Summary
Mouse embryonic stem cells (ESCs) cultured in defined medium resemble the pre-implantation epiblast in the ground state, with full developmental capacity including the germline. β-Catenin is required to maintain ground state pluripotency in mouse ESCs, but its exact role is controversial. Here, we reveal a Tcf3-independent role of β-catenin in restraining germline and somatic lineage differentiation genes. We show that β-catenin binds target genes with E2F6 and forms a complex with E2F6 and HMGA2 or E2F6 and HP1γ. Our data indicate that these complexes help β-catenin restrain and fine-tune germ cell and neural developmental potential. Overall, our data reveal a previously unappreciated role of β-catenin in preserving lineage differentiation integrity in ground state ESCs.
Keywords: mouse embryogenesis, ground state, naive pluripotency, ESC ground state maintenance, germline, Wnt signaling, lineage differentiation
Highlights
-
•
β-Catenin depletion irreversibly compromised lineage development of ground state ESCs
-
•
TCF3-independent role of β-catenin in determining lineage differentiation potential
-
•
E2F6, HP1γ, and HMGA2 are β-catenin interaction partners and co-bound to target genes
-
•
β-Catenin and protein partners fine-tune germline and neural development potential
In this article, Tao and colleagues show that β-catenin functions beyond derepressing TCF3 targeted pluripotency network in ESCs. β-Catenin physically interacts with protein partners, including transcription factor and chromatin modulators. Together, they preserve the integrity of lineage differentiation of ground state ESCs by restraining and fine-tuning their germline and neural development potential.
Introduction
Pre-implantation epiblast cells from the inner cell mass (ICM) can give rise to all cell lineages, including the germline (Nichols and Smith, 2009; Silva and Smith, 2008). This naive pluripotent or ground state is captured in vitro by self-renewing mouse embryonic stem cells (ESCs) grown in defined media with two inhibitors (2i), which inhibit MEK and GSK3 (Evans and Kaufman, 1981; Ying et al., 2008). The ground state is established and maintained by a core transcription program controlled by the transcription factors (TFs) Oct4, Sox2, Nanog, and others (Chambers et al., 2007; Chen et al., 2008; Dunn et al., 2014; Hackett and Surani, 2014). After implantation, the epiblast becomes primed for somatic germ layer specification marked by the upregulation of Fgf5, Otx2, and Pou3f1 (Oct6) (Boroviak et al., 2015; Kojima et al., 2014). Exit from naive pluripotency involves disassembly of the pluripotency machinery and acquisition of a post-implantation transcription program (Hackett and Surani, 2014). Transition to the primed state has been studied in vitro (Hayashi et al., 2011); however, it is unclear how signaling pathways orchestrate the transition from pluripotency to lineage specification. In particular, Wnt signaling's role is puzzling (Anton et al., 2007; Lyashenko et al., 2011; Raggioli et al., 2014; Sokol, 2011).
Wnt proteins are required to prevent ESCs from exiting ground state pluripotency and entering the primed state (ten Berge et al., 2011). The Wnt effector T cell factor-3 (Tcf3) helps maintain the ground state and prevents differentiation, despite being a repressor (Cole et al., 2008). β-Catenin is critical for keeping ESCs in the ground state (Wray et al., 2011) but also promotes cell fate specification and differentiation of ESCs, including neural development (Okumura et al., 2013; Rudloff and Kemler, 2012). But while β-catenin-null ESCs have defective ectoderm formation (Lyashenko et al., 2011), blocking Wnt signaling/β-catenin promoted neuronal differentiation, and overexpression of β-catenin inhibited neuronal differentiation (Cajanek et al., 2009; Kelly et al., 2011). It was proposed that β-catenin might mediate some of its functions independently of Tcf3, e.g., via enhancing Oct4 activity to reinforce pluripotency of ESCs (Kelly et al., 2011), but this hypothesis has not been sufficiently explored. The β-catenin interactome has been widely studied (Mosimann et al., 2009; Zhurinsky et al., 2000), but it is not known how interacting components contribute to β-catenin function in vivo.
Here, we investigated how β-catenin and associated proteins regulate ground state maintenance and lineage commitment of ESCs. We revealed a role of β-catenin in repressing differentiation by interacting with E2F6, HP1γ, and HMGA2. Genome-wide binding of β-catenin and associated protein partners indicates that they act as transcriptional repressors of key lineage development genes. Knockdown of β-catenin-associated protein partners leads to exit from the ground state and premature differentiation into the germline and neuronal lineages.
Results
β-Catenin Depletion Leads to Ground State Exit before Core Pluripotency TF Downregulation
To confirm β-catenin function in maintaining mouse ESCs, we knocked down β-catenin expression (Figure S1A). In conventional media, growth of β-catenin-depleted ESCs was reduced (Figure S1B), likely due to cell death (Raggioli et al., 2014), but cell morphology and alkaline phosphatase (AP) activity remained normal (Figure S1C). In contrast, under culture with two inhibitors (2i), downregulation of β-catenin resulted in disrupted ESC morphology with reduced AP activity, which indicated differentiation (Figure 1A). This is consistent with a previous report (Wray et al., 2011) and confirms that β-catenin is required for ESC ground state maintenance. Unexpectedly, Oct4 and Sox2 were still detected at 72 h and Nanog was slightly reduced by 10% and 20% at 48 h and 72 h, respectively, after β-catenin knockdown (Figures 1B–1D and S1D).
Figure 1.
β-Catenin Knockdown in 2i Culture
(A) Morphology of β-catenin-knockdown ESCs at 48 h. Differential interference contrast microscopy with AP staining. Scale bar, 100μm.
(B) Western blot of β-catenin, Oct4, Sox2, and β-actin (loading control) at 24, 48, and 72 h in Cre−β-cateninfl/fl and Cre+β-cateninfl/fl ESCs in 2i media.
(C) IF staining of β-catenin, OCT4, SOX2, and NANOG at 48 h after β-catenin knockdown compared with mock control ESCs. Scale bar, 25μm.
