Abstract
Abnormal epigenetic patterns correlate with effector T cell malfunction in tumors1–4. However, their causal link is unknown. Here, we show that tumor cells disrupt methionine metabolism in CD8+ T cells, thereby lowering intracellular methionine levels and the methyl donor S-adenosylmethionine (SAM), resulting in loss of H3K79me2. Consequently, loss of H3K79me2 led to low STAT5 expression and impaired T cell immunity. Mechanistically, tumor cells avidly consumed and outcompeted T cells for methionine via high expression of SLC43A2, a methionine transporter. Genetic and biochemical inhibition of tumor SLC43A2 rescued T cell H3K79me2 levels, boosting spontaneous and checkpoint-induced tumor immunity. Moreover, we found that methionine supplementation improved expression of H3K79me2 and STAT5 in T cells, accompanied by increased T cell immunity in tumor bearing models and colon cancer patients. Clinically, tumor SLC43A2 negatively correlated with T cell histone methylation and functional gene signatures. Our work reveals a novel mechanistic connection between methionine metabolism, histone patterns, and T cell immunity in the tumor microenvironment. Thus, cancer methionine consumption is an unappreciated immune evasion mechanism, and targeting cancer methionine signaling may provide an immunotherapeutic approach.
Keywords: Methionine, SAM, H3K79me2, STAT5, SLC43A2, CD8+ T cell, cancer, immunotherapy, metabolism, epigenetic, anti-PD-L1, checkpoint blockade
Main
Immune checkpoint blockade therapies have demonstrated unprecedented clinical efficacy in cancer treatment, however, harnessing this strategy is largely encumbered by therapeutic resistance5. CD8+ T cells mediate anti-tumor immunity. Unfortunately, tumor infiltrating CD8+ T cells are often dysfunctional (exhaustion)1,6. T cell differentiation and activation are associated with epigenetic landscape changes at gene loci encoding effector molecules, including interferon (IFN) and granzyme B4,7. However, these dynamic epigenetic changes in T cells may be subject to disruption by tumor cells2,4,8 via metabolic regulation in the tumor microenvironment9–12. Tumor intrinsic mechanisms, particularly oncogenic signaling, may contribute to abnormal tumor metabolism13,14. However, it is unknown whether amino acid metabolism could affect the T cell epigenetic landscape and in turn alter T cell function in tumors.
Tumor cells outcompete T cells for methionine to impair T cell function
Exhausted T cells exhibit distinct histone profiles and limit tumor immunotherapy2,3. To test if abnormal amino acid metabolism was related to T cell histone alteration and dysfunction, we cultured CD8+ T cells without individual amino acids and found methionine omission resulted in the most dramatic T cell death and dysfunction, as shown by Annexin V+, and IFNγ+ and TNFα+ cells (Fig. 1a–c). Thus, ready access to methionine is critical for T cell survival and function.
Fig.1. Tumor cells outcompete T cells for methionine to impair T cell function.

a-c, Effect of amino acids on T cell apoptosis and effector cytokines. Activated mouse CD8+ T cells were cultured with complete medium (CM) or media with individual amino acid omission for 36 hours. d, e, Effect of tumor supernatants on T cell apoptosis. CD8+ T cells were cultured for 36 hours with ID8 (d) and B16F10 (e) supernatants. f, Mass spectrometry (MS) detection of amino acids consumption by tumor culture. g, h, Effect on T cell apoptosis and cytokines by amino acid supplementation in tumor supernatant. CD8+ T cells were cultured with B16F10 supernatants supplemented with amino acids for 36 hours. i, j, Apoptosis of T cells and tumor cells by methionine competition. B16F10 cells and CD8+ T cells were cultured at different ratios for 72 hours in Transwell with 30 or 100 μM methionine. k, l, Effect of methionine on CD8+ T cell (k) and tumor cell (l) viability. Data are mean ± s.e.m. Sample sizes (n), P values, statistical tests and number of times experiments were replicated are listed in ‘Statistics and reproducibility’.
Next, we tested whether tumor cells impaired CD8+ T cell function through altering methionine levels. We cultured ID8 (Fig. 1d) and B16F10 (Fig. 1e) cells with media containing 20–100 μM methionine. Regardless of methionine concentrations, fresh media had minimal impact on T cell apoptosis (Fig. 1d, e). However, both ID8 (Fig. 1d) and B16F10 (Fig. 1e) supernatants induced CD8+ T cell apoptosis when < 100 μM methionine was originally added. Similar results were obtained in mouse CD8+ T cells cultured with the supernatants from MC38 and CT26 colon cancer cells (Extended Data Fig. 1a, b), as well as in human CD8+ T cells cultured with the supernatants from A375 melanoma cells (Extended Data Fig. 1c). Moreover, supernatants resulted in more death and dysfunction in cultured ID8 tumor infiltrating T cells (Extended Data Fig. 1d, e). Thus, tumor cells restrain methionine access to T cells, and impair their survival and function.
The physiological concentration of methionine in human serum is ~30 μM15,16. Blood methionine were lower in cancer patients than healthy donors (Extended Data Fig. 1f, g). To evaluate methionine consumption within the physiological range, we cultured B16F10 with 30 μM methionine, and analyzed amino acids abundance in the supernatants. Tumor cells consumed the majority of amino acids, including methionine and tryptophan (Fig. 1f, Extended Data Fig. 1h). We subsequently supplemented individual amino acids, and cultured with CD8+ T cells. Among all amino acids, methionine supplementation prevented T cell apoptosis and rescued IFNγ+TNFα+ cytokine (Fig. 1g, h). Similar results were obtained with human CD8+ T cells (Extended Data Fig. 1i, j). Tumor glycolysis regulates T cell function17, we cultured T cells in supernatant supplemented with glucose or methionine. Methionine, but not glucose, recovered T cell survival and cytokine production (Extended Data Fig. 1k–m). Moreover, simultaneous supplementation of glucose and methionine didn’t additionally enhance this recovery effect. Thus, tumor methionine consumption impairs T cell survival and function.
We cultured B16F10 and CD8+ T cells in a Transwell system (Extended Data Fig. 1n). High concentration of methionine (100 μM) had minimal effect on both tumor and CD8+ T cell apoptosis. However, low concentration of methionine (30 μM) caused CD8+ T cell (Fig. 1i), but not tumor cell (Fig. 1j) apoptosis. Then, we evaluated the half maximal effective concentrations (EC50) of methionine to maintain CD8+ T and tumor cell viabilities. Both mouse and human CD8+ T cells were more sensitive than tumor cells to methionine deprivation, as shown by their different EC50s (Fig. 1k, l and Extended Data Fig. 1o, p). Thus, tumor cells outcompete T cells for methionine, thereby impairing T cell survival and function.
Methionine depletion decreases SAM and H3K79me2 in CD8+ T cells
To explore the mechanism by which tumor cells affected CD8+ T cells through methionine deprivation, we performed RNA-sequencing on CD8+ T cells cultured with fresh medium (FM), B16F10 supernatants (Sup), and supernatants plus methionine (Sup+Met) (Extended Data Fig. 2a). Network grouping analysis revealed that the pathways related to metabolism, function, and survival were affected by tumor supernatants (Extended Data Fig. 2b). Correspondingly, GSEA showed an enrichment of T cell apoptosis signature and poor T cell receptor signaling in the presence of tumor supernatants (Fig. 2a), while methionine supplementation largely rescued this phenotype (Extended Data Fig. 2c). Moreover, one carbon metabolic process and the methionine cycle were defective in CD8+ T cells cultured with supernatants (Fig. 2b), which were recovered by methionine addition (Extended Data Fig. 2d, e).
Fig. 2. Tumor alters CD8+ T cell methionine metabolism to diminish H3K79me2.
