Summary
Mutations and loss of activity in PARKIN, an E3 ubiquitin ligase, play a role in the pathogenesis of Parkinson's disease (PD). PARKIN regulates many aspects of mitochondrial quality control including mitochondrial autophagy (mitophagy) and mitochondrial biogenesis. Defects in mitophagy have been hypothesized to play a predominant role in the loss of dopamine (DA) neurons in PD. Here, we show that although there are defects in mitophagy in human DA neurons lacking PARKIN, the mitochondrial deficits are primarily due to defects in mitochondrial biogenesis that are driven by the upregulation of PARIS and the subsequent downregulation of PGC-1α. CRISPR/Cas9 knockdown of PARIS completely restores the mitochondrial biogenesis defects and mitochondrial function without affecting the deficits in mitophagy. These results highlight the importance mitochondrial biogenesis versus mitophagy in the pathogenesis of PD due to inactivation or loss of PARKIN in human DA neurons.
Keywords: PARKIN, PARIS, ZNF746, PGC-1α, dopamine, Parkinson’s disease, isogenic, human IPSC, mitochondrial biogenesis, mitophagy
Graphical Abstract

Highlights
-
•
Human DA neuron loss of PARKIN leads to increased PARIS and decreased PGC-1α
-
•
Human DA neuron PARKIN loss leads to reduced mitochondrial autophagy and biogenesis
-
•
Decreased mitochondrial biogenesis drives the mitochondrial defects in DA neurons
-
•
PARIS is a key mediator of the mitochondrial defects in DA neurons lacking PARKIN
In this article, Dawson and colleagues show that human DA neurons lacking PARKIN exhibit mitochondrial defects that are driven by the upregulation of PARIS, downregulation of PGC-1α, and decreased mitochondrial biogenesis. While mitophagy is impaired in human DA neurons lacking PARKIN, it does not appear to contribute to the mitochondrial deficits.
Introduction
The progressive loss of dopamine (DA) neurons is one of the hallmark features of Parkinson's disease (PD) (Savitt et al., 2006). The death of DA neurons in PD is a complex process, and the exact causes of their demise are still under active investigation. Mutations in specific genes, including PARKIN, PTEN-induced kinase 1 (PINK1), DJ-1, SNCA and LEUCINE RICH REPEAT KINASE 2 (LRRK2) among others, have been linked to PD (Domingo and Klein, 2018; Martin et al., 2011). PARKIN is an ubiquitin E3 ligase that is mutated in autosomal recessive early-onset familial PD (Domingo and Klein, 2018; Kitada et al., 1998) leading to loss of its E3 ligase activity (Panicker et al., 2017). Its E3 ligase activity is also compromised in sporadic PD (for review, see Dawson and Dawson, 2014; Panicker et al., 2017). Genetic interaction studies in Drosophila indicate that Parkin and PINK1 function in the same pathway to regulate mitochondrial quality control (Clark et al., 2006; Park et al., 2018).
Early overexpression studies of GFP-tagged PARKIN in mitotic cancer cell lines coupled with gain and loss of function studies with PINK1 indicated that PINK1 regulates the ability of overexpressed PARKIN to translocate to the mitochondria and clear damaged mitochondria after treatment with mitochondrial toxins that simultaneously depolarize mitochondria (Narendra et al., 2010; Vives-Bauza et al., 2010). These initial observations led to tremendous insight into the underlying mechanisms of how PARKIN and PINK1 coordinate the removal of damaged mitochondria via mitochondrial autophagy (mitophagy) (Pickles et al., 2018; Pickrell and Youle, 2015). Loss of PARKIN and PINK1 function leads to defects in mitophagy in a variety of cultured cell types, including neurons (Cummins and Gotz, 2018; Whitworth and Pallanck, 2017). This has led to the hypothesis that defective mitophagy accounts for neurodegeneration of dopamine neurons in PD due to PARKIN or PINK1 mutations.
Despite the evidence that PARKIN and PINK1 coordinate mitophagy in cell culture systems, there is very little evidence of PINK1- and PARKIN-mediated mitophagy in vivo and in particular whether the loss of DA neurons is due to defects in mitophagy (Whitworth and Pallanck, 2017). There is evidence that the accumulation of Parkin Interacting Substrate (PARIS), also known as Zinc Finger Protein 746 (ZNF746), drives the loss of dopamine neurons in adult mice (Lee et al., 2017; Shin et al., 2011; Siddiqui et al., 2015; Stevens et al., 2015) or Drosophila (Pirooznia et al., 2020) lacking Parkin or due to pathologic α-SYNUCLEIN in sporadic PD (Brahmachari et al., 2019; Siddiqui et al., 2015, 2016) through downregulating PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA, COACTIVATOR 1α (PGC-1α), a master regulator of mitochondrial biogenesis (Scarpulla, 2011), suggesting that defects in mitochondrial biogenesis may be important in the loss of DA neurons. Since it is difficult to simultaneously study mitophagy, biogenesis, and mitochondrial function in DA neurons in vivo, we employed human DA neurons derived from human embryonic stem cells (hESCs) in which Exon 7 of the PARKIN gene was deleted, as well as induced pluripotent stem cells (iPSCs) from PD patients with PARKIN mutations. Using these human DA neurons lacking PARKIN, we explored the role of defects in mitophagy versus mitochondrial biogenesis in the mitochondrial dysfunction due to the absence of PARKIN.
Results
Generation and Characterization of Isogenic PARKIN KO Human Embryonic Stem Cells and Inducible Pluripotent Stem Cells Lacking p
PARKIN exon 7 was deleted from H1 hESCs via zinc finger nuclease (ZFN) technology (Figure S1A). ZFN nucleofected cells were selected with puromycin, and 18 clones were picked for PCR analysis (Figure S1B). Ten PCR positive clones were analyzed via Southern blot. Nine homozygous and one heterozygous PARKIN exon 7 human stem cell (HSC) clones were successfully generated (Figure S1C). Two clones (7.23.13 and 7.23.14), PARKIN KO1 and KO2 were confirmed by RT-PCR (Figures S1A and S1D) and by immunoblot (Figure S1E). Pluripotency markers, SSEA4, TRA-1-60, OCT4, and NANOG were positive in the isogenic parental PARKIN KO1 and KO2 lines (Figure S1F). No major alterations in chromosomal structure occurred as determined by karyotyping (Figure S1G). iPSCs were generated from two PD patients (PD1 and PD2) containing PARKIN mutations and two control patients (CTL1 and CTL2) (Figure S1). PD1 contains a 459 bp deletion spanning exons 4 to 7, and PD2 contains a 363 bp deletion spanning exons 3 and 4. The pluripotency markers, SSEA4, TRA-1-60, OCT4, and NANOG were positive in the two control and two PARKIN mutations lines (Figure S1H). The two control lines and two PD patients containing PARKIN mutations exhibited all three germ layers (Figure S1I). Karyotyping indicates that no major alterations in chromosomal structure occurred during the generation of the iPSCs (Figure S1J). Sequencing of PD1 and PD2 iPSCs confirmed the exon deletions (Figures S1K and S1L). RT-PCR using primers from exon 2 and exon 9 revealed a 1,039 bp product in both control iPSCs and a 580 bp product in PD1 and 676 bp product in PD2 (Figure S1M). Immunoblot analysis confirmed that PARKIN was deleted from both PD1 and PD2 iPSCs (Figure S1).