(D) Flow cytometric analysis of Nanog expression at 48 and 72 h after β-catenin knockdown in 2i culture (n = 4–5 biological replicates).
(E) MA plot of RNA-seq expression data from GFP mock control and β-catenin knockdown ESCs (n = 2–6 biological replicates).
(F–I) Gene set enrichment analysis and examples of relative expression of two selected groups of signature genes: ICM (F–G) and post implantation epiblast (Postimp epi) (H and I) in β-catenin-knockdown ESCs compared with mock control (n = 2–6 biological replicates).
(J) IF staining of OTX2 and PRDM1 at 48 h after β-catenin knockdown compared with mock control ESCs and epiLCs. Scale bar, 20 μm.
(K) Image quantification of OTX2 signal intensity per ESC from (J) (n value indicates number of cells, n = 1,939 shGFP ESCs, n = 1,279 shβcat ESCs).
(L) Image quantification of PRDM1 signal intensity out of DAPI background from (J) (n value indicates number of ESC colonies, n = 12 shGFP ESC colonies, n = 13 shβcat ESC colonies).
(M) Distribution of β-catenin binding sites at transcription starting site (TSS, −1 to +1 kb), gene body, downstream, and upstream of the nearest genes (n = 2 independent experiments).
(N) Integration of the β-catenin ChIP-seq dataset with DE genes in β-catenin-knockdown ESCs (RNA-seq) identified 515 direct target genes of β-catenin (p = 6.67566 × 10−66 of overlapped genes with hypergeometric test) (n = 2 biological replicates).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Error bars, SEM.
Altered morphology in β-catenin-depleted ESCs prior to core TF downregulation indicates that β-catenin has a direct role in maintaining the ground state. RNA-sequencing (RNA-seq) analysis comparing β-catenin-knockdown cells with controls after 48 h identified 391 and 862 upregulated and downregulated genes, respectively (Figure 1E). Surprisingly, expression of pluripotency genes such as Pecam1, Nr0b1, and Stat3 were unchanged after β-catenin knockdown (Figure S1E). However, genes predominantly expressed in the ICM were downregulated, whereas post-implantation genes, including markers for primed epiblast cells such as Fgf5, Otx2, and Oct6, were significantly increased (Figures 1F–1I). An increase in OTX2 protein was confirmed by immunofluorescent (IF) staining: 428 ± 7 in β-catenin-knockdown ESCs compared with 373 ± 4 in the mock control (Figures 1J and 1K). We also observed a marked increase of the DNA methylase Dnmt3l, which is often associated with hypermethylation, indicating loss of ground state (Leitch et al., 2013). Intriguingly, signature genes downregulated in epiLCs such as Zfp42 (Hayashi et al., 2011) were not changed in β-catenin-knockdown ESCs (Figure S1E).
To clarify whether β-catenin-knockdown ESCs are similar to epiLCs, we performed RNA-seq time-course experiments using mock control and β-catenin knockdown cells with epiLCs induction. β-catenin-depleted cells showed similar morphological changes to those of control cells (Figure S1F) and stalled cell growth (Figure S1G). Overall, we observed differences between β-catenin-knockdown and control cells as shown by principal component analysis at 48 h (Figure S1H). However, β-catenin-knockdown cells after 24 and 48 h in induction media strongly correlated with control cells after 96 h in induction media when cells resemble the epiLC state. This indicates that loss of β-catenin drives cells more rapidly toward epiLC fate. Indeed, β-catenin-knockdown cells showed signs of further lineage differentiation. For example, Prdm1, which is required for specification of primordial germ cells (PGCs) (Vincent et al., 2005), was upregulated >2-fold by IF staining (Figures 1J and 1L). Furthermore, neurogenesis was upregulated at 48 h after knockdown (Figure S1I). These data indicate that β-catenin inhibits ESC transition from the ground state.
To understand the molecular mechanism, we identified genomic targets bound by β-catenin using a knockin ESC line with C-terminal FLAG-tagged endogenous β-catenin (Zhang et al., 2013). We identified 18,469 β-catenin binding regions, which mapped to 11,178 genes (Figures 1M and 1N). Among these genes, 160 were upregulated and 355 were downregulated in the absence of β-catenin, suggesting that β-catenin both activates and represses target genes. Gene ontology (GO) analysis showed upregulated genes enriched for processes indicating differentiation, while downregulated genes were consistent with β-catenin's feedback regulation of Wnt signaling and in anterior/posterior axis formation (Huelsken et al., 2000) (Figures 1N, S1I, and S1J). Both upregulated and downregulated genes were enriched for reproduction-related processes, suggesting that β-catenin regulates priming germ cell development in ground state ESCs.
