Abstract
Airway neutrophilia is correlated with disease severity in a number of chronic and acute pulmonary diseases, and dysregulation of neutrophil chemotaxis can lead to host tissue damage. The gene Zfp30 was previously identified as a candidate regulator of neutrophil recruitment to the lungs and secretion of CXCL1, a potent neutrophil chemokine, in a genome-wide mapping study using the Collaborative Cross. ZFP30 is a putative transcriptional repressor with a KRAB domain capable of inducing heterochromatin formation. Using a CRISPR-mediated knockout mouse model, we investigated the role that Zfp30 plays in recruitment of neutrophils to the lung using models of allergic airway disease and acute lung injury. We found that the Zfp30 null allele did not affect CXCL1 secretion or neutrophil recruitment to the lungs in response to various innate immune stimuli. Intriguingly, despite the lack of neutrophil phenotype, we found there was a significant reduction in the proportion of live Zfp30 homozygous female mutant mice produced from heterozygous matings. This deviation from the expected mendelian ratios implicates Zfp30 in fertility or embryonic development. Overall, our results indicate that Zfp30 is an essential gene but does not influence neutrophilic inflammation in this particular knockout model.
Keywords: Zfp30, ZFP30, neutrophil, inflammation, innate immunity
Introduction
Neutrophils are key participants in the innate immune system’s response to pathogens, but the mechanisms by which these cells respond to immune challenge are prone to generating collateral host tissue damage (Nathan 2006). This makes regulation of neutrophil chemotaxis particularly important in defending against outside insults and preventing unwanted organ damage. This signaling balance is particularly vital in the lungs due to the continual exposure to pathogens, allergens, and other environmental exposures that are introduced through respiration.
Dysregulation of the chemokines that attract neutrophils into tissues may tip the scale from appropriate innate immune response to unwanted damage. The CXC chemokine family members CXCL8 (IL-8), CXCL1 (KC/Gro-α), CXCL2 (MIP-2/Gro-β), and CXCL5 (LIX/ENA-78) are all hallmark neutrophil recruitment molecules that signal through the chemokine receptor CXCR2 (Charo and Ransohoff 2006). Pharmaceutical targeting of CXCR2 has proven effective in reducing airway neutrophilia in early chronic pulmonary disease trials (Kirsten et al. 2015; Moss et al. 2013; Nair et al. 2012; Rennard et al. 2015; Todd et al. 2016; Watz 2017). CXCR2 antagonism also reduced airway neutrophils in mild atopic asthmatics and patients with severe or persistent neutrophilic asthma (Nair et al. 2012; Todd et al. 2016; Watz et al. 2017).
Previously, we identified the murine gene Zfp30 as novel regulator of neutrophil recruitment to the airways in a mouse model of asthma (Rutledge et al. 2014). Our approach involved use of incipient lines of Collaborative Cross (CC), a multiparental genetics reference population (Srivastava et al. 2017), that were treated with the house dust mite allergen Der p 1 (Rutledge et al. 2014). In that study, we mapped quantitative trait loci (QTL) to proximal chromosome (Chr) 7 for both neutrophil number and the neutrophil chemokine CXCL in bronchoalveolar lavage fluid. Using expression QTL (eQTL) mapping of whole lung RNA levels, we identified a cis-eQTL for Zfp30 that co-localized with the neutrophil/CXCL1 QTL. Zfp30 expression was strongly and negatively corrrelated with neutrophil number and CXCL1 concentration, and in subsequent in vitro experiments we showed that decreasing Zfp30 expression (by siRNA knockdown) lead to increased CXCL1 secretion by airway epithelial cells, consistent with the in vivo data. Thus, we concluded that Zfp30 expression, which we subsequently showed is largely determined by rs51434084 genotype (Laudermilk et al 2018), is a key determinant of CXCL1 and neutrophil recruitment to the airways in response to house dust mite allergen exposure in the CC population. Additionally, because previous studies identified QTL for endotoxin response (Matesic et al. 1999) and Streptococcus pneumoniae infection (Denny et al. 2003) at the same location on Chr 7, we reasoned that ZFP30 may be involved in response to multiple innate immune stimuli.
ZFP30 is a C2H2 zinc finger protein with a KRAB domain. The C2H2 domains allow ZFP30 to bind to DNA in a sequence specific manner, and the KRAB domain recruits KAP1, a well-studied transcriptional repressor to these binding sites (Friedman et al. 1996). This transcriptional repression proceeds through recruitment of HP1, SETDB1, and histone deacetylases that induce heterochromatin formation and silence nearby genes (Groner et al. 2010; Medugno et al. 2005; Ryan et al. 1999; Schultz et al. 2002;). There is precedent for zinc finger proteins to regulate immune signaling through heterochromatin domains, including ZNF160 which down-regualtes TLR4 expression in intestinal epithelia to ameliorate immune response to the host microbiome (Takahashi et al. 2009).
In this study, we further examined Zfp30 and its role in neutrophil recruitment to the airways through generation of a CRISPR-Cas9 Zfp30 knockout mouse strain. Given that we previously found that low Zfp30 expression was associated with higher neutrophil counts in the airways after exposure to house dust mite allergen, we hypothesized that Zfp30 knockout mice would exhibit heightened levels of neutrophilic inflammation after treatment with allergen and other innate immune stimuli. Additionally, because a recent report described a regulatory effect of ZFP30 on Pparg2 expression and adipogenesis (Chen 2019), we also assessed metabolic phenotypes in Zfp30 wildtype and knockout mice. Somewhat surprisingly, we observed a significant depletion of homozygous Zfp30 knockout mice in matings of Zfp30+/− x Zfp30+/− mice, implicating ZFP30 in fertility or development. However, contrary to expectation, we found that Zfp30 knockout mice did not exhibit differences in neutrophil recruitment to the airways after exposure to innate immune stimuli, nor did we observe any differences in metabolic phenotypes or Pparg2 expression in adipose tissue from Zfp30 knockout mice.
