SUMMARY
The xbp-1 mRNA encodes the XBP-1 transcription factor, a critical part of the unfolded protein response. Here we report that an RNA fragment produced from xbp-1 mRNA cleavage is a biologically active non-coding RNA (ncRNA) essential for axon regeneration in Caenorhabditis elegans. We show that the xbp-1 ncRNA acts independently of the protein-coding function of the xbp-1 transcript, as part of a dual output xbp-1 mRNA stress response axis. Structural analysis indicates that the function of the xbp-1 ncRNA depends on a single RNA stem; this stem forms only in the cleaved xbp-1 ncRNA fragment. Disruption of this stem abolishes the non-coding but not coding function of the endogenous xbp-1 transcript. Thus, cleavage of the xbp-1 mRNA bifurcates it into a coding and a non-coding pathway; modulation of the two pathways may allow neurons to fine-tune their response to injury and other stresses.
Graphical Abstract

eTOC BLURB
Liu et al. show that the mRNA encoding the XBP-1 transcription factor has an unexpected second function in C. elegans. Cytoplasmic cleavage of xbp-1 mRNA activates a ncRNA sequestered within the mRNA. This ncRNA is critical for axon regeneration, and acts independently of the protein-coding function of the xbp-1 mRNA.
INTRODUCTION
The IRE1/XBP1 branch of the unfolded protein response (XBP-UPR) is conserved from yeast to human and is a critical component of the cellular response to protein stress in the ER (Walter and Ron, 2011). The output of the XBP-UPR is activity of the XBP-1 protein, a DNA-binding transcription factor encoded by the xbp-1 locus. Translation of active XBP-1 protein requires processing of xbp-1 mRNA by a non-canonical cytoplasmic RNA splicing event. This splicing event removes a short central sequence from the xbp-1 mRNA, resulting in a frame shift that brings the functional C-terminal half of the XBP-1 protein into frame. Mechanistically, cytoplasmic xbp-1U mRNA is first cleaved twice by the endonuclease IRE-1(Calfon et al., 2002; Kawahara et al., 1998; Sidrauski and Walter, 1997; Yoshida et al., 2001). Next, the 5’ and 3’ fragments are ligated by the RNA ligase RtcB to generate the spliced xbp-1S mRNA that encodes the XBP-1 protein (Figure 1A)(Jurkin et al., 2014; Kosmaczewski et al., 2014; Lu et al., 2014; Sidrauski et al., 1996). The XBP-UPR is crucial for cellular protein homeostasis, and dysregulation of xbp-1 splicing is implicated in inflammatory diseases, metabolic disease and several types of cancer (Jiang et al., 2015). In the nervous system, the function of the XBP-UPR has been associated with a wide range of neurodegenerative diseases including Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), Huntington’s disease, and Parkinson’s disease (Hetz and Mollereau, 2014; Jiang et al., 2015).
Figure 1. A processing intermediate of the xbp-1 mRNA promotes axon regeneration.

(A) Diagram of the xbp-1 mRNA splicing pathway. (B) RNA-seq analysis identifies non-canonical RNA junctions that are enriched in RtcB mutant animals compared to non-mutant controls. The Venn diagram plots genes with such RtcB-dependent junctions identified under normal condition (blue) or with neuronal injury (orange) at the indicated fold change cutoff. See also Table S1 and Table S2. (C) Animals with uncleavable xbp-1 fail to mount the UPR either in control conditions or upon tunicamycin treatment (5 μg/mL, 24 h). Scale bar, 50 μm. (D) Scheme of axotomy in C. elegans GABA neurons. DNC, dorsal nerve cord. VNC, ventral nerve cord. (E) Animals deficient of the RNA ligase RtcB show significantly higher regeneration. n = 37 and 44 from left to right. (F) Axon regeneration is eliminated in animals with the xbp-1(uncleavable) allele (see also Figure S1 for details about this allele). n = 44 and 43 from left to right. In (E) and (F), black bar represents the median. ***P<0.001, ****P<0.0001, 2-tailed K-S test.
We previously found that RtcB, the RNA ligase that is required for xbp-1 splicing and the XBP-UPR, has a very strong effect on axon regeneration in C. elegans neurons. Neurons can respond to axon injury by initiating axon regeneration to restore structure and function. Both an unbiased functional screen (Nix et al., 2014) and detailed genetic analysis (Kosmaczewski et al., 2015) indicated that RtcB mutants have extremely high regeneration, among the strongest effects seen. This result was surprising, since neuronal injury is a form of cellular stress, and RtcB mutants completely lack the XBP-UPR and die quickly when treated with tunicamycin to disrupt ER protein homeostasis (Kosmaczewski et al., 2014). Further experiments showed that the effect of RtcB required its ligase activity, but was independent of the XBP-UPR. It was also independent of the ligation of tRNAs, the other RNA substrate of RtcB (Kosmaczewski et al., 2015). These data indicate that RtcB affects axon regeneration by an unknown mechanism involving RNA ligation.
