Summary
The Wnt/β-catenin signaling pathway is a key regulator of embryonic stem cell (ESC) self-renewal and differentiation. Constitutive activation of this pathway has been shown to increase mouse ESC (mESC) self-renewal and pluripotency gene expression. In this study, we generated a novel β-catenin knockout model in mESCs to delete putatively functional N-terminally truncated isoforms observed in previous knockout models. We showed that aberrant N-terminally truncated isoforms are not functional in mESCs. In the generated knockout line, we observed that canonical Wnt signaling is not active, as β-catenin ablation does not alter mESC transcriptional profile in serum/LIF culture conditions. In addition, we observed that Wnt signaling activation represses mESC spontaneous differentiation in a β-catenin-dependent manner. Finally, β-catenin (ΔC) isoforms can rescue β-catenin knockout self-renewal defects in mESCs cultured in serum-free medium and, albeit transcriptionally silent, cooperate with TCF1 and LEF1 to inhibit mESC spontaneous differentiation in a GSK3-dependent manner.
Keywords: mESCs, Wnt, β-catenin, Ctnnb1, CRISPR, self-renewal, pluripotency, TCF, LEF, embryonic stem cells
Highlights
-
•
N-terminally truncated β-catenin isoforms are produced in mESCs upon inducible knockout
-
•
β-Catenin is fully deleted upon CRISPR/Cas9 whole-gene knockout
-
•
Wnt/β-catenin prevents differentiation without affecting pluripotency genes
-
•
β-Catenin/TCF/LEF functions are required to prevent spontaneous differentiation
In this article, Cosma and colleagues show that previously published inducible β-catenin knockout models produce N-terminally truncated isoforms in mouse embryonic stem cells (mESCs). Complete β-catenin knockout was obtained by CRISPR/Cas9 whole-gene deletion. With the newly generate knockout ESC model it was observed that β-catenin/TCF/LEF functions are required to prevent spontaneous differentiation of mESCs.
Introduction
β-Catenin regulates different cellular processes spanning from development to cancer progression. In addition to its central role in adherens junctions, β-catenin is the key effector of the canonical Wnt signaling pathway. Exposure to canonical Wnt ligands, such as WNT3A or small-molecule inhibitors of GSK3 activity, triggers β-catenin stabilization and its nuclear translocation. In the nucleus, β-catenin acts as a scaffolding protein for transcriptional co-factors such as TCF/LEF family members, thereby activating the expression of Wnt target genes. Over the past years, accumulating evidence highlighted a key role for Wnt/β-catenin signaling in sustaining self-renewal, pluripotency, and cell-cycle progression of mouse embryonic stem cells (mESCs) (De Jaime-Soguero et al., 2017; Sato et al., 2004; Ten Berge et al., 2011) and in regulating somatic cell reprogramming (Aulicino et al., 2014; Lluis et al., 2008, 2011; Marucci et al., 2014). Despite its importance in mESC physiology, β-catenin knockout mESC lines developed so far show no defects in self-renewal or pluripotency marker expression when cultured in a medium containing serum plus the leukemia inhibitory factor (LIF), but display a strict LIF requirement for pluripotency maintenance and fail to efficiently differentiate in vitro (Lyashenko et al., 2011; Wray et al., 2011). Surprisingly, LIF dependency can be rescued by transcriptionally defective β-catenin isoforms (ΔC mutants), challenging the hypothesis that β-catenin transcriptional activity could be relevant for mESC pluripotency and self-renewal (Lyashenko et al., 2011; Wray et al., 2011). More recently, however, it has been demonstrated that the most widely used inducible β-catenin knockout alleles lead to the production of uncharacterized N-terminally truncated β-catenin isoforms (ΔN β-cat) during pre-implantation embryo development (Messerschmidt et al., 2016).
Here, we confirmed the production of ΔN β-cat isoforms in mESCs and generated a novel β-catenin full knockout model in mESCs using CRISPR/Cas9. We found that complete β-catenin deletion produces similar phenotypes observed in the previously described knockout models retaining ΔN β-cat fragments, suggesting that N-terminally truncated isoforms are biologically inactive. We furthermore analyzed the impact of β-catenin loss at transcriptomic level, in presence or absence of GSK3 chemical inhibition. Our results show that the Wnt/β-catenin pathway is not transcriptionally active in mESCs cultured in serum/LIF. However, upon GSK3 inhibition, we observed β-catenin-dependent inhibition of differentiation markers, while the expression of pluripotency genes remained unchanged. Finally, we showed that transcriptionally impaired C-terminally truncated β-catenin rescue isoforms (ΔC β-cat) can inhibit mESC differentiation in the absence of LIF when GSK3 is inhibited, as their full-length counterpart. However, this phenotype is impaired upon silencing of TCF1/LEF1, suggesting that ΔC β-cat isoforms are not entirely transcriptionally silent and their nuclear function could depend on TCF/LEF factors.
Results
Inducible β-Catenin Knockout Alleles Generate N-Terminally Truncated Isoforms in mESCs
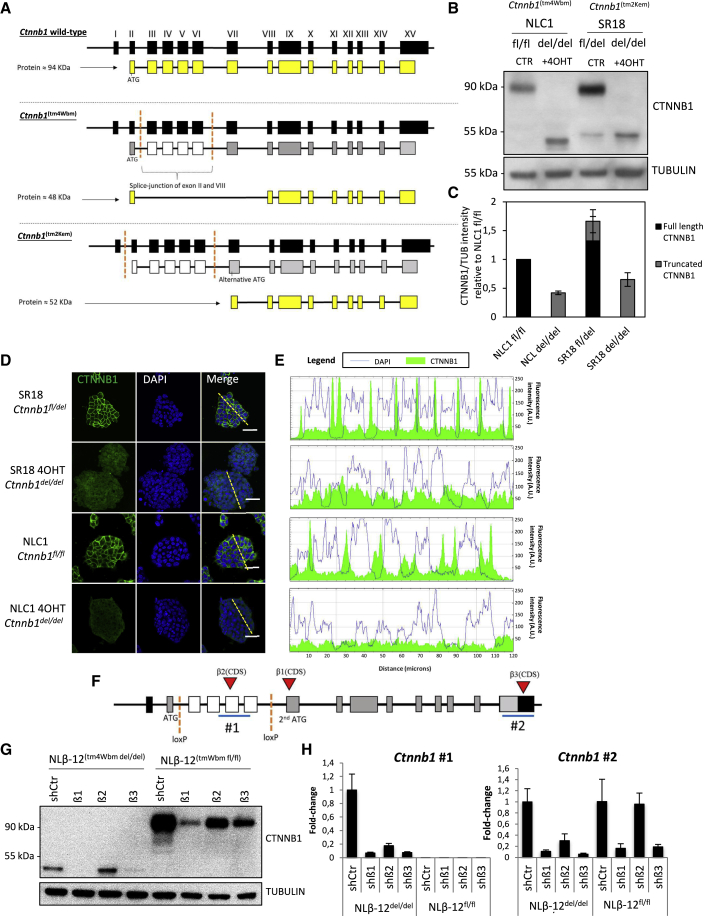
β-Catenin (Ctnnb1) mRNA includes 15 exons, with an open reading frame (ORF) spanning from exon 2 to exon 15. Previously reported β-catenin knockout models in mESCs were generated using CRE-mediated excision of a DNA fragment flanked by two LoxP sites encompassing exons 3–6 (Ctnnb1tm4Wbm) or 2–6 (Ctnnb1tm2Kem). Three groups have independently studied both alleles in mESCs and concluded that β-catenin is dispensable for mESC self-renewal (Lyashenko et al., 2011; Raggioli et al., 2014; Wray et al., 2011).
A recent study reported that, during pre-implantation embryo development, both these β-catenin knockout alleles are flawed by the production of N-terminally truncated (ΔN) proteins, possibly generated by alternative splicing (Ctnnb1tm4Wbm) or by a secondary ATG within a Kozak consensus sequence downstream of the excised region (Ctnnb1tm2Kem) (Figure 1A) (De Vries et al., 2004; Messerschmidt et al., 2016). Remarkably the production of these ΔN β-cat isoforms in mESCs has not been previously reported (Anton et al., 2007; Lyashenko et al., 2011; Wagner et al., 2010; Wray et al., 2011). We therefore tested the expression of ΔN β-cat isoforms by using an antibody raised against the C-terminal portion of β-catenin in protein extracts of NLC1 and SR18 mESCs, which harbor the Ctnnb1tm4Wbm fl/fl and Ctnnb1tm2Kem fl/del alleles, respectively, and stably express the CRE-ERT2. Full-length β-catenin was successfully excised upon 4-hydroxytamoxifen (4OHT) treatment in both cell lines, and ΔN β-cat isoforms with a molecular weight of approximately 48 and 52 kDa, respectively, were detected upon CRE recombination (Figures 1B and 1C). Immunofluorescence staining did not show any clear subcellular localization of ΔN isoforms, which instead appeared distributed among cytoplasm and nuclei showing little or no membrane localization (Figures 1D and 1E).