Human DA Neurons Lacking PARKIN Have Reduced Levels of DA Markers Compared with Isogenic Controls
PARKIN KO1, KO2, and the isogenic control (iCont) hESCs line were differentiated into DA neurons (Figure 1A) (Kriks et al., 2011). There was a significant 36.5% reduction of the percentage of tyrosine hydroxylase (TH) positive neurons in both PARKIN KO1 and PARKIN KO2 lines compared with the iCont line (Figure 1B), while there was only a 19.2% reduction in overall neuronal number as assessed by TUJ1 immunoreactivity (Figure 1C). Of the TUJ1 positive cells, there was a 40% reduction in the number of TH-positive neurons in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figure 1D). Immunoblot analysis confirmed the loss of TH, TUJ1, and relative TH/TUJ1 levels (Figures 1E–1H). To determine whether the loss of TH was due to cell death or a differentiation defect, staining with propidium iodide (PI), a marker of cell death, and Ki67, a proliferation marker, was performed. In both PARKIN KO1 and KO2 lines compared with the iCont line, there was an increase of PI (13.2% ± 3.7%) and Ki67 (3.32% ± 0.68%) staining indicative of cell death in the setting of enhanced proliferation (Figures 1I–1K). To confirm cell death in TH-positive neurons, annexin V staining was performed by co-labeling with the TH immunoreactivity, and there was a significant increase in the percentage of Annexin V staining in the TH-positive neurons consistent with enhanced cell death (Figures 1L and 1M). Similar to previous reports (Chung et al., 2016), no significant abnormality in DA neuron number was observed in the PD1 and PD2 iPSCs derived DA neurons compared with the iPSC control DA neurons (Figures S2A and S2B).
Figure 1.
Differentiation and Characterization of iCont and PARKIN KO hESCs to Dopamine Neurons
(A) Immunofluorescence staining of DA neuron differentiated cultures at 60 days from iCont and PARKIN KO lines. Neuron-specific class IIIβ-tubulin (TUJ1) is indicated in green and tyrosine hydroxylase (TH) is indicated in red. DAPI (blue) staining represents the nucleus. Scale bar, 50 μm.
(B–D) Percentage of (B) TH, (C) TUJ1, and (D) TH/TUJ1 immunofluorescence staining. n = 5 independent experiments.
(E) Representative immunoblots of TH and TUJ1 normalized to actin 60 days post differentiation to DA neurons.
(F–H) Quantification of immunoblots of (F) TH, (G) TUJ1, and (H) TH/TUJ1. n = 3 independent experiments.
(I) Immunofluorescence staining of Ki67 (red) and PI staining (red), nuclei are stained with DAPI (blue) 60 days post differentiation to DA neurons. Scale bar, 50 μm.
(J and K) Quantification of immunofluorescence staining of (J) PI and (K) Ki67staining. n = 5 independent experiments.
(L) Immunofluorescence staining of Annexin V (red) TH (green), nuclei are stained with DAPI (blue) 60 days post differentiation to DA neurons. Scale bar, 50 μm.
(M) Quantification of immunofluorescence staining of Annexin V. n = 5 independent experiments.
Error bar represents mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figures S1 and S2.
Loss of PARKIN Leads to Mitochondrial Respiratory Decline
Mitochondrial respiration was measured in the human DA neuron cultures using a Seahorse XF24 Flux Analyzer (Agilent Technologies, USA). There was a 27% reduction in basal respiration, a 54% reduction in spare respiratory capacity, and a 45% reduction in carbonyl cyanide m-chlorophenyl hydrazine (CCCP)-induced maximal respiration in both PARKIN KO1 and KO2 culture compared with the iCont culture (Figures 2A and 2B). In addition, there was a 24% reduction in basal respiration, a 78% reduction in spare respiratory capacity, and an 87% reduction in CCCP-induced maximal respiration in the PD1 and PD2 iPSC-derived DA neurons compared with the iPSC control DA neurons (Figures S2C–S2H). Mitochondrial membrane potential assessment using MitoCMXRos revealed a 47% reduction in both PARKIN KO1 and KO2 cultures compared with the iCont cultures (Figures 2C and 2D). Similarly, a 41% reduction in membrane potential was found in both PD1 and PD2 iPSC-derived DA neurons compared with the iPSCs control DA neurons (Figures S2G and S2H). Taken together, these results indicate there is a deficit in mitochondrial respiration in PARKIN KO-derived DA neurons.
Figure 2.
Characterization of Mitochondrial Function Defects in PARKIN KO Neurons
(A) Mitochondrial oxygen consumption rate (OCR) using the Seahorse platform in iCont and PARKIN KO neuronal cultures 55 days post differentiation. The blue line represents iCont, green and red represents PARKIN KO-1and KO-2 respectively.
(B) Quantification of basal respiration, reserve capacity, and maximal respiration in iCont and PARKIN KO neuronal cultures in the presence of oligomycin, CCCP, and rotenone, respectively. n = 3 independent experiments.
(C) Immunofluorescence staining of mitochondrial marker Tom20 (green), TH (blue) positive neurons, and MitoTracker Red CMXRos 60 days post differentiation to DA neuron iCont and PARKIN KO neuronal cultures. Scale bar, 25 μm.
(D) Quantification of Tom20 and MitoTracker Red CMXRos intensity within TH-positive neurons. n = 3 independent experiments.
Error bar represents the mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S2.
Reduced Mitochondrial Autophagy in PARKIN-Deficient Human DA Neuron Culture
Mitophagy was measured in human DA neuron cultures by utilizing Mito-Keima (Katayama et al., 2011). Human DA neuron cultures were transduced with a lentivirus expressing Mito-Keima, a pH-sensitive marker of mitophagy that provides a measure of the status of mitochondria where mitochondria that are localized to cytoplasm are green and those mitochondria that are being degraded via autophagy are red due to the acidic environment of the lysosome (Katayama et al., 2011). The ratio of the lysosomal red signal over the mitochondrial green signal within the neuronal body was calculated as the mitophagy index. There was a 50%–70% reduction in the mitophagy index in both PARKIN KO1 and KO2 cultures compared with the iCont cultures (Figures 3A and 3B) and a 30%–35% reduction in the mitophagy index in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron culture (Figures S3A and S3B). We evaluated LC3I/II, an integral structural protein of the autophagasome for autophagy. The LCII to LCI ratio was decreased by 50% in both PARKIN KO1 and KO2 cultures compared with the iCont cultures (Figures 3C and 3D), and there was a 50%–30% reduction in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron cultures (Figures S3C and S3D). The p62 protein significantly accumulates in both PARKIN KO1 and KO2 cultures compared with the iCont cultures (Figures 3C and 3E) and in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron cultures (Figures S3C and S3E). These results taken together suggest that there are defects in mitophagy in DA neuron cultures lacking PARKIN.