β-Catenin Depletion in ESCs Irreversibly Compromises Lineage Development
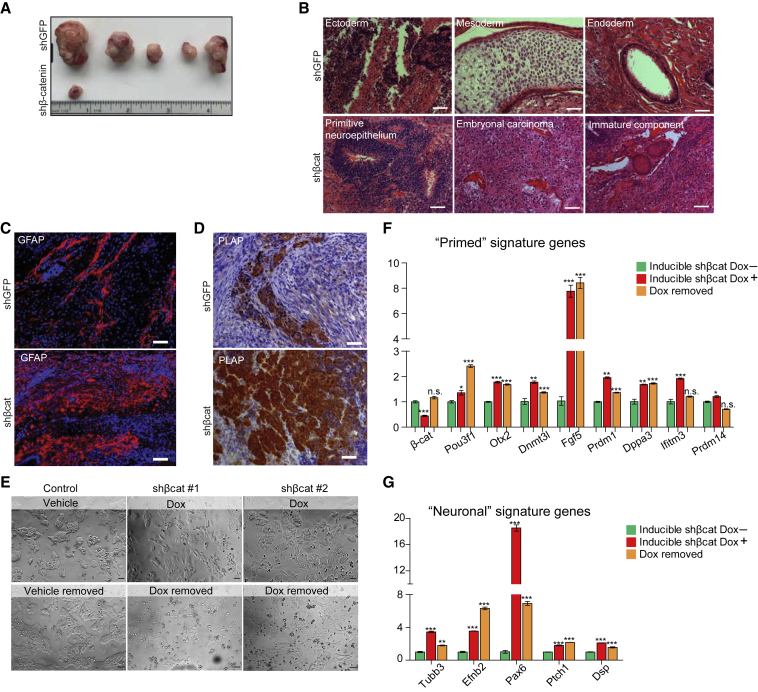
To better understand lineage differentiation potential, we assessed β-catenin knockdown ESCs in vivo. One month after transplantation, 5/5 control mice developed teratomas (0.5–2 cm) compared with 1/5 (0.25 cm) that received β-catenin-depleted ESCs (Figure 2A). Control ESCs gave rise to all three germ layers; however, β-catenin-depleted teratomas were largely embryonal carcinoma, which corresponds to an immature teratoma of grade 3 of the Norris system in humans. Specifically, they are characterized by central nucleoli with clear cytoplasm containing elements of primitive neuroepithelia undergoing mitosis, although foci of well-differentiated epithelium and smooth muscle were also present (Figure 2B). Consistently, we found substantially stronger signals of both neuroectoderm marker GFAP and embryonal carcinoma marker PLAP in β-catenin-depleted ESC-derived teratoma than in control ESCs (Figures 2C and 2D).
Figure 2.
β-Catenin Depletion in ESCs Irreversibly Compromises Lineage Development
(A–D) Teratoma formation assay (A), H&E staining (B), and IHC staining of embryonal carcinoma marker PLAP (C) and neuroectoderm marker GFAP (D) in teratoma tissue sections from control and β-catenin-depleted ESCs. Scale bar, 50 μm.
(E) Live images of ESCs in 2i culture with Dox (1 μg/mL) induced β-catenin knockdown for 72 h (upper panels) and Dox removal for 48 h (lower panels). Scale bar, 50 μm.
(F and G) qPCR showing expression of primed (F) and neuronal (G) signature genes in 2i cultured ESCs with Dox-induced β-catenin knockdown for 72 h and Dox removal for 48 h (n = 3 biological replicates for all 3 groups).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Error bars, SEM.
β-catenin is indispensable for mesoderm specification (Lindsley et al., 2006). Indeed, mesoderm gene expression was reduced after β-catenin knockdown, and β-catenin bound directly to Cdx1 and Wnt8 promoters or the 3′ region of Hoxa1 mesoderm genes (Figures S2A and S2B), providing candidate enhancer regions by which β-catenin promotes mesoderm development.
To test whether β-catenin depletion in ground state ESCs led to transient or permanent effects on developmental trajectory, we established a doxycycline (Dox)-inducible β-catenin knockdown ESC line, in which transcription is restored by Dox removal. Differentiation was observed 72 h post Dox induction, but removing Dox did not rescue ESC cultures (Figure 2E). Primed state signature genes were de-repressed after Dox induction and retained higher expression levels after Dox removal (Figure 2F). Restoring β-catenin did not reverse the neuronal lineage differentiation phenotype caused by β-catenin knockdown (Figure 2G), showing that β-catenin knockdown irreversibly changes ESC differentiation potential.
Tcf3-Independent Functions of β-Catenin in Determining Lineage Differentiation Potential
To decipher the relationship between β-catenin and Tcf3 in ESCs, we performed TCF3 chromatin immunoprecipitation sequencing (ChIP-seq) and intersected target regions with β-catenin (Figure 3A). Co-occupied regions included canonical Wnt signaling components (Axin2), stem cell maintenance (Oct4 and Zfp42), trophectoderm and endoderm formation (Cdx2, Foxa2), and anti-apoptosis (Mcl1) (Figures S3A–S3C). These data are consistent with previous reports of the Tcf3-dependent role of β-catenin in ESC maintenance by directly binding to pluripotency factors and developmental regulators (Cole et al., 2008). We reasoned if the sole function of β-catenin is to counteract the repressive effect of Tcf3, one might expect that the additional knockdown of Tcf3 would rescue the phenotype of the β-catenin depletion phenotype. We found double knockdown of β-catenin and Tcf3 partially alleviated the ESC differentiation phenotype (Figures S3D and S3E); however, morphological features were still similar to β-catenin-depleted ESCs (Figure S3F). Because partial rescue could be due to incomplete knockdown of Tcf3 (Figure S3D), we tested quadruple-knockout (QKO) mouse ESCs lacking all full-length TCF/LEFs (Moreira et al., 2017). QKO ESCs remained indistinguishable at 48 h after β-catenin knockdown (Figure 3B), indicating that loss of all TCFs rescues the ESC phenotype morphologically. However, RNA-seq analysis revealed 429 differentially expressed (DE) genes solely bound by β-catenin and 149 genes bound by both β-catenin and TCF3 (Figure 3C). Annotation of β-catenin-only bound genes showed circulatory and skeletal developmental regulation and stem cell differentiation (Figure 3D). Altogether, the phenotypic rescue confirms that β-catenin counteracts the repressive effect of Tcf3 in ground state ESCs but supports a Tcf3-independent function lineage differentiation. To test whether ESC differentiation was compromised in β-catenin-deficient QKO cells, we performed teratoma assays. β-catenin-deficient QKO cells gave rise to 721-fold higher CD30+ cells, which represent germ cell-derived embryonal carcinoma, but 1.5-fold fewer GFAP+ (glial fibrillary acidic protein) neuronal lineage cells (Figures 3E and 3F). Thus, despite normal morphology, β-catenin-deficient QKO cells have altered developmental potential.
Figure 3.
Tcf3-Independent Functions of β-Catenin in Determining Lineage Differentiation Potential
(A) Heatmap of β-catenin binding loci showing β-catenin and TCF3 co-bound and β-catenin-only bound regions (n = 2 independent experiments).