Figure 1.
Inducible β-Catenin Knockout Alleles Produce N-Terminally Truncated Isoforms in mESCs
(A) Schematic representation of murine β-catenin (Ctnnb1) locus and the two loxP alleles used for β-catenin studies in mESCs. Black boxes represent exons, yellow boxes coding exons, dashed red lines indicate loxP sites, and white boxes represent exons excised upon CRE-mediated recombination of loxP sites.
(B and C) Western blot (B) and relative quantification (C) of NLC1 and SR18 cell lines upon 72-h 4′-hydroxytamoxifen treatment (+4OHT) and respective untreated controls (CTRs). SR18 untreated cell line is heterozygous for full-length β-catenin deletion. Western blot band intensities (C) are normalized on NLC1 full-length CTTNB1.
(D) β-Catenin immunofluorescence staining on fixed SR18 or NLC1 parental cell lines or upon 72-h +4OHT treatment. A primary antibody raised against the C-terminal portion of β-catenin was used. DAPI was used to counterstain nuclei. Scale bar represents 50 μm.
(E) Multichannel fluorescence intensity measurement of immunofluorescence images in (D). Image quantification has been performed across the dashed yellow line depicted in (D), merge panel.
(F) Schematic representation of short-hairpin targeted regions (red triangles, β1, β2, and β3) and qRT-PCR amplicons (blue lines, #1 and #2) along the Ctnnb1Tm4Wbm allele.
(G) Western blot of β-catenin of mESCs harboring the Birchmeier β-catenin allele after (Ctnnb1Tm4Wbmdel/del; left) or before (Ctnnb1Tm4Wbm fl/fl; right) CRE-mediated recombination of the loxP sites. Cells were transduced with a control short hairpin (shCtr) or three different short hairpins against β-catenin mRNA (β1, β2, or β3).
(H) qRT-PCR on total mRNA extracts of Ctnnb1Tm4Wbm fl/fl or Ctnnb1Tm4Wb del/del cells transduced with the short-hairpin constructs used in Figure 1D. Two different amplicons were amplified to monitor deleted region (Ctnnb1 #1) or 3′ UTR (Ctnnb1 #2). GAPDH was used as housekeeping control. Error bars represents standard deviation of technical triplicates.
To assess if N-terminally truncated isoforms were a product of the Ctnnb1 genomic locus and not a mere technical artifact, we designed three different short hairpins against three different regions of β-catenin mRNA (β1-3) (Figure 1F) to silence β-catenin in both knockout (del/del) and parental (fl/fl) mESCs (NLβ-12tm4Wbm background). A scrambled short hairpin (shSCR) was used as control. All the short hairpins successfully induced knockdown of β-catenin at protein level (Figure 1G) in wild-type cells, while N-terminally truncated isoforms were depleted efficiently by β1 and β3 but not by β2, which targets the portion of mRNA excised upon CRE-mediated recombination. The same results were confirmed analyzing β-catenin mRNA levels by qRT-PCR using two different primer pairs targeting the CRE-excised region (Figure 1H, left panel), and the 3′ UTR (Figure 1H, right panel). The oligonucleotides targeting the excised region failed to detect β-catenin mRNA in knockout cells, while the ones designed on the 3′ UTR revealed that mRNA regions downstream of the excision had comparable expression levels in wild-type (NLβ-12 fl/fl shSCR) and knockout (NLβ-12 fl/fl del/del shSCR) cells and were efficiently silenced only by β1 and β3 but not, as expected, by β2 (Figure 1H). Importantly, we never observed ΔN isoforms in wild-type cells, including cells expressing short hairpins targeting β-catenin, indicating that they are only produced upon recombination of Ctnnb1 locus.
Next, we characterized SR18 cells (Ctnnb1tm2Kem fl/del) upon β-catenin CRE-mediated deletion (Raggioli et al., 2014). Upon 4OHT treatment and consequent β-catenin deletion, clone morphology and alkaline phosphatase (AP) expression (Figure S1A), as well as Nanog and Oct4 expression patterns (Figure S1B), remained unaltered, confirming earlier results (Lyashenko et al., 2011; Wray et al., 2011). The lack of morphological changes upon β-catenin deletion was likely due to compensatory effects by the upregulation of PLAKOGLOBIN (Figure S1C), as reported by Lyashenko et al. (2011) and Wray et al. (2011). Furthermore, no changes were detected in the protein levels of pluripotency markers (NANOG, OCT4, SOX2) (Figures S1D and S1E). Although these results show that β-catenin loss does not affect morphology and pluripotency markers (confirming results in Lyashenko et al., 2011; Wray et al., 2011), we asked whether the ΔN isoforms, produced by Ctnnb1tm2Kem or the Ctnnb1tm4Wbm alleles, could be responsible for the maintenance of the observed mESC phenotype.
Generation of a New β-catenin Knockout Model in mESCs Using CRISPR/Cas9 Technology
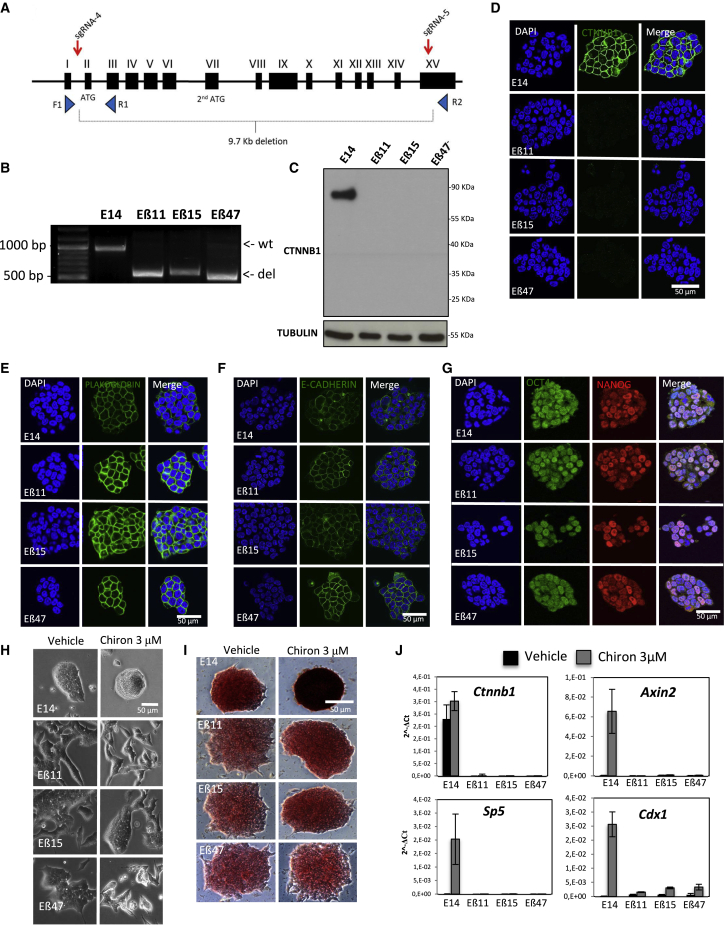
We generated a full β-catenin knockout mESC line via CRISPR/Cas9 technology. As the presence of ΔN β-catenin isoforms could be due either to an alternative splicing or to the presence of a secondary ATG on exon VII (Figure 1A), we designed two different single-guide RNA (sgRNA) pairwise combinations (sgRNA1+sgRNA3 and sgRNA2+sgRNA3, resulting in 5,287-bp and 4,970-bp deletion, respectively, Figure S1F) to induce deletions spanning both alternative splicing sites and the entire exon VII. While the editing efficiency of sgRNA2+sgRNA3 was low, sgRNA1+sgRNA3 induced the deletion in a high percentage of cells (Figure S1G) with a significant loss of full-length β-catenin protein in the pool of transfected cells (Figures S1H and S1I). The deletion generated by sgRNA1+sgRNA3 resulted in the production of a new ΔN isoform with a lower molecular weight (40 kDa) with respect to the N-terminally truncated isoforms produced by Ctnnb1tm2Kem (48 kDa) or the Ctnnb1tm4Wbm (52 kDa) alleles (Figure S1H). The generation of this new ΔN isoform could only be explained by the presence of alternative downstream ATG sites, which, however, were not predictable a priori.