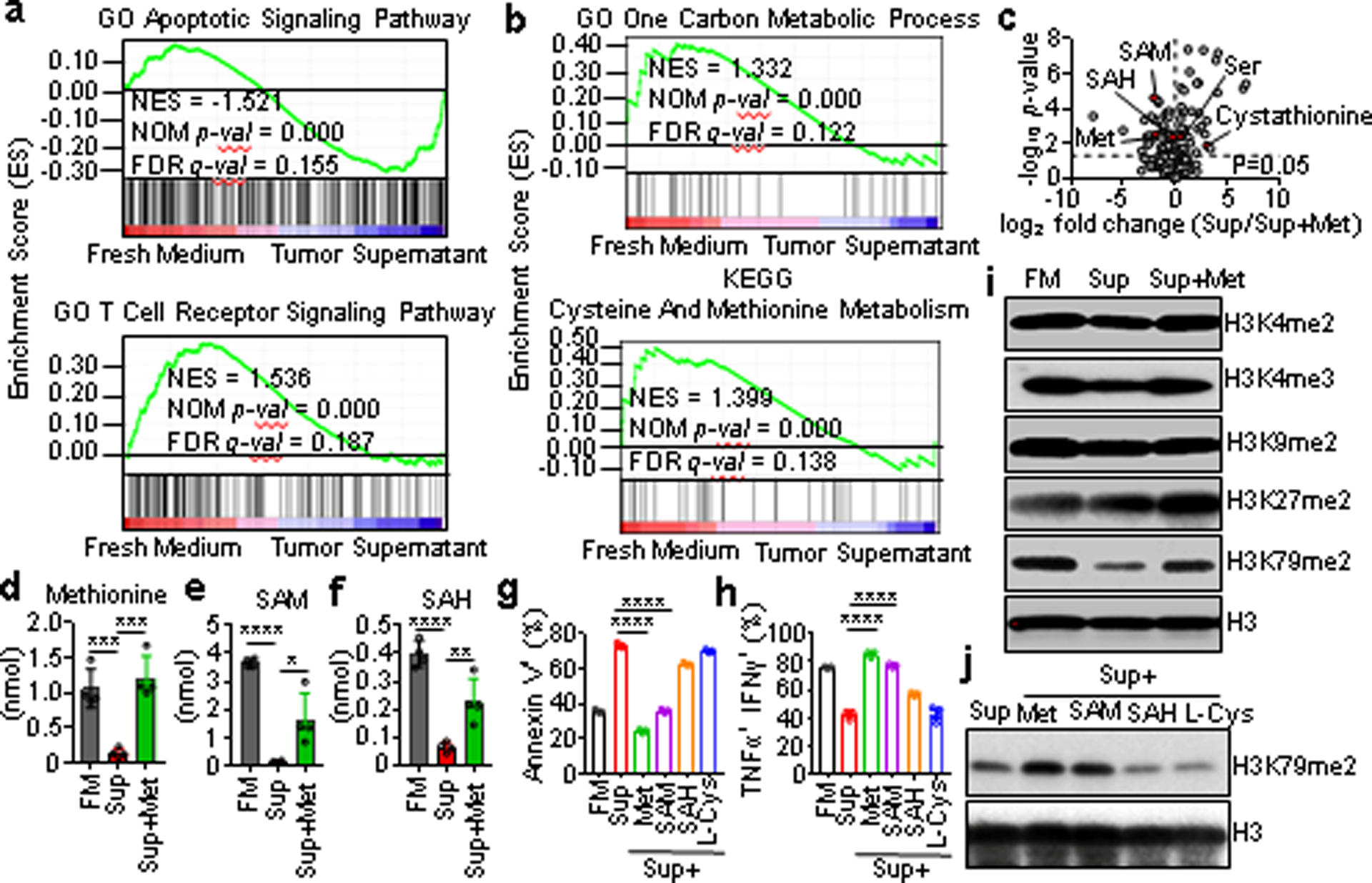
a, b, GSEA plot showed enriched apoptotic and TCR signaling pathways (a), and defective methionine metabolism signaling (b) in tumor supernatant (Sup) cultured CD8+ T cells. c-f, Methionine pathway metabolic changes in CD8+ T cells cultured with FM, Sup and Sup+Met. Volcano showed metabolites changes between groups from Sup and Sup+Met (c). Intracellular methionine (d), SAM (e), and SAH (f) were detected by MS. g, h, Effect of metabolites supplementation on CD8+ T cell apoptosis (g) and cytokines (h). i, Effect of tumor supernatants on CD8+ T cell histone methylation. j, Role of methionine metabolites supplementation in CD8+ T cell H3K79 methylation. Data are mean ± s.e.m. Sample sizes (n), P values, statistical tests and number of times experiments were replicated are listed in ‘Statistics and reproducibility’.
Next, we performed a metabolomics analysis of parallel CD8+ T cells. We observed obvious metabolite changes in T cells cultured with the tumor supernatants, and these too were rescued by methionine addition (Extended Data Fig. 2f). We specifically examined one carbon process and methionine cycle related metabolites (Fig. 2c, Extended Data Fig. 2g), and found that tumor supernatant cultured CD8+ T cells showed a dramatic decrease of intracellular methionine, SAM and S-adenosyl-homocysteine (SAH) (Fig. 2d–f). Supplementation of methionine resulted in recovery of intracellular methionine, SAM, and SAH (Fig. 2d–f), and a decrease in serine and L-cystathionine (Extended Data Fig. 2h, i). To test which metabolite was the key factor, we cultured CD8+ T cells with supernatants and supplemented with methionine, SAM, SAH, or L-cystathionine. Supplementation of methionine or SAM prevented CD8+ T cell apoptosis, and rescued T cell cytokine profile (Fig. 2g, h).
Intracellular methionine is converted to SAM, the donor for epigenetic methylation18,19. Thus, we tested T cell histone marks and found supernatants induced a dramatic decrease in H3K79 methylation, but not in other marks (Fig. 2i). Similar results were obtained in mouse CD8+ T cells cultured with CT26 and MC38 supernatants, and human CD8+ T cells with A375 supernatants (Extended Data Fig. 2j, k). Moreover, the reduced H3K79 methylation could be recovered by supplementation of methionine or SAM, but not by SAH or L-cystathionine (Fig. 2j). Thus, methionine restriction by tumor cells reduces the methyl donor SAM, and in turn, impairs H3K79me2 in CD8+ T cells.
Loss of H3K79me2 impairs T cell immunity through STAT5 pathway
Disruptor of telomeric silencing 1-like (DOT1L) is the specific and sole methyltransferase for H3K7920,21. We cultured CD8+ T cells with EPZ004777, a DOT1L inhibitor. EPZ004777 inhibited H3K79me2, induced CD8+ T cell apoptosis, and suppressed CD8+ T cell cytokine expression in a dose dependent manner (Fig. 3a–c). To genetically explore the role of H3K79me2 in T cell function, we crossed conditional Dot1l allele (Dot1l flox/flox, also as Dot1l+/+) mice22 with CD4-Cre transgenic mice to delete Dot1l specifically in T cells (Extended Data Fig. 3a, referenced as Dot1l−/−). DOT1L deletion led to a loss of H3K79me2 in CD8+ T cells (Fig. 3d, Extended Data Fig. 3b) and resulted in higher apoptosis, especially upon activation (Fig. 3e). Moreover, intracellular cytokine staining showed an impaired function in Dot1l−/− CD8+ T cells (Fig. 3f). We performed RNA array on Dot1l+/+ and Dot1l−/− CD8+ T cells (Extended Data Fig. 3c), and found it was comparable to that of T cells exposed to methionine deficiency (Extended Data Fig. 2b). For instance, similar to methionine deficient T cells (Fig. 2a), Dot1l−/− T cells showed enriched apoptotic gene signature and impaired T cell receptor functional gene signature (Extended Data Fig. 3d, e). The data suggest a mechanistic correlation in T cell malfunction caused by methionine deficiency and by impaired Dot1l-dependent histone methylation.
Fig. 3. Loss of H3K79me2 impairs T cell anti-tumor immunity through STAT5.

a-c, CD8+ T cells were treated with EPZ004777 for 48 hours. Western Blot showed H3K79me2 in CD8+ T cells (a). FACS demonstrated CD8+ T cell apoptosis (b) and cytokines (c). d, Western Blot showed H3K79me2 in Dot1lf/f and Dot1l−/− CD8+ T cells. e, f, Effect of DOT1L deletion on CD8+ T cells apoptosis (e) and cytokines (f). g-i, Effect of T cell DOT1L deficiency on MC38 growth (g, h) and T cell viability (i). j, k, Effect of methionine supplementation on Dot1lf/f and Dot1l−/− CD8+ T cells apoptosis (j) and cytokines (k). l, GSEA plot showed enriched apoptotic pathway in Dot1l−/− CD8+ T cells. m, Heat map showed Jak-Stats mRNA levels in mouse Dot1l−/− vs Dot1lf/f CD8+ T cells. n, Western blot showed STAT5 and p-STAT5 in Dot1lf/f (f/f) and Dot1l−/− (−/−) CD8+ T cells. o, Western blot showed STAT5 and p-STAT5 in CD8+ T cells cultured with fresh medium (FM), Supernatant (Sup), and Sup supplemented with different metabolites (Sup+). p, ChIP assay showed H3K79me2 occupancy on the Stat5b promoter in CD8+ T cells. q, ChIP assay showed H3K79me2 occupancy on the Stat5b promoter in CD8+ T cells cultured with FM, Sup, or Sup+Met. Data are mean ± s.e.m. Sample sizes (n), P values, statistical tests and number of times experiments were replicated are listed in ‘Statistics and reproducibility’.
We assessed T cell DOT1L in tumor immunity and found MC38 tumor grew faster in Dot1l−/− mice (Fig. 3g, h). Correspondingly, we detected an increase in CD8+ T cell apoptosis in tumor draining lymph node (dLN) and tumor tissues (Fig. 3i), as well as a decrease in tumor infiltrating CD8+ T cell TNFα, IFNγ, and granzyme B (Extended Data Fig. 3f). In addition, PD-L1 blockade resulted in tumor inhibition in Dot1l+/+ mice, but not in Dot1l−/− mice (Extended Data Fig. 3g). We obtained similar results in B16F10 tumors (Extended Data Fig. 3h, i). To confirm the positive role of methionine in T cells is dependent on DOT1L, we cultured Dot1l+/+ and Dot1l−/− T cells with methionine supplementation in the presence of tumor medium. Methionine supplementation failed to prevent Dot1l−/− T cells from apoptosis (Fig. 3j) and to rescue their impaired cytokine production (Fig. 3k). Thus, loss of DOT1L, which mediates H3K79me2, weakens anti-tumor immunity.