Here we show that the xbp-1 locus, in addition to encoding the XBP-1 protein, also encodes a biologically active non-coding RNA (ncRNA). The xbp-1 ncRNA is the 3’ fragment produced by cleavage of the cytoplasmic xbp-1U mRNA. The effect of RtcB on axon regeneration is due in large part to modulation of the xbp-1 ncRNA function, rather than the canonical output of the XBP-1 protein. We show that the function of the xbp-1 ncRNA is context-dependent, and that it is not functional before cleavage, when it is still part of the xbp-1U mRNA. We identify a rearrangement of secondary structure that correlates with xbp-1 ncRNA activity, and show that a single base pair within this structure is essential for ncRNA function.
ncRNAs are produced from various origins and biogenesis pathways. In addition to being generated from distinct regions from the coding RNAs, ncRNAs can also be transcribed from genomic regions that are closely linked to coding genes. For example, many miRNAs and circRNAs are processed from intronic regions of pre-mRNAs (Fu, 2014). However, it is less clear whether ncRNAs can be produced from functional, mature mRNAs. Only until recently has it been postulated that coding RNAs can also have non-coding functions (Crerar et al., 2019; Kumari, 2015; Sampath and Ephrussi, 2016). By characterizing the non-coding function of the xbp-1 locus, our study provides the first example of a ncRNA directly derived from the cleavage of a mature mRNA. Discovery of a ncRNA pathway that coexists with the coding function of the xbp-1 locus opens a new window into the molecular mechanisms neurons use to respond to cellular conditions.
RESULTS
Blocking xbp-1 cleavage versus ligation results in opposite phenotype in axon regeneration
RtcB mutant animals, which lack the XBP-UPR completely because the xbp-1 RNA fragments cannot be ligated to produce the protein-coding xbp-1S, have extremely high axon regeneration (Kosmaczewski et al., 2015). However, the high regeneration phenotype in these mutants is not caused by the loss of the XBP-UPR, as either inactivating or activating the XBP-UPR does not phenocopy or rescue RtcB mutants and overall has relatively minor effects on regeneration (Kosmaczewski et al., 2015). The high regeneration phenotype is also not due to lack of tRNA maturation, despite the clear role of RtcB in ligating tRNAs (Englert et al., 2011; Popow et al., 2011; Tanaka and Shuman, 2011).
We considered the possibility that an unidentified RNA substrate mediates the effect of RtcB ligation on axon regeneration. We performed RNA-seq comparing RtcB mutant animals and non-mutant controls, and searched for RtcB-dependent RNA junctions that did not have the canonical sequences used by the spliceosome (see Methods). To account for the possibility that neuron stress was required, we sequenced both groups with and without neuronal injury, using a mutation in β-spectrin (unc-70) to trigger spontaneous axon breaks (Hammarlund et al., 2007). The analysis (Table S1) confirmed that RtcB is required for production of the xbp-1S mRNA, as the RtcB-dependent junction was >500 fold enriched in presence of RtcB compared to RtcB mutant animals. Further, this enrichment was observed in both normal conditions and in the β-spectrin mutant background, suggesting that xbp-1 splicing occurs constitutively and is not significantly altered by neuronal injury. However, when comparing all junctions using a fold-change cutoff at 10, we identified only 3 genes when we compared the two groups with neuronal injury, and 8 genes when without. Other than xbp-1, there was no overlap between the two sets of genes (Figure 1B, Table S2). The effect of RtcB on axon regeneration is neuron-autonomous (Kosmaczewski et al., 2015), but many of the genes we identified are not expressed in neurons (Figure 1B, Table S2). Further, many of these junctions were located in non-coding regions near the 5’ or 3’ ends of transcripts, and likely have little effect on gene function (Table S2). Overall, we did not identify strong candidates other than xbp-1 for RtcB-mediated ligation.