Figure 3.
Mitochondrial Autophagy Defects in PARKIN KO Neurons
(A) Confocal live imaging of neurons at 60 days post differentiation to DA neurons transduced with lentivirus encoding for Mito-Keima in iCont and PARKIN KO lines. Scale bar, 25 μm.
(B) Quantification of Mito-Keima in iCont and PARKIN KO neuronal cultures. n = 10 neurons in each condition.
(C) Immunoblot of analysis of LC3I/II and autophagy marker P62.
(D and E) (D) Quantification of LC3 I/II and (E) P62 60s day post differentiation to DA neurons. n = 5 independent experiments.
Error bar represents the mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S3.
Elevation of PARIS and Reduction of PGC-1α in PARKIN-Deficient Human DA Neuron Cultures
The level of PARIS was elevated 3-fold in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 4A and 4B) along with an 80% reduction in TH expression via immunoblot analysis (Figures 4A and 4C). In addition, there was a 4-fold increase in PARIS levels in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron cultures (Figures S4A and S4B). In the PD1 and PD2 iPSC-derived DA neurons, there was no substantial difference in TH immunoreactivity compared with the iPSC control DA neurons (Figures S2A and S2B). PARIS is a transcriptional repressor that regulates the expression of peroxisome proliferator-activated receptor gamma, coactivator 1α, PGC-1α, a master coregulator of mitochondrial biogenesis, oxidative stress management, and function (Scarpulla, 2011). There was a 50%–60% reduction in PGC-1α levels in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 4D and 4E). There was a 55% reduction in PGC-1α levels in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron cultures (Figures S4A and S4C). Transmission electron microscopy of mitochondria from the human DA neuronal cultures revealed that the mitochondria were significantly less in size and number. There was also an increase in abnormal mitochondrial cristae in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 4F–4I). The abnormal cristae were defined by the presence of hollow cristae and a reduction in the mitochondrial matrix. Mitochondrial copy number was assessed by measuring the mitochondrial genes, tRNA Leu (MT), NADH-ubiquinone oxidoreductase chain 1(ND1), mitochondrially encoded ATP synthase membrane subunit 6 (ATP6), and mitochondrially encoded cytochrome c oxidase I (COX1), normalized to the nuclear-encoded β2-microglobulin gene. There was a 20%–30% reduction in tRNA Leu (MT), ND1, ATP6, and COX1 levels in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figure 4J) and a 40%–60% reduction in tRNALeu (MT), ND1, ATP6, and COX1 levels in the PD1 and PD2 iPSC-derived DA neurons compared with the iPSC control DA neuron cultures (Figure S4D). There was also a significant reduction in the integral mitochondrial proteins, cytochrome c (Cyt C), and CoxIV levels in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 4K, 4L, and 4M). Taken together, these results indicate that there was a reduction in mitochondrial mass in human DA neurons lacking PARKIN.
Figure 4.
Mitochondrial Biogenesis Defect in PARKIN KO Neurons
(A) Immunoblot analysis of PARIS, TH, PARKIN, and in PARKIN iCont and PARKIN KO, 60 days post differentiation to DA neurons.
(B and C) Quantification of immunoblot of (B) PARIS and (C) TH normalized to actin in iCont and PARKINKO neurons.
(D) Immunoblot analysis of PGC-1α in iCont and PARKIN KO lines, 60 days post differentiation to DA neurons.
(E) Quantification of immunoblots of PGC-1α normalized to actin in iCont and PARKIN KO neuronal cultures. n = 3 independent experiments.
(F) Transmission electron microscopy images of neurons derived from iCont and PARKIN KO hESCs, 60 days post differentiation to DA neurons. Scale bar, 500 nm.
(G–I) Quantification of (G) abnormal mitochondria cristae, (H) mitochondria size, and (I) mitochondria number/cell body in iCont and PARKIN KO hESC neurons. n = 16–17 cells for each condition.
(J) Mitochondrial DNA quantification of tRNALeu (MT), ND1, ATP6, and COX1, 60 days post differentiation to DA neurons in iCont and PARKIN KO hESCs, normalized to nuclear β2-microglobulin gene. n = 6 independent experiments.
(K) Immunoblot analysis of CytC and CoxIV in iCont and PARKIN KO hESCs, 60 days post differentiation to DA neurons.
(L and M) Quantification of immunoblots of (L) CytC and (M) CoxIV in iCont and PARKIN KO neuronal cultures. n = 5 independent experiments.
Error bar represents the mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figures S2 and S4.
Mitochondrial Biogenesis Defect in PARKIN-Deficient Human DA Neurons
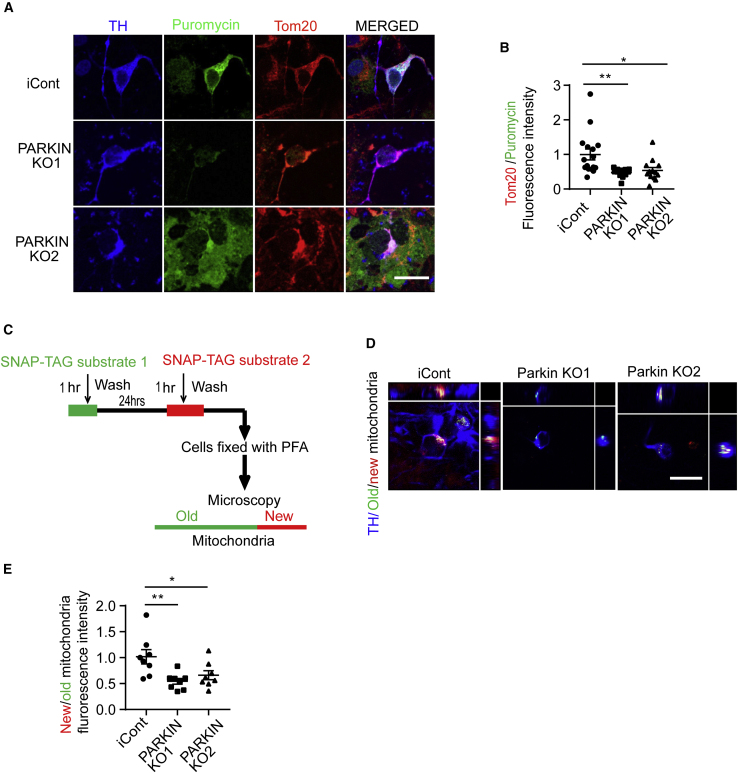
A modified non-radioactive protein puromycin labeling called surface sensing of translation (SUnSET) assay (Schmidt et al., 2009) was performed by assessing TOM20 and puromycin immunofluorescence levels within TH-positive neurons and used to assess mitochondrial biogenesis (Figures 5A and 5B). There was a 50%–60% reduction of puromycin labeling in TOM20-labeled mitochondria in human DA neurons (Figures 5A and 5B). Next, a (SNAP-TAG) self-protein labeling tag covalently fused with a suitable ligand targeted to the mitochondria via fusion to the integral mitochondrial protein Cox8A was transduced into human DA neuron cultures via lentivirus to monitor mitochondrial turnover (Figure 5C) (Gautier et al., 2008a; Hussain et al., 2013). Old versus new mitochondria were identified by using a green (Oregon Green) SNAP-TAG and a red (TMR-Star) SNAP-Tag substrate in TH-positive neurons and demonstrated that there was significant 50% reduction in the formation of new mitochondria within DA neurons in both PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 5D and 5E). In non-DA neurons, no significant defect in mitochondrial biogenesis was observed using both the SUnSET and SNAP-TAG assays (Figures S5A–S5D).