In order to eliminate any possible undesired gene-editing product, we sought to induce a 10-kb deletion encompassing the whole gene body. For this aim, we used an additional couple of sgRNAs (sgRNA4+sgRNA5), targeting the Ctnnb1 locus upstream of the canonical ATG and within a portion of the last coding exon (XV), respectively (Figure 2A). Successful gene editing was confirmed at pool levels by PCR genotyping (Figure S2A). The pool of transfected cells did not show any expansion defect or cell detachment. Single-cell clones were picked, expanded, and finally three independent clones were isolated with homozygous deletions, Eβ11, Eβ15, and Eβ47. All three clones carried homozygous deletion of a 9.7-kb region as assessed by PCR genotyping and Sanger sequencing (Figures 2B and S2B and Supplemental DNA sequences) encompassing the whole β-catenin coding sequence (CDS), therefore preventing Ctnnb1 locus rearrangement.
Figure 2.
CRISPR/Cas9-Mediated Excision of Whole Ctnnb1 Locus Results in a Complete β-Catenin Knockout Model in mESCs
(A) Schematic representation of sgRNA design for CRISPR/Cas9-mediated excision of whole β-catenin coding sequence. Red arrows indicate sgRNAs target sites. Blue triangles indicate position and orientation of oligonucleotides used for PCR genotyping.
(B) PCR genotyping of three homozygous β-catenin knockout clones (Eβ11, Eβ15, and Eβ47) and parental E14 mESCs. Expected amplicon size is 951 bp for wild-type (wt) alleles and 551 bp for knockout alleles (del).
(C) Western blot of total protein extracts from Eβ11, Eβ15, Eβ47, and wild-type E14 cells. Protein extracts were probed for β-catenin (using a C-terminally raised antibody), stripped and re-probed for TUBULIN as loading control.
(D–G) Immunofluorescence in fixed parental E14, Eβ11, Eβ15, and Eβ47 cells for β-catenin (D), PLAKOGLOBIN (E), E-CADHERIN (F), OCT4 and NANOG (G). DAPI was used to counterstain nuclei. Scale bar represents 50 μm.
(H) Phase contrast pictures of Eβ11, Eβ15, Eβ47, and parental E14 clones upon 72-h vehicle (0.3% DMSO, left) or Chiron 3 μM treatment (right) in serum/LIF. Cells were seeded at 4 × 105 cells/well density in 6-well plates. Scale bar represents 50 μm.
(I) Phase contrast pictures of AP staining on E14, Eβ11, Eβ15, and Eβ47 cells cultured in serum/LIF in presence of vehicle (0.3% DMSO, left) or Chiron 3 μM (right) for 5 days; 300 cells were seeded in each well of a 6-well plate. Scale bar represents 50 μm.
(J) Histogram of qRT-PCR data on total RNA extracts of E14, Eβ11, Eβ15, and Eβ47 cells exposed to vehicle (0.3% DMSO, black bars) or Chiron 3 μM; 2−ΔCt are represented, and GAPDH was used as internal control. Error bars represents standard deviations of three technical replicates.
Western blot analysis with an antibody raised against the C-terminal portion of β-catenin revealed the absence of any ΔN isoform in all the analyzed clones (Figure 2C); this, together with the absence of any detectable signal by immunofluorescence (Figure 2D), confirmed the complete β-catenin depletion in the newly generated clones.
These data demonstrate that, regardless of the gene-editing technology used (CRE-mediated excision or CRISPR/Cas9), the gene-editing by-product cannot be easily predicted, and that complete locus ablation is a viable alternative to avoid undesired protein rearrangements.
Complete β-Catenin Loss Does Not Affect Self-Renewal and Pluripotency Marker Expression in mESCs Cultured in Serum/LIF
In order to characterize the impact of complete β-catenin ablation in mESCs, we analyzed the newly generated cell lines for proliferation, self-renewal, and pluripotency marker expression defects under serum/LIF culturing conditions. β-Catenin is normally found at the plasma membrane, where it physically interacts with E-CADHERIN and Α-CATENIN connecting adherens junctions to the actin cortex. As in Lyashenko et al. (2011) and Wray et al. (2011), Eβ11, Eβ15, and Eβ47 clones did not show major morphological defects; PLAKOGLOBIN levels were upregulated in response to β-catenin loss (Figures 2E, S2C, and S2D), while E-CADHERIN localization and expression levels remained unchanged (Figures 2F and S2E). We did not detect any difference in the cell-cycle (Figures S2F and S2G) or proliferation rate (Figure S2H) in all three knockout clones with respect to the parental cell lines, which displayed overall comparable population doubling times (Figure S2I).
Similarly to previously reported knockout models (Lyashenko et al., 2011; Raggioli et al., 2014; Wray et al., 2011), complete β-catenin loss did not show any additional defects of NANOG and OCT4 pluripotency marker expression as their localization (Figure 2G) and expression levels (Figure S2C and S2D) remained similar to the parental E14 cell line in all the analyzed clones.
The addition of small-molecule inhibitors of GSK3 activity has been reported to promote mESC pluripotency and self-renewal, while inhibiting spontaneous differentiation, by increasing β-catenin levels and activating the expression of Wnt target genes (Sato et al., 2004).
We therefore monitored the morphological and transcriptional response of β-catenin knockout clones to CHIR99021 (Chiron), a selective small-molecule inhibitor of GSK3. Wild-type and knockout cells were cultured in serum/LIF supplemented with 3 μM Chiron or vehicle (DMSO 0.3 μL/mL). After 72 h of Chiron treatment, wild-type cells acquired a homogeneous round morphology with tight colony boundaries, while Eβ11, Eβ15, and Eβ47 mESCs did not show any major morphological changes with respect to vehicle (Figure 2H), confirming that morphological changes induced by GSK3 inhibition are mediated by β-catenin (Wray et al., 2011). Morphological defects were only evident when cells were reaching >50% confluency (Figure 2H, vehicle), while β-catenin knockout clones were indistinguishable from wild-type E14 if plated at clonal density (Figure 2I, vehicle).
These data suggest that ΔN β-cat isoforms are biologically inactive as complete β-catenin depletion does not result in any additional phenotypic defect with respect to previously characterized knockout models.
Next, we assessed the colony formation capacity and AP expression of wild-type and β-catenin knockout cells in response to GSK3 inhibition. Cells were plated at clonal density and, while cultured in serum/LIF in presence of vehicle, wild-type E14 and β-catenin knockout clones were all positive for AP expression, although β-catenin knockout cells displayed a slightly lower AP staining intensity (Figures 2I, S2J, and S2K). In presence of Chiron, however, wild-type cells dramatically increased AP staining intensity. By contrast, AP staining intensity in β-catenin knockout cells was only moderately increased in response to Chiron treatment, probably as the result of slightly increased colony compaction (Figures 2I, S2J, and S2K). In addition, the AP transcript (Alpl) was upregulated upon Chiron treatment only in wild-type cells (Table S1, WTC/WTV differentially expressed genes [DEGs], row 89).
We then assessed the transcriptional response to GSK3 inhibition in wild-type and β-catenin knockout clones upon 72 h of Chiron treatment through qRT-PCR. As expected, β-catenin mRNA levels were undetectable in all knockout clones and remained unchanged upon Chiron treatment in wild-type cells. Instead, canonical Wnt targets such as Axin2 and Sp5 showed comparable levels between wild-type and knockout cells, while their expression was activated only in wild-type in response to Chiron treatment (Figure 2J). Of note, slightly higher levels of Cdx1 mRNA were found in all knockout clones in basal conditions with respect to wild-type cells. In addition, although the absence of β-catenin severely impaired Cdx1 upregulation upon Chiron treatment, it did not completely abrogate it, suggesting that downstream targets of GSK3 contribute to partially regulate its expression (Figure 2J).