Thus, we considered the possibility that another function of the xbp-1 gene, independent of the XBP-UPR, might affect axon regeneration. To better characterize xbp-1 function, we sought to block xbp-1U mRNA processing at the cleavage step, rather than at the ligation step (as in the RtcB mutants). Cleavage of xbp-1U mRNA is mediated by the IRE-1 endonuclease. However, IRE-1 has many RNA targets (Hollien and Weissman, 2006; Hollien et al., 2009). Further, unlike xbp-1 mutants, ire-1 mutant animals are visibly sick. These data suggest that IRE-1’s additional targets are biologically important, and might confound the study of the XBP-UPR. Therefore, to block specifically the cleavage of xbp-1U mRNA, we used CRISPR techniques to generate a novel allele, xbp-1(uncleavable) (Figure S1A). Cleavage of the xbp-1U mRNA by IRE-1 requires a conserved RNA motif at the two cleavage sites (Gonzalez et al., 1999). We introduced mutations into the endogenous xbp-1 locus that alter these motifs, but that do not affect amino acid coding in either the xbp-1U or the xbp-1S frame (Figure S1B).
Both cleavage and ligation are required for the production of the xbp-1S transcript that encodes the XBP-1 protein (Figure 1A). Consistent with this, xbp-1(uncleavable) animals could neither produce xbp-1S (Figure S1C) nor activate the XBP-UPR (Figure 1C), similar to RtcB ligase mutants (Kosmaczewski et al., 2014). Interestingly, however, we found that these two mutants have completely opposite phenotypes in axon regeneration. The RtcB ligase mutants have extremely high axon regeneration (Figure 1D, E), while the xbp-1(uncleavable) allele eliminates regeneration (Figure 1F). These data suggest that an xbp-1 mRNA processing intermediate between the cleavage step and the ligation step functions in axon regeneration.
The xbp-1 3’ fragment promotes axon regeneration independently of the UPR
During xbp-1U mRNA processing, IRE-1 cleaves the mRNA twice, generating three fragments: a 5’ fragment, a central fragment, and a 3’ fragment. The 5’ and 3’ fragments are then ligated by RtcB to generate xbp-1S mRNA. Blocking cleavage would be expected to result in depletion of all three fragments, while blocking ligation would be expected to cause relative accumulation of the 5’ and 3’ fragments. Thus, the opposite regeneration phenotypes in the cleavage and ligation mutants could be due to a novel function for these intermediate fragments. To determine which intermediate fragment is responsible, we generated rtcb-1; xbp-1(zc12) double mutant animals. We found that this allele of xbp-1, which contains an early stop codon, suppresses the high regeneration phenotype in RtcB mutant animals (Figure 2A, ‘control’), likely because nonsense-mediated decay reduces overall xbp-1 mRNA abundance, affecting all outputs of the xbp-1 locus. We expressed transgenic xbp-1 in this double mutant background, with the expectation that expressing the form of xbp-1 that mediates regeneration would result in the rtcb-1 phenotype of high regeneration. Expressing xbp-1 coding sequence along with its native 3’ UTR, either under its native promoter or under a GABA-specific promoter, restored regeneration back to the high rtcb-1 single mutant level. These data confirm that the function of the xbp-1 locus in axon regeneration is cell-autonomous, consistent with previous results from RtcB (Kosmaczewski et al., 2015), and indicate that the function of the xbp-1 locus is mediated by some aspect of the xbp-1 mRNA. However, when the endogenous xbp-1 3’ UTR was swapped with the commonly-used unc-54 3’ UTR, expression of xbp-1 was no longer able to restore regeneration (Figure 2A). These data indicate that the xbp-1 3’ UTR is required to promote axon regeneration in the rtcb-1; xbp-1(zc12) context. We concluded that the xbp-1 3’ fragment, which contains the 3’ UTR, is likely responsible for the high regeneration phenotype in the RtcB ligase mutant animals. In support of this, we detected with northern blot an accumulation of xbp-1 3’ fragments in vivo in RtcB mutant animals compared to control (Figure 2B).
Figure 2. The xbp-1 3’ fragment is necessary and sufficient to promote axon regeneration.