Materials and Methods
Generation of Zfp30 Knockout Mice by CRISPR/Cas9 Embryo Microinjection
A CRISPR/Cas9 guide RNA (5’-GAATCCAGATACAGCAGTAA(CGG)-3’) was designed to target mouse (NCBI Taxon ID: 10090) Zfp30 gene (NCBI Gene ID: 22693) near the 5’ end of exon 5. While targeting earlier exons would have been preferable, exon 5 was the only region that could be targeted with specificity owing to high homology across ZFP family members. Exon 5 encodes the C2H2 zinc finger domains of ZFP30 that are required for DNA binding. The guide RNA was produced by T7 in vitro transcription and validated in vitro by incubating guide RNA, Cas9 enzyme, and plasmid harboring the guide RNA target site. This was followed by gel electrophoresis to determine the extent of in vitro cleavage activity. A donor oligonucleotide (“Zfp30-H1-T”: 5’-GTTTTTCTTCTTTTTGCTTTCAGATCTGGAATCCAGATACAGC[TGA][TAG]GATCC[T AG]ACCGGTAACGGGTTACTTCCAGAAAAGAATACTTACGAAATTAATCTATCT-3’) was used for homologous recombination to insert stop codons (brackets) and a BamHI restriction site (underlined) at the target site (Supplementary Figure 1).
C57BL/6J females were then superovulated by injection with pregnant mare’s serum gonadotropin (PMSG) and human chorionic gonadotropin and then mated with C57BL/6J stud males for zygote production. One-cell embryos were collected from the ampulla oviducts the morning after mating and microinjected with either “low” or “high” mix, containing, respectively, 20 ng/μl or 100 ng/μl in vitro transcribed Cas9 mRNA, 20 ng/μl or 50 ng/μl Zfp30 guide RNA and 100 ng/μl Zfp30-H1-T donor oligonucleotide. The microinjected embryos were then implanted into pseudopregnant recipients.
Fourteen live pups born from microinjected embryos were screened by polymerase chain reaction (PCR) amplification of the Zfp30 target site followed by digestion of the PCR product with BamHI restriction enzyme. The BamHI restriction site was detected in nine animals. Two founders with apparent biallelic insertion of the BamHI restriction site were mated to C57BL/6J animals for germline transmission of the targeted allele.
The founder animals harboring the intended Zfp30 mutant allele were screened for mutations at 10 potential off-target sites (Supplementary Table 1). Each potential off-target site was PCR amplified and products were analyzed by T7endo1 assay. Founders chosen for line establishment were further analyzed by Sanger sequencing of PCR products for all 10 off-target sites. A single founder line was subsequently backcrossed to C57BL/6J again to remove detected off-target mutations. The mutant strain, C57BL/6J-Zfp30em1Snpk/Mmnc (hereafter referred to simply as Zfp30−/−), has been deposited into the Mutant Mouse Research and Resource Center at UNC (https://www.mmrrc.org/catalog/sds.php?mmrrc_id=50629).
In the initial stages of breeding, genotyping of Zfp30−/− mice was performed using allele-specific PCR. Primer sets were designed to specifically target either the Zfp30+/+ (Fwd: GGGCTGCTAAGTCCATTCAG; Rev: GGAAGTAACCCGTTACTGCTG) or Zfp30−/− (Fwd: GGGCTGCTAAGTCCATTCAG; Rev: CGTTACCGGTCTAGGATCCT) allele. We later transitioned to a proprietary qPCR-based genotyping protocol through Transnetyx (Cordova, TN).
qPCR for Zfp30 Quantification in the Knockout Strain
To quantify Zfp30 gene expression level in Zfp30+/+ and Zfp30−/− mice, we designed primer sets that specifically quantify the Zfp30+/+ allele (Fwd: TGTTGGAACAAGGGAAGGAG; Rev: GTAACCCGTTACTGCTGTAT) or specifically quantify the Zfp30−/− allele (Fwd: TGTTGGAACAAGGGAAGGAG; Rev: CGGTCTAGGATCCTATCAGCT). qPCR reactions were carried out using iTaq Universal SYBR Green Supermix (Bio-Rad; Hercules, CA USA).
qPCR for Mouse Tracheal Epithelial Cell cultures and adipose tissue
To quantify expression of selected genes in mouse tracheal epithelial cell cultures, Taqman assays for the following genes were used:
Krt5: (Assay ID: Mm00503549_m1, Thermo Fisher)
Muc5ac: (Assay ID: Mm01276718_m1, Thermo Fisher)
Foxj1: (Assay ID: Mm01267279_m1, Thermo Fisher)
Scgb1a1: (Assay ID: Mm00442046_m1, Thermo Fisher)
Tgm2: (Assay ID: Mm00436987_m1, Thermo Fisher)
Lyz2: (Assay ID: Mm01612741_m1, Thermo Fisher)
Lif: (Assay ID: Mm00434762_g1, Thermo Fisher)
Abhd6: (Assay ID: Mm00481199_m1, Thermo Fisher)
To quantify expression of Pparg in adipose tissue harvested from WT and KO mice, the following assays were used:
Pparg: (Assay ID: Mm00440945_m1, Thermo Fisher)
Rps20 (Normalization gene): (Mm02342828_g1, Thermo Fisher)
Reactions were carried out using Thermo Fisher Taqman Universal PCR Master Mix (Thermo Fisher). Fold changes were determined using the ΔΔCt method, and significance was calculated using two-tailed Welch’s t-tests.
Complete Blood Count Assays
For complete blood counts, blood was collected in EDTA tubes and stored on ice for a minimal amount of time before processing via a ProCyte Dx Hematology Analyzer.