β-Catenin Ablation Promotes a Weak Upregulation of Primitive Endoderm Genes
According to previously published works, the canonical Wnt pathway is active in mESCs (Ten Berge et al., 2011) and β-catenin nuclear translocation reinforces the pluripotency network by upregulating pluripotency genes such as Nanog, Esrrb, or Tcfp2l1 (Martello et al., 2012; Pereira et al., 2006; Qiu et al., 2015) mainly by inhibiting TCF3 repressive activity on their promoters (Pereira et al., 2006). Based on this model, a reasonable consequence of β-catenin ablation would be the increase in TCF3 repressive activity on core-pluripotency network genes with an expected destabilization of self-renewal and pluripotency features. Complete β-catenin loss did not, however, affect Tcf3 levels or its subcellular localization pattern (Figure S2L), suggesting that, in absence of exogenous WNT3A or GSK3 inhibitors, β-catenin does not control TCF3 activity. On the other hand, a previous β-catenin knockout model did not reveal major transcriptomic defects (Lyashenko et al., 2011); due to the lack of characterization of truncated isoforms, a possible explanation for this apparently controversial phenotype could be that ΔN β-cat isoforms retain some of the functions of the full-length counterpart, as recently proposed in pre-implantation embryo development (Messerschmidt et al., 2016).
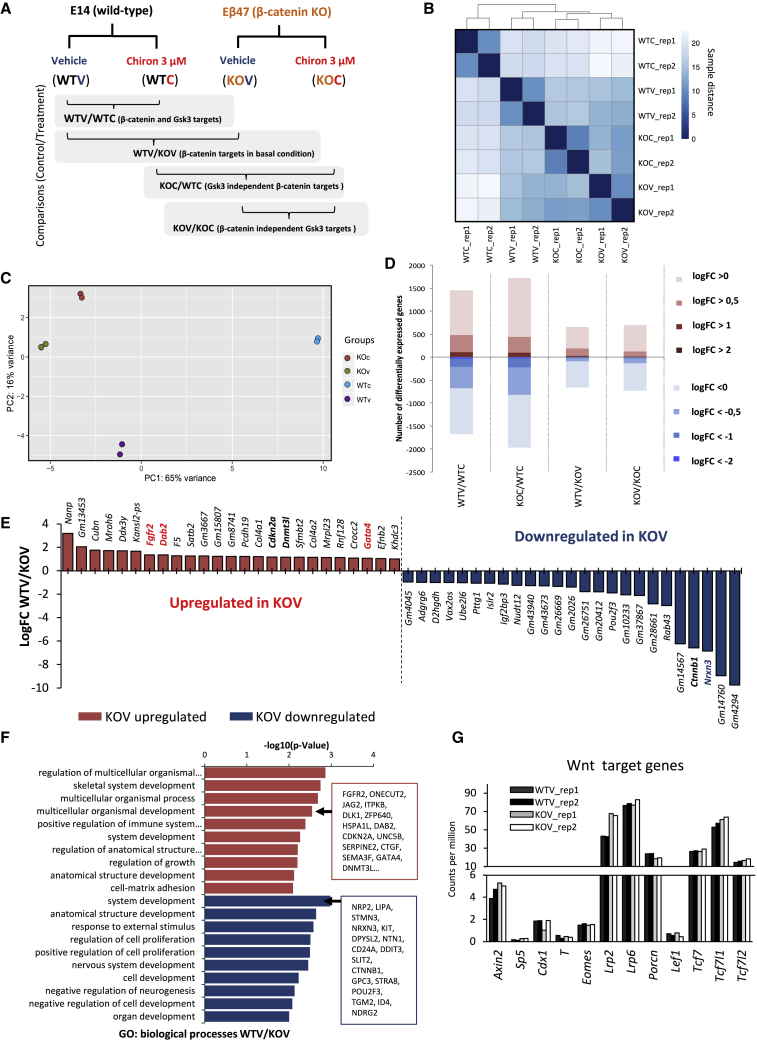
We therefore sought to analyze the expression profile of E14 parental cell line and the Eβ47 knockout clone through high-throughput RNA sequencing (RNA-seq) in cells cultured in serum/LIF culture conditions upon treatment with either vehicle (0.3% DMSO) or 3 μM Chiron for 72 h (Figure 3A). Since the impact of complete β-catenin loss in mESCs has never been assessed before at transcriptomic level, we designed an assay to study, through different comparisons between samples, the β-catenin and GSK3-dependent transcriptional changes in mESCs (Figure 3A). As expected, β-catenin was not detectable at transcriptional level in Eβ47 clone (Figure S3A), confirming the difference with previous knockout models in which β-catenin mRNA was still detectable (Figure 1F).
Figure 3.
β-Catenin Depletion Produces Minor Changes at Transcriptomic Level
(A) Schematic representation of experimental design for RNA-seq analysis. E14 parental cells (WT) or Eβ47 cells (knockout) were cultured in serum/LIF upon 72-h vehicle (0.3% DMSO, V) or Chiron 3 μM (C) treatment. Two biological replicates were analyzed for each sample. Pairwise sample comparisons are indicated as control/treatment.
(B) Sample distance matrix and hierarchical clustering of biological replicates (rep_1 and rep_2) for WTV, WTC, KOV, and KOC samples.
(C) PCA plot of indicated samples.
(D) Histogram of differentially expressed genes across pairwise comparisons as indicated in Figure 3A. Shades of red indicate overexpressed genes; shades of blue indicate downregulated genes. Shade intensity represents log fold-change cutoff from >0 (no fold-change cutoff, light), to absolute log fold change >2 (dark). Adjusted p value cutoff is 0.05.
(E) Top differentially expressed genes in WTV/KOV comparison ranked for log fold change. Adjusted p value <0.05.
(F) GO analysis of biological processes enriched in differentially expressed genes in WTV/KOV comparison (adjusted p value <0.05, absolute logFC >0.5). Upregulated features are shown in red, downregulated features are shown in blue.
(G) Histogram of RNA level (counts per million reads [CPM]) of canonical Wnt target genes and components across WTV (black and dark gray) or KOV (light gray and white) samples. Individual replicates are shown for each sample.
The transcriptomic profile of E14 wild-type cells cultured in serum/LIF (WTV), was overall very similar to Eβ47, independently if cultured in presence of vehicle (KOV) or Chiron (KOC), while E14 wild-type cultured in presence of 3 μM Chiron (WTC) clustered away from all the other samples in the samples distance matrix (Figure 3B). Principal component analysis (PCA) showed that the highest amount of variance was due to GSK3 inhibition in presence of β-catenin (WTC samples), while only subtle differences could be spotted between KOC, KOV, and WTV samples (Figure 3C). These results confirmed that, similarly to what was observed in β-catenin knockout models producing N-terminally truncated isoforms (Lyashenko et al., 2011), β-catenin absence does not significantly alter the transcriptional profile of serum/LIF cultured mESCs.
We then analyzed differential expressed genes (DEGs) across different samples/treatments, considering a threshold for adjusted p value <0.05 and absolute log2 fold change >0.5 (logFC, defined as the log ratio of a transcript’s expression values in two different conditions). Upon β-catenin depletion, 286 genes were differentially expressed with respect to E14 cells cultured in serum/LIF + vehicle (WTV/KOV comparison, of which 192 upregulated and 94 downregulated; Figures 3D and S3B and Table S1), and only 11 genes displayed a logFC higher than 2. Genes such as Fgfr2, Dab2, Pdgfra, and Gata4 were slightly upregulated upon β-catenin depletion, although with very low fold changes (between 1.2- and 2-fold enrichment), suggesting a partial priming toward primitive endoderm (PrE), while Neurexin 3 (Nrxn3) was downregulated (Figure 3E). We then performed Gene Ontology (GO) analysis on WTV/KOV DEGs (adjusted p value <0.05, logFC > 0.5); both upregulated and downregulated DEGs were enriched for developmental categories such as multicellular organism development and system development (Figure 3F and Table S2). Surprisingly, β-catenin depletion did not alter the expression of canonical Wnt target genes such as Axin2, Sp5, Cdx1, or T/Brachyury (Figure 3G), suggesting that canonical Wnt pathway is not transcriptionally active in mESCs cultured in serum/LIF, although β-catenin is highly expressed (Figure S3A). These results are in line with previous reports that failed to identify activity of the TOP/FOP reporter in serum/LIF cultured mESCs (Aulicino et al., 2014; De Jaime-Soguero et al., 2017; Faunes et al., 2013), while they stand at odds with the evidence that the Wnt/β-catenin pathway is active in the inner cell mass of the blastocyst, and the canonical Wnt signaling is required for mESC self-renewal in serum/LIF (Ten Berge et al., 2011).