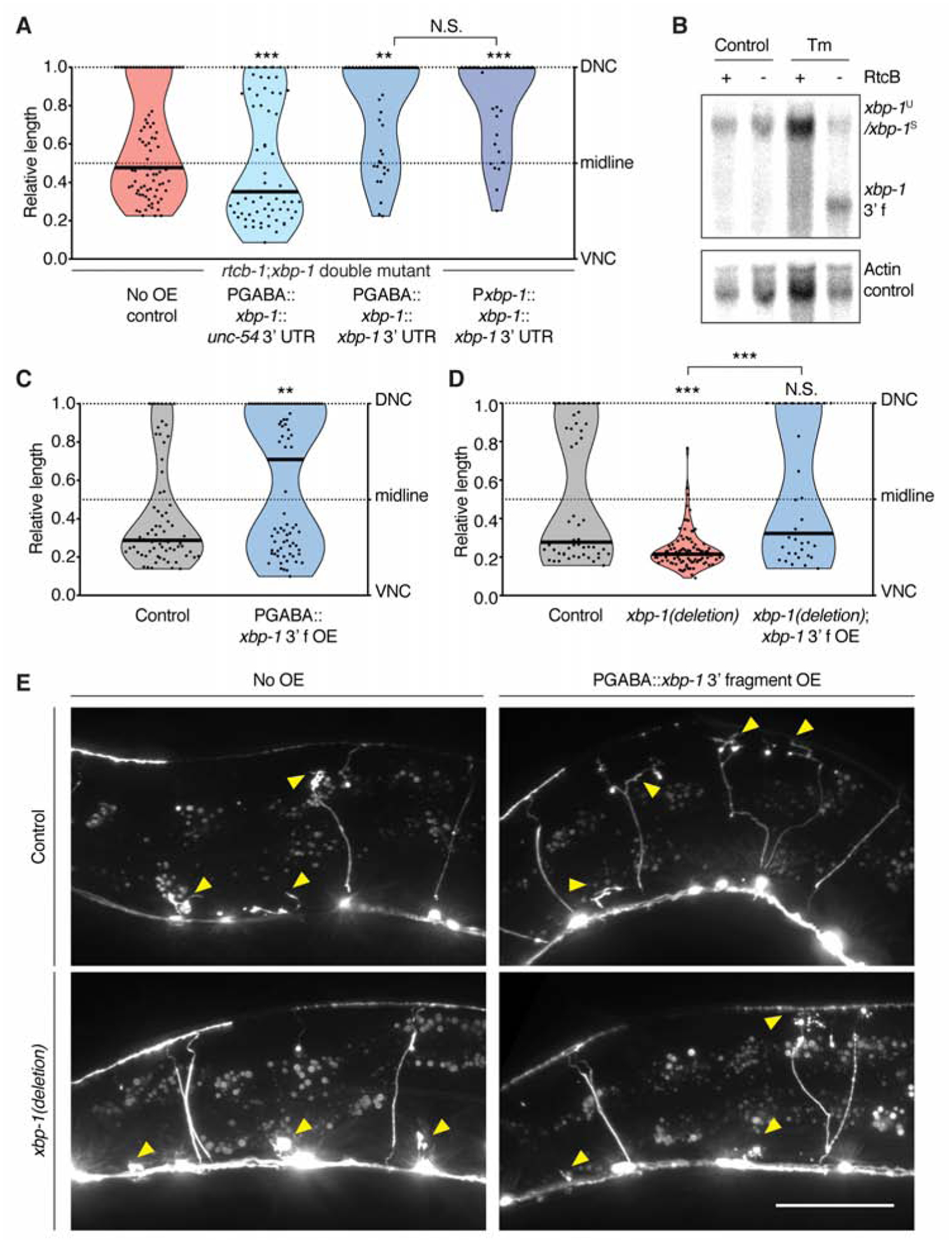
(A) The xbp-1 3’ UTR is required to increase regeneration in the rtcb-1(gk451); xbp-1(zc12) double mutant background. n = 62, 69, 40, and 31 from left to right. (B) Northern blot showing xbp-1 3’ fragment accumulation in RtcB mutant animals. (C) The xbp-1 3’ fragment is sufficient to increase regeneration cell-autonomously. n = 70 and 79 from left to right. (D) Deletion of the genomic xbp-1 locus eliminates regeneration, which is rescued by GABA-specific expression of the xbp-1 3’ fragment. n = 52, 92 and 34 from left to right. (E) Representative micrographs of animals of the indicated genotype 24 h post axotomy. Arrows indicate cut axons. Scale bar, 50 μm. In (A), (C) and (D), black bar represents the median. N.S., not significant, **P<0.01, ***P<0.001, 2-tailed K-S test.
To further test the function of the xbp-1 3’ fragment, and to determine whether it can promote axon regeneration outside the RtcB mutant context, we overexpressed the 3’ fragment specifically in GABA neurons in a non-mutant background. We found that 3’ fragment overexpression increased axon regeneration (Figure 2C, E). To further characterize this effect in relation to the XBP-UPR, we used CRISPR techniques to completely delete the endogenous xbp-1 gene. These xbp-1(deletion) animals had decreased regeneration (Figure 2D, E), similar to xbp-1(uncleavable) (Figure 1F). In this xbp-1(deletion) background, we expressed the xbp-1 3’ fragment specifically in GABA neurons. We found that 3’ fragment overexpression restored regeneration to normal levels (Figure 2D, E). Thus, overexpression of the xbp-1 3’ fragment is sufficient to increase regeneration in animals with a normal XBP-UPR and also in animals that completely lack conventional xbp-1 transcripts and have no XBP-UPR. Further, preventing the production of endogenous xbp-1 3’ fragments (in xbp-1(uncleavable) (Figure 1F), and xbp-1(deletion) (Figure 2D)) reduces regeneration below control levels, and this is rescued by the expression of the 3’ fragment (Figure 2D). These data indicate that the xbp-1 3’ fragment is a critical part of the endogenous regeneration response.