Hereafter, we explored how H3K79me2 loss results in T cell dysfunction. We observed apoptosis gene signature enrichment in Dot1l−/− CD8+ T cells (Fig. 3l, Extended Data Fig. 3c, d). The JAK-STAT pathway regulates T cell survival and effector function. Among the JAK-STATs, Stat5 were mostly affected by H3K79me2 deficiency (Fig. 3m, Extended Data Fig. 3j). We confirmed a decrease in total STAT5 and phosphorylated STAT5 (p-STAT5), but not other STATs, in Dot1l−/− T cells (Fig. 3n). Then, we cultured mouse CD8+ T cells with B16F10 supernatants. Supernatants induced a decrease in Stat5 transcripts (Extended Data Fig. 3k), total STAT5, and p-STAT5 (Fig. 3o). These effects were rescued by supplementation of methionine or SAM, but not SAH or L-cystathionine (Fig. 3o). Moreover, the RNA-seq data from human CD8+ T cells treated with a DOT1L inhibitor, SGC094623, showed reduced STAT5, enriched apoptotic gene signatures, and impaired T cell signaling (Extended Data Fig. 3l–n). Altogether, tumor cells outcompete T cells for methionine, resulting in a reduction of H3K79me2 and defective STAT5 signaling in CD8+ T cells.
H3K79me2 is an active gene mark in mammalian cells and occurs on the promoter and 5’ regions within the coding regions of transcriptionally active genes24,25. ChIP-seq data26,27 revealed high H3K79me2 occupancies in the key regulatory regions of the STAT5b promoter in mouse and human (Extended Data Fig. 3o, p). ChIP assay demonstrated a high H3K79me2 abundance binding to the Stat5 promoter (Fig. 3p, Extended Data Table 1). This binding was diminished in T cells cultured with B16F10 supernatants and recovered by methionine supplementation (Fig. 3q). Thus, H3K79me2 is involved in the direct regulation of STAT5 transcription in CD8+ T cells.
Methionine supplementation restores T cell immunity
To demonstrate the relevance of methionine competition between tumor cells and T cells in vivo, we conducted 4 complementary studies. First, we detected a decrease in H3K79me2 and STAT5 in tumor infiltrating CD8+ T cells, compared to T cells in dLN and spleen (Extended Data Fig. 4a–d). Second, compared to peripheral T cells, we validated a decrease in H3K79me2 and STAT5 in human tumor infiltrating CD8+ T cells from ovarian carcinoma omentum, malignant ascites, and several other cancers (Extended Data Fig. 4e–i). To examine the effect of methionine on human tumor infiltrating T cells, we cultured human colorectal cancer infiltrating T cells with or without methionine. Addition of methionine enhanced expression of T cell effector cytokines, H3K79me2, and STAT5 (Extended Data Fig. 4j–m). Third, we supplemented methionine by intratumor injection in B16F10 bearing mice. Methionine supplementation delayed tumor growth, along with enhanced H3K79me2 and STAT5 expression in tumor infiltrating CD8+ T cells, and increased T cell survival and polyfunctional cytokines (Fig. 4a–e). We performed similar studies in ID8 tumor bearing mice. Again, methionine supplementation slowed down tumor progression (Fig. 4f), enhanced T cell (but not tumor cell) survival (Extended Data Fig. 4n) and effector cytokine in tumor ascites and tumor infiltrating CD8+ T cells (Fig. 4g, h). As a confirmation, after methionine supplementation, we detected high levels of methionine in ID8 ascites (Extended Data Fig. 4o). Furthermore, we treated mice bearing CT26 tumor with methionine, anti-PD-L1, and their combination. We observed a synergistic anti-tumor effect mediated by methionine plus anti-PD-L1, compared to single agent therapy. This was accompanied with increased T cell tumor infiltration and reduced T cell apoptosis (Extended Data Fig. 4p–r). Fourth, we recruited and provided methionine supplementation to patients with colorectal cancer (Extended Data Table 2). Methionine supplementation resulted in an increase in H3K79me2 and p-STAT5 in CD8+ T cells (Fig. 4i), and enhanced T cell IL-2 (Fig. 4j), CD8+ T cell polyfunctional cytokine expression (Fig. 4k), and decreased CD8+ T cell apoptosis (Fig. 4l). Altogether, our data suggest that methionine deficiency impairs T cell H3K79me2 and STAT5 expression and function.
Fig. 4. Methionine supplementation in tumor restores T cell immunity.

a-e, Methionine supplementation restored T cell immunity in B16F10-bearing mice. Tumor growth (a), tumor infiltrating CD8+ T cell H3K79me2 (b) and STAT5 (c) were monitored. FACS showed intratumor CD8+ T cell apoptosis (d) and cytokines (e). f-h, Methionine supplementation restored T cell immunity in ID8-bearing mice. Tumor growth was monitored by bioluminescence imaging (f). FACS showed ascites CD8+ T cell (g) and intratumor CD8+ T cell cytokines (h). i-l, Studies on colorectal cancer patients treated with methionine. Western blot showed p-STAT5 and H3K79me2 in peripheral CD8+ T cells prior and post methionine treatment (i). FACS showed IL-2+ T cells (j), CD8+ T cell effector cytokines (k) and apoptosis (l) in patients prior and post methionine treatment. Data are mean ± s.e.m. Sample sizes (n), P values, statistical tests and number of times experiments were replicated are listed in ‘Statistics and reproducibility’.
Tumor impairs tumor immunity through SLC43A2
Methionine is transported into cells by the solute carrier family (SLC), including system L-type and A-type transporters28. We cultured B16F10 cells with BCH (a system L transporter inhibitor) or MeAIB (a system A transporter inhibitor)28. Then, we cultured CD8+ T cells with these corresponding tumor supernatants and analyzed CD8+ T cells. BCH, but not MeAIB, prevented T cell apoptosis and rescued the impaired cytokine profile (Extended Data Fig. 5a, b). Thus, system L transporters may be responsible for tumor methionine consumption.
Next, we compared the SLC transcripts in effector CD8+ T cells and tumor cells, and found SLC7A5 and SLC43A2 (two system L transporters) were relatively highly expressed on tumor (Extended Data Fig. 5c). Western blots revealed minimal SLC43A2 in effector CD8+ T cells and comparable SLC7A5 in effector CD8+ T cells and several tumor cells (Extended Data Fig. 5d). In line with this, we detected minimal SLC43A2 in human CD8+ T cells, compared to several tumor cells (Extended Data Fig. 5e). The differential SLC43A2 in tumor and CD8+ T cells suggests that tumor cells may be well-positioned to outcompete T cells for methionine via SLC43A2.
To test this possibility, we knocked down SLC43A2 in B16F10 (Extended Data Fig. 5f), which induced a decrease in methionine consumption (Extended Data Fig. 5g). Then, we cultured CD8+ T cells with the tumor supernatants from shSLC43A2, and compared to scrambled tumor cells. We observed reduced T cell apoptosis and enhanced polyfunctional cytokine in CD8+ T cells cultured with the supernatants from shSLC43A2 cells (Fig. 5a, b). Furthermore, by using the Transwell (Extended Data Fig. 1n), we found a reduction in T cell apoptosis and an increase in cytokine by T cells cultured with shSLC43A2 cells (Fig. 5c, d). Moreover, high H3K79me2 were detected in T cells cultured with shSLC43A2 cells (Fig. 5e). Thus, tumor cells outcompete T cells for methionine via SLC43A2, and affect T cell histone methylation and function.
Fig. 5. Tumor cells outcompete T cells for methionine via SLC43A2.

a, b, Effects of supernatants from sh-SLC43A2 tumors on T cell apoptosis (a) and cytokines (b). c-e, Effects of tumor SLC43A2 knockdown on T cell apoptosis (c), cytokines (d) and H3K29me2 modification (e). CD8+ T cells were co-cultured with wild type and sh-SLC43A2 B16F10 cells in Transwell in media containing with 30 μM methionine. CD8+ T cell apoptosis (c), cytokines (d) and H3K79me2 (e) were determined by FACS and Western Blot after 72 hours. f, g, Effect of tumor SLC43A2 knockdown on tumor growth (f) and tumor T cells function (g). h-l, Effect of the combination of BCH and anti-PD-L1 on B16F10 (h, i) and ID8 (j-l) bearing mice. Tumor growth (h, j, k), and TNFα+, IFNγ+ and granzyme B+ CD8+ T cells (i, l) were compared. m, SLC43A2 transcripts in tumors and paired adjacent normal tissues in several types of tumor in TCGA. Data are mean ± s.e.m. Sample sizes (n), P values, statistical tests and number of times experiments were replicated are listed in ‘Statistics and reproducibility’.
We next inoculated shSLC43A2 B16F10 cells into Dot1l+/+ and Dot1l−/− C57BL/6 mice, and observed slower tumor growth in Dot1l+/+ mice bearing shSLC43A2 B16F10 (Fig. 5f). However, SLC43A2 knockdown failed to affect tumor progression in Dot1l−/− mice (Extended Data Fig. 5h). The data suggest a functional connection between tumor SLC43A2 and T cell DOT1L in anti-tumor immunity. Furthermore, tumor growth was comparable in Rag1−/− mice (Extended Data Fig. 5i). Thus, tumor immunity contributed to tumor control in shSLC43A2 tumor. Consistent with this, we showed increased tumor T cell infiltration (Extended Data Fig. 5j) and effector molecule in shSLC43A2 tumor (Fig. 5g). Furthermore, anti-PD-L1 additionally inhibited shSLC43A2 B16F10 growth (Extended Data Fig. 5k). We also studied shSLC43A2 ID8 model (Extended Data Fig. 5l). Tumor growth was similar in Rag1−/− mice (Extended Data Fig. 5m), however, we observed slower tumor growth and enhanced CD8+ T cells infiltration in WT mice (Extended Data Fig. 5n, o). The data suggest that pharmacologically targeting SLC43A2 may promote anti-tumor immunity. Given that specific SLC43A2 inhibitor is not available, we treated B16F10-bearing mice with BCH with or without anti-PD-L1 (Fig. 5h). Single treatment with BCH or anti-PD-L1 partially inhibited tumor growth, and the combination manifested a synergistic effect (Fig. 5h). Moreover, the combination treatment induced the highest T cell infiltration (Extended Data Fig. 5p), and highest effector molecule in tumor infiltrating CD8+ T cells (Fig. 5i). We also tested the combination therapy in ID8 model. Again, the combination synergistically inhibited tumor growth and enhanced tumor infiltrating CD8+ T cell cytokine (Fig. 5j–l). These results suggest that targeting tumor SLC43A2 in combination with checkpoint blockade may be an effective anti-cancer approach.