(B) Live culture images of QKO cells at 48 h after β-catenin knockdown compared with mock control. Scale bar, 50 μm.
(C) Differentially expressed (DE) genes at 48 h in β-catenin-knockdown QKO cells. DE genes were shown as bound by β-catenin only or by both β-catenin and TCF3.
(D) GO analysis (Metascape) of 429 DE genes bound by β-catenin only in β-catenin-knockdown QKO cells.
(E and F) Representative images of IF staining (E) and quantification (F) of pan-Cytokeratin, CD30, and GFAP at 48 h after β-catenin knockdown in QKO cells (n = 4–6 biological replicates). Scale bar, 50 μm.
(G) Venn diagram showing overlap between DE genes in β-catenin-depleted ESCs and genes bound by β-catenin only (ChIP-seq, n = 2 independent experiments; RNA-seq, n = 2–6 biological replicates).
(H) Putative transcription factor regulatory motif analysis of the 429 genes in (G).
∗p < 0.05. Error bars, SEM.
To identify Tcf3-independent functions of β-catenin, we analyzed DE genes of β-catenin-knockdown ESCs bound by β-catenin only (Figures 3G and 3H). We found TF DNA motifs including Sp1, Klf4/5 (early embryonic development and pluripotency (Marin et al., 1997)), and Zic3 (transition from naive to primed pluripotency (Yang et al., 2019)). Furthermore, we found enriched motifs of Ap-2γ (Tfap2c) and BCL6B, which are implicated in PGC development (Ishii et al., 2012), adding more evidence to a role of β-catenin in regulating germ cell development (Figures S1I and S1J). Overall, these data further show that β-catenin regulates the differentiation potential of ground state ESCs independently of Tcf3.
E2F6, HP1γ, and HMGA2 Are β-Catenin Interaction Partners
To identify proteins mediating TCF3-independent binding of β-catenin, we affinity-purified β-catenin-associated proteins from FLAG-β-catenin expressing 293T cells (Figure 4A). Multiple proteins immunoprecipitated with FLAG-β-catenin compared with controls (Figure 4B). MudPIT analysis found enrichment of nine known β-catenin interacting proteins such as LEF1, TCF7L1, TCF7L2, TLE3, and SMARCA4, and the β-catenin destruction complex of AXIN2, GSK3β, and APC (Figure 4C). We also identified three candidates with higher or similar dNSAF (distributed normalized spectral abundance factor) values: E2F6, L3MBTL2, and HMGA2. E2F6 and L3MBTL2 are part of the polycomb repressive complex 1 (PRC1)-like 4 (PRC1L4), which establishes repressive chromatin regulating gene expression in ESCs (Trojer et al., 2011). As E2F6 interacts with HP1γ as a part of the PRC1L4 complex (Qin et al., 2012), we also included HP1γ in our analysis.
Figure 4.
E2F6, HP1γ, and HMGA2 Are β-Catenin Interaction Partners
(A) Schematic diagram of affinity purification of FLAG-β-catenin. FLAG-tagged β-catenin induced for 48 h in HEK293FRT cells was isolated by FLAG affinity chromatography from both nuclear and chromatin bound extract. The resulting protein complexes were analyzed by mass spectrometry.
(B) FLAG affinity-purified elutes were analyzed by silver staining. Asterisk indicates FLAG-tagged β-catenin.
(C) List of proteins from MudPIT analyses with dNSAF values showing their relative abundance (n = 4 biological replicates).
(D-G) Coimmunoprecipitation in ESCs with β-catenin (D), E2F6 (E), HP1γ (F) or HMGA2 (G) as bait showing the interaction among endogenouse β-catenin, E2F6, HP1γ, or HMGA2.
We next tested whether endogenous β-catenin complexes with the identified partners in 293FRT cells. From size exclusion chromatography with crude nuclear extract, HMGA2 coeluted with β-catenin and E2F6 in fractions 7–8, whereas HP1γ and E2F6 were more abundant and coeluted with β-catenin in fractions 13–14 (Figure S4A). Interaction between β-catenin and E2F6 was confirmed in 293T cells (Figure S4B). These results show that β-catenin can be part of a complex that contains E2F6 and HMGA2 and another complex that contains HP1 γ and E2F6. We then analyzed ground state ESCs and found E2F6, HMGA2, and HP1γ, but not L3MBTL2, immunoprecipitated with endogenous β-catenin in ESCs (Figure 4D). HP1γ was not initially observed in our MudPIT analysis in 293FRT cells, indicating that the interaction between HP1γ and β-catenin is cell-type specific. Reverse immunoprecipitation using E2F6, HP1γ or HMGA2 as baits further confirmed interaction with β-catenin and each other (Figures 4E–4G). Taken together, we identified β-catenin-interacting proteins with repressive functions, including E2F6, HP1γ and HMGA2.
β-Catenin and Interacting Partners Regulate Germ Cell Genes and Neurogenesis
To understand the role of β-catenin-associated proteins in regulating ESC ground state, we first tested E2F6 by shRNA knockdown (Figure 5A). No morphological defects were observed in E2F6-depleted ESC colonies in 2i culture, suggesting that E2F6 is not essential for ground state pluripotency. However, although teratoma formation using E2F6-depleted ESCs was as efficient as control, the former showed foci of hemorrhage and necrosis that are common features of embryonal carcinoma (Figure 5B). Microscopic examination revealed mixed embryonal carcinoma containing primitive neuroepithelial, mesenchymal, and mature epithelial elements (Figure 5C). Although these teratomas do not lack mesodermal elements, the embryonal nature resembles β-catenin-depleted teratomas. The similarities between the teratomas support that β-catenin and E2F6 function together in ESCs to influence later cell fate decisions. RNA-seq of the E2F6-depleted ESCs revealed increased expression of meiosis-related genes Stag3, Dazl, Tex12, Tex15, and Ddx4 (Figure 5D). GO analysis showed significant enrichment for genes involved in meiosis, spermatogenesis, and chromosome organization (Figure 5E), which is consistent with the role of E2F6 in restricting germ-cell-specific gene expression (Pohlers et al., 2005).