The xbp-1 3’ fragment functions as a ncRNA
The xbp-1 3’ fragment contains some of the xbp-1 coding sequence (Figure S2A), and the overexpression experiments (Figure 2C, D) included a start codon in the plasmid at the beginning of the xbp-1 3’ fragment sequence. We asked whether the xbp-1 3’ fragment acts as a non-coding RNA molecule, or whether it encodes a functional peptide. We generated an xbp-1 3’ fragment construct with the plasmid start codon frame-shifted (Figure S2A) and found that this construct retained its ability to promote regeneration (Figure S2B and 2C), suggesting the coding potential of the xbp-1 3’ fragment is dispensable for function in regeneration. Next, we attached only the predicted non-coding (UTR) portion of the xbp-1 3’ fragment to the GFP coding sequence, after the GFP stop codon, and expressed this construct (GFP-UTR) specifically in GABA neurons. Expression of this hybrid construct resulted in GFP expression and also increased regeneration. By contrast, expression of GFP with a control 3’ UTR did not affect regeneration, although similar levels of GFP fluorescence were observed (Figure 3A, S2C and S2D). Of note, the experiments in Figure 3A were performed in a completely wild-type background, as neurons were visualized using GFP produced from these arrays, rather than from the integrated GFP marker oxIs12. Thus, these data indicate that the regeneration-enhancing effect of the xbp-1 3’ fragment is not dependent on oxIs12. Most importantly, these experiments also indicate that the regeneration-promoting activity of the xbp-1 3’ fragment is contained within the 3’ UTR, suggesting that the xbp-1 3’ fragment acts as a ncRNA.
Figure 3. The xbp-1 3’ UTR promotes axon regeneration as a ncRNA.

(A) The xbp-1 3’ UTR increases regeneration even when fused to GFP coding sequence. See also Figure S2D for representative micrographs showing GFP expression. To visualize GFP fluorescence, these constructs were expressed in a wild-type N2 background, as opposed to the oxIs12 background (GABA-specific GFP marker) used in other axotomy experiments. Regeneration was scored 14 h (instead of 24 h) after axotomy. n = 42 and 50 from left to right. See also Figure S2C and S2D. (B) When exported from the nucleus into the cytoplasm with the help of CTE, the xbp-1 3’ UTR increases regeneration without any coding sequence. n = 35 and 39 from left to right. (C) Diagrams of xbp-1 3’ fragment deletion constructs. (D) Axotomy results of animals expressing constructs in (C). n = 148, 39, 36, 57, and 48 from left to right. (E) Predicted secondary structure of the xbp-1 3’ fragment based on SHAPE-MaP results. (F) The xbp-1 3’ fragment with the RNA stem S134–209 deleted no longer promotes axon regeneration. n = 47 and 34 from left to right. In (A), (B), (D) and (F), black bar represents the median. N.S., not significant, *P<0.05, **P<0.01, ***P<0.001, 2-tailed K-S test.
The endogenous xbp-1 3’ fragment is produced in the cytoplasm by cleavage of the xbp-1U transcript on the ER membrane. On the other hand, the overexpressed xbp-1 3’ fragment is transcribed in the nucleus. Proper pre-mRNA processing and mRNP assembly is required for mRNA stability and efficient export into the cytoplasm (Maniatis and Reed, 2002). Consistent with this idea, we found that the xbp-1 3’ fragment only increases regeneration if it contains at least one intron, which is likely to promote nuclear export (Maniatis and Reed, 2002)(Figure S2E and S2F). Deletion of the start codon (rather than frame-shifting it) also abolished the overexpressed construct’s ability to promote regeneration (Figure S2B), perhaps due to an effect on RNA stability or export. To construct a minimal functional sequence without any introns, start codons, or coding potential, we fused the xbp-1 3’ UTR with the retrovirus-derived constitutive transport element (CTE). The CTE RNA sequence hijacks the cellular mRNA nuclear export machinery and is exported independently of mRNA processing (Ernst et al., 1997; Grüter et al., 1998). We found that expression of this minimal construct (CTE-UTR) increased axon regeneration (Figure 3B), similar to expression of the entire xbp-1 3’ fragment (Figure 2C). Together, these data support the idea that the xbp-1 3’ fragment acts as a ncRNA to promote regeneration, and that cytoplasmic localization is essential for its function.