Finally, when analyzing the expression levels of key lineage markers, we could not detect any change in pluripotency genes, while a slight overexpression of PrE lineage markers (Gata4, Gata6, Foxa2, Dab2, Ihh, Cerberus, and Pdgfra) was observed independently of Chiron treatment in absence of β-catenin (Figure S3C, bottom).
As colony formation and AP staining phenotypes are slightly ameliorated in β-catenin knockout mESCs in presence of Chiron with respect to vehicle (Figures 2I and S2J), we asked whether GSK3 inhibition per se could alter the transcriptome of mESCs independently of β-catenin. To address this question, we compared Eβ47 cells cultured in presence of vehicle (KOV) or 3 μM Chiron (KOC) for 72h (KOV/KOC comparison). Chiron treatment only resulted in minor changes as the transcriptome of KOV and KOC were overall very similar (Figures 3B and 3C). Only 254 genes were differentially expressed in cells lacking β-catenin and exposed to GSK3 chemical inhibition, again with overall low fold change (p value adjusted <0.05 and logFC >0.5, of which 124 upregulated and 130 downregulated) (Figures 3D and S3B and Table S1). Upregulated genes were enriched for biological processes such as stress response and metabolic features, while downregulated genes were enriched for metabolic and developmental processes (Figure S3D and Table S3). Nevertheless, the slight transcriptional changes induced by Chiron in absence of β-catenin revealed a certain degree of overlap with canonical Wnt target genes (Figure S3E). Canonical target genes such as Myc or Axin2 were slightly inhibited by Chiron treatment in absence of β-catenin as well as Tcf3 (Tcf7l1) mRNA levels (Figure S3F). These data confirm, as previously demonstrated (Doble et al., 2007), that GSK3 inhibition has little or no effect on mESC transcriptional landscape independently of β-catenin. Of note PLAKOGLOBIN can partially mimic β-catenin function (Mahendram et al., 2013), but, although PLAKOGLOBIN protein levels are upregulated in β-catenin knockout cells (Figures 2E, S2C, and S2D), Plakoglobin mRNA was not found among the differentially expressed genes upon β-catenin loss, suggesting the existence of a post-translational regulation mechanism for PLAKOGLOBIN that relies on β-catenin levels. In addition, our data demonstrate that Plakoglobin cannot replace nuclear β-catenin functions in response to Chiron in mESCs as no major transcriptional changes were observed in KOV/KOC comparison and, among the canonical Wnt/β-catenin targets, inhibition rather than activation was observed at best (Figures S3E and S3F).
Canonical Wnt Signaling Inhibits Differentiation of mESCs Toward Ectoderm
Although we assessed that the transcriptional effects of Chiron in absence of β-catenin are negligible, the DEGs in the KOC/WTC comparison were dependent solely on the presence of β-catenin. A similar approach has been used previously (Zhang et al., 2013) by comparing mESCs treated with Chiron or XAV (a small-molecule inhibitor of the Wnt pathway) without considering possible off-target effects of both drugs. Our experimental approach, instead, allowed us to decouple β-catenin-dependent targets (DEGs in KOC/WTC) from GSK3-only-dependent targets (DEGs in KOV/KOC).
The highest number of DEGs between comparisons represented as control/treatment was found in the WTV/WTC (1,157 DEGs, 476 upregulated, 681 downregulated) and in the KOC/WTC (1,259 DEGs, 437 upregulated, 822 downregulated) comparisons (Figures 3D and S3B and Table S1). Wild-type mESCs treated with Chiron clustered far apart from all the other conditions, as assessed by sample distance matrix PCA (Figures 3B and 3C). Numerous reports associated Wnt activation with transcriptional activation of Nanog, Klf4, Esrrb, and Tcfp2l1 in mESCs (Ai et al., 2016; Martello et al., 2012; Pereira et al., 2006; Qiu et al., 2015). However, with the exception of a slight Tcfp2l1 upregulation and Dppa3 downregulation, we were not able to identify any change at transcriptional levels of pluripotency marker genes upon Chiron treatment in wild-type cells (Figure S3C). Accordingly, we previously showed that Nanog, Oct4, and Rex1 levels do not change in mESCs even upon prolonged (up to 8 passages) exposure to Chiron (De Jaime-Soguero et al., 2017). Nevertheless, lineage markers such as Pax6, Fgf1, Nes, Otx2, Lefty1, and Pou3f1 were strongly inhibited by Chiron treatment in presence of β-catenin, indicating an overall reduction of differentiation commitment toward the ectoderm lineage (Figure S3C). Conversely, canonical Wnt target genes required for trophectoderm and mesoderm specification, including Cdx1, T/Brachyury, and Eomes, were strongly upregulated upon Chiron treatment.
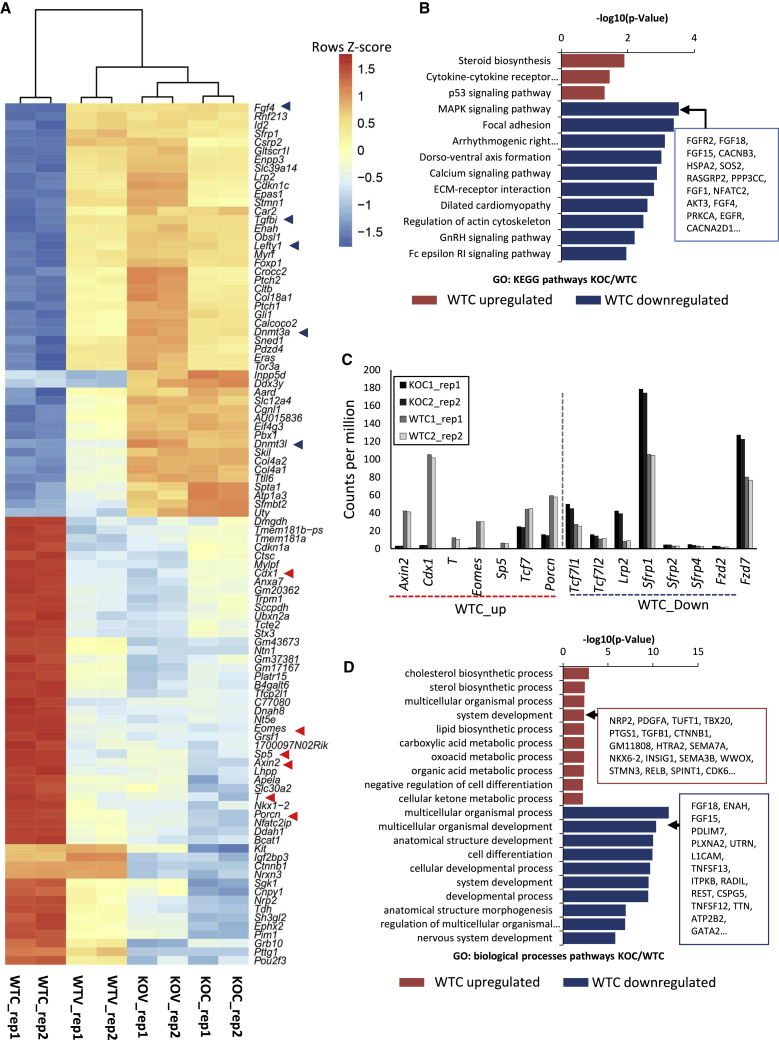
We then clustered the top 100 DEGs in the KOC/WTC comparison across the various samples (Figure 4A). As expected, sample clustering did not change with respect to previous analysis (Figures 3B and 3C) and canonical Wnt targets such as Axin2, Cdx1, and T/Brachyury were only activated in wild-type cells treated with Chiron, while few or no differences were observed in the other samples (Figures 4A and 4C). Furthermore, this subset of genes was transcriptionally perturbed only in presence of both Chiron and β-catenin, once again confirming that the canonical Wnt pathway is not active in mESCs cultured in serum/LIF unless an external stimulus (i.e., chemical GSK3 inhibition) is applied (Figure 4A).
Figure 4.
RNA-Seq Analysis of β-Catenin-Dependent Differentially Expressed Genes
(A) Heatmap clustering of the top 100 differentially expressed genes in KOC/WTC comparison across KOV, KOC, WTV, and WTC samples (p value adjusted <0.05, absolute logFC >0.5). Minimum and maximum are scaled across conditions on single genes; the heatmap represents logCPM rescaled on each gene for their Z scores (average of the values is the center of a normal distribution; color codes represent positive or negative deviations from the average).
(B) GO analysis of KEGG pathway categories enriched in differentially expressed genes in the KOC/WTC comparison (p value adjusted <0.05, absolute logFC >0.5).