Figure 5.
Mitochondrial Biogenesis Defects in PARKIN KO DA Neurons
(A) Puromycin labeling SUnSET assay in iCont and PARKIN KO cells 60 days post differentiation to DA neurons. Anti-Puromycin (green), -TH (blue), and -Tom20 (red) immunofluorescence staining. Scale bar, 25 μm.
(B) Quantification of immunofluorescence staining for puromycin, TH, and Tom20 within the TH-positive neurons. n = 13–16 for each condition.
(C) Schematic diagram showing SNAP-tag, a protein labeling technique targeting the mitochondria. Old mitochondria are labeled with substrate 1 (green), and after 24 h substrate 2 (red) is added to label new mitochondria.
(D) Confocal Z stacked images of SNAP-tag Cox8a labeled mitochondria along with anti-TH (blue) immunostained neurons. Green (old) and new (red) mitochondria are labeled. Scale bar, 25 μm.
(E) Quantification of the fluorescence intensity of old (green) and new (red) Cox8a labeled mitochondria in TH (blue)-positive neurons. n = 8 in each condition.
Error bar represents the mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01. See also Figure S5.
PARKIN-Associated Mitophagy Defects and Reduction in PGC-1α Can Be Rescued by Expressing WT PARKIN in PARKIN KO and PARKIN Mutant iPSC Neuronal Cultures
Loss of PARKIN-induced defects were restored by introducing WT PARKIN in PARKIN KO hESCs and PARKIN mutant iPSC neuronal cultures. Mature neurons from PARKIN KO and PARKIN mutant iPSC neuronal cultures were transduced with a lentivirus expressing WT PARKIN. High levels of PARKIN were expressed in the PARKIN KO hESCs and PARKIN mutant iPSC neuronal cultures (Figures S5E and S5F). Lentiviral-mediated expression of WT PARKIN in PARKIN KO hESCs (Figures S5G and S5H) and PARKIN mutant iPSC (Figures S5I and S5J) neurons restored the mitophagy defects. Immunoblot analysis for PGC-1α also revealed that the levels of PGC-1α were restored following lentiviral-mediated expression of WT PARKIN in PARKIN KO hESCs and PARKIN mutant iPSC neuronal cultures (Figures S5K–S5N).
Reducing PARIS Levels Restores PGC-1α Levels without Restoring the Defects in Mitophagy
PARIS levels were reduced in human DA neuron cultures via CRISPR/Cas9 using lentiviral transduction of a guide RNA to PARIS. The levels of PARIS were reduced by 90%–95% in PARKIN KO1/PARIS KD and KO2/PARIS KD compared with the PARKIN KO1 and KO2 neuronal cultures (Figures 6A and 6B), and there was a 50%–80% reduction in the PD1/PARIS KD and PD2/PARIS KD iPSC-derived DA neurons compared with the PD1 and PD2 iPSCs DA neurons (Figures S6A and S6B). Accompanying the reduction in PARIS levels, PGC-1α was restored in the PARKIN KO1 and KO2 neuronal cultures compared with the iCont neuronal cultures (Figures 6C and 6D) and in the PD1 and PD2 iPSC-derived DA neuron cultures compared with the iPSC control DA neuron cultures (Figures S6C and S6D). In the PARKIN KO1 and KO2 neuronal cultures with PARIS knockdown, TH levels were also restored (Figures 6A, 6E, 6F, and 6G). The mitophagy index in both PARKIN KO1 and KO2 neuronal cultures remained reduced in the setting of PARIS knockdown (Figures 6H and 6I). The p62 and LC3 I/II levels also remain unchanged following PARIS knockdown (Figures 6J, 6K, and 6L).
Figure 6.
Generation and Characterization of PARKIN/PARIS Knockdown (PARKIN/PARIS KD) hESCs Lines
(A) Representative immunoblot of PARIS, TH, and PARKIN protein levels in iCont, PARKIN KO, and PARKIN/PARIS KD cells, 60 days post differentiation to DA neurons.
(B) Quantification of the immunoblot of PARIS protein levels. n = 3–4 independent experiments.
(C) Representative immunoblot for PGC-1α protein levels in iCont, PARKIN KO, and PARKIN KO/PARIS KD DA neurons in (A).
(D) Quantification of immunoblots for PGC-1α. n = 3–4 independent experiments.
(E) Quantification of the immunoblots of TH protein levels in iCont, PARKIN KO, and PARKIN/PARIS KD neuronal cultures in (A). n = 3–4 independent experiments.
(F) Immunofluorescence staining of TH-positive neurons (green) and nuclei (blue) iCont, PARKIN KO, and PARKIN/PARIS KD cells, 60s day post differentiation to DA neurons.
(G) Quantification of TH-positive neurons in (F). n = 5 independent experiments.
(H) Confocal live imaging of neurons transduced with lentivirus encoding for Mito-Keima in iCont, PARKIN KO, and PARKIN/PARIS KD cells, 60 days post differentiation to DA neurons.
(I) Quantification of Mito-Keima (mitophagy index) in (H). n = 19–25 neurons in each condition.
(J) Representative immunoblot of P62 and LC3 I/II in iCont, PARKIN KO, and PARKIN/PARIS KD cells, 60 days post differentiation to DA neurons.
(K and L) Quantification of (K) P62 and (L) LC3 I/II. n = 3–4 independent experiments.
Error bar represents the mean ± SEM. p values were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S6 and Table S1.
Rescuing Mitophagy Does Not Restore PGC-1α Levels in PARKIN KO Neurons
In the absence of PARKIN, mitophagy occurs at a low level, and this process can be enhanced by exposing neurons to the mitochondrial ionophore CCCP to induce mitophagy. We analyzed mitophagy in human DA neuron cultures by utilizing Mito-Keima in PARKIN KO neurons by treating with CCCP for 2 h at 10 μM. We observed that mitophagy was significantly induced in PARKIN KO neurons following treatment with CCCP (Figures S6E and S6F). Treatment of PARKIN KO neurons with CCCP does not change the level of PGC-1α (Figures S6G and S6H). These results suggest that inducing mitophagy in the absence of PARKIN is unable to rescue the reduction in the levels of PGC-1α.