The function of the xbp-1 ncRNA depends on an RNA stem
To determine which region of the xbp-1 3’ fragment is required for its function in regeneration, we tested a series of truncations of the 3’ fragment (Figure 3C) expressed under a GABA-specific promoter. The results were consistent with our findings that the coding sequence is dispensable, as deletions in this region preserved function (Figure 3D, Δ1–189 and Δ190–378). By contrast, we found that a 189nt region in the predicted 3’ UTR is required for the 3’ fragment to increase regeneration (Figure 3D, Δ379–567). In silico folding by Vienna RNAfold of this region indicated that it contains a high-probability RNA stem, which was confirmed by SHAPE-MaP (Siegfried et al., 2014) performed on in vitro transcribed and folded RNAs (Figure 3E). We hypothesized that this stem (nucleotides 134–209 from the start of the xbp-1 3’ UTR, S134–209 hereafter) could be a functional element of the xbp-1 3’ fragment. In support of this, deleting S134–209 was sufficient to abolish the function of the xbp-1 3’ fragment (compare Figure 3F and 2C).
To further explore the function of S134–209, we began by observing that the xbp-1 3’ UTR (which contains S134–209) does not function in every condition. The xbp-1 3’ UTR promotes regeneration when expressed as part of the xbp-1 3’ fragment (Figure 2C), or when attached to the GFP coding sequence (GFP-UTR, Figure 3A), or when fused to CTE (CTE-UTR, Figure 3B). By contrast, we found that it does not have a significant effect on regeneration when it is part of the full-length xbp-1 transcript. The xbp-1(uncleavable) allele has poor regeneration even though the 3’ UTR is present (Figure 1F), and overexpression of either wild-type or uncleavable full-length xbp-1 with the 3’ UTR under a GABA-specific promoter does not increase regeneration (Figure 4A). These data suggest that the xbp-1 3’ UTR, and perhaps the key S134–209 region, is in an inactive structural conformation when part of the full-length xbp-1 transcript.
Figure 4. A single base pair within S134–209 is essential for xbp-1 ncRNA function.

(A) Overexpression of the full-length xbp-1 transcript does not increase regeneration. n = 50, 41, and 45 from left to right. (B) Base-by-base comparisons of SHAPE reactivity difference across the length of S134–209. Arrows point to C159. (C) Predicted secondary structures of partial S134–209 based on SHAPE-MaP results. Left, the active forms (xbp-1 3’ fragment, CTE-UTR, and GFP-UTR). Right, the inactive form (full-length xbp-1). Box insert, diagram of point mutations used in (D-F). (D) Base pairing at C159 is essential for the overexpressed CTE::xbp-1 3’ UTR to increase regeneration. n = 67, 48 and 55 from left to right. (E) C159G mutation at the endogenous xbp-1 locus decreases regeneration. n = 49 and 50 from left to right. (F) The endogenous xbp-1 ncRNA impacts animal life span. n = 91, 93 and 116 from top to bottom. ****P<0.0001, log rank Mantel-Cox test. In (A), (D) and (E), black bar represents the median. N.S., not significant, *P<0.05, **P<0.01, 2-tailed K-S test. See also Figure S3.
We used SHAPE-MaP to compare the RNA structures of the active RNAs (xbp-1 3’ fragment, CTE-UTR, GFP-UTR) with the inactive RNA (full-length xbp-1). We focused on the S134–209 sequence, since our analysis demonstrated that it is necessary for function (Figure 3F). SHAPE-MaP identified a single nucleotide within S134–209 whose reactivity differences correlated with the different function of the RNAs (Figure 4B and S3A). Specifically, C159 shows low SHAPE reactivity and is predicted to be paired in a stem structure in the three active RNAs (Figure 4C, left, and Figure S3B). On the other hand, in the inactive full-length xbp-1 RNA, C159 is highly accessible, indicating that the stem containing it does not form (Figure 4C, right, and Figure S3B). These in vitro SHAPE data suggest that pairing at C159 may be critical in vivo for the function of the xbp-1 ncRNA. To test the functional importance of C159 pairing, we mutated it to G (C159G, Figure 4C, box) in the CTE-UTR construct. This single mutation completely abolished the ability of the CTE-UTR construct to promote regeneration (compare Figure 4D and 3B). Next, we introduced a compensatory mutation (G177C, Figure 4C, box) that our structural analysis predicted would restore base pairing and stem formation. This second mutation restored the regeneration-promoting activity of the CTE-UTR construct (Figure 4D). Together, these results confirm that the CTE-UTR construct acts as a ncRNA, and demonstrate that pairing at C159 is required for function.