(C) Histogram of CPMs of canonical Wnt target genes and components differentially expressed in KOC/WTC comparison. Individual biological replicates are shown.
(D) GO analysis of biological processe categories enriched in differentially expressed genes in KOC/WTC comparison (p value adjusted <0.05, absolute logFC >0.5).
We next asked about the nature of transcriptional changes induced by GSK3 inhibition in presence of β-catenin. We focused once again on the KOC/WTC comparison and performed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways enrichment and gene ontology (DEGs, KOC/WTC, adjusted p value 0.05, logFC > 0.5, Figures 4B and 4D and Table S5). Interestingly, Chiron treatment led to a perturbation of Tcf/Lefs levels, with the downregulation of Tcf3 (Tcf7l1) and the upregulation of Tcf1 (Tcf7) (Figure 4C). Furthermore, downregulated genes were strongly enriched for the mitogen-activated protein kinase (MAPK) signaling pathway (Figure 4B) and for focal adhesion. Both upregulated and downregulated genes were enriched for biological process categories associated with development. Nevertheless, the enrichment was stronger among DEGs downregulated upon Chiron treatment (Figure 4D and Table S5), confirming the general inhibition of spontaneous differentiation induced by the coupled action of GSK3 inhibition and β-catenin.
Transcriptional β-Catenin Activity Is Required to Inhibit Differentiation in Absence of LIF
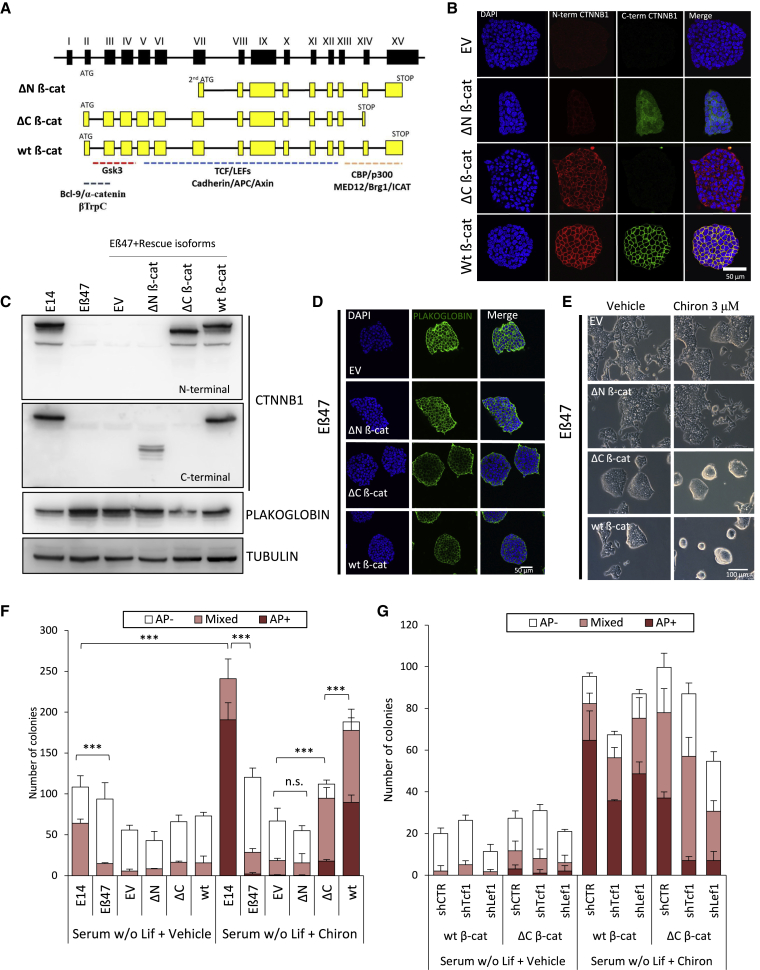
Previous reports have shown that transcriptional activity of β-catenin is dispensable for mESC self-renewal (Lyashenko et al., 2011; Wray et al., 2011). In order to validate or disprove these findings, we generated lentiviruses carrying different β-catenin isoforms. In addition to the full-length β-catenin (wild-type [wt] βcat), an N-terminally truncated (ΔN β-cat) and a C-terminally truncated isoform (ΔC β-cat) were generated. While ΔN β-cat isoforms mimic the isoforms generated from previous knockout models (Messerschmidt et al., 2016), ΔC β-cat carries a deletion of the transactivation domain, which results in an impaired transcriptional activity (Figure 5A).
Figure 5.
Canonical β-catenin Functions Are Required for Inhibition of Differentiation
(A) Schematic representation of β-catenin isoforms used for rescue experiments. N-terminally (ΔN β-cat) truncated β-catenin isoform mimics N-terminally truncated β-catenin isoforms obtained in previously published knockout models.
(B) Immunofluorescence of Eβ47 cells transduced with lentiviral vectors encoding empty vector (EV), wild-type (wt β-cat), ΔN β-cat, and C-terminally (ΔC β-cat) truncated β-catenin isoforms. Cells were stained with N-terminally (red) and C-terminally (green) β-catenin antibodies. Nuclei were counterstained with DAPI.
(C) Western blot of total protein extracts from E14 and Eβ47 untransduced cells and Eβ47 transduced with EV, ΔN β-cat, ΔC β-cat, and wt β-cat encoding lentiviruses. Membranes were probed with N-terminally or C-terminally raised β-catenin antibodies and anti-PLAKOGLOBIN. TUBULIN was used as loading control. Scale bar represents 50 μm.
(D) Immunofluorescence of Eβ47 cells transduced with EV, ΔN β-cat, ΔC β-cat, or wt β-cat encoding lentiviruses. Cells were stained for PLAKOGLOBIN. DAPI was used to counterstain nuclei (Scale bar represents 50 μm).
(E) Phase contrast pictures of Eβ47 cells transduced with EV, ΔN β-cat, ΔC β-cat, or wt β-cat encoding lentiviruses and cultured in serum/LIF in presence of 3 μM Chiron or vehicle (0.3% DMSO). Scale bar represents 100 μm.
(F) AP staining quantification of E14 and Eβ47 untransduced cells or Eβ47 transduced with EV, ΔN β-cat, ΔC β-cat, or wt β-cat encoding lentiviruses. Cells were plated in serum without LIF and supplemented with 3 μM Chiron (right) or vehicle (0.3% DMSO, left). Error bars represent standard error of three biological replicates. Student's t test was used to measure statistical significance as indicated, stars indicate p value (n.s. = not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
(G) AP staining of Eβ47 cells transduced with either wt βcat or ΔC β-cat encoding lentiviruses. Cells were further transduced with lentivirus encoding short hairpins against Lef1 (shLef1), Tcf1 (shTcf1), or shCTR. Cells were cultured in serum without LIF in presence of 3 μM Chiron or vehicle (0.3% DMSO) for 1 week and stained for AP expression. Error bars represent standard error of three biological replicates.
Eβ47 cells were transduced with a lentivirus carrying the different β-catenin isoforms under the constitutive EF1α promoter, or an empty lentivirus as a control (EV). Upon isoform expression, we studied protein subcellular localization and expression levels using N-terminal and C-terminal antibodies against β-catenin. N-terminally truncated β-catenin isoforms were only detected with the C-terminal raised antibody, and their subcellular localization recapitulated the one observed in previous knockout models (Figures 5B and 1C). Furthermore, as in ΔN β-cat isoforms emerging from previous knockout models, the overall ΔN β-cat levels were reduced, as compared to total β-catenin levels in wild-type cells (Figure 5C).
The expression and localization of ΔC β-cat and wt β-cat rescue isoforms were overall comparable, with intense membrane localization and similar expression levels (Figures 5B and 5C). Interestingly, expression of ΔC and wt β-cat, but not ΔN β-cat, restored normal Plakoglobin levels (Figures 5C, 5D, and S4A), indicating that impaired membrane functions, but not the transcriptional ones, are responsible for the increased Plakoglobin levels in β-catenin knockout cells. Similarly, ΔN β-cat isoforms could not rescue the loss of morphological changes induced by Chiron treatment, while both ΔC and wt rescued Eβ47 cells exhibited a round morphology (Figure 5E) indistinguishable from E14 wild-type cells upon Chiron addition (Figure 2H). Upon GSK3 inhibition, however, eGFP expression under the control of the synthetic Wnt reporter was only detectable in wild-type rescued Eβ47, confirming that both ΔN and ΔC β-cat isoforms are transcriptionally impaired for canonical Wnt signaling (Figure S4B).