Reducing PARIS Levels Restores Mitochondrial Respiratory and Biogenesis Defects in Human DA Neuron Cultures Lacking PARKIN
The reduction in basal respiration, spare respiratory capacity, and maximal respiration was rescued in the PARKIN KO1 (Figures 7A and 7B) and PARKIN KO2 (Figures S7A and S7B) neuronal cultures by knocking down PARIS. Basal respiration, spare respiratory capacity, and maximal respiration was also rescued in the in the PD1 and PD2 iPSC-derived DA neuron cultures (Figures S7C–S7F). Mitochondrial copy number as assessed by tRNA Leu (MT), ND1, ATP6, and COX1 levels normalized to the nuclear-encoded β2-microglobulin gene was restored in the PARKIN KO1 line (Figure 7C) and the PARKIN KO2 line (Figure S7G) and in the PD1 and PD2 iPSC-derived DA neuron cultures (Figures S7H and S7I). Knockdown of PARIS restores the mitochondrial biogenesis defect in DA neurons as determined by the puromycin labeling with the SUnSET (Figures 7D and 7E) and SNAP-tag assays in the PARKIN KO1 and KO2 neuronal cultures (Figures 7F and 7G). The status of healthy and unhealthy mitochondria was assessed based on mitochondrial morphology where elongated and tubular mitochondrial were counted as healthy and round and swollen mitochondria were counted as unhealthy. The reduction in size and number as well as the reduction in the percentage of healthy mitochondria was significantly restored by PARIS knockdown in the PARKIN KO1 and KO2 neuronal cultures (Figures 7H–7K). At the same time, the number of healthy mitochondria increased significantly following PARIS knockdown in the PARKIN KO1 and KO neuronal cultures (Figure 7L). The reduction in levels of the outer mitochondrial protein TOM20 in TH-positive neurons was significantly restored by PARIS knockdown in both PARKIN KO1 and KO2 neuronal cultures (Figures 7M and 7N). Mitochondrial membrane potential, as assessed via MitoCMXRos, was restored by PARIS knockdown in both PARKIN KO1 and KO2 neuronal cultures (Figures S7J and S7K). Taken together, these results indicate that a reduction in PARIS restored the mitochondrial respiratory and biogenesis defects due to the absence of PARKIN in human DA neurons.
Figure 7.
Rescue of Mitochondrial Function Defects in PARKIN/PARIS KD Lines
(A) Mitochondrial oxygen consumption rate (OCR) by the Seahorse platform in iCont, PARKIN KO1, and PARKIN/PARIS KD cells, 55 days post differentiation to DA neurons. The green line represents iCont, the red and blue lines represent PARKIN KO-1 and PARKIN/PARIS KD neurons, respectively.
(B) Quantification of basal respiration, reserve capacity, and maximal respiration in iCont, PARKIN KO1, and PARKIN/PARIS KD neurons. n = 3 independent experiment.
(C) Mitochondrial DNA quantification of tRNALeu (MT), ND1, ATP6, and COX1, 60 days post differentiation to DA neurons in iCont, PARKIN KO, and PARKIN/PARIS KD cells normalized to nuclear β2-microglobulin gene. n = 4 independent experiments.
(D) Puromycin labeling SUnSET assay in iCont, PARKIN KO neuronal cultures, and PARKIN/PARIS KD, 60 days post differentiation to DA neurons. Anti-Puromycin (green), anti-TH (blue), and anti-Tom20 (red) immunofluorescence staining.
(E) Quantification of the SUnSET assay. n = 10–15 TH neurons in each condition.
(F) Confocal Z stacked images of SNAP-tag Cox8a labeled mitochondria along with anti-TH (blue) immunostained neurons. New mitochondria are labeled in red and old mitochondria are labeled in green. Scale bar, 50 μm.
(G) Quantification of the fluorescence intensity of new (red) and old (green) Cox8a labeled mitochondria in TH (blue) positive neurons. n = 16 TH neurons in each condition.
(H) Transmission electron microscopy images of neurons from iCont and PARKIN KO and PARKIN/PARIS KD hESC lines from 60-day-old cultures. Scale bar, 500 nm.
(I–L) Quantification of mitochondrial (I) abnormal cristae, (J) size, (K) number/cell, and (L) number of healthy and unhealthy mitochondria in iCont and PARKIN KO and PARKIN/PARIS KD hESC-derived neurons, 60 days post differentiation. n = 15–35 cells in each condition.
(M) Immunofluorescence staining of Tom20 (red), TH (green), and nuclei stained with DAPI (blue) from iCont, PARKIN KO, and PARKIN/PARIS KD TH neurons, 60 days post differentiation. Colocalized Tom20 and TH are in white. Scale bar, 50 μm.
(N) Quantification of colocalized Tom20and TH in iCont, PARKIN KO, and PARKIN/PARIS KD TH neurons. n = 7–15 TH neurons in each condition.
Error bar represents the mean ± SEM. p value were determined using one-way ANOVA followed by Bonferroni's multiple comparisons test. Asterisks indicate the level of statistical significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S7 and Table S1.
Discussion
The major finding of this manuscript is the observation that human DA neurons lacking PARKIN have defects in mitochondrial respiratory capacity and biogenesis that are due in large part to elevation of the PARKIN substrate, PARIS. Moreover, the mitochondrial deficits are not likely to be due to defects in mitophagy since reducing PARIS levels fails to reverse the mitophagy defects, while it restores the defects in mitochondrial respiratory capacity and biogenesis.
Mitochondria play important roles in maintaining cellular health and metabolism. Alterations in mitochondrial quality control are thought to play a pivotal role in PD (Pickles et al., 2018; Scarffe et al., 2014). There are many aspects to mitochondrial quality control, but mitochondrial number is, in large part, governed by removal of damaged mitochondria (mitophagy) and production of new mitochondria (mitochondrial biogenesis) (Scarffe et al., 2014). PARKIN plays an important role in regulating both mitophagy (Ordureau et al., 2014; Panicker et al., 2017; Pickles et al., 2018) and biogenesis (Mouton-Ligeri et al., 2017; Scarffe et al., 2014) through processes that are regulated by PINK1. Consistent with this notion, we show that there are defects in both mitophagy and mitochondrial biogenesis in human DA neurons lacking PARKIN.
The use of human DA neurons provided an opportunity to dissect out the relative contribution of mitophagy versus mitochondrial biogenesis in the pathogenesis of PD due to the loss of PARKIN. Reducing PARIS levels by CRISPR/Cas9 restored PGC-1α levels without affecting the defects in mitophagy as assessed via Mito-Keima, while enhancing mitophagy failed to restore PGC-1α levels in DA neurons lacking PARKIN. Thus, although there are defects in mitophagy as assessed by Mito-Keima in human DA neurons, the major driver of the mitochondrial respiratory and mitochondrial depolarization defects appears to be decreased mitochondrial biogenesis. These mitochondrial respiratory and depolarization defects are likely occurring through the elevation of PARIS and the reduction of PGC-1α.