Metabolic phenotyping
Magnetic Resonance Imaging (MRI)I
MRIs scans were performed using an EchoMRI-3n1–100TM analyzer prior to glucose tolerance tests to accurately determine body mass composition.
Glucose Tolerance Test
At 13 weeks of age, mice were fasted overnight and dosed with glucose (2g per kg lean body mass) via intraperitoneal injection. Blood glucose was measured at baseline and 15, 30, 45, 60, and 120 minutes after injections, using an Accu-Chek Performa glucometer and test strips (Roche, Basel Switzerland).
Neutrophil Recruitment Models
Lipopolysaccharide (LPS) challenge
Intratracheal instillation of LPS from E. coli (LIST Biologicals Campbell, CA) into lungs of Zfp30+/+ and Zfp30−/− mice was carried out at a dose of 0.3 mg per kg of body weight using previously described methods (Limjunyawong et al. 2015, Mock et al. 2020). Bronchoalveolar lavage fluid was collected between 8 and 48 hours after exposure, and differential cell counts in bronchoalveolar lavage fluid were performed. Aliquots of BALF were saved for cytokine quantification. Oropharyngeal aspiration of LPS from E. coli into lungs of Zfp30+/+ and Zfp30−/− mice was carried out with 5μg LPS in 40μl PBS.
Dermatophagoides pteronyssinus house dust mite allergen (Der p 1): We used a model of allergic inflammation involving Der p 1 that we previously showed induces predominantly eosinophilic but also neutrophilic inflammation (Kelada et al. 2011). Zfp30+/+ and Zfp30−/− mice were sensitized with 10 μg Der p 1 (Indoor Biotechnologies, Charlottesville, VA) administered through intraperitoneal injection (in 100 μl of PBS) on days 0 and 7 of the experiment, and a 50 μg Der p 1 challenge was administered on day 15 of the experiment (Kelada et al. 2011). Mice were sacrificed 48–72 hours after challenge, and differential cell counts in bronchoalveolar lavage fluid were performed. Aliquots of BALF were saved for cytokine quantification.
Ozone exposure
Zfp30+/+ and Zfp30−/− mice were exposed to filtered air, 1 ppm ozone, or 2 ppm ozone for three hours as previously described (Smith et al. 2019). Bronchoalveolar lavage fluid was collected 24 hours after exposure, and differential cell counts in bronchoalveolar lavage fluid were performed.
Mouse tracheal epithelial cell (MTEC) culture model
MTEC cultures were generated and cultured according to a previously established protocol (You and Brody 2013). Tissues isolated from 4 week old male and female mice were grown using PluriQ differentiation media and plated in 12 well plates with Transwell inserts. Cells were maintained at air-liquid interface for a minimum of three weeks to allow for differentiation. LPS, a TLR4 ligand that induces strong CXCL1 secretion, exposures were carried out at 10μg/mL in 100μl of PBS added to the apical surface of MTECs for 24h.
Luminex Assays
Cytokines in bronchoalveolar lavage fluid or PBS used in MTEC LPS exposures were measured using Milliplex assays (Millipore, Billerica, MA) according to manufacturer’s instructions.
Histology
Histological preparation and analysis of lungs were carried out using previously described methods (Donoghue et al. 2017). Briefly, left lung lobes were fixed in formalin and cut in cross-section starting at the hilum and 2mm apart along the main stem bronchus. Sections were embedded in paraffin and stained with Hematoxylin and eosin (H&E) stain. Images were captured on an Olympus BX605F microscope with CellSens Standard software.
Statistical Analysis
We tested for departures from expected mendelian genotype ratios in offspring from matings of Zfp30+/− x Zfp30+/− mice using two approaches. First, we used standard X2 goodness of fit tests to examine whether the observed genotype frequencies deviated from the expected genotype ratios. Second, because we observed a depletion of Zfp30−/− mice specifically, we used the binomial distribution formula to test whether the observed number was less than the expected value assuming a 25% genotype frequency. Significance tests for qPCR data and quantitative traits were determined by two-tailed Welch’s t-test between WT and KO groups within an experiment unless otherwise noted.
Results
We sought to investigate the function of Zfp30 using a CRISPR-Cas9 generated in vivo knockout model. Of fourteen pups born from microinjected embryos, nine had successful insertion of the donor oligonucleotide harboring the BamHI restriction site, and two founders with biallelic insertion were mated to C57BL/6J for germline transmission of the targeted allele. These founder animals were screened for ten potential off-target mutations. Bi-allelic insertion/deletion (indel) mutations were identified at a single off-target site in both founders. N1 backcross animals from a single founder were subsequently backcrossed once more to C57BL/6J to remove the off-target indel mutation and establish our Zfp30+/− colony.
Throughout the maintenance of this colony, we repeatedly observed a relative depletion of homozygous knockout mice produced from matings of heterozygous dams and sires. Initially, we sought to overcome this through selective breeding of mice that produced litters with normal genotype distributions. However, after several generations of breeding, we again observed less than expected number of homozygous mutant mice (Table 1). On the whole (n=1,270 mice), the difference in genotype frequencies compared to Mendelian ratios was only suggestive (X2=3.88, p-value = 0.14) because heterozygous mice were present at 51%. However, only 289 (22.8%) offspring were homozygous knockouts. The cumulative probability of observing 289 or fewer homozygous mutant mice in this population size is 0.03, according the binomial distribution, i.e., a significant depletion of homozygous knockout mice. Further examination revealed that this effect was only apparent among female pups (Table 1). More specifically, of 648 female mice produced from Zfp30+/− x Zfp30+/− matings, only 21% were homozygous knockout genotypes (p=0.01 by binomial distribution test). Likewise, among all female offspring, the overall genotype distributions were significantly different from Mendelian expectations.