We next assessed the colony formation capacity and AP expression of rescued β-catenin knockout cells in different culture conditions. In serum/LIF, only ΔC β-cat and wt β-cat rescued Eβ47 cells recapitulated the parental E14 cells phenotype, while ΔN β-cat overexpression did not alter the phenotype with respect to empty vector transduced cells (EV, Figure S4C, top panel). Eβ47 showed impair clonogenicity and self-renewal, in line with previously published knockout models. In serum-free media, Eβ47 could not be expanded in PD + LIF, Chiron + LIF, or PD + Chiron (2i), but could self-renew in 2i/LIF; again, ΔN β-cat rescued Eβ47 cells did not show any improvement of AP expression or self-renewal (Figure S4C, bottom panel). As previously reported, both ΔC β-cat and wt β-cat isoforms restored self-renewal and AP staining intensity defects (Figure S4C, bottom panel), suggesting that canonical Wnt pathway activity is dispensable for self-renewal in these culture conditions.
However, nuclear β-catenin activity appears to be required for inhibition of differentiation in serum and in absence of LIF (Ogawa et al., 2006; Sato et al., 2004). Therefore, we performed clonal assay and AP staining of Eβ47 rescued cells in serum without LIF. While all the cell lines quickly differentiated in absence of LIF, Chiron treatment maintained AP staining in ΔC β-cat and wt β-cat rescued cells, but not in ΔN β-cat rescued Eβ47 (Figures 5F and S4D). Furthermore, ΔC β-cat expression only partially rescued AP staining, indicating that canonical Wnt activity is required to counteract spontaneous differentiation upon LIF withdrawal.
These results suggest that the inhibition of spontaneous mESC differentiation in the absence of LIF could be due to combined nuclear and membrane-associated β-catenin functions. However, while the ΔC β-cat isoform fails to activate the synthetic Wnt reporter (Figure S4B), it has previously been observed that the ΔC β-cat isoform exhibit a certain degree of transcriptional activity, despite lacking the transactivation domain (Wray et al., 2011). A possible explanation could be that ΔC β-cat isoforms still interact with TCF3 and, like the full-length β-catenin, alleviate TCF3-mediated repression on a subset of target genes (Wray et al., 2011). On the other hand, it has been reported that ΔC β-cat isoforms can still form a transcriptionally active complex in association with LEF1 (Hsu et al., 1998) or TCF1. We therefore silenced Lef1 or Tcf1 in Eβ47 cells rescued with either full-length or ΔC β-cat (Figure S4E) and assessed AP staining intensity upon exposure to vehicle or Chiron in absence of LIF. Tcf1 or Lef1 depletion efficiently reduced the number of AP colonies formed in response to Chiron treatment in ΔC β-cat rescued cells and, to a lesser extent, also in wild-type β-catenin mESCs (Figures 5G and S4F).
These results suggest that ΔC β-cat could still interact with TCF1 and LEF1, and that nuclear β-catenin/TCF/LEF functions could be required to inhibit mESC spontaneous differentiation in the absence of LIF, while the membrane-associated β-catenin functions may have little or no role in this phenotype.
Discussion
In this work, we showed that the inducible β-catenin knockout models currently available result in the production of N-terminally truncated isoforms in mESCs. While ΔN β-cat isoforms are not physiologically expressed, their appearance is a consequence of rather unpredictable gene rearrangement upon genetic manipulation. By using CRISPR-Cas9 technology, we isolated three mESC clones with no detectable β-catenin expression. Of note, only a paired sgRNA approach generating a deletion encompassing the whole β-catenin gene body was successful in generating knockout alleles (10-kb genomic DNA deletion), while standard strategies (ORF shifting and microdeletion) only produced ΔN-truncated isoforms at best. As CRISPR-Cas9 is boosting genetic engineering, the production of undesired isoforms with potential gain of functions, which could undermine all the conclusion of a study, should be carefully monitored and avoided. For this reason, we foresee that whole-gene deletion using CRISPR-Cas9 will be an advantageous approach for generating knockout models free of undesired side products.
We assessed that the appearance of ΔN β-cat isoforms in previously published β-catenin knockout mESC models is not detrimental for their phenotype, as no compensatory or overlapping effects were to be attributed to the truncated proteins. In addition, none of the impaired phenotypes observed upon complete β-catenin loss could be rescued by a ΔN β-cat isoform, thus recapitulating the phenotype observed in the previously generated knockout models (Lyashenko et al., 2011; Wray et al., 2011).
β-Catenin depletion does not alter self-renewal and pluripotency marker expression of mESCs under serum/LIF culturing conditions, with expected morphological changes probably rescued by Plakoglobin stabilization. Moreover, β-catenin depletion also does not alter the expression of known canonical Wnt targets in basal culturing conditions, suggesting that the Wnt/β-catenin pathway is not constitutively active in mESCs cultured in feeder-free media supplemented with serum/LIF. Accordingly, β-catenin loss does not globally alter the transcriptional profile of mESCs but induces a mild activation of PrE marker genes. The latter phenotype, coupled to the absence of self-renewal and proliferation defects in serum/LIF, could either not be significant or could potentially highlight the presence of a rare population of terminally differentiated cells committed toward PrE that cannot withstand β-catenin absence.
Furthermore, only chemical inhibition of GSK3 activates a transcriptional Wnt response in wild-type cells, while eliciting little or no response on β-catenin knockout mESCs, suggesting that Plakoglobin upregulation cannot recapitulate nuclear β-catenin functions. Nevertheless, activation of the canonical Wnt pathway shields mESCs from spontaneous epiblast/ectoderm differentiation by suppressing the expression of lineage-specific genes and not by enhancing the expression levels of pluripotency factors. This effect is particularly evident when cells are deprived of LIF, suggesting overlapping functions between LIF and Wnt signaling in mESC self-renewal. With respect to these functions, Chiron treatment can prolong, but not indefinitely sustain, mESC self-renewal in the absence of LIF. This phenotype is abrogated by β-catenin loss but can be rescued by wild-type β-catenin and, to a lesser extent, by ΔC β-cat isoforms, posing the question of whether nuclear β-catenin functions are truly required for differentiation inhibition. We demonstrated that TCF1 and LEF1 are involved in the observed phenotypes, as their depletion further impairs the colony formation capacity and AP staining of both wild-type and ΔC rescued β-catenin knockout cells. These results suggest that ΔC β-cat isoforms are not completely transcriptionally silent and can still interact not only with TCF3 (as previously suggested by Wray et al., 2011) but also with other TCF/LEFs; we cannot, however, exclude that other regulatory mechanisms might be involved. Finally, we proved that activation of the canonical Wnt pathway sustains mESC self-renewal through inhibition of spontaneous differentiation; nuclear β-catenin functions, in association with TCF/LEF family members, could play a role in this phenotype.
In the future, it will be particularly interesting to further investigate the transcriptional response of ΔC-rescued β-catenin knockout cells to Chiron, as it could pinpoint a subset of powerful transcriptional targets truly responsible for Wnt-mediated self-renewal enhancement. In addition, since most of the observed pluripotency phenotypes are inevitably dependent on the specific culture conditions we used (mostly, serum based and feeder free using gelatin coating), it might be of interest to extend the results presented in this work to study pluripotency phenotypes upon β-catenin deletion in serum-free media and MEK1/2 and GSK3β inhibitors (e.g., 2i/LIF), which can confer ground-state cell pluripotency to mESCs (Ying et al., 2008); notably, 2i/LIF cultured mESCs should better resemble in vivo features observed in the inner cell mass in the pre-implantation embryo, where Wnt signaling is active (De Jaime-Soguero et al., 2018; Ten Berge et al., 2011).
Experimental Procedures
Detailed experimental procedures are provided in Supplemental Information.
Cell Culture
mESCs E14 (129/Ola strain) were cultured on gelatin-coated plates in ESC medium: DMEM supplemented with 15% FBS (Hyclone), 1× non-essential amino acids, 1× GlutaMax, 1× penicillin/streptomycin, 1× 2-mercaptoethanol, and 1,000 U/mL LIF ESGRO (Chemicon). mESCs cultured in serum + LIF medium were re-plated every 3 days at a split ratio from 1:30 following dissociation with trypsin 0.05% EDTA (Gibco). mESCs containing floxed alleles of β-catenin were a kind gift of Prof. Rolf Kemler (SR18 and NLC1 cell lines, stably expressing CRE-ERT2) and Prof. Christine Hartmann (NLβ-12tm4Wbm fl/fl and del/del cells).