Figure 5.
E2F6 Regulates Germline Lineage and Co-binds Genomic Loci with β-Catenin
(A) Western blot of E2F6 and β-actin (loading control) at 48 h after E2F6 knockdown in ESCs in 2i culture.
(B) Gross examination of teratoma derived from control and E2F6-depleted ESCs.
(C) H&E staining of teratoma sections derived from control ESCs or E2F6-depleted ESCs. Scale bar, 20μm.
(D) MA plot of RNA-seq of control and E2F6-depleted ESCs in 2i culture (n = 2–6 biological replicates).
(E) GO analysis of upregulated genes at 48 h after E2F6 knockdown. False discovery rate <0.05, log2(fold-change) >0.
(F) Venn diagram showing genes nearest to FLAG-β-catenin and E2F6 binding regions.
(G) Heatmap showing β-catenin peaks sorted by E2F6 with co-occupancy of TCF3, as well as bivalency histone marks H3K4me3 and H3K27me3 (n = 2 independent experiments).
We next performed E2F6 ChIP-seq in ESCs to test whether E2F6 co-binds β-catenin. Among 5,945 regions bound by E2F6, ∼44% were shared with β-catenin (Figure 5F). Strikingly, co-bound regions showed weaker TCF3 occupancy, whereas β-catenin-only regions had stronger TCF3 occupancy, suggesting that TCF3 and E2F6 were inversely correlated at β-catenin-bound regions (Figure 5G). The pattern of β-catenin bound with either TCF3 or E2F6 at target regions in ESCs suggests that E2F6 collaborates with β-catenin independently of TCF3. Histone modifications H3K4me3 and H3K27me3 were also differentially enriched at the E2F6- or TCF3-bound targets. Almost no H3K27me3 signal was present in the β-catenin-only occupied regions (Figure 5G), but β-catenin and E2F6 co-bound regions tended to be bivalently marked by H3K4me3 and H3K27me3, indicating a poised state regulated by polycomb repression (Bernstein et al., 2006).
Co-bound target genes of β-catenin and E2F6 were mainly at or near the transcription starting site (Figure 6A). MsigDB (molecular signatures database) analysis showed that these genes were related to meiotic recombination, H3K27 methylation, DNA methylation, and chromatin organization (Figure 6B), indicating that germ cell differentiation in ESCs, particularly meiosis-related genes, is co-regulated by E2F6 and β-catenin.
Figure 6.
β-Catenin and Its Interacting Partners Regulate Germ Cell Genes and Neurogenesis
(A) Genomic regions associated genes bound by β-catenin only or bound by both β-catenin and E2F6 (n = 2 independent experiments).
(B) MsigDB (molecular signatures database) pathway analysis of genes co-bound by E2F6 and β-catenin.
(C) Normalized gene expression of germline development genes including early PGC, PGC migration and late meiotic genes from RNA-seq of control and β-catenin-depleted ESCs (n = 2–6 biological replicates).
(D and E) Early PGC genes co-bound by β-catenin and HMGA2 (D) and meiotic genes co-bound by β-catenin, E2F6, and HP1γ (E).
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Error bars, SEM.
To further analyze the role of β-catenin in restricting germ cell differentiation in ESCs, we analyzed expression of key genes related to early PGC specification, migration, and meiosis entry in β-catenin-depleted ESCs (Lin et al., 2008; Saitou et al., 2002). RNA-seq revealed that 5/7 PGC specification genes (Prdm1, Ifitm3, Bmpr1b, Lin28a, and Lin28b) and 2/3 PGC migration genes (Ifitm1 and Kitl) were upregulated, but all meiosis-related genes (Stra8, Dazl, Ddx4, and Tex19.1) were downregulated after loss of β-catenin (Figure 6C). These results confirm that depletion of β-catenin unleashed PGC lineage differentiation when exiting ground state.
Regarding the possible function of HMGA2, our ChIP-seq results showed that β-catenin and HMGA2 co-localized at promoter or gene body of PGC genes (Figure 6D). Although knocking down HMGA2 in ESCs did not cause morphological changes (data not shown), teratoma size was reduced (Figures S5A and S5B). HMGA2-deficient teratomas showed only 26.8% decrease in GFAP staining (ectoderm) but no changes in pan-cytokeratin or α-smooth muscle actin (Figure S5C). These data suggest that HMGA2 does not substantially influence the composition of the somatic cell lineages but does affect their proliferation.
In addition to PGC genes, we found that β-catenin and E2F6 also co-occupied promoters of meiotic genes, such as Ddx4, Dazl, and Tex19.1, expressed during late germline development (Figure 6E). These genes were also bound by HP1γ, suggesting that HP1γ might be involved in meiotic gene regulation together with β-catenin and E2F6. We knocked down HP1γ in ESCs grown in 2i culture (Figure S5D). ESC colonies showed similar features of differentiation as in β-catenin-depleted ESCs (Figure S5E). Furthermore, astrocyte-related genes (Slc4a4, Slc1a3, and Fzd2) were de-repressed in HP1γ or β-catenin-knockdown ESCs, and HP1γ knockdown upregulated a broad panel of genes of neurogenesis (Cahoy et al., 2008) (Figure S5F). Additional neuronal genes were upregulated in β-catenin-depleted ESCs (Figure S5G). The de-repressed genes might be due to E2F6, as β-catenin and E2F6 co-occupied the promoter of Pax6 and Dsp and the distal region of Tubb3 (Figure S5H). HP1γ was located >10 kb from the β-catenin and E2F6 co-bound regions, suggesting that HP1γ might play an indirect role of inhibiting neuronal gene expression. Taken together, these data suggest that HP1γ is critical for restraining neuronal differentiation of ESCs together with β-catenin.