Table 1.
Genotype counts and ratios of all mice born from heterozygous matings
| Zfp30+/+ | Zfp30+/− | Zfp30−/− | Total | |
|---|---|---|---|---|
| Male | 152 (24.4%) | 318 (51.1%) | 152 (24.4%) | 622 |
| Female | 185 (28.6%) | 326 (50.3%) | 137 (21.1%) | 648 |
| Total | 337 (26.5%) | 644 (50.7%) | 289 (22.8%) | 1270 |
X2 for both sexes = 3.88, 2 d.f., p-value = 0.14.
X2 for males = 0.32, 2 d.f., p-value = 0.85.
X2 for females = 7.14, 2 d.f., p-value = 0.03.
It is unclear if Zfp30−/− female mice died during embryonic development or if the knockout allele was selected against through some other mechanism. However, mean litter size for the colony is 7.15, which is highly comparable to the reported values for C57BL/6J (Silver 1995), and we did not observe any evidence that Zfp30−/− mice died after birth. Recently, the International Mouse Phenotyping Consortium generated a Zfp30 knockout strain on a C57BL/6N background and also found that among litters from heterozygous parents there was a significant depletion of both male and female homozygous knockout mice (www.mousephenotype.org, Dickinson et al. 2016), providing a partial replication of our result.
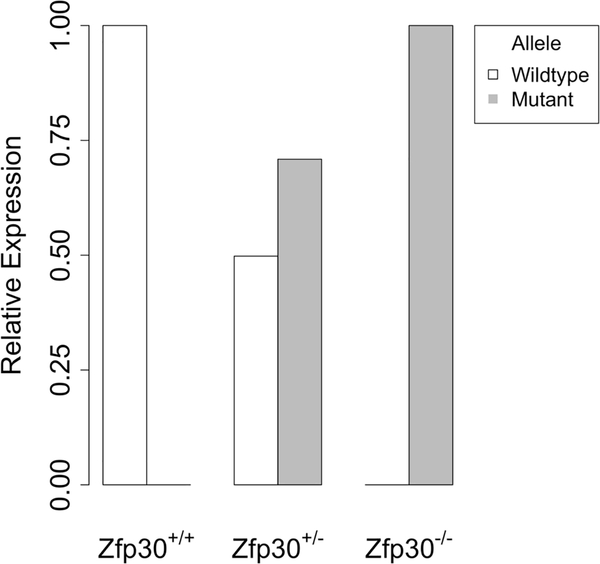
Because no reliable antibody for mouse ZFP30 exists, we verified the Zfp30 knockout at the RNA level in whole lung tissue from 9–10 week old mice. We designed a qPCR-based approach to specifically quantify expression of either the Zfp30 wildtype allele or the CRISPR-Cas9 modified Zfp30 mutant allele. We detected expression of only the Zfp30 wildtype allele in WT mice, only the Zfp30 mutant allele in KO mice, and intermediate expression of the two alleles in heterozygous mice (Figure 1).
Figure 1. Allele-specific qPCR confirms genotype-dependent expression of Zfp30 wildtype and mutant alleles in the lung.
Primer pairs that specifically amplify either the wildtype or CRISPR-Cas9 modified allele were used to confirm the status of Zfp30 expression in whole lung tissue from 4–6 mice per genotype. Mice were 9–10 weeks of age at harvest.
Baseline Phenotyping
As a family, the KRAB domain-containing C2H2 zinc finger proteins are thought to play an important role in differentiation and development (Lupo et al. 2013; Urrutia 2003). To assess the impact that a whole-body knockout of Zfp30 might have on development, we assayed the weights of lungs, pancreases, spleens, and livers in Zfp30+/+ and Zfp30−/− mice, and we detected no significant differences in these phenotypes (Supplementary Table 2). Complete blood count (CBC) assays did not reveal any significant differences in red blood cell or circulating leukocyte phenotypes between Zfp30+/+ and Zfp30−/− samples (Supplementary Table 3). Because ZFP30 has been reported to play a role in adipogenesis via regulation of Pparg2 expression (Chen 2019), we tested for metabolic impacts of Zfp30 knockout in fasting glucose and glucose tolerance (Figure 2A) but saw no significant differences. We did, however, observe a significant difference in the body weights of Zfp30+/+ and Zfp30−/− male mice (Figure 2B) and a marginally significant difference in the lean body weights among male mice (Figure 2C). We did not, however, observe a substantial impact of Zfp30 knockout on fat mass (Figure 2D) or Pparg2 expression in white adipose tissue (Fold change (KO vs WT)= 0.82; p-value = 0.62; WT n=11, KO n=10).
Figure 2. Zfp30−/− mice exhibit no differences in glucose tolerance but have lower body weight.
A. Thirteen week old mice were fasted overnight and dosed with glucose (2 g per kg lean body mass) via intraperitoneal injection. Blood glucose was then measured at baseline and 15, 30, 45, 60, and 120 minutes after injections. B. Bodyweight at harvest. C. Lean body mass determined by MRI. D. Fat mass determined by MRI normalized to body weight. N=10–11 per genotype. * P < 0.05; ^ P < 0.1.
Finally, we carried out histological analysis of lungs from Zfp30+/+ and Zfp30−/− lungs to investigate any obvious differences in the airways, alveoli, or vasculature and detected no striking differences (Supplementary Figure 2).