The use of isogenic PARKIN KO lines compared with the isogenic controls also indicates that the absence of PARKIN leads to a reduction in the number of DA neurons, which is restored by reducing PARIS levels. These results in human DA neurons are consistent with the observation that reducing PARIS levels in adult conditional Parkin KO mice prevents mitochondrial biogenesis defects and the loss of DA neurons through restoration of PGC-1α levels (Shin et al., 2011; Stevens et al., 2015). Similar to our results, a reduction in mitochondrial volume and TH neuronal counts was observed in isogenic PARKIN KO DA neurons compared with isogenic controls (Shaltouki et al., 2015). In iPSC DA neurons from patients lacking PARKIN, we also observed elevations of PARIS and accompanying defects in mitochondrial biogenesis, respiration, and depolarization that were restored by reducing PARIS levels without affecting the defects in mitophagy. However, there was no alteration in TH, which is in contrast to what was observed in the isogenic human PARKIN KO DA neurons reported here and by Shaltouki et al. (2015). These results highlight the importance of using isogenic controls to uncover phenotypic differences due to a genetic mutation in human-derived cultures (Soldner and Jaenisch, 2018). Lentiviral-mediated expression of PARKIN restores the defects in mitophagy and the reduction in PGC-1α levels, indicating the key phenotypes described in the human PARKIN KO and iPSC DA neurons are due to the loss of PARKIN and not some non-specific background effect. Since the number of iPSC DA neurons from patients lacking PARKIN was equal to the number of control iPSC DA neurons, and the defects in mitochondrial biogenesis and mitochondrial respiratory capacity and the defects in mitophagy were equivalent to those observed in the isogenic PARKIN KO DA neurons, it is unlikely that the defects in the isogenic lines are due to a reduction in TH number.
Our results, which indicate that there are defects in mitophagy in human DA neurons as assessed by a reduction in Mito-Keima, reduced LC3A, and increased p62 levels, strengthens the emerging data that PARKIN and by inference, PINK1, regulate mitophagy in DA neurons. These findings are consistent with the evidence that PINK1 and PARKIN regulate mitophagy in cancerous cell lines and rodent and Drosophila neurons (Cummins and Gotz, 2018; Whitworth and Pallanck, 2017). Our results contrast with in vivo studies in Drosophila expressing Mito-Keima, which uses a mitochondrial matrix-targeted pH-sensitive variant of GFP (Katayama et al., 2011), or mito-QC, which uses a tandem GFP-mCherry fusion protein targeted to the outer mitochondrial membrane (McWilliams et al., 2016) where defects in basal mitophagy were not observed in Drosophila lacking Parkin or PINK1 in a variety of cell types including DA neurons despite the loss of DA neurons in these models (Lee et al., 2018). PINK1 knockout mice expressing mito-QC also failed to show defects in basal mitophagy (McWilliams et al., 2018). On the other hand, combining Mito-Keima imaging with correlative light and electron microscopy in Drosophila indicates that mitophagy can occur in vivo and that the absence of Parkin or PINK1 reduced basal mitophagy (Cornelissen et al., 2018). However, 1-week-old Drosophila lacking PINK1 or Parkin exhibited mitochondrial abnormalities (Greene et al., 2003) but failed to exhibit a significant reduction in mitophagy (Cornelissen et al., 2018), suggesting that the mitochondrial deficits are separate from the mitophagy defects similar to the results in human DA neurons lacking PARKIN. The Drosophila PARIS homolog (Merzetti and Staveley, 2016) also drives the mitochondrial defects as it does in mouse and human DA neurons (Pirooznia et al., 2020).
Studies on both Drosophila and mice lacking either PINK1 or Parkin have failed to observe an increase in mitochondrial mass, which would be expected if defects in mitophagy were driving the mitochondrial defects. For instance, mass spectrometry revealed that mitophagy might not control mitochondrial turnover in the heads of PINK1 and Parkin KO files (Vincow et al., 2013). Mitochondria do not accumulate in PINK1 KO mice (Gautier et al., 2008b). Despite evidence of elevated phospho-ubiquitin, a marker of PINK1 activation and mitophagy, and loss of DA neurons, no increase in mitochondrial mass was observed in germline Parkin KO mice crossed to the DNA polymerase subunit gamma (POLG) 257A mitochondrial mutator mouse (Pickrell et al., 2015). An increase in mitochondrial mass or mitophagy was not observed in germline Parkin KO crossed to the PD-mito-PstI mouse despite mitochondrial DNA damage and enhanced DA neuron degeneration (Pinto et al., 2018). Indices of impaired mitophagy were not observed in germline Parkin KO mice crossed to mitochondrial transcription factor A (Tfam) KO mice (Sterky et al., 2011). Taken together, these reports suggest that defective mitophagy is not driving the alterations in mitochondrial function or loss of DA neurons (Cummins and Gotz, 2018; Whitworth and Pallanck, 2017). On the other hand, careful examination of mitochondrial content via mass spectrometry revealed decreased mitochondrial proteins (Palacino et al., 2004) and reduced respiratory capacity (Damiano et al., 2014) in germline Parkin KO mice. In human DA neurons lacking PARKIN, we observed a reduction in mitochondrial size, mass, and number, which is consistent with a defect in mitochondrial biogenesis. Reducing PARIS levels restores the defects in mitochondrial biogenesis and the indices of mitochondrial function without restoring the Mito-Keima defects. On the other hand, while enhancing mitophagy in human DA neurons lacking PARKIN restored the Mito-Keima defects, it was unable to restore the key defect in the reduction in PGC-1α levels. These results are consistent with the notion that increased PARIS levels and reduced mitochondrial biogenesis due to decreased PGC-1α levels is driving the mitochondrial phenotype, not mitophagy. In adult conditional Parkin KO mice, there was also a reduction in mitochondrial size, mass, and number, which was reduced by lowering PARIS levels (Stevens et al., 2015). There is emerging data that there are forms of mitophagy independent of PARKIN and PINK1, which is consistent with our observation that CCCP is able to induce mitophagy in human DA neurons lacking PARKIN (Di Rita et al., 2018; Kageyama et al., 2014; Villa et al., 2017). We cannot exclude the possibility that these other mitophagy pathways play a role in the survival of DA neurons in PD.
In summary, our findings highlight that mitochondrial mitophagy and mitochondrial biogenesis are impaired in human DA neurons lacking PARKIN and that the impairment in mitochondrial biogenesis is an important driving force in the reduction of mitochondrial function and the loss of DA neurons in PD due to PARKIN deficiency or inactivation. Strategies focused on enhancing mitochondrial biogenesis may offer new therapeutic approaches to preventing the loss of DA neurons in PD.
Experimental Procedures
iPSC Generation and Characterization
hiPSCs were generated using a CytoTune -iPS Sendai Reprogramming Kit (Thermo Scientific) following the manufacturer's instruction. Established hiPSC clones were karyotyped and characterized for stemness by evaluating the expression level of stem cell markers using immunocytochemistry. Pluripotency was tested by an in vivo teratoma assay. Gene deletion was confirmed using RT-PCR and immunoblots.
Maintenance and Differentiation of hESCs and hiPSCs to DA Neurons
All procedures involving human embryonic and inducible pluripotent stem cells were approved by and conformed to the guidelines of the Johns Hopkins Medicine Institution Review Board and Johns Hopkins University Institutional Stem Cell Research Oversight Committee. Differentiation of hESCs and hiPSCs to dopamine neurons was done as described by Kriks et al. (2011). Full details are provided in Supplemental Experimental Procedures.