In summary, we found that independent of Tcf3, β-catenin in ground state ESCs cooperates with E2F6 and HMGA2 to restrain germline differentiation and with E2F6 and HP1γ to inhibit neuronal differentiation (Figure S5I and Table S1).
Discussion
Given the conflicting evidence regarding β-catenin's role in ESC self-renewal, here we provide insight into β-catenin's target genes and developmental functions in ESCs. While our findings confirm many previous results, we also show that β-catenin has functions in ESCs beyond Tcf3 targets of the pluripotency network. Genomic regions bound by β-catenin, but not TCF3, were strongly bound by E2F6. Furthermore, β-catenin physically associated with E2F6 in ESCs, in a complex with either HMGA2 or HP1γ, showing that β-catenin engages with transcriptional repressors other than TCF3.
Notably, functional analysis of the β-catenin complex components showed that individual knockdowns were insufficient to completely recapitulate the β-catenin-deficient ESC phenotype in 2i culture. Instead, they shared some phenotypes with β-catenin. For example, β-catenin or E2F6 knockdown resulted in embryonal carcinoma teratomas, β-catenin or HMGA2 knockdown showed decreased teratoma size, and β-catenin or HP1γ knockdown led to defective morphology of ESC colonies and upregulation of neural development genes.
β-catenin and the new interaction partners play a role in regulating competence of ESCs for germline development and neurogenesis, a critical characteristic of ground state pluripotency (Marks et al., 2012; Ying et al., 2008). Furthermore, β-catenin has been implicated in neuronal development (Brault et al., 2001; Hirabayashi et al., 2004) but with conflicting evidence on its exact function (Cajanek et al., 2009; Kelly et al., 2011; Lyashenko et al., 2011). Our data help clarify the role of β-catenin in germline development and neurogenesis.
First, we found that β-catenin functions together with E2F6 and HMGA2 in restraining germline differentiation. Depleting either β-catenin or E2F6 in ESCs disrupted germ cell development genes, leading to preferential differentiation toward germ cell tumors in vivo. However, β-catenin depletion and E2F6 depletion had the opposite effect on the expression of meiotic genes, suggesting that β-catenin has an activating role, while E2F6 has a repressing role. We propose that β-catenin alleviates repression by E2F6, similar to β-catenin acting on TCFs, allowing the timing and expression levels of meiotic target genes to be tightly controlled. Meiosis-related and PGC genes may also be regulated by β-catenin/E2F6/HMGA2, including Prdm1, a critical TF for the specification of PGCs (Vincent et al., 2005). Prdm1 may be induced through a Lin28b-let-7-HMGA2 regulatory axis, as found in fetal hematopoietic stem cells (Cai et al., 2013; Copley et al., 2013).
Second, we found that β-catenin functions together with E2F6 and HP1γ in restraining neural development. Neural development genes are bound by β-catenin, HP1γ, and E2F6, and both β-catenin and HP1γ knockdown led to their upregulation. This suggests that these proteins form a repressive complex. This complex is likely different from the PRC1L4 complex (Qin et al., 2012; Trojer et al., 2011), since we did not detect interaction between β-catenin and L3mbtl2 in ESCs.
Third, our results corroborate the idea that both embryonic and adult stem cell self-renewal depends on inhibition of lineage differentiation (Perry et al., 2011; Zhang and Li, 2005). β-Catenin not only regulates pluripotency genes but also has specific roles in restricting lineage development to reinforce the ground state. This role is to some extent independent of the pluripotency regulatory network since β-catenin knockdown led to transcriptional changes toward a primed fate before pluripotency TFs were downregulated. This suggests that β-catenin has a direct repressive role in preventing the transition from the ground state toward the primed state.
Experimental Procedures
Cell Lines
The Flp-In T-REx 293 cell line (Invitrogen R78007) and V6.5 mouse ESCs (Novus Biologicals NBP1-41162) were maintained by Stowers Institute for Medical Research (SIMR) Tissue Culture Core (TCC) facility. The Ctnnb1-3xFLAG mouse ESC line was a gift from Dr. Andrew McMahon (University of South California). The β-cateninfl/fl ESCs were derived from B6.129-Ctnnb1tm2Kem/KnwJ (B-cateninflox) and maintained by the SIMR TCC facility. Cre+β-cateninfl/fl ESCs were from β-cateninfl/fl ESCs transduced with EF1α-Cre-SV40-Puro packaged in lentivirus particles and selected with puromycin (2 μg/mL). The stable ESC line with Dox-inducible β-catenin knockdown was generated by transducing ESCs with SMARTvector-inducible lentiviral shRNA constructs (Horizon Discovery; V3SM11253–234566878 and V3SM11253-236512822) and maintained in the SIMR TCC Facility.
Alkaline Phosphatase Assay
Six-well plates with ESCs were fixed and stained with VECTOR Red Alkaline Phosphatase Substrate Kit (Vector Laboratories, SK-5100) following the manufacturer's instructions.
Flow Cytometry
Intracellular staining of NANOG was conducted with a BD Cytofix/Cytoperm Kit (catalog no. 554714) following the manufacturer's instructions. Flow cytometry data were collected with a ZE5 Cell Analyzer (Bio-Rad).
Affinity Purification of FLAG-β-Catenin
Approximately 2 L of 293T cells from 10 × 800 cm2 roller bottles at 100% confluency for each assay was collected, and the soluble nuclear extract was prepared with a final NaCl concentration of 0.3 M. For each assay, 150 μL of packed FLAG M2 affinity gel (Sigma A2220) was mixed with the nuclear extract and incubated for at least 4 h or overnight at 4°C. The gel was washed with the wash buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100). The protein complex was eluted two times with the elution buffer (10 mM HEPES [pH 7.9], 0.1 M NaCl, 1.5 mM MgCl2, 0.05% Triton X-100, 10% glycerol, 200 μg/mL FLAG peptide; Sigma F3290, 5 mg/mL).