Ex vivo Mouse Tracheal Epithelial Cell Cultures
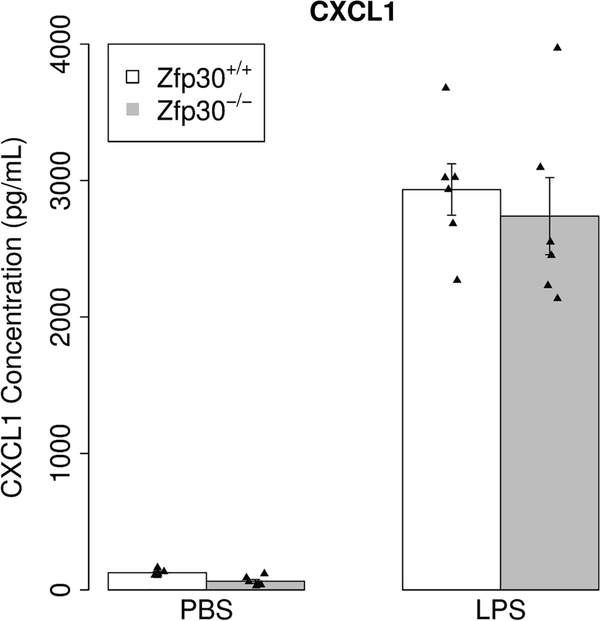
Given our previous results indicating a correlation between Zfp30 expression and innate immune response in the lung (Rutledge et al. 2014), we tested innate immune responses in mouse tracheal epithelial cultures (MTECs) from Zfp30+/+ and Zfp30−/− mice. This system was particularly well suited to study the impact of Zfp30 knockout on neutrophil recruitment, because recent single-cell RNA sequencing (RNA-seq) data suggests expression of Zfp30 across a broad array of cell types in the airway epithelium (Plasschaert et al. 2018). Additionally, we previously showed that MTEC cultures have high expression of Zfp30 and that perturbation of Zfp30 expression with siRNAs results in increased CXCL1 production following LPS exposure in a mouse airway epithelial cell line (Rutledge et al. 2014). After establishing MTEC cultures, we tested whether there were differences in the proportions of airway epithelial cell types using qRT-PCR for markers of ciliated cells (Foxj1), club cells (Scgb1a1), goblet cells (Muc5ac), and basal cells (Krt5). We detected a marginally significant doubling of Muc5ac expression in Zfp30−/− MTECs (Table 2), suggesting elevated goblet cell numbers in the knockout. We then stimulated Zfp30+/+ and Zfp30−/− MTECs with LPS, but did not detect significant differences in CXCL1 secretion in response (Figure 3).
Table 2.
Airway epithelial cell marker gene expression in primary tracheal epithelial cell cultures from Zfp30−/− vs. Zfp30+/+ mice.
| Sample | Foxj1 | Krt5 | Scgb1a1 | Muc5ac | Tgm2 | Lyz2 | Lif |
|---|---|---|---|---|---|---|---|
| Fold Change* | 0.95 | 1.24 | 1.29 | 2.09 | 1.18 | 1.12 | 0.82 |
| p-value | 0.48 | 0.48 | 0.44 | 0.07 | 0.25 | 0.76 | 0.28 |
Fold change for Zfp30−/− vs. Zfp30+/+ mice using Abhd6 for normalization.
Statistical significance was calculated using a Welch’s corrected t-test on Ct values of target genes in WT and KO cultures. Fold changes were determined using the ΔΔ Ct method.
Figure 3. No difference in CXCL1 secretion by genotype in primary mouse tracheal epithelial cells after LPS exposure.
Mouse tracheal epithelial cell cultures were generated from Zfp30+/+ and Zfp30−/− mice and maintained at air-liquid interface for 21 days. Cells were then exposed to 100 μM LPS for 24 hours, and supernatants were assayed for CXCL1 concentrations using Luminex assays. N= 6 per condition per genotype.
In vivo Lung Inflammation Phenotypes in Zfp30−/− mice
Zfp30 was identified as a candidate regulator of CXCL1 levels and neutrophils in bronchoalveolar lavage fluid in the context of a model of allergic airway disease (Rutledge et al. 2014). As a direct follow-up to these experiments, we utilized the same house dust mite model of allergic airway disease in Zfp30+/+ and Zfp30−/− mice to further probe the connection between Zfp30 and neutrophil recruitment. We saw no significant differences in neutrophil counts or CXCL1 in bronchoalveolar lavage fluid (BALF) 48 or 72 hours post-challenge (Figure 4A–B).
Figure 4. No difference in neutrophil count or CXCL1 concentration in BALF after house dust mite allergen challenge in Zfp30+/+ and Zfp30−/− mice.
Zfp30+/+ and Zfp30−/− mice were sensitized to and challenged with HDM allergen, and effects on neutrophil counts (A) and CXCL1 concentration (B) in BALF were monitored 48 and 72 hours after challenge. Cell count results are the combined data of two experiments and were assayed via differential cell counts. Cytokine were measured using multiplex assays. N= 9–11 mice per condition per genotype.