Finally, we examined a potential relationship between tumor SLC43A2, T cell signature, and clinical outcome in cancer patients. Based on available TCGA database, we found that SLC43A2 transcripts were higher in tumors than matched normal tissues (Fig. 5m). Moreover, high tumor SLC43A2 were associated with poor survival (Extended Data Fig. 5q–s). Based on the single cell RNA-seq in tumor infiltrating T cells and melanoma cells in patients29, we found higher SLC43A2 transcripts in tumor cells than in T cells (Extended Data Fig. 5t). We divided the patients into high and low tumor SLC43A2 groups. GSEA showed that methionine metabolic signaling genes were enriched in melanoma cells with high tumor cell SLC43A2 (Extended Data Fig. 5u). Tumor SLC43A2 negatively correlated with CD8 and IFNγ transcripts in the same tumors (Extended Data Fig. 5v). Furthermore, compared to patients with low tumor SLC43A2, T cells in patients with high tumor SLC43A2 exhibited weak methionine metabolic signatures, histone methylation signatures, and poor effector genes (Extended Data Fig. 5w–y). Thus, high tumor SLC43A2 negatively correlates with T cell immune responses in patients with cancer.
Discussion
Recent studies have started to explore amino acids in T cell activation and epigenetic reprogramming30–32. Methionine is an essential amino acid, and is converted to SAM for methyltransferases to yield methylated substrates, including histone methylation18,19. Hence, SAM provides a link between methionine metabolism and epigenetic regulation. Dysfunctional T cells exhibit a distinct epigenetic landscape including histone alteration2,3. Thus, abnormal methionine metabolism may lead to particular histone alteration in T cells and contribute to their dysfunction in the tumor microenvironment.
We demonstrate a direct competition between tumor cells and T cells for methionine, which results in a decrease of a series of substrates in one-carbon metabolism including SAM. DOT1L is the only methyltransferase for H3K7920,21, and has a relatively low Km for SAM33. These characteristics contribute to the H3K79 methylation sensitivity to methionine and SAM deprivation, and explain why T cell H3K79me2 is predominantly sensitive to tumor-altered methionine metabolism. The mechanistic connection of methionine, H3K79me2, and STAT5 has additionally been validated in human and murine tumor infiltrating T cells. Notably, as substantial methionine is required for abnormal tumor cell proliferation and metabolism, dietary methionine restriction in tumor growth has been tested in immune deficient systems34. Our work indicates that both human and murine effector T cells are sensitive to methionine. Thus, tumor specific methionine restriction is essential to maintain T cell immunity in patients with cancer.
H3K79me2 is an active transcriptional histone mark21. Biochemical and genetic inhibition of DOT1L has abolished H3K79me2 and STAT5, resulting in T cell apoptosis and dysfunction. Mechanistically, H3K79me2 controls STAT5 transcription in CD8+ T cells. Thus, we have identified a causal and biological link between a particular histone alteration (H3K79me2) and a critical transcription factor (STAT5) in defining T cell phenotype. Moreover, tumor cells outcompete T cells for methionine via SLC43A2, a major methionine transporter. Given that SLC43A2 is highly expressed on multiple human and mouse tumor cells with different genetic backgrounds, abnormal tumor SLC43A2 and its related methionine metabolism may be unlikely driven by shared key oncogenes. Inhibition of tumor SLC43A2 can normalize methionine metabolism in effector T cells and rescue their function, as well as improve spontaneous and checkpoint blockade-induced anti-tumor immunity in preclinical models. Our work has not only generated novel insights into SLC biology in T cells, but also identified tumor SLC43A2 as an immunotherapy resistance mechanism in patients with cancer. Indeed, we have established a negative correlation between tumor cell SLC43A2, tumor infiltrating T cell histone methylation, and effector functional signatures in the same tumor tissues in cancer patients.
In summary, we have demonstrated that tumor cells metabolically and epigenetically impair T cell function and tumor immunity by outcompeting T cells for methionine via SLC43A2 (Extended Data Fig. 6). Our work demonstrates a long-thought crosstalk between metabolism, histone pattern, and functional profile in tumor infiltrating T cells. On this basis, selectively targeting tumor methionine metabolism may be a novel approach for cancer immunotherapy.
Methods
Mice
Six- to eight-week-old female wild-type C57BL/6, BALB/c, and Rag1 knock out (KO) mice were from the Jackson Laboratory (Bar Harbor, ME, USA). Dot1lflox/flox mice were bred with CD4-Cre mice to generate mice with specific DOT1L deletion in T cells. All mice Dot1lf/f or Dot1l−/− mice were utilized at the age of 6–12 weeks unless specified in the text. Mice were housed under specific pathogen-free conditions and handled according to the guidelines of the University Committee on the Use and Care of Animals at the University of Michigan.
Clinical studies
Colorectal cancer patients were recruited for the methionine supplementation study. Eligible patients were of Eastern Cooperative Oncology Group performance status 0/1 with adequate organ and bone marrow function. Patients were excluded from this study if they had received or were receiving any concurrent chemotherapy, immunotherapy, biologic, and/or hormonal therapy for cancer. All patients took two capsules of methionine (500 mg/capsule, NOW Foods, Bloomingdale, IL, USA) daily for two weeks. This study was conducted according to the Declaration of Helsinki and approved by the institutional review board (IRB) of the Medical University of Lublin, with written informed consent obtained from all patients. Study participants were not compensated.
Human specimens
Plasma, peripheral blood mononuclear cells (PBMCs), and tumor infiltrating T cells were isolated from healthy donors and cancer patients, respectively. Plasma from patients diagnosed with high-grade serous ovarian carcinomas were collected for this study. Human specimens were collected with informed consent and procedures approved by the IRB of the University of Michigan.
Reagents
Amino acids, including L-isoleucine, L-leucine, L-lysine, L-methionine, L-phenylalanine, L-threonine, L-tryptophan, L-valine, L-histidine, L-arginine, L-cystine, L-tyrosine, and MEM non-essential amino acid solution (100×, including L-alanine, L-aspartic acid, L-asparagine, L-glutamic acid, L-glycine, L-proline and L-serine) were from Sigma (Saint Louis, MO, USA). L-glutamine (100x), 2-Mercaptoethanol and Dialyzed Fetal Bovine Serum (FBS) were from GIBCO (Carlsbad, CA, USA). RPMI 1640 medium without amino acids and sodium phosphate powder (#R8999–04A) were from the US Biological (Salem, MA, USA). 1x RPMI without L-glutamine, L-cysteine, L-cystine, and L-methionine was from MP Biomedicals (Solon, OH, USA). Methionine Assay Kit (Fluorometric) was from Abcam (ab234041, Cambridge, UK). Anti-mouse CD3 and anti-CD28, anti-human CD3 and anti-CD28 monoclonal antibodies (mAbs) were from eBioscience (San Diego, CA, USA). Mouse and human interleukin 2 (IL-2) was from R&D Systems, Inc. (Minneapolis, MN, USA). S-(5-Adenosyl)-L-methionine iodide (SAM), S-(5-Adenosyl)-L-homocysteine (SAH), and L-cystathionine, as well as SLC transporter inhibitors, including α-(Methylamino) isobutyric acid (MeAIB) and 2-Amino-2-norbornanecarboxylic acid (BCH) were from Sigma. DOT1L Inhibitor, EPZ004777 (#1338466–77-5) were from Millipore (Burlington, MA, USA). Anti-mouse PD-L1 (Clone: 10F.9G2) and rat IgG2B isotype (Clone: LTF-2) were from Bioxcell (West Lebanon, NH, USA).
Cell Separation and Culture
Human cells (including A375, CHL-1, SK-MEL-2, 293T cells) and mouse tumor cells (including B16F10 and CT26 cells) were obtained from ATCC (Manassas, VA. USA). Mouse ID8-luc and MC38 cells were previously reported35,36. Human primary high grade serous ovarian carcinoma cells (OC8) were generated in our laboratory36. All cell lines in our laboratory are routinely tested for mycoplasma contamination and cells used in this study are negative for mycoplasma. None of our cell lines are on the list of commonly misidentified cell lines (International Cell Line Authentication Committee). Tumor cells were maintained in RPMI1640 (HyClone SH30255, GE healthcare, Chicago, IL, USA) containing 10% (v/v) FBS (Alkali Scientific, Fort Lauderdale, FL, USA) and 1% (v/v) pen/strep (GIBCO).
