulin mutant isoforms transiently expressed in MDCK cells.
Supplementary Methods. Genetic evaluation and UMOD mutational sequencing, rs4293393 genotyping, rs4293393 and UMOD mutation phase determination, and in vitro score determination
Supplementary References.
Contributor Information
Luca Rampoldi, Email: rampoldi.luca@hsr.it.
Anthony J. Bleyer, Email: ableyer@wakehealth.edu.
Supplementary Materials
References
- 1.Bleyer AJ, Hart PS, Kmoch S. Autosomal dominant tubulointerstitial kidney disease, UMOD-related. 2016. GeneReviews [Internet]. Seattle, WA: University of Washington, Seattle, 1993-2020.
- 2.Bleyer A.J., Woodard A.S., Shihabi Z. Clinical characterization of a family with a mutation in the uromodulin (Tamm-Horsfall glycoprotein) gene. Kidney Int. 2003;64:36–42. doi: 10.1046/j.1523-1755.2003.00081.x. [DOI] [PubMed] [Google Scholar]
- 3.Bollee G., Dahan K., Flamant M. Phenotype and outcome in hereditary tubulointerstitial nephritis secondary to UMOD mutations. Clin J Am Soc Nephrol. 2011;6:2429–2438. doi: 10.2215/CJN.01220211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moskowitz J.L., Piret S.E., Lhotta K. Association between genotype and phenotype in uromodulin-associated kidney disease. Clin J Am Soc Nephrol. 2013;8:1349–1357. doi: 10.2215/CJN.11151012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devuyst O., Olinger E., Weber S., Eckardt K.U. Autosomal dominant tubulointerstitial kidney disease. Nat Rev Dis Primers. 2019;5:60. doi: 10.1038/s41572-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 6.Vylet’al P., Kublova M., Kalbacova M. Alterations of uromodulin biology: a common denominator of the genetically heterogeneous FJHN/MCKD syndrome. Kidney Int. 2006;70:1155–1169. doi: 10.1038/sj.ki.5001728. [DOI] [PubMed] [Google Scholar]
- 7.Scolari F., Caridi G., Rampoldi L. Uromodulin storage diseases: Clinical aspects and mechanisms. Am J Kidney Dis. 2004;44:987–999. doi: 10.1053/j.ajkd.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Bernascone I., Vavassori S., Di P.A. Defective intracellular trafficking of uromodulin mutant isoforms. Traffic. 2006;7:1567–1579. doi: 10.1111/j.1600-0854.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 9.Rampoldi L., Caridi G., Santon D. Allelism of MCKD, FJHN and GCKD caused by impairment of uromodulin export dynamics. Hum Mol Genet. 2003;12:3369–3384. doi: 10.1093/hmg/ddg353. [DOI] [PubMed] [Google Scholar]
- 10.Williams S.E., Reed A.A., Galvanovskis J. Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum Mol Genet. 2009;18:2963–2974. doi: 10.1093/hmg/ddp235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahan K., Devuyst O., Smaers M. A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol. 2003;14:2883–2893. doi: 10.1097/01.asn.0000092147.83480.b5. [DOI] [PubMed] [Google Scholar]
- 12.Serafini-Cessi F., Malagolini N., Hoops T.C., Rindler M.J. Biosynthesis and oligosaccharide processing of human Tamm-Horsfall glycoprotein permanently expressed in HeLa cells. Biochem Biophys Res Commun. 1993;194:784–790. doi: 10.1006/bbrc.1993.1890. [DOI] [PubMed] [Google Scholar]
- 13.Raffi H., Bates J.M., Laszik Z., Kumar S. Tamm-Horsfall protein knockout mice do not develop medullary cystic kidney disease. Kidney Int. 2006;69:1914–1915. doi: 10.1038/sj.ki.5000411. [DOI] [PubMed] [Google Scholar]
- 14.Trudu M., Janas S., Lanzani C. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med. 2013;19:1655–1660. doi: 10.1038/nm.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vyletal P., Bleyer A.J., Kmoch S. Uromodulin biology and pathophysiology - an update. Kidney Blood Press Res. 2010;33:456–475. doi: 10.1159/000321013. [DOI] [PubMed] [Google Scholar]
- 16.Bleyer A.J., Kidd K., Robins V. Outcomes of patient self-referral for the diagnosis of several rare inherited kidney diseases. Genet Med. 2019;22:142–149. doi: 10.1038/s41436-019-0617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith G.D., Robinson C., Stewart A.P. Characterization of a recurrent in-frame UMOD indel mutation causing late-onset autosomal dominant end-stage renal failure. Clin J Am Soc Nephrol. 2011;6:2766–2774. doi: 10.2215/CJN.06820711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczewski K.J.F., Francioli L.C., Tiao G. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding regions. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.