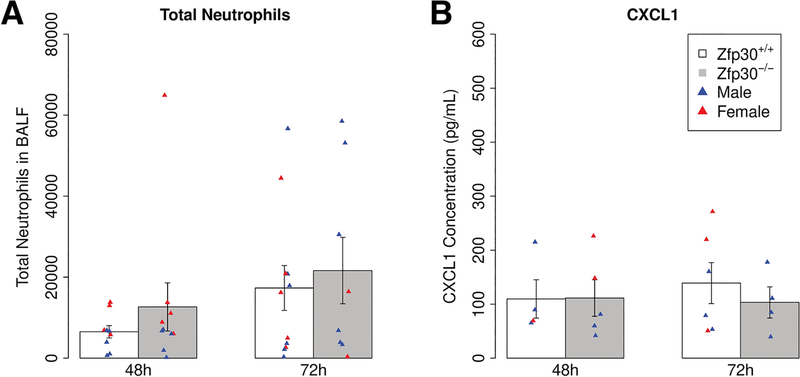
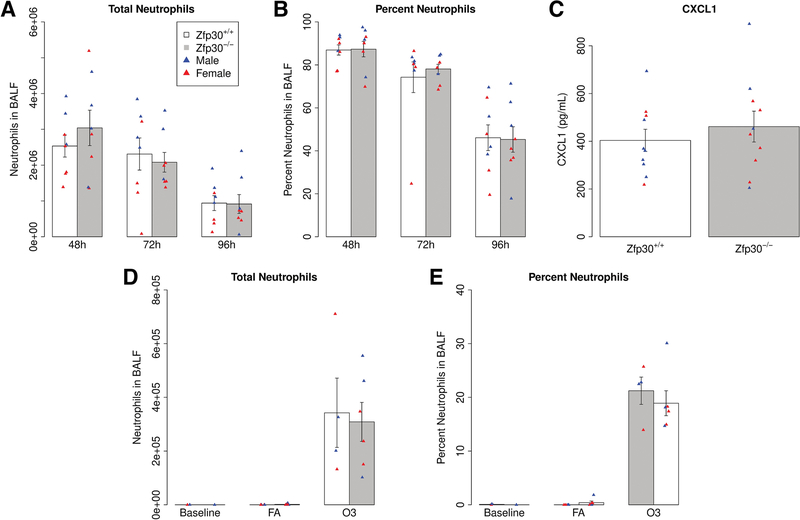
The allergic airway disease model we employed is dominated by eosinophilia. To test for differences in CXCL1 secretion or neutrophil recruitment into the lungs in the context of neutrophil-dominated immune responses, we employed LPS and ozone exposure models. These models induce a much more robust neutrophilic airway recruitment than the HDM model, so differences in chemotactic signaling may be more apparent. We found that intratracheal instillation of LPS did not cause a significant difference in neutrophil or CXCL1 levels in BALF of Zfp30−/− vs. Zfp30+/+ mice (Figure 5A–C). Additionally, there were no significant differences in neutrophilia by genotype in a model of sterile inflammation induced by the air pollutant ozone (1 and 2 parts per million concentration).
Figure 5. No differences in neutrophil chemotaxis by Zfp30 genotype in two models of neutrophilic inflammation.
(A-C) Zfp30+/+ and Zfp30−/− mice were given LPS by intratracheal administration and phenotyped 48, 72, or 96 hours later Neutrophils counts (A) and neutrophils as a percentage of total cell count in bronchoalveolar lavage (B) were we measured at indicated time points. CXCL1 was measured at 48 hours post-challenge (C). Neutrophil counts reported are from a single intratracheal administration and are representative of four experiments that span collections 8–96 hours after challenge. Neutrophil levels were assayed via differential cell counts. N= 8 per genotype per condition. Cytokine measurements were carried out via Luminex assays. N= 10 per genotype. (D-E) Zfp30+/+ and Zfp30−/− mice were exposed to 2 ppm ozone for three hours and phenotyped 21 hours later. Total (D) and percent neutrophils (E) are shown. N= 4–6/genotype/group.
Discussion
Based on previous data that implicates Zfp30 in the regulation of neutrophil chemotaxis, we developed a Zfp30 knockout mouse strain to test for potential impacts on in vivo neutrophil recruitment. We accomplished this through CRISPR-Cas9 targeting of the DNA binding domains within Zfp30. This strategy was chosen to disrupt wiltype DNA binding activity of ZFP30, a crucial component of this transcription factor family’s sequence-specific chromatin remodeling activity. Though no suitable antibody for ZFP30 exists, allele-specific qPCR revealed a total loss of wildtype allele expression in homozygous mutant mice. Contrary to expectation based on our previous work (Rutledge 2014), the results shown here indicate that loss of wiltype ZFP30 function does not significantly affect CXCL1 secretion or neutrophil chemotaxis in response to various innate immune stimuli.
One potential explanation for why ZFP30 loss did not have the predicted effects on neutrophil chemotaxis is that genetic background, i.e., strain, could have altered the impact of Zfp30 genotype. We generated the Zfp30 knockout on the C57BL/6J genome, which is one of the Collaborative Cross founder strains. This inbred strain, however, was recently shown to harbor a mutation in Nlrp12 that impacts neutrophil recruitment (Hornick 2017; Ulland et al. 2016). Introducing a knockout into an already-mutated cytokine secretion pathway may have masked the effects of the knockout. Hence, it is possible that generating a Zfp30 knockout on another genetic background could reveal different phenotypic consequences, as has been observed in other cases (Sittig et al. 2016). Zfp30 is more highly expressed in the lungs of 129S1/SvImJ, A/J, NOD/ShiLtJ, and NZO/HILtJ laboratory mouse strains, so they may be good candidates for further analysis. Since we have generated a full-body, non-conditional knockout, it is also possible that in Zfp30−/− mice, other genes compensated for Zfp30, thereby masking any effects of ZFP30 absence (Rossi 2015).
Our analysis of MTEC cultures revealed a marginally significant difference in expression of Muc5ac among Zfp30−/− cultures, suggesting a possible difference in goblet cell abundance between Zfp30+/+ and Zfp30−/− mice. Given the important role of MUC5AC is mucus hypersecretion and airway obstruction (Evans et al. 2015, Ordoñez et al. 2001), this finding may merit further investigation. For example, determining whether loss of ZFP30 affects airway epithelial progenitor cell differentiation, or whether Muc5ac expression was increased due to some inflammatory process in these cells.