Mouse lymphocytes were isolated from spleen and lymph nodes. CD8+ T cells were separated by the EasySep™ Mouse CD8+ T Cell Isolation Kit (STEMCELL Technologies Inc., Cambridge, MA, USA). Human PBMCs were isolated from blood by using Lymphoprep™ (STEMCELL Technologies Inc.). Human CD8+ T cells were separated through the EasySep™ Human CD8+ T Cell Isolation Kit (STEMCELL Technologies Inc.). CD8+ T cells were re-suspended (106 cells/mL) and activated with anti-CD3 and anti-CD28 mAbs for 48 hours. Activated CD8+ T cells were maintained with IL-2 (10ng/mL) and 2-mercaptoethanol, and cultured with fresh complete media, media with/without individual amino acids, or the tumor supernatants for 36–48 hours. Media with/without amino acids were formulated with RPMI1640 (US Biological, #R8999–04A) by supplementation or omission of individual amino acids. The tumor supernatants were collected from media initially cultured with tumor cells in RPMI absence of L-glutamine, L-cysteine, L-cystine, and L-methionine (MP Biomedicals #1646454), and subsequently supplemented with L-glutamine (GIBCO), L-cystine 2HCl (Sigma), and different concentrations of L-methionine (Sigma), as specified in different experiments. The following amino acids were used to supplement tumor culture supernatant: Iso 380 μM/L, Leu 380 μM/L, Lys 220 μM/L, Met 30 μM/L, Phe 90 μM/L, Thr 17 0μM/L, Trp 20 μM/L, Val 170 μM/L, His 100 μM/L, Arg 1160 μM/L, Cys 200 μM/L, Gln 2000 μM/L, Tyr 110 μM/L, and other amino acids (MEM Non-essential Amino Acid Solution 100x, a mix of Ala, Asp, Asn, Glu, Gly, Pro, and Ser (Sigma)). The following metabolites were used to supplement tumor culture supernatant: Met 30 μM/L, SAM 50 μM/L, SAH 50 μM/L or L-Cystathionine 100 μM/L.
Intratumor CD8+ T cells from mice and human were isolated as follows: mononuclear cells from the whole tumor or ascites suspension were first enriched by density gradient centrifugation using Lymphoprep™ (STEMCELL). CD8+ T cells were further separated a negative-positive two steps isolation. First, the enriched cells were isolated by the EasySep™ Mouse/Human CD8+ T Cell Isolation Kit (negative selection, STEMCELL), then further enriched by the EasySep™ Mouse/human CD8a Positive Selection Kit II (positive selection, STEMCELL). The purity of CD8+ T cell were further determined by FACS staining.
Generation of knockdown cells
SLC43A2 knockdown cells were generated by using the MISSION® shRNA (Sigma) and the GIPZ Lentiviral shRNA system (Dharmacon, Inc. Lafayette, CO, USA). 293T cells were co-transfected with lentiviral shRNA system together with plasmids psPAX2 and pMD2.G using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) for lentivirus package. 48 hours after transfection, the supernatant was collected. Tumor cells were infected with the virus supernatant for 24 hours and then selected with 2 μg/mL puromycin (Santa Cruz Biotechnology, Dallas, TX, USA) for additional 48 hours. Knockdown efficiency was validated by immunoblotting.
Flow cytometry analysis (FACS)
For FACS staining, cells were stained with a combination of fluorescence-conjugated mAbs from BD Biosciences (Franklin Lakes, NJ, USA) or Thermo Fisher Scientific (Waltham, MA, USA). Mouse samples were stained with FITC-Annexin V, 7-AAD, PE-Texas Red-anti mouse CD45 (30-F11), FITC-anti-Mouse CD90 (53–2.1), APC-Cy7-anti-mouse CD4 (RM4–5), AF700-anti-mouse CD8 (53–6.7), APC-anti-mouse IL-2 (JES6–5H4), BV786-anti-mouse IFNγ (XMG1.2), PE-Cy7-anti-mouse TNFα (MP6-XT22) and PE-anti-mouse granzyme B (NGZB) mAbs. Human samples were stained with FITC-Annexin V, 7-AAD, Pacific Blue-anti-human IFNγ (4S.B3), and APC-anti-human TNFα (MAb11). For apoptosis staining, cells were washed with 1x binding buffer (BD Biosciences) and stained with Annexin V and 7-AAD in 1x binding buffer at dark for 10 minutes. For surface staining, the cells were dark incubated with antibodies for 30 minutes. For intracellular staining, the cells were fixed in Fix/Perm solution (BD Biosciences). After being washed with Perm/Wash buffer (BD Biosciences), the cells were stained intracellularly for 30 minutes under dark. For STAT5, cells were stained with APC-anti-STAT5 (REA549, Miltenyi Biotec Inc., Bergisch Gladbach, Germany). For DOT1L and H3K79me2 intracellular staining, the cells were first stained with DOT1L or H3K79me2 antibodies (Abcam), and then stained using a FITC-conjugated goat anti-rabbit IgG (H+L) secondary antibody (Invitrogen). All samples were acquired on BD LSRFortessa™ (BD Biosciences) and were analyzed with FACSDiva (BD Biosciences) or FlowJo (FlowJo LLC, USA).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed according to the protocol (Upstate, Millipore). Briefly, crosslinking was performed with 1% formaldehyde or 1% paraformaldehyde for 10 minutes. To enhance cell lysis, we ran the lysate through a 27g needle three times and flash froze it in −80°C. Sonication was then performed with the Misonix 4000 water bath sonication unit at 15% amplitude for 20 minutes. Protein/DNA complex was precipitated by specific antibodies against H3K79me2 (Abcam) and IgG control (Millipore, Burlington, MA, USA). DNA was purified using DNA Purification Kit (Qiagen, Hilden, Germany). ChIP-enriched chromatin was used for real-time PCR. Relative expression levels were normalized to Input. Specific primers are listed in Extended Data Table 1.
Real-time PCR and Western Blotting
CD8+ T cells were incubated in fresh media or specific tumor supernatant for 24–48 hours. Tumor cells were maintained with complete media. The cells were washed and harvested. RNA was isolated from these cells using Direct-zolTM RNA miniprep Plus kit (ZOMO research, Irvine, CA, USA), and then subjected to reverse transcription with first-strand cDNA Synthesis for Quantitative RT-PCR kit (OriGene, Rockville, MD, USA). Real-time PCR was performed using SYBR green chemistry (Applied Biosystems, Foster City, CA, USA). Reactions were run on a real-time PCR system (StepOnePlus Real-Time PCR System, Applied Biosystems, Foster City, CA, USA). Specific primers are listed in Extended Data Table 3.
For Western blotting, CD8+ T and tumor cells were washed and lysed in a modified RIPA buffer with 1× protease inhibitor cocktail (Roche, Basel, Switzerland). The lysates were stored at −80 °C until immunoblot analysis. For histone isolation, CD8+ T cells were first lysed with PBS with 0.5% Triton X-100. The lysates were then incubated on ice for 10 minutes and cleared by centrifugation at 5000g for 15 minutes. The precipitate was dissolved with 0.2 N HCl. Protein concentration was quantified using a BCA protein assay kit (Thermo Fisher Scientific) and denatured at 95°C for 5 minutes. The lysates samples were stored at −80 °C for immunoblot analysis. Briefly, the proteins were separated electrophoretically using a 12% SDS-polyacrylamide gel and transferred onto an PVDF membrane (Millipore). The membranes were blocked in 5% fat-free milk for 1 hour, and then incubated with a specific primary antibody at 4°C overnight. Blots were probed with rabbit anti-H3K79me2, H3K4me2, H3K4me3, H3K9me2, H3K27me2, total H3, STAT5, p-STAT5, STAT1, STAT3, SLC43A2, SLC7A5 and β-actin. All antibodies were from Abcam or Cell Signaling Technology (Danvers, MA, USA). After hybridization with HRP-conjugated secondary antibody (Life Technologies), protein bands were visualized using a chemiluminescence detection kit (Bio-Rad Laboratories, Hercules, CA, USA).
Tumor inoculation and treatments
For the in vivo tumor growth experiments, the animals were inoculated subcutaneously (s.c.) with 2 × 105 B16F10 cells or 5 × 105 MC38 cells, or intraperitoneally (i.p.) with 2 × 106 ID8-Luc ovarian cancer cells. The B16F10 and MC38 tumor volumes were measured along three orthogonal axes (a, b, and c) and were calculated as follows: tumor volume = a*b*c/2. The ID8-luc tumor growth was monitored by using the Xenogen IVIS Spectrum In Vivo Bioluminescence Imaging System (PerkinElmer, Waltham, MA, USA). Tumor load was calculated based on the total flux (photons per second [p/s]). Anti–PD-L1 and IgG1 isotype mAbs (Bioxcell) were given intraperitoneally at a dose of 100 μg per mouse on day 7 after tumor cell inoculation, then every 3 days for the duration of the experiment. 2-Amino-2-norbornanecarboxylic acid (BCH) was given intravenously at a dose of 180 mg/kg per mouse on day 7 after tumor inoculation, then every 2 days for the duration of the experiment. Methionine was given by intratumor (B16F10 model) or intraperitoneal (ID8 model) injection at a dose of 40 mg/kg per mouse on day 7 after tumor inoculation, then every 2 days for the duration of the experiment. Animal studies were conducted under the approval of the University of Michigan Committee on Use and Care of Animals. In none of the experiments, tumour size surpasses 2 cm in any dimension. No animal had severe abdominal distension (≥10% original body weight increase). Sample size was chosen at the basis of preliminary data. After tumour inoculation mice were randomized and assigned to different groups for treatment.