xbp-1 ncRNA is essential for in vivo axon regeneration
The endogenous xbp-1 transcript encodes the XBP-1 protein and has a well-established role in the XBP-UPR. Our experiments show that overexpressed fragments of this transcript can also act as ncRNAs. But does the endogenous xbp-1 transcript give rise to a ncRNA in addition to its coding function? To determine the potential ncRNA function of the xbp-1 3’ fragment in its natural context, we used CRISPR to engineer the C159G (Figure 4C, box) equivalent change into the endogenous xbp-1 locus (xbp-1(no-stem)). In contrast to the xbp-1(uncleavable) and xbp-1(deletion) alleles, xbp-1(no-stem) animals have a normal XBP-1 protein-mediated UPR (Figure S3C). Thus, the xbp-1(no-stem) allele does not affect the coding output of the endogenous xbp-1 transcript, as is expected from a single nucleotide change in the 3’ UTR. However, xbp-1(no-stem) animals have significantly impaired regeneration compared to control animals (Figure 4E). The loss of regeneration is nearly as strong as in the xbp-1(uncleavable) and xbp-1(deletion) alleles (Figure 1F, 2D), indicating that C159 is required for the function of the endogenous, non-coding xbp-1 3’ fragment in vivo. Next, we CRISPR engineered the G177C (Figure 4C, box) equivalent change into the xbp-1(no-stem) allele to restore the predicted base pairing of endogenous xbp-1 ncRNA (xbp-1(restored-stem)). In the RtcB mutant background, in which the ncRNA fragment accumulates and results in high regeneration, we observed a strikingly significant increase in regeneration in xbp-1(restored-stem) animals compared to xbp-1(no-stem) animals (Figure S3D). Restoring the stem did not have a significant effect in the RtcB wild-type background (Figure S3E), although some neurons were able to regenerate fully to the dorsal nerve cord, which was never observed in xbp-1(no-stem) animals (P = 0.058, 2-tailed Fisher’s exact test). Together, these data indicate that the endogenous xbp-1 3’ fragment acts as a ncRNA, and support the idea that the stem structure is a key functional element. In addition to the structure of the stem, sequence elements in the stem appear to play an auxiliary role in certain contexts.
The non-coding output of the endogenous xbp-1 transcript impacts more than just axon regeneration. We found that xbp-1(no-stem) animals have a significantly longer life span compared to control animals (Figure 4F). Similar to regeneration, this effect is also dependent on base pairing at the C159 site, because the long life span was fully restored back to normal in xbp-1(restored-stem) animals. Taken together, our data indicate that in its natural context, xbp-1 mediates a non-coding pathway that is separable from its coding-dependent function, and that the endogenous xbp-1 ncRNA affects both axon regeneration and animal life span.
DISCUSSION
RNA processing intermediates with unidentified functions
The xbp-1U transcript undergoes a well-conserved non-canonical splicing pathway, where an endonuclease and a ligase act sequentially to generate the protein-coding xbp-1S transcript. While previous studies of the output of the xbp-1 pathway focused on the function of the xbp-1S transcript, which encodes the critical UPR mediator XBP-1, our work reveals an unexpected role of a processing intermediate fragment of this pathway. We report that this intermediate RNA fragment functions as a ncRNA, entirely independently of the XBP-UPR.
More broadly, our data support the concept that RNA processing events like RNA cleavage can produce functional intermediate fragments in addition to the final product. This phenomenon has been observed previously, the most well-documented example being tRNA fragments (Hanada et al., 2013; Sobala and Hutvagner, 2011; Thompson and Parker, 2009). However, prior to this study, it was not known whether mRNA cleavage would also result in RNA fragments with non-coding functions. mRNA cleavage is a widespread cellular event. For example, during the regulated IRE1-dependent decay of mRNA (RIDD), numerous mRNAs are cleaved by the endonuclease IRE-1 (Hollien et al., 2009). Our study widens the potential landscape of ncRNAs produced by cleavage to include those produced from mature mRNAs.