Immunostaining
For IF staining, the ESCs were seeded onto 0.1% gelatin-coated chamber slides (Nunc Lab-Tek II 154852) and stained after 48 h. The ESC culture was fixed with 4% paraformaldehyde, washed with PBST (0.05% Tween 20), and permeabilized in PBS with 0.8% Triton X-100. Cells were blocked with universal blocking reagent (BioGenex HK0855K) and incubated overnight at 4°C with primary antibodies. Images were taken with a PerkinElmer UltraVIEW spinning-disk confocal microscope. Primary and secondary antibodies used in this study are shown in Table S2.
For immunohistochemistry staining, tumor samples were fixed in 100% zinc formalin, embedded in paraffin, and cut into 4 mm sections.
Microscopic Imaging
Fluorescence imaging was performed on an Olympus VS120 slide scanner, equipped with an X-Cite 120 LED light source and a Hamamatsu Flash 4.0 sCMOS camera or a spinning-disk confocal microscope (UltraVIEW; PerkinElmer), comprising an inverted stand (Axiovert 200 M; Carl Zeiss Microimaging, Jena, Germany) and a spinning-disk confocal head (CSU-X1; Yokogawa Corporation of America) with Velocity acquisition software (PerkinElmer) and dual cameras: a Hamamatsu C9100-23B EMCCD camera and a Hamamatsu Orca-R2 camera. Fluorescence channels were acquired sequentially. DAPI fluorescence was acquired with a 405 nm diode laser and a 445 (W60), 615 (W70) dual bandpass emission filter. Red (Alexa 546) fluorescence a 561 nm diode laser and a 445 (W60), 615 (W70) dual bandpass emission filter.
Western Blot
Protein extracts were resolved by SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was incubated with 5% BSA in TBST (0.1% Tween 20), washed with TBST, and incubated at 4°C overnight with primary antibodies. Membranes were washed with TBST and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma). Blots were developed with Luminata Forte western HRP substrate (Millipore WBLUF0100). Primary and secondary antibodies used in this study are shown in Table S2.
Coimmunoprecipitation
ESCs were resuspended in 5× packed cell volume of isotonic Dignam A buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT). Cells were lysed by passing through a 27-gauge needle. The crude nuclei pellet was resuspended in nuclear extraction buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 420 mM NaCl) and incubated at 4°C for 30 min. The lysate was centrifuged for 10 min at 20,000 × g, and the supernatant was mixed with antibody or IgG-coated protein G dynabeads (Invitrogen 10003D) overnight at 4°C at a ratio of 5 μL per 1 μg of antibody listed in Table S2. The resulting protein complexes were washed with the washing buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100) and eluted with 1× Laemmli buffer (Bio-Rad160747).
Lentivirus-Based RNAi in ESCs
The shRNA targeting sequence in Table S3 was cloned into the PLKO.1 TRC vector (Addgene10878). The mock control shRNA targeting GFP was provided by Ali Shilatifard (Northwestern University). Briefly, 10 μg of the shRNA plasmid, 7.5 μg of PsPAX2, and 2.5 μg of pMD2.G were co-transfected into 293T cells. The medium was filtered through 0.45 μm filters and concentrated at 18,000 rpm with a SW32Ti rotor for 2 h at 4°C. ESCs were infected with concentrated lentiviral particles with 8 μg/mL Polybrene (Sigma) and selected 24 h after infection with puromycin (2 μg/mL) for an additional 48 h.
Teratoma Assay
ESCs (100 μL at 5 × 106 cells/mL) in PBS were injected subcutaneously into the dorsal flank of NSG mice with a 25-gauge needle. Tumors were dissected 4 weeks after injection. Samples were fixed in 100% formalin, embedded in paraffin and subjected to IF or H&E staining. All mice used in this study were housed in the animal facility at SIMR and were handled according to SIMR and National Institutes of Health (NIH) guidelines. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the SIMR.
Quantitative RT-PCR and Total RNA-Seq Analysis
Cells were homogenized in 1 mL of TRIzol (Invitrogen15596026) with a 27-gauge needle followed by RNA extraction with phenol-chloroform. The RNA was treated with DNase (Promega M6101) and the cDNA was synthesized with a High Capacity RNA-to-cDNA Kit (Applied Biosystems 4387406). qPCR was carried out with Fast SYBR Green Master Mix (Applied Biosystems 409155) on StepOne RT-PCR Systems (Life Technologies) with the primers listed in Table S4.
Statistical Analysis
Statistical parameters, including the exact value of n, the definition of center, dispersion and precision measures (mean ± SEM), and statistical significance are reported in the figures and figure legends. Data are judged to be statistically significant when p < 0.05 by two-tailed Student's t test. In figures, asterisks denote statistical significance as calculated by Student's t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). All statistical analyses were generated using GraphPad Prism 5.
Other Materials and Methods
Other materials and methods are described in Supplemental Experimental Procedures.
Data and Code Availability
Original data repository: https://www.stowers.org/research/publications/LIBPB-1343. The accession number for the data reported in this paper is GEO: GSE108620.
Author Contributions
F.T. designed and performed experiments, analyzed data, and wrote the manuscript. J.S., D.H., D.T., and Y.Z. performed part of the experiments. S.C. and X.G. performed bioinformatics analysis. C.Z. and T.P. optimized cell culture system. D.Z., S.E.S., J.R.U., and Z.L contributed to data analysis. A.V., M.Z., P.Q., X.C.H., M.W., L.F., J.W., and J.Z. provided training and intellectual discussion. J.M.P. collaborated and supported the project. L.L. directed the overall project and co-wrote the manuscript. All authors contributed to reading and editing the manuscript.
Acknowledgments
We thank Dr. Andrew P. McMahon and Dr. Bradley W. Doble for kindly providing the FLAG-β-catenin knockin stable mouse ESC line and QKO mouse ESC line; M. Mohan, K. Chen, and W. Shao for training and scientific discussion; M. Miller for scientific illustration; K. Tannen for proofreading and editing; M. Hembree, K. Maria, Y. Han, J. Smith, Q. Jiang, O. Kenzior, A. Murrey, K. Zapien, D. Dukes, A. Moran, M. Durnin, A. Box, J. Park, Y. Wang, A. Perera, E. Rhonda, and A. Peak for technical support. We are grateful to L.L.’s lab members for scientific discussion and critical reading of manuscript. This work was supported by Stowers Institute for Medical Research funding to L.L. (SIMR-1004).