RNA-seq and bioinformatics analysis
CD8+ T cells were cultured in complete fresh medium (FM), tumor supernatant (Sup), and tumor supernatant plus methionine (Sup+Met) for 24 hours. CD8+ T cells from Dot1l−/− and littermates (Dot1lf/f) were isolated and sorted. The RNA was isolated by using Direct-zolTM RNA miniprep Plus kit (ZOMO research). RNA-seq and RNA-array were conducted in CD8+ T cells by the DNA sequencing core at the University of Michigan. The data were processed by the Bioinformatic Core at the University of Michigan, and analyzed with ClueGo37 and GSEA software v. 3.038. RNA sequencing data that support the findings of this study have been deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE150887. Public RNA-seq data were from GSE108694 and GSE72056. For single cell RNA-seq data in patients with melanoma, the expression levels of genes were quantified as Ei,j=log2(TPM i,j/10+1), where TPM i,j refers to transcript-per-million (TPM) for gene i in sample j29. We then evaluated the average Ei,j values of tumor cell SLC43A2 transcripts39–41. Based on median of average Ei,j values, we divided the patients into high (average SLC43A2 Ei,j >0.056, including patients #53, 79, 81, 82, 84, and 94), low (average SLC43A2 Ei,j < 0.056, including patients #60, 65, 71, 80, 88, and 89) groups. GSEA analysis for infiltration T cells were characterized and compared between these patients with high and low levels of SLC43A2 expression.
Metabolomics
Metabolomics and sample collection were performed as previously reported42,43. Briefly, CD8+ T cells were collected and transferred to a 15 mL tube to centrifuge at 300 g for 5 minutes at 4°C. Cells were then washed with cold PBS. After centrifugation at 300 g for 5 minutes at 4°C, 1 mL of 80% cold methanol was added and vigorously vortexed to ensure the cell pellet was completely disrupted. The samples were placed on dry ice and moved to a −80°C freezer for 10 minutes, followed by vigorous vortex. The samples were again centrifuged at maximum speed for 10 minutes at 4°C. The supernatant was collected in new tubes and normalized by protein concentration. Samples were kept in −80°C until measured. We used liquid chromatography–mass spectrometry to detect intracellular metabolites in CD8+ T cells and amino acids in human sera from healthy donors and patients with ovarian cancer. Intracellular SAM and amino acids in B16F10 supernatants were measured in Creative-proteomics (Shirley, NY, USA).
Statistical Analysis
Statistical analysis was performed in GraphPad Prism statistical software (version 7, GraphPad Software Inc., San Diego, CA, USA). Error bars in data represent mean ± standard error of the mean (SEM). Inter-group data were analyzed using an unpaired or paired two-tailed t test. The tumor growth was analyzed by using two-way analysis of variance (ANOVA). Survival functions were estimated by the Kaplan-Meier methods. Log-rank test was used to calculate the statistical differences. The correlations between tumor SLC43A2 and immune associated genes were analyzed using Person correlation test. A value of p<0.05 was considered statistically significant.
Extended Data
Extended Data Fig. 1. Tumor cells outcompete T cells for methionine to impair T cell function.

a-c, Effect of tumor cells on T cell apoptosis. Tumor supernatants were collected from MC38 (a), CT26 (b), and human melanoma A375 (c) tumor cells cultured for 48 hours with media containing different concentrations of methionine (Met). Then, CD8+ T cells were cultured for 36 hours with these tumor supernatants (Sup) or fresh medium (FM). Apoptosis was determined by Annexin V staining. d, e, Effect of methionine on ID8 tumor infiltrating cells. T cells were cultured with fresh medium (FM), ID8 supernatant (Sup), and supernatant plus methionine (Sup+Met). T cell apoptosis (d) and cytokine production (e) were determined by FACS. f, g, Amino acid levels in ovarian cancer patient plasma. Amino acids were detected in healthy donor and ovarian cancer patient plasma by liquid chromatography mass spectrometry (LC-MS). (f) Volcano showed plasma free amino acid changes. Red dot showed methionine (Met). (g) Plasma methionine in ovarian cancer patients vs healthy controls. h, Methionine concentration in pre- and post- tumor cultured medium. i, j, Effect of amino acid supplementation on human T cell function. CD8+ T cells were cultured with A375 supernatants (Sup) supplemented with different amino acids for 36 hours. FACS analysis showed T cell apoptosis (i) and effector cytokines (j). k-m, Effect of glucose supplementation on the role of methionine-affected T cell apoptosis and function. n, Schematic figure showing tumor and T cell co-culture in the Transwell system. o, p, Effect of methionine on human CD8+ T cell (o) and tumor cell (p) viability, EC50 was determined by nonlinear regression (log (agonist) vs. response). FM: fresh medium. Sup: tumor supernatant. EAA: essential amino acid, NEAA: non-essential amino acid. Data are mean ± s.e.m. Information on sample sizes, experimental number, times, biological replicates, statistical tests, and P values is available in ‘Statistics and reproducibility’.
Extended Data Fig. 2. Tumor alters CD8+ T cell methionine metabolism to diminish H3K79me2.

a, Gene profile changes in CD8+ T cells. Mouse CD8+ T cells were cultured with fresh medium (FM), B16F10 tumor supernatant (Sup), or tumor supernatants plus methionine (Sup+Met) for 36 hours. Gene profile changes were analyzed by RNA-seq. b, Gene signatures were compared between groups from FM and Sup. Functionally grouped network of enriched categories was generated for the hub genes and their regulators using ClueGO. Visualization has been carried out using Cytoscape 3.7.1.c-e, GSEA plot showed recovery of TCR signaling pathway (c) and methionine metabolism signaling (d, e) in CD8+ T cells cultured with Sup+Met compared to Sup. f, Metabolites changes in CD8+ T cells cultured with FM, Sup and Sup+Met. Upper panel: Metabolites induced upon methionine supplementation. Lower panel: Metabolites suppressed upon methionine supplementation. g, The diagram of methionine cycle is shown. h, i, CD8+ T cells were cultured with FM, Sup, or Sup+Met for 36 hours. Metabolites related to the methionine cycle, including intracellular serine (h) and L-cystathionine (i), were detected by MS. j, k, Effect of tumor supernatants on CD8+ T cell histone methylation. Mouse (j) or human (k) CD8+ T cells were cultured with or without methionine (Met) for 36 hours with fresh medium (FM), CT26 and MC38 tumor supernatants (j), or human A375 tumor supernatants (Sup) (k). T cell histone marks were determined by Western blots. Data are mean ± s.e.m. Information on sample sizes, experimental number, times, biological replicates, statistical tests, and P values is available in ‘Statistics and reproducibility’.
Extended Data Fig. 3. Loss of H3K79me2 impairs T cell anti-tumor immunity through STAT5.
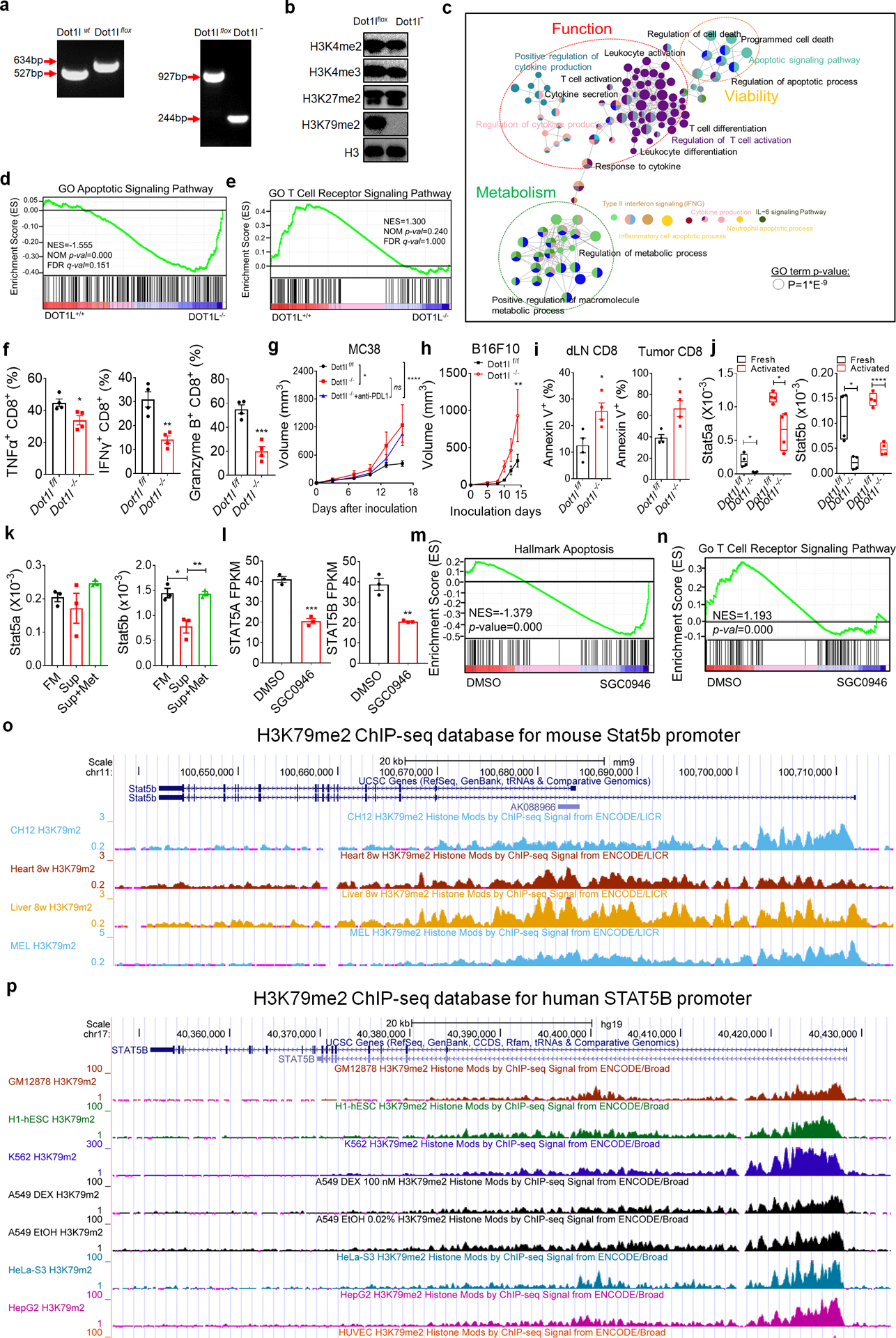
a, Genotyping for Dot1lf/f and Dot1l−/− mice by PCR. b, Effect of Dot1l knockout on histone marks in T cells. c-e, Gene signature comparison between Dot1l−/− and Dot1lf/f CD8+ T cells. Functionally grouped network of enriched categories was generated for the hub genes and their regulators using ClueGO. Visualization has been carried out using Cytoscape 3.7.1. (c). GSEA plot showed enriched apoptotic gene pathway (d) and impaired TCR signaling pathway (e) in Dot1l−/− CD8+ T cells. f, Effect of DOT1L deficiency on T cell function in MC38 tumor. MC38 cells were inoculated into Dot1lf/f and Dot1l−/− mice. Expression of TNFα, IFNγ, and granzyme B in tumor infiltrating CD8+ T cells was determined by FACS. g, Effect of anti-PD-L1 on tumor growth in Dot1l+/+ and Dot1l−/− mice. h, i, B16F10 cells were inoculated into Dot1l−/− and Dot1lf/f mice. Effect of T cell DOT1L deficiency on tumor growth (h) and T cell viability (i) were monitored. j, Real-time PCR showed Stat5a and Stat5b transcripts in fresh or anti-CD3/CD28 activated Dot1lf/f and Dot1l−/− CD8+ T cells. k, Real-time PCR showed Stat5a and Stat5b transcripts in activated CD8+ T cells cultured with fresh media (FM), B16F10 tumor supernatants (Sup), or supernatants plus methionine (Sup+Met) for 24 hours. l-n, RNA-seq showed the effect of DOT1L inhibitor (SGC0946) on human CD8+ T cells (Database: GSE108694). STAT5A and STAT5B (l) transcripts were quantified in human CD8+ T cells treated with DOT1L inhibitor SGC0946. GSEA enrichment plot showed enrichment of apoptotic gene pathway (m) and defects in T cell receptor related pathways (n) in human CD8+ T cells treated with DOT1L inhibitor. o, p, H3K79me2 ChIP-seq in ENCODE database showing Stat5b promoter in mice (o) and humans (p). Data are mean ± s.e.m. Information on sample sizes, experimental number, times, biological replicates, statistical tests, and P values is available in ‘Statistics and reproducibility’.