ZFN Design and Generation of PARKIN KO hESCs
The ZFN expression and targeting vector were purchased from Transposagen. ZFNs were designed against the human PARKIN gene locus targeting exon 7. Individual targeted colonies were selected and screened. Full details are provided in Supplemental Experimental Procedures.
hESC and hiPSC Analysis
Immunoblot and immunohistochemistry were performed using previously described methods (Xu et al., 2016). Genomic DNA was extracted using QIAGEN kit and was separated on a 0.7% agarose gel after restriction digest with appropriate restriction enzyme for Southern blots. Total RNA was extracted from the cells using a QIAGEN RNA isolation kit for RT-PCR. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature for microscopy and imaging with an LSM7 or Airy scan (Carl Zeiss) confocal laser scanning microscope under a 20× or 40× oil objective. The oxygen consumption rate was assessed via a Sea Horse instrument. For CRISPR Cas9, knockdown guide RNAs were designed using the online tool from http://crispr.mit.edu/. Lentivirus was used to transduce human cells. Transmission electron microscopy was performed on neurons that were fixed in 2.5% glutaraldehyde, 3 mM MgCl2, in 0.1 M sodium cacodylate buffer, 60 days post differentiation. Images were captured with an AMT XR80 high-resolution (16-bit) 8 M pixel camera. Full details are provided in Supplemental Experimental Procedures.
Antibodies
Anti-mouse parkin, Cell Signaling, catalog no. 4211; anti-mouse PARIS, NeuroMab, catalog no. 75-195; anti-rabbit TH, EMD Millipore, catalog no. AB152; anti-rabbit P62, Cell Signaling, catalog no. 8025; anti-rabbit LC3A, Cell Signaling, catalog no. 4599; anti-mouse TUJ1, Covance, catalog no. MMS-435P; anti-rabbit PGC1α, Novus, catalog no. NBP1-04676; anti-rabbit Annexin V, Abcam, catalog no. ab14196; anti-mouse Puromycin, EMD Millipore, catalog no. MABE343; anti-mouse Tom20, Santacruz, catalog no. SC-17764; anti-rabbit CytC, Cell Signaling, catalog no. 4280; anti-rabbit COXIV, Cell Signaling, catalog no. 4850; anti-mouse Actin, Sigma, catalog no. A3854; anti-rabbit Ki76, Abcam, catalog no. ab15580; goat anti-mouse IgG, Thermo Fisher, catalog no. A-11029; StemLight Pluripotency Antibody Kit, Cell Signaling, catalog no. 9656; goat anti-rabbit IgG, Thermo Fisher, catalog no. A-11036; goat anti-rabbit IgG, Thermo Fisher, catalog no. A-21068.
Quantification and Statistical Analysis
Details of the statistical analyses performed can be found in Supplemental Experimental Procedures and in the figure legends of the respective figures.
Author Contributions
Conceptualization, V.L.D. and T.M.D.; Methodology, M.K., J.A.-C., A.J., H.W., S.A.A., S.E., S.S.K., S.B., R.C., H.K., H.S.K., V.L.D., and T.M.D.; Formal Analysis, M.K.; Investigation, M.K., J.A.-C., A.J., H.W., S.A.A., S.E., S.S.K., S.B., R.C., H.K., and H.S.K; Writing – Original Draft, M.K., V.L.D., and T.M.D.; Writing – Review & Editing, M.K., J.A.-C., A.J., S.A.A., S.E., S.S.K. S.B., R.C., H.K., H.S.K., V.L.D., and T.M.D.; Funding Acquisition, H.S.K., V.L.D., and T.M.D.; Supervision, H.S.K., V.L.D., and T.M.D.
Conflicts of Interest
The value of patents owned by Valted, LLC, could be affected by the study described in this article. Also, T.M.D. and V.L.D. are founders of Valted, LLC, and hold an ownership equity interest in the company. This arrangement has been reviewed and approved by The Johns Hopkins University in accordance with its conflict of interest policies.
Acknowledgments
This work was supported by grants from the NIH/NINDS NS38377, and the JPB Foundation. The authors acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation and the Diana Helis Henry Medical Research Foundation through their direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation's Parkinson's Disease Programs M-1, M-2, H-2014. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. We thank the Dawson lab personnel for helpful suggestions.
Published: August 13, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.07.013.
Contributor Information
Valina L. Dawson, Email: vdawson@jhmi.edu.
Ted M. Dawson, Email: tdawson@jhmi.edu.
Supplemental Information
References
- Brahmachari S., Lee S., Kim S., Yuan C., Karuppagounder S.S., Ge P., Shi R., Kim E.J., Liu A., Kim D. PARKIN interacting substrate zinc finger protein 746 is a pathological mediator in Parkinson's disease. Brain. 2019;142:2380–2401. doi: 10.1093/brain/awz172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.Y., Kishinevsky S., Mazzulli J.R., Graziotto J., Mrejeru A., Mosharov E.V., Puspita L., Valiulahi P., Sulzer D., Milner T.A. PARKIN and PINK1 patient iPSC-derived midbrain dopamine neurons exhibit mitochondrial dysfunction and alpha-synuclein accumulation. Stem Cell Rep. 2016;7:664–677. doi: 10.1016/j.stemcr.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with PARKIN. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Cornelissen T., Vilain S., Vints K., Gounko N., Verstreken P., Vandenberghe W. Deficiency of PARKIN and PINK1 impairs age-dependent mitophagy in Drosophila. eLife. 2018;7:e35878. doi: 10.7554/eLife.35878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N., Gotz J. Shedding light on mitophagy in neurons: what is the evidence for PINK1/PARKIN mitophagy in vivo? Cell Mol. Life Sci. 2018;75:1151–1162. doi: 10.1007/s00018-017-2692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano M., Gautier C.A., Bulteau A.L., Ferrando-Miguel R., Gouarne C., Paoli M.G., Pruss R., Auchere F., L'Hermitte-Stead C., Bouillaud F. Tissue- and cell-specific mitochondrial defect in PARKIN-deficient mice. PLoS One. 2014;9:e99898. doi: 10.1371/journal.pone.0099898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson T.M., Dawson V.L. PARKIN plays a role in sporadic Parkinson's disease. Neuro Degener. Dis. 2014;13:69–71. doi: 10.1159/000354307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rita A., Peschiaroli A., P D.A., Strobbe D., Hu Z., Gruber J., Nygaard M., Lambrughi M., Melino G., Papaleo E. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKalpha. Nat. Commun. 2018;9:3755. doi: 10.1038/s41467-018-05722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo A., Klein C. Genetics of Parkinson disease. Handb Clin. Neurol. 2018;147:211–227. doi: 10.1016/B978-0-444-63233-3.00014-2. [DOI] [PubMed] [Google Scholar]
- Gautier A., Juillerat A., Heinis C., Correa I.R., Jr., Kindermann M., Beaufils F., Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Gautier C.A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J.C., Whitworth A.J., Kuo I., Andrews L.A., Feany M.B., Pallanck L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila PARKIN mutants. Proc. Natl. Acad. Sci. U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A.F., Amoury M., Barth S. SNAP-tag technology: a powerful tool for site specific conjugation of therapeutic and imaging agents. Curr. Pharm. Des. 2013;19:5437–5442. doi: 10.2174/1381612811319300014. [DOI] [PubMed] [Google Scholar]
- Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Hoke A., Dawson V.L., Dawson T.M. PARKIN-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33:2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Kogure T., Mizushima N., Yoshimori T., Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the PARKIN gene cause autosomal recessive juvenile PARKINsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., Sanchez-Martinez A., Zarate A.M., Beninca C., Mayor U., Clague M.J., Whitworth A.J. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or PARKIN. J. Cell Biol. 2018;217:1613–1622. doi: 10.1083/jcb.201801044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Stevens D.A., Kang S.U., Jiang H., Lee Y.I., Ko H.S., Scarffe L.A., Umanah G.E., Kang H., Ham S. PINK1 primes PARKIN-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 2017;18:918–932. doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin I., Dawson V.L., Dawson T.M. Recent advances in the genetics of Parkinson's disease. Annu. Rev. Genomics Hum. Genet. 2011;12:301–325. doi: 10.1146/annurev-genom-082410-101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams T.G., Prescott A.R., Allen G.F., Tamjar J., Munson M.J., Thomson C., Muqit M.M., Ganley I.G. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams T.G., Prescott A.R., Montava-Garriga L., Ball G., Singh F., Barini E., Muqit M.M.K., Brooks S.P., Ganley I.G. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 2018;27:439–449.e5. doi: 10.1016/j.cmet.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzetti E.M., Staveley B.E. Identifying potential PARIS homologs in D. melanogaster. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr15048934. [DOI] [PubMed] [Google Scholar]
- Mouton-Ligeri F., Jacoupy M., Corvol J.C., Corti O. PINK1/PARKIN-dependent mitochondrial surveillance: from pleiotropy to Parkinson's disease. Front Mol. Neurosci. 2017;10:120. doi: 10.3389/fnmol.2017.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate PARKIN. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., Sarraf S.A., Duda D.M., Heo J.M., Jedrychowski M.P., Sviderskiy V.O., Olszewski J.L., Koerber J.T., Xie T., Beausoleil S.A. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacino J.J., Sagi D., Goldberg M.S., Krauss S., Motz C., Wacker M., Klose J., Shen J. Mitochondrial dysfunction and oxidative damage in PARKIN-deficient mice. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Panicker N., Dawson V.L., Dawson T.M. Activation mechanisms of the E3 ubiquitin ligase PARKIN. Biochem. J. 2017;474:3075–3086. doi: 10.1042/BCJ20170476. [DOI] [PubMed] [Google Scholar]
- Park J.S., Davis R.L., Sue C.M. Mitochondrial dysfunction in Parkinson's disease: new mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. 2018;18 doi: 10.1007/s11910-018-0829-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S., Vigie P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018;28:R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell A.M., Huang C.H., Kennedy S.R., Ordureau A., Sideris D.P., Hoekstra J.G., Harper J.W., Youle R.J. Endogenous PARKIN preserves dopaminergic substantia nigral neurons following mitochondrial DNA mutagenic stress. Neuron. 2015;87:371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell A.M., Youle R.J. The roles of PINK1, PARKIN, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto M., Nissanka N., Moraes C.T. Lack of PARKIN anticipates the phenotype and affects mitochondrial morphology and mtDNA levels in a mouse model of Parkinson's disease. J. Neurosci. 2018;38:1042–1053. doi: 10.1523/JNEUROSCI.1384-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia S.K., Yuan C., Khan M.R., Karuppagounder S.S., Wang L., Xiong Y., Kang S.U., Lee Y., Dawson V.L., Dawson T.M. PARIS induced defects in mitochondrial biogenesis drive dopamine neuron loss under conditions of PARKIN or PINK1 deficiency. Mol. Neurodegener. 2020;15:17. doi: 10.1186/s13024-020-00363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitt J.M., Dawson V.L., Dawson T.M. Diagnosis and treatment of Parkinson disease: molecules to medicine. J. Clin. Invest. 2006;116:1744–1754. doi: 10.1172/JCI29178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe L.A., Stevens D.A., Dawson V.L., Dawson T.M. PARKIN and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Shaltouki A., Sivapatham R., Pei Y., Gerencser A.A., Momcilovic O., Rao M.S., Zeng X.M. Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Rep. 2015;4:847–859. doi: 10.1016/j.stemcr.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A., Bhaumik D., Chinta S.J., Rane A., Rajagopalan S., Lieu C.A., Lithgow G.J., Andersen J.K. Mitochondrial quality control via the PGC1alpha-TFEB signaling pathway is compromised by PARKIN Q311X mutation but independently restored by rapamycin. J. Neurosci. 2015;35:12833–12844. doi: 10.1523/JNEUROSCI.0109-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A., Rane A., Rajagopalan S., Chinta S.J., Andersen J.K. Detrimental effects of oxidative losses in PARKIN activity in a model of sporadic Parkinson's disease are attenuated by restoration of PGC1alpha. Neurobiol. Dis. 2016;93:115–120. doi: 10.1016/j.nbd.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Soldner F., Jaenisch R. Stem cells, genome editing, and the path to translational medicine. Cell. 2018;175:615–632. doi: 10.1016/j.cell.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F.H., Lee S., Wibom R., Olson L., Larsson N.G. Impaired mitochondrial transport and PARKIN-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. U S A. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.A., Lee Y., Kang H.C., Lee B.D., Lee Y.I., Bower A., Jiang H., Kang S.U., Andrabi S.A., Dawson V.L. PARKIN loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc. Natl. Acad. Sci. U S A. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E., Proics E., Rubio-Patino C., Obba S., Zunino B., Bossowski J.P., Rozier R.M., Chiche J., Mondragon L., Riley J.S. PARKIN-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep. 2017;20:2846–2859. doi: 10.1016/j.celrep.2017.08.087. [DOI] [PubMed] [Google Scholar]
- Vincow E.S., Merrihew G., Thomas R.E., Shulman N.J., Beyer R.P., MacCoss M.J., Pallanck L.J. The PINK1-PARKIN pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci. U S A. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S. PINK1-dependent recruitment of PARKIN to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth A.J., Pallanck L.J. PINK1/PARKIN mitophagy and neurodegeneration-what do we really know in vivo? Curr. Opin. Genet. Dev. 2017;44:47–53. doi: 10.1016/j.gde.2017.01.016. [DOI] [PubMed] [Google Scholar]
- Xu J.C., Fan J., Wang X., Eacker S.M., Kam T.I., Chen L., Yin X., Zhu J., Chi Z., Jiang H. Cultured networks of excitatory projection neurons and inhibitory interneurons for studying human cortical neurotoxicity. Sci. Transl. Med. 2016;8:333ra348. doi: 10.1126/scitranslmed.aad0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.