Published: August 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.07.018.
Supplemental Information
References
- Anton R., Kestler H.A., Kuhl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. doi: 10.1016/j.febslet.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Lombard P., Okahara J., Behr R., Sasaki E., Nichols J., Smith A., Bertone P. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell. 2015;35:366–382. doi: 10.1016/j.devcel.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault V., Moore R., Kutsch S., Ishibashi M., Rowitch D.H., McMahon A.P., Sommer L., Boussadia O., Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W.Y., Wei T.Z., Luo Q.C., Wu Q.W., Liu Q.F., Yang M., Ye G.D., Wu J.F., Chen Y.Y., Sun G.B. The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J. Cell Sci. 2013;126:2877–2889. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- Cajanek L., Ribeiro D., Liste I., Parish C.L., Bryja V., Arenas E. Wnt/beta-catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells. 2009;27:2917–2927. doi: 10.1002/stem.210. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cole M.F., Johnstone S.E., Newman J.J., Kagey M.H., Young R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley M.R., Babovic S., Benz C., Knapp D.J., Beer P.A., Kent D.G., Wohrer S., Treloar D.Q., Day C., Rowe K. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat. Cell Biol. 2013;15:916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- Dunn S.J., Martello G., Yordanov B., Emmott S., Smith A.G. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y., Itoh Y., Tabata H., Nakajima K., Akiyama T., Masuyama N., Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Kanatsu-Shinohara M., Toyokuni S., Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139:1734–1743. doi: 10.1242/dev.076539. [DOI] [PubMed] [Google Scholar]
- Kelly K.F., Ng D.Y., Jayakumaran G., Wood G.A., Koide H., Doble B.W. β-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Kaufman-Francis K., Studdert J.B., Steiner K.A., Power M.D., Loebel D.A., Jones V., Hor A., de Alencastro G., Logan G.J. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Gill M.E., Koubova J., Page D.C. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322:1685–1687. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- Lindsley R.C., Gill J.G., Kyba M., Murphy T.L., Murphy K.M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- Lyashenko N., Winter M., Migliorini D., Biechele T., Moon R.T., Hartmann C. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat. Cell Biol. 2011;13:753–761. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M., Karis A., Visser P., Grosveld F., Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira S., Polena E., Gordon V., Abdulla S., Mahendram S., Cao J., Blais A., Wood G.A., Dvorkin-Gheva A., Doble B.W. A single TCF transcription factor, regardless of its activation capacity, is sufficient for effective trilineage differentiation of ESCs. Cell Rep. 2017;20:2424–2438. doi: 10.1016/j.celrep.2017.08.043. [DOI] [PubMed] [Google Scholar]
- Mosimann C., Hausmann G., Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Okumura N., Akutsu H., Sugawara T., Miura T., Takezawa Y., Hosoda A., Yoshida K., Ichida J.K., Yamada M., Hamatani T. β-catenin functions pleiotropically in differentiation and tumorigenesis in mouse embryo-derived stem cells. PLoS One. 2013;8:e63265. doi: 10.1371/journal.pone.0063265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.M., He X.C., Sugimura R., Grindley J.C., Haug J.S., Ding S., Li L. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. 2011;25:1928–1942. doi: 10.1101/gad.17421911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers M., Truss M., Frede U., Scholz A., Strehle M., Kuban R.J., Hoffmann B., Morkel M., Birchmeier C., Hagemeier C. A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr. Biol. 2005;15:1051–1057. doi: 10.1016/j.cub.2005.04.060. [DOI] [PubMed] [Google Scholar]
- Qin J., Whyte W.A., Anderssen E., Apostolou E., Chen H.H., Akbarian S., Bronson R.T., Hochedlinger K., Ramaswamy S., Young R.A., Hock H. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–332. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggioli A., Junghans D., Rudloff S., Kemler R. Beta-catenin is vital for the integrity of mouse embryonic stem cells. PLoS One. 2014;9:e86691. doi: 10.1371/journal.pone.0086691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudloff S., Kemler R. Differential requirements for beta-catenin during mouse development. Development. 2012;139:3711–3721. doi: 10.1242/dev.085597. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Silva J., Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.Y. Maintaining embryonic stem cell pluripotency with Wnt signaling. Development. 2011;138:4341–4350. doi: 10.1242/dev.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R.K., Nusse R. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P., Cao A.R., Gao Z., Li Y., Zhang J., Xu X., Li G., Losson R., Erdjument-Bromage H., Tempst P. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell. 2011;42:438–450. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S.D., Dunn N.R., Sciammas R., Shapiro-Shalef M., Davis M.M., Calame K., Bikoff E.K., Robertson E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Andrabi M., Biss R., Murtuza Baker S., Iqbal M., Sharrocks A.D. ZIC3 controls the transition from naive to primed pluripotency. Cell Rep. 2019;27:3215–3227.e6. doi: 10.1016/j.celrep.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li L. BMP signaling and stem cell regulation. Dev. Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Zhang X., Peterson K.A., Liu X.S., McMahon A.P., Ohba S. Gene regulatory networks mediating canonical Wnt signal-directed control of pluripotency and differentiation in embryo stem cells. Stem Cells. 2013;31:2667–2679. doi: 10.1002/stem.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J., Shtutman M., Ben-Ze'ev A. Plakoglobin and beta-catenin: protein interactions, regulation and biological roles. J. Cell Sci. 2000;113:3127–3139. doi: 10.1242/jcs.113.18.3127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data repository: https://www.stowers.org/research/publications/LIBPB-1343. The accession number for the data reported in this paper is GEO: GSE108620.