Extended Data Fig. 4. Methionine supplementation promotes T cell anti-tumor immunity.

a, b, H3K79me2 (a) and STAT5 (b) levels in CD8+ T cells from tumor draining lymph node (dLN) and tumor in B16F10 bearing mice. c, d, H3K79me2 (c) and STAT5 (d) levels in CD8+ T cells from spleen and tumor ascites in ID8 bearing mice. e, H3K79me2 levels in CD8+ T cells from healthy peripheral blood and human ovarian cancers ascites. f, g, H3K79me2 (f) and STAT5 (g) levels in CD8+ T cells from healthy human blood and human ovarian cancer omentum tissues. h, i, FACS showed H3K79me2 and STAT5 levels in human tumor infiltrating CD8+ T cells. j-m, Effect of methionine on human tumor infiltrating CD8+ T cells. Human colorectal cancer infiltrating CD8+ T cells were cultured with or without methionine. T cell cytokine production (j, k), H3K79me2 (l), and STAT5 (m) were analyzed by FACS. One representative of four is shown. n, Effect of methionine supplementation on apoptosis of tumor infiltrating CD8+ T cells and ID8 tumor cells in vivo. ID8 tumor bearing mice were treated with methionine or PBS. T cell and tumor cell apoptosis was determined by FACS. o, Methionine levels in ID8 tumor after methionine or PBS treatment. p-r: Effect of anti-PD-L1 on methionine-affected CT26 tumor progression. Mice bearing CT26 tumor were treated with anti-PD-L1, methionine, and their combination. Tumor volume (p), T cell tumor infiltration (q) and apoptosis (r) were assessed. Data are mean ± s.e.m. Information on sample sizes, experimental number, times, biological replicates, statistical tests, and P values is available in ‘Statistics and reproducibility’.
Extended Data Fig. 5. Tumor SLC43A2 correlates to poor T cell immunity.

a, b, Effects of SLC inhibitors (BCH or MeAIB) on tumor cell affected CD8+ T cell apoptosis (a) and cytokine production (b). c, Real-time PCR showed SLC transporter transcripts in activated CD8+ T cells and B16F10 tumor cells. d, Western Blot showed SLC43A2 and SLC7A5 proteins in activated CD8+ T cells and tumor cells. e, Western Blot showed SLC43A2 protein in human CD8+ T cells and human tumor cells. f, Western Blot showed SLC43A2 knockdown efficiency in B16F10 cells. g, Effect of tumor cell SLC43A2 knockdown on methionine consumption. WT (scramble) and sh-SLC43A2 tumor cells were cultured with fresh medium containing 30 μM methionine for 24 hours. Methionine concentration was measured by MS in fresh medium and supernatants. h, Wild type and sh-SLC43A2 B16F10 tumor growth in Dot1l−/− mice. i, Wild type and SLC43A2 knockdown B16F10 tumor growth in Rag1−/− mice. j, Effect of tumor SLC43A2 knockdown on T cell tumor infiltration in WT or sh-SLC43A2 B16F10 bearing mice. k, Effect of SLC43A2 knockdown and the combination of anti-PD-L1 on B16F10 bearing mice. l, Western Blot showed SLC43A2 knockdown efficiency in ID8-luc cells. m, Wild type and SLC43A2 knockdown ID8-luc tumor growth in Rag1−/− mice. n, o, Effect of tumor SLC43A2 knockdown on ID8 growth (n) and T cell tumor infiltration in WT or sh-SLC43A2 ID8 bearing mice. p, T cell tumor infiltration in B16F10 bearing mice treated with BCH, anti-PD-L1, or their combination. q-s, Kaplan-Meier survival curves showed the prognostic values of SLC43A2 expression in different types of tumor: Cholangiocarcinoma (CHOL, q), low grade glioma (LGG, r), and lung squamous cell carcinoma (LUSC, s). The raw data was from TCGA. t-y, The analysis was based on single cell RNA-seq data (GSE72056). t, SLC43A2 transcripts were compared in tumor cells versus tumor infiltrating T cells from the same human melanoma tissues. u, GSEA plots showed methionine metabolic process genes in tumor cells expressing high versus low SLC43A2. v, Correlation was analyzed between CD8A, CD8B, IFNG transcripts in T cells and SLC43A2 transcripts in tumor cells in the same human melanoma tissues. w-y, GSEA enrichment plot analysis showed defective pathways in tumor infiltrating T cells in melanoma patients with high tumor SLC43A2 compared to low tumor SLC43A2. The pathways included T cell methionine metabolic process (w), histone methylation (x), and IFNγ production (y). Data are mean ± s.e.m. Information on sample sizes, experimental number, times, biological replicates, statistical tests, and P values is available in ‘Statistics and reproducibility’.
Extended Data Fig. 6. Graphical model.

Extended Data Table 1.
ChIP primers for mouse Stat5b.
| Site | CHIP primer region | Primer Sequence |
|---|---|---|
| 1 | Stat5b promoter #1 (−186~−369) | F: 5’-TCATTCAGTCAGGATACGGGC-3’ |
| R: 5’-GAATTCCCCAGCTGAAAAGGC-3’ | ||
| 2 | Stat5b promoter #2 (−790~−930) | F: 5’-AAAGGCGAAGAACAAACGGC-3’ |
| R: 5’-TACAAGTTCCGACCCACAGC-3’ | ||
| 3 | Stat5b promoter #3 (−1072~−1240) | F: 5’-GCTTGAATGTGTGGTGGTGG-3’ |
| R: 5’-AGACAGCTCTCCTTCCGACT-3’ | ||
| 4 | Stat5b promoter #4 (−1072~−1240) | F: 5’- CGTGCTCCTGCTGTCTAGAAGCTGGG-3’ |
| R: 5’- GGGATCGGCTCTGTCGGCGTC-3’ | ||
| 5 | Stat5b promoter #5 (−3148~−3308) | F: 5’-AGGCCAGGAGTGTGTTTCTG-3’ |
| R: 5’-TGGAAATCAGCAGCTCTGGG-3’ | ||
| 6 | Stat5b promoter #6 (−3886~−3940) | F: 5’-ATAGTGGGTGGCAGGGTTTG-3’ |
| R: 5’-CTGTCTACCTCATGGCGTCC-3’ |
Extended Data Table 2.
Characteristics of patients with colorectal cancer.
| Number | Gender | Age | Tumor histology | Grade | Primary tumor location | Stage | TNM |
|---|---|---|---|---|---|---|---|
| 1 | M | 62 | Adenocarcinoma tubulare | G2 | Sigmoid colon | III | T4N1M0 |
| 2 | M | 84 | Adenocarcinoma tubulare | G2 | Transverse colon (Hepatic flexure) | II | T3N0M0 |
| 3 | F | 62 | Adenocarcinoma tubulare | G2 | Sigmo-rectal flexure | III | T3NxM0 |
| 4 | M | 66 | Adenocarcinoma | G2 | Ceacum | IV | T4N1M1 |
| 5 | M | 59 | Adenocarcinoma tubulare | G3 | Sigmoid colon | IV | T4N1M1 |
| 6 | M | 81 | Adenocarcinoma tubulare | G2 | Sigmoid colon | II | T3N0M0 |
| 7 | F | 65 | Adenocarcinoma | G2 | Sigmoid | III | T2N0M0 |
Extended Data Table 3.