Zfp30 was recently shown to affect adipogenesis and Pparg2 expression in vitro (Chen 2019), and human ZFP30 is differentially expressed in the pancreatic beta cells of type-2 diabetes patients (Lawlor 2017). Additionally, previous results suggest that ZFP148 regulates Zfp30 expression (Laudermilk et al. 2018), and Zfp148 was recently implicated in glucose tolerance and insulin secretion from pancreatic islets in mice (Keller et al. 2019). However, follow-up studies on insulin sensitivity here revealed no metabolic impact of ZFP30 loss, nor did we observe a significant difference in Pparg2 expression in the white adipose tissues of our mice. We did, however, observe a significant difference in body weight among male Zfp30−/− mice. In contrast to our results, male Zfp30 knockout mice generated on a C57BL/6N genetic background by the International Mouse Phenotyping Consortium exhibited decreased fasting glucose concentrations compared to wildtype mice. Differences in genetic background may underlie this difference, as our colony was generated on and backcrossed to a C57BL/6J background.
One of the most intriguing findings generated here was that there was a significant depletion of homozygous knockout offspring from Zfp30+/− x Zfp30+/− matings, a finding that was independently reproduced by the International Mouse Phenotyping Consortium. These results implicate ZFP30 in fertility or embryonic development and demonstrate that Zfp30 is an essential gene. The underlying cause of this effect and its sex-specific effect remains unclear, especially in light of the fact that litter sizes were not obviously affected. That said, there is precedent for similar effects of other zinc finger protein knockout mice to display some degree of embryonic lethality. Knockout of Zfp57, a C2H2 ZFP with a KRAB domain, affects the establishment and maintenance of critical DNA methylation imprints, and disruption of this imprinting leads to death (Li et al. 2008). Further studies will be required to explore the mechanisms underlying this phenotype, including an assessment of whether the Zfp30 knockout allele is associated with transmission ratio distortion per se.
To conclude, we demonstrate here that a knockout of Zfp30 on a C57BL/6J genetic background does not affect CXCL1 secretion or neutrophil recruitment to the lungs in mouse models of asthma or acute lung injury. Zfp30−/− male mice do differ from their wildtype littermates in body weight but not in baseline metabolic phenotypes assayed here. Finally, we provide initial evidence that Zfp30 is an essential gene based on the observation that significantly fewer homozygous mutant mice were produced from Zfp30+/− x Zfp30+/− matings.
Supplementary Material
Acknowledgments
The authors would like to thank: Gregory J. Smith, Ph.D. for his assistance with in vivo ozone exposures; Larry Ostrowski, Ph.D. and Ximena Bustamante, Ph.D. for their assistance with MTEC isolation and culture; Kim Burns for her assistance with histology; Max Lowman for technical assistance with qPCR work; Autumn Sanson for her assistance with in vivo data generation; Gang Chen for his suggestions regarding mouse tracheal epithelial cell qPCR; Praveen Sethupathy, Ph.D. and Yu-Han Hung, Ph.D. for their consultation on metabolic phenotypes; and David Aylor, Ph.D., for input on statistical analysis related to transmission ratio distortion. The authors would additionally like to thank the UNC CGIBD Advanced Analytics core for their work on cytokine multiplex assays, the Animal Histopathology and Laboratory Medicine core for their work in processing complete blood count assays,and the UNC NORC Animal Metabolism Phenotyping core for their work on mouse MRIs.
Funding:
This work was supported by NIH grants ES024965 and HL122711. The UNC NORC Animal Metabolism Phenotyping core is supported by DK056350. The UNC CGIBD Advanced Analytics core is supported by DK034987. The UNC Animal Histopathology Core is supported in part by an NCI Center Core Support Grant (5P30CA016086-41) to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Declarations
Conflicts of interest:
Dale Cowley is employed by, has equity ownership in and serves on the board of directors of TransViragen, the company which has been contracted by UNC-Chapel Hill to manage its Animal Models Core Facility.
Ethics approvals:
Not applicable
Consent to participate:
Not applicable
Consent for publication:
Not applicable
Availability of data and material:
The datasets generated during and/or analysed during the current study will be made available as a data supplement if/when the manuscript is accepted.
Code availability:
Not applicable
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- Charo IF, and Ransohoff RM, 2006. The Many Roles of Chemokines and Chemokine Receptors in Inflammation. N. Engl. J. Med 354: 610–621. [DOI] [PubMed] [Google Scholar]
- Chen W, Schwalie PC, Pankevich EV, et al. ZFP30 promotes adipogenesis through the KAP1-mediated activation of a retrotransposon-derived Pparg2 enhancer. Nat Commun. 2019;10(1):1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P, Hopes E, Gingles N, Broman K, McPheat W et al. , 2003. A major locus conferring susceptibility to infection by Streptococcus pneumoniae in mice. Mamm. Genome 14: 448–453. [DOI] [PubMed] [Google Scholar]
- Dickinson M, Flenniken A, Ji X et al. , 2016. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue LJ, Livraghi-Butrico A, McFadden KM, Thomas JM, Chen G et al. , 2017. Identification of trans protein QTL for secreted airway mucins in mice and a causal role for Bpifb1. Genetics 207: 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman D, Aksoy E, Agca MC, Kocak ND, Ozmen I et al. , 2015. The utility of inflammatory markers to predict readmissions and mortality in COPD cases with or without eosinophilia. Int. J. Chron. Obstruct. Pulmon. Dis 10: 2469–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CM, Raclawska DS, Ttofali F, Liptzin DR, Fletcher AA, et al. , 2015. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat. Commun. 6: 6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP et al. , 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10: 2067–2078. [DOI] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G et al. , 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick EE, Banoth B, Miller AM, Zacharias ZR, Jain N et al. , 2017. Nlrp12 Mediates Adverse Neutrophil Recruitment during Influenza Virus Infection. J. Immunol ji1700999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelada SNP, Wilson MS, Tavarez U, Kubalanza K, Borate B et al. , 2011. Strain-dependent genomic factors affect allergen-induced airway hyperresponsiveness in mice. Am. J. Respir. Cell Mol. Biol 45: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Rabaglia M, Schueler K, Stapleton D, Gatti D, Vincent M, … Attie A (2019). Gene loci associated with insulin secretion in islets from nondiabetic mice. Journal of Clinical Investigation, 129(10), 4419–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten AM, Förster K, Radeczky E, Linnhoff A, Balint B et al. , 2015. The safety and tolerability of oral AZD5069, a selective CXCR2 antagonist, in patients with moderate-to-severe COPD. Pulm. Pharmacol. Ther 31: 36–41. [DOI] [PubMed] [Google Scholar]