For serum-free cultures, mESCs were cultured without feeders or serum in pre-formulated N2B27 medium (NDiff N2B27 base medium, Stem Cell Sciences Ltd, cat. no. SCS-SF-NB-02) supplemented with small-molecule inhibitors PD0325901 (PD, 1 μM, Selleck) and CHIRON99021 (CH, 3 μM, Selleck) and 1000 U/mL LIF (ESGRO, Millipore). Cells were routinely propagated on 0.1% gelatin-coated plastic and re-plated every 3 days at a split ratio of 1 in 10 following dissociation with Accutase (Gibco) as previously reported (Wray et al., 2011).
Human embryonic kidney 293t (HEK293t) were purchased from ATCC (293T; ATCC CRL-3216) and cultured in DMEM supplemented with 10% FBS (Hyclone), 1× penicillin/streptomycin. HEK293t were re-plated every 3 days at a split ratio of 1 in 6 following dissociation with trypsin 0.05% EDTA (Gibco, Life technologies).
RNA-Seq and Data Analysis
Total RNA from mESCs was extracted wit RNAeasy kits (Qiagen) following manufacturer's instructions. RNA integrity check, Poly-A pulldown, and library preparation were performed by CRG genomic facility. Samples were sequenced in two biological replicates to a depth of 30 million reads (100 bp) per sample using Illumina HiSeq. Reads were mapped to a reference transcriptome (mouse transcriptome from Ensembl v80) using kallisto (v0.42.5) (Bray et al., 2016) to generate counts and transcripts per million (TPM) values. We tested for differentially expressed genes for all pairwise conditions using edgeR (Robinson et al., 2010) from raw counts, filtering for genes with adjusted p value <0.05 and logFC > 0.5. Table S1 reports the list of all DEGs across pairwise comparisons regardless of their logFC. GO and KEGG analyses were performed using DAVID (version 6.7) (Huang da et al., 2009; Huang et al., 2007) using differentially expressed genes with adjusted p value <0.05 and logFC > 0.5. For each pairwise comparison, the list of expressed genes in control and analyzed samples was obtained by filtering out all the genes with zero counts in at least one sample. Expressed genes for each pairwise comparison were set as background universe to estimate enriched categories among differentially expressed genes in DAVID (Timmons et al., 2015). A p value threshold of 0.05 was set to determine enrichment of gene ontology categories. Multiple test correction was not performed.
Data and Code Availability
RNA-seq data that support the findings of this study have been deposited in the NCBI/GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143340.
Author Contribution
F.A., L.M., and M.P.C. wrote the manuscript. F.A. and M.P.C. designed the experiments. M.P.C. supervised the project. F.A., F.S., E.P., and F.L.L. performed the experiments.
Acknowledgments
SR18 and NLC1 mESCs cell lines were a kind gift from Professor Rolf Kemler. NLβ-12 (Ctnnb1tm4Wbm fl/fl and Ctnnb1tm4Wbm del/del) were a kind gift from Professor Christine Hartmann.
The authors would like to thank the Genomics Unit at the CRG for assistance with the mRNA-seq (library prep and sequencing), and CRG and the Bristol University flow-cytometry facilities. This work was supported by the European Union's Horizon2020 Research and Innovation Programme (CellViewer No 686637 to M.P.C.), Ministerio de Ciencia e Innovación, grant BFU2017-86760-P (AEI/FEDER, UE), and an AGAUR grant from Secretaria d’Universitats i Recerca del Departament d’Empresa I Coneixement de la Generalitat de Catalunya (2017 SGR 689 to M.P.C.). We acknowledge the support of the Spanish Ministry of Science and Innovation to the EMBL partnership, the Centro de Excelencia Severo Ochoa, and the CERCA Programme/Generalitat de Catalunya and Ministerio de Ciencia e Innovación FPI (to F.A.), La Caixa international PhD fellowship (to F.S.); KU Leuven C1 funds (C14/16/078) and FWO (G097618N) funds to F.L.L. We acknowledge the support of the Medical Research Council (grant MR/N021444/1) to L.M., and of the Engineering and Physical Sciences Research Council (grants EP/R041695/1 and EP/S01876X/1) to L.M.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: August 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.07.019.
Supplemental Information
References
- Ai Z., Shao J., Wu Y., Yu M., Du J., Shi X., Zhang Y., Guo Z. CHIR99021 enhances Klf4 expression through beta-catenin signaling and miR-7a regulation in J1 mouse embryonic stem cells. PLoS One. 2016;11:e0150936. doi: 10.1371/journal.pone.0150936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton R., Kestler H.A., Kuhl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Aulicino F., Theka I., Ombrato L., Lluis F., Cosma M.P. Temporal perturbation of the Wnt signaling pathway in the control of cell reprogramming is modulated by TCF1. Stem Cell Reports. 2014;2:707–720. doi: 10.1016/j.stemcr.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- De Jaime-Soguero A., Abreu de Oliveira W.A., Lluis F. The pleiotropic effects of the canonical Wnt pathway in early development and pluripotency. Genes. 2018;9:93. doi: 10.3390/genes9020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jaime-Soguero A., Aulicino F., Ertaylan G., Griego A., Cerrato A., Tallam A., Del Sol A., Cosma M.P., Lluis F. Wnt/Tcf1 pathway restricts embryonic stem cell cycle through activation of the Ink4/Arf locus. PLoS Genet. 2017;13:e1006682. doi: 10.1371/journal.pgen.1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries W.N., Evsikov A.V., Haac B.E., Fancher K.S., Holbrook A.E., Kemler R., Solter D., Knowles B.B. Maternal beta-catenin and E-cadherin in mouse development. Development. 2004;131:4435–4445. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- Doble B.W., Patel S., Wood G.A., Kockeritz L.K., Woodgett J.R. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S.M., Chatterjee S.S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A.K., Dasgupta R. A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.C., Galceran J., Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F., Ombrato L., Pedone E., Pepe S., Merrill B.J., Cosma M.P. T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proc. Natl. Acad. Sci. U S A. 2011;108:11912–11917. doi: 10.1073/pnas.1017402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluis F., Pedone E., Pepe S., Cosma M.P. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493–507. doi: 10.1016/j.stem.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Lyashenko N., Winter M., Migliorini D., Biechele T., Moon R.T., Hartmann C. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendram S., Kelly K.F., Paez-Parent S., Mahmood S., Polena E., Cooney A.J., Doble B.W. Ectopic gamma-catenin expression partially mimics the effects of stabilized beta-catenin on embryonic stem cell differentiation. PLoS One. 2013;8:e65320. doi: 10.1371/journal.pone.0065320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the GSK3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marucci L., Pedone E., Di Vicino U., Sanuy-Escribano B., Isalan M., Cosma M.P. β-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 2014;8:1686–1696. doi: 10.1016/j.celrep.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Messerschmidt D., de Vries W.N., Lorthongpanich C., Balu S., Solter D., Knowles B.B. β-catenin-mediated adhesion is required for successful preimplantation mouse embryo development. Development. 2016;143:1993–1999. doi: 10.1242/dev.133439. [DOI] [PubMed] [Google Scholar]
- Ogawa K., Nishinakamura R., Iwamatsu Y., Shimosato D., Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem. Biophys. Res. Commun. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- Pereira L., Yi F., Merrill B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D., Ye S., Ruiz B., Zhou X., Liu D., Zhang Q., Ying Q.L. Klf2 and Tfcp2l1, two Wnt/beta-catenin targets, act synergistically to induce and maintain naive pluripotency. Stem Cell Reports. 2015;5:314–322. doi: 10.1016/j.stemcr.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggioli A., Junghans D., Rudloff S., Kemler R. Beta-catenin is vital for the integrity of mouse embryonic stem cells. PLoS One. 2014;9:e86691. doi: 10.1371/journal.pone.0086691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A.H. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons J.A., Szkop K.J., Gallagher I.J. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol. 2015;16:186. doi: 10.1186/s13059-015-0761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.T., Xu X., Yi F., Merrill B.J., Cooney A.J. Canonical Wnt/beta-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells. 2010;28:1794–1804. doi: 10.1002/stem.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.-L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Peterson K.A., Liu X.S., McMahon A.P., Ohba S. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells. 2013;31:2667–2679. doi: 10.1002/stem.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study have been deposited in the NCBI/GEO database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143340.