Primers for RT-PCR and Dot1l mouse genotyping.
| Gene | Forward Sequence (5’–3’) | Reverse Sequence (5’–3’) |
|---|---|---|
| Slc3a2 | GAGCGTACTGAATCCCTAGTCAC | GCTGGTAGAGTCGGAGAAGATG |
| Slc7a5 | GGTCTCTGTTCACGTCCTCAAG | GAACACCAGTGATGGCACAGGT |
| Slc7a6 | TCTACCTTCGCTGGAAAGAGCC | GCCACCAGAAACAAGGAGCAGA |
| Slc7a7 | AAGGTGTTGGCGCTGATTGCAG | AGAGTGCCAGAGCAATGTCACC |
| Slc7a9 | GGATTCCTCTGGTGACCGTATG | CAAGATGCTGGATAGAGAACGCG |
| Slc38a1 | TACCAGAGCACAGGCGACATTC | ATGGCGGCACAGGTGGAACTTT |
| Slc38a2 | GCGTTGGCATTCAATAGCACCG | TCGTAGATGGGAAGAACAGCGG |
| Slc38a4 | CTCTTCACAGCAATGGCGTGGA | GACCTCAGGGTGGCAGACAAAA |
| Slc43a1 | TTCCTGTGGAGCCTTGTCACCA | CTCCACCTTCTGTCTCTGCTCA |
| Slc43a2 | CAGCATCCTTGAGTTCCTGGTC | TGATGTAGCCGATGACAGGAGC |
| Stat5a | CCTGTTTGAGTCTCAGTTCAGCG | TGGCAGTAGCATTGTGGTCCTG |
| Stat5b | CACAGTTCAGCGTCGGTGGAAA | CTGTGGCATTGTTGTCCTGGCT |
| Actb | CATTGCTGACAGGATGCAGAAGG | TGCTGGAAGGTGGACAGTGAGG |
| Dot1 L-genotype-Dot1l alleles | GCCTACAGCCTTCATCATTC | GATAGTCTCAATAATCTCA |
| Dot1 L-genotype-confirming excision | GAAGTTCCTATTCCGAAGTT | GAACCACAGGATGCTTCAG |
Supplementary Material
Acknowledgments
We thank Philip King for scientific input for this work. We acknowledge support from the Advanced Genomics Core and Bioinformatics Core of the University of Michigan Medical School’s Biomedical Research Core Facilities. This work was supported in part by the research grants from the U.S. NIH/NCI R01 grants (W.Z) (CA217648, CA123088, CA099985, CA193136 and CA152470) and the NCI Cooperative human tissue network (CHTN). C.A.L. was supported by a 2017 AACR NextGen Grant for Transformative Cancer Research (17-20-01-LYSS) and an ACS Research Scholar Grant (RSG-18-186-01). Metabolomics studies performed at the University of Michigan were supported by the NIH grant DK097153, the Charles Woodson Research Fund, and the University of Michigan Pediatric Brain Tumor Initiative. C.A.L. and W.Z. were supported by the NIH through the University of Michigan Rogel Cancer Center Grant (P30 CA046592).
Footnotes
Data availability
RNA sequencing data that support the findings of this study have been deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE150887. All other data that supported the findings of this study are available from the corresponding author upon request.
Declaration of Interests
The authors declare no competing interests.
References:
- 1.Wherry EJ T cell exhaustion. Nature Immunology 12, 492–499, doi: 10.1038/ni.2035 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Pauken KE et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science (New York, N.Y.) 354, 1160–1165, doi: 10.1126/science.aaf2807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen DR et al. The epigenetic landscape of T cell exhaustion. Science (New York, N.Y.) 354, 1165–1169, doi: 10.1126/science.aae0491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philip M et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456, doi: 10.1038/nature22367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou W, Wolchok JD & Chen L PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Science translational medicine 8, 328rv324, doi: 10.1126/scitranslmed.aad7118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton BL, Williams JB, Cabanov A, Spranger S & Gajewski TF Intratumoral CD8(+) T-cell Apoptosis Is a Major Component of T-cell Dysfunction and Impedes Antitumor Immunity. Cancer Immunol Res 6, 14–24, doi: 10.1158/2326-6066.cir-17-0249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoenborn JR et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-γ. Nature Immunology 8, 732, doi: 10.1038/ni1474 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan O et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218, doi: 10.1038/s41586-019-1325-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao E et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nature Immunology 17, 95, doi: 10.1038/ni.3313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song M et al. IRE1α–XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562, 423–428, doi: 10.1038/s41586-018-0597-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W et al. Aerobic Glycolysis Controls Myeloid-Derived Suppressor Cells and Tumor Immunity via a Specific CEBPB Isoform in Triple-Negative Breast Cancer. Cell Metabolism 28, 87–103.e106, doi: 10.1016/j.cmet.2018.04.022 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maj T et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nature Immunology 18, 1332, doi: 10.1038/ni.3868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levine AJ & Puzio-Kuter AM The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 330, 1340, doi: 10.1126/science.1193494 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Cairns RA, Harris IS & Mak TW Regulation of cancer cell metabolism. Nature Reviews Cancer 11, 85, doi: 10.1038/nrc2981 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Guttormsen AB, Solheim E & Refsum H Variation in plasma cystathionine and its relation to changes in plasma concentrations of homocysteine and methionine in healthy subjects during a 24-h observation period. The American Journal of Clinical Nutrition 79, 76–79, doi: 10.1093/ajcn/79.1.76 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Schmidt JA et al. Plasma concentrations and intakes of amino acids in male meat-eaters, fish-eaters, vegetarians and vegans: a cross-sectional analysis in the EPIC-Oxford cohort. European Journal of Clinical Nutrition 70, 306–312, doi: 10.1038/ejcn.2015.144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu J et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell reports 27, 2063–2074.e2065, doi: 10.1016/j.celrep.2019.04.022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mentch SJ et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab 22, 861–873, doi: 10.1016/j.cmet.2015.08.024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraki N et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19, 780–794, doi: 10.1016/j.cmet.2014.03.017 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Min J, Feng Q, Li Z, Zhang Y & Xu R-M Structure of the Catalytic Domain of Human DOT1L, a Non-SET Domain Nucleosomal Histone Methyltransferase. Cell 112, 711–723, doi: 10.1016/S0092-8674(03)00114-4 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Nguyen AT & Zhang Y The diverse functions of Dot1 and H3K79 methylation. Genes Dev 25, 1345–1358, doi: 10.1101/gad.2057811 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo SY, Granowicz EM, Maillard I, Thomas D & Hess JL Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood 117, 4759–4768, doi: 10.1182/blood-2010-12-327668 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kagoya Y et al. DOT1L inhibition attenuates graft-versus-host disease by allogeneic T cells in adoptive immunotherapy models. Nature Communications 9, 1915, doi: 10.1038/s41467-018-04262-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubeler D et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18, 1263–1271, doi: 10.1101/gad.1198204 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barski A et al. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 129, 823–837, doi: 10.1016/j.cell.2007.05.009 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Kent WJ et al. The human genome browser at UCSC. Genome research 12, 996–1006, doi: 10.1101/gr.229102 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenbloom KR et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research 41, D56–63, doi: 10.1093/nar/gks1172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander SPH et al. THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology 176, S397–S493, doi: 10.1111/bph.14753 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tirosh I et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189, doi: 10.1126/science.aad0501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma EH et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metabolism 25, 345–357, doi: 10.1016/j.cmet.2016.12.011 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Geiger R et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842.e813, doi: 10.1016/j.cell.2016.09.031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy DG et al. Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metabolism 31, 250–266.e259, doi: 10.1016/j.cmet.2020.01.006 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Richon VM et al. Chemogenetic analysis of human protein methyltransferases. Chemical biology & drug design 78, 199–210, doi: 10.1111/j.1747-0285.2011.01135.x (2011). [DOI] [PubMed] [Google Scholar]
- 34.Gao X et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401, doi: 10.1038/s41586-019-1437-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roby KF et al. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis 21, 585–591, doi: 10.1093/carcin/21.4.585 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Wang W et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell 165, 1092–1105, doi: 10.1016/j.cell.2016.04.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindea G et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093, doi: 10.1093/bioinformatics/btp101 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102, 15545, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bacher R & Kendziorski C Design and computational analysis of single-cell RNA-sequencing experiments. Genome biology 17, 63, doi: 10.1186/s13059-016-0927-y (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner GP, Kin K & Lynch VJ Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory in biosciences = Theorie in den Biowissenschaften 131, 281–285, doi: 10.1007/s12064-012-0162-3 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Hwang B, Lee JH & Bang D Single-cell RNA sequencing technologies and bioinformatics pipelines. Experimental & Molecular Medicine 50, 96, doi: 10.1038/s12276-018-0071-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H-J, Kremer DM, Sajjakulnukit P, Zhang L & Lyssiotis CA A large-scale analysis of targeted metabolomics data from heterogeneous biological samples provides insights into metabolite dynamics. Metabolomics 15, 103, doi: 10.1007/s11306-019-1564-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan M et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC–MS/MS. Nature Protocols 14, 313–330, doi: 10.1038/s41596-018-0102-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