- Laudermilk LT, Thomas JM, Kelada SN. Differential Regulation of Zfp30 Expression in Murine Airway Epithelia Through Altered Binding of ZFP148 to rs51434084. G3 (Bethesda). 2018;8(2):687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor N, George J, Bolisetty M, Kursawe R, Sun L et al. , 2017. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type – specific expression changes in type 2 diabetes. Genome Res. 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X et al. , 2008. A maternal-zygotic effect of gene Zfp57 maintains both maternal and paternal imprints. Dev Cell. 15: 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limjunyawong N, Mock J, and Mitzner W, 2015. Instillation and Fixation Methods Useful in Mouse Lung Cancer Research. J. Vis. Exp i: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P et al. , 2013. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr. Genomics 14: 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesic LE, De Maio A, and Reeves RH, 1999. Mapping lipo-polysaccharide response loci in mice using recombinant inbred and congenic strains. Genomics 62: 34–41. [DOI] [PubMed] [Google Scholar]
- Medugno L, Florio F, De Cegli R, Grosso M, Lupo A et al. , 2005. The Krüppel-like zinc-finger protein ZNF224 represses aldolase a gene transcription by interacting with the KAP-1 co-repressor protein. Gene 359: 35–43. [DOI] [PubMed] [Google Scholar]
- Mock JR, Tune MK, Dial CF, Torres-Castillo J, Hagan RS, & Doerschuk CM (2020). Effects of IFN-γ on immune cell kinetics during the resolution of acute lung injury. Physiological reports, 8(3), e14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss RB, Mistry SJ, Konstan MW, Pilewski JM, Kerem E et al. , 2013. Safety and early treatment effects of the CXCR2 antagonist SB-656933 in patients with cystic fibrosis. J. Cyst. Fibros 12: 241–248. [DOI] [PubMed] [Google Scholar]
- Nair P, Gaga M, Zervas E, Alagha K, Hargreave FE et al. , 2012. Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: A randomized, placebo-controlled clinical trial. Clin. Exp. Allergy 42: 1097–1103. [DOI] [PubMed] [Google Scholar]
- Nathan C, 2006. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol 6: 173–182. [DOI] [PubMed] [Google Scholar]
- Ordoñez CL, Khashayar R, Wong HH, Ferrando R, Wu R, et al. , 2001. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am. J. Respir. Crit. Care Med 163: 517–523. [DOI] [PubMed] [Google Scholar]
- Plasschaert LW, Žillionis R, Choo-Wing R, Savova V, Knehr J et al. , 2018. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560: 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennard SI, Dale DC, Donohue JF, Kanniess F, Magnussen H et al. , 2015. CXCR2 antagonist MK-7123 a phase 2 proof-of-concept trial for chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 191: 1001–1011. [DOI] [PubMed] [Google Scholar]
- Rutledge H, Aylor DL, Carpenter DE, Peck BC, Chines P et al. , 2014. Genetic Regulation of Zfp30;, CXCL1, and Neutrophilic Inflammation in Murine Lung. Genetics 198: 735 LP–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, & Stainier DYR (2015). Genetic compensation induced by deleterious mutations but not gene [dummy_no space]knockdowns.Nature, 524(7564), 230–233. [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR et al. , 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol 19: 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, and Rauscher FJ, 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16: 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver Lee M. 1995. Mouse genetics: concepts and applications. New York: Oxford University Press. [Google Scholar]
- Sittig LJ, Carbonetto P, Engel KA, Krauss KS, Barrios-Camacho CM, & Palmer AA (2016). Genetic Background Limits Generalizability of Genotype-Phenotype Relationships. Neuron, 91(6), 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Walsh L, Higuchi M, Kelada SNP. Development of a large-scale computer-controlled ozone inhalation exposure system for rodents. Inhal Toxicol. 2019;31(2):61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Morgan AP, Najarian ML, et al. , 2017. Genomes of the Mouse Collaborative Cross. Genetics.; 206(2):537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Sugi Y, Hosono A, and Kaminogawa S, 2009. Epigenetic Regulation of TLR4 Gene Expression in Intestinal Epithelial Cells for the Maintenance of Intestinal Homeostasis. J. Immunol 183: 6522–6529. [DOI] [PubMed] [Google Scholar]
- Todd CM, Salter BM, Murphy DM, Watson RM, Howie KJ et al. , 2016. The effects of a CXCR1/CXCR2 antagonist on neutrophil migration in mild atopic asthmatic subjects. Pulm. Pharmacol. Ther 41: 34–39. [DOI] [PubMed] [Google Scholar]
- Ulland TK, Jain N, Hornick EE, Elliott EI, Clay GM et al. , 2016. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat. Commun 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia R, 2003. KRAB-containing zinc-finger repressor proteins. Genome Biol. 4: 231.1–231.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watz H, Uddin M, Pedersen F, Kirsten A, Goldmann T et al. , 2017. Effects of the CXCR2 antagonist AZD5069 on lung neutrophil recruitment in asthma. Pulm. Pharmacol. Ther 45: 121–123. [DOI] [PubMed] [Google Scholar]
- You Y, Brody S, 2013. Culture and differentiation of mouse tracheal epithelial cells. Epithelial Cell Culture Protocols: Second Edition 123–143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.