Roles of xbp-1 in regulating the neuronal injury response
The xbp-1 locus has been shown to function in neuronal injury across species (Hu et al., 2012; Kosmaczewski et al., 2015; Ohtake et al., 2018; Oñate et al., 2016; Song et al., 2015; Ying et al., 2015). It has been linked to axon regeneration itself, as well as to related cellular activities such as cell survival, myelin removal, and microphage infiltration (Hu et al., 2012; Oñate et al., 2016). Our work identifies a novel mechanism by which the xbp-1 locus functions in neuronal injury: via a ncRNA generated by xbp-1 mRNA cleavage. The cellular mechanisms that mediate the non-coding function of the xbp-1 3’ fragment remain to be determined. In general, ncRNAs can function via diverse mechanisms, including acting as enzymes, ligands, structural scaffolds, and molecular sponges (Fu, 2014; Ponting et al., 2009). For the xbp-1 3’ fragment, one or more of these mechanisms might impinge on the cellular pathways that mediate axon regeneration, for example the well-characterized DLK-1 pathway (Hammarlund et al., 2009; Yan et al., 2009), which has a similar effect size on regeneration as the xbp-1 ncRNA. In addition, the xbp-1 3’ fragment influences lifespan, perhaps via independent cellular pathways. Further experiments will be necessary to describe how the xbp-1 ncRNA functions and to identify the cellular mechanisms it affects.
The endogenous xbp-1 transcript has coding/non-coding dual outputs
RNA molecules are generally divided into two worlds with little overlap, coding and non-coding. Our data identify xbp-1 as the first ncRNA that is directly derived from the cleavage of a mature mRNA. Thus, the endogenous xbp-1 transcript has coding/non-coding dual outputs. The coding output mediates the XBP-UPR. The non-coding output is mediated by the xbp-1 3’ fragment. It impacts both axon regeneration and animal life span. Our work thus places the xbp-1 RNA into the small group of RNAs that act both as coding and non-coding RNAs, sometimes called “coding and non-coding RNAs” (cncRNAs)(Crerar et al., 2019; DeJesus-Hernandez et al., 2011; Ji et al., 2015; Kumari, 2015; Mori et al., 2013; Sampath and Ephrussi, 2016). Such RNAs blur the line between the coding and non-coding RNA worlds and raise the question of how their dual roles are regulated.
In the case of xbp-1, the coding/non-coding dual outputs may be regulated by modulating the balance between cleavage and ligation. While regulation of cleavage of xbp-1 mRNA by IRE-1 has been extensively characterized, less is known about regulation of ligation by RtcB. However, multiple potential avenues for ligation regulation exist. First, although in vitro RtcB exhibits RNA ligase activity on its own, in vivo it is thought to act as the catalytic subunit of a complex that includes archease and DDX1 (Desai et al., 2014, 2015; Popow et al., 2014). RtcB has also been reported to complex in vivo with multiple other accessory proteins like FAM98B and hCLE/C14orf166/RTRAF (Kanai et al., 2004; Pazo et al., 2019; Pérez-González et al., 2014). These interactions may affect ligase function and provide a mechanism for regulation. Second, RNA ligation by RtcB requires specific chemistry at the RNA ends to be joined; a number of factors, including the cyclic phosphodiesterase CNP, the RNA cyclase RtcA (Unlu et al., 2018), as well as cytoplasmic capping machinery, could potentially alter this chemistry. Such modifications could also affect the stability of the cleaved fragments, as the 5’ fragment has been shown in yeast to be degraded by exonuclease if not ligated (Peschek and Walter, 2019). Third, RtcB can be transported by kinesin along microtubules in neurons (Kanai et al., 2004). We previously observed that RtcB translocates to severed axon tips after injury (Kosmaczewski et al., 2015). Thus, ligation could be modulated by subcellular localization. Injury and other cellular stresses might act via one or more of these regulatory mechanisms to shift the cleavage-ligation balance and regulate xbp-1 ncRNA function.
Our study indicates that cleavage and ligation modulate the function of the xbp-1 ncRNA because they have a secondary effect on RNA structure. We hypothesize that suppression of ncRNA function in the full length xbp-1 transcript is due to interaction with specific sequences within the 5’ fragment sequence. This interaction would inhibit ncRNA function in both the uncleaved mRNA (xbp-1U) and the ligated mRNA (xbp-1S), but allow ncRNA function in the unligated 3’ fragment or when the xbp-1 ncRNA is placed in the context of a different mRNA (encoding GFP). We find that cleavage-and presumably removal of inhibition-causes a structural switch in the xbp-1 3’ fragment, activating its non-coding function. Specifically, cleavage favors formation of a stem-loop structure within the xbp-1 3’ UTR, by base pairing at the C159G site. This may in turn allow the ncRNA to interact with downstream effectors. Stem-loop structures are also predicted in the 3’ UTR of the xbp-1 homologues in other organisms, but such structures are common in UTR regions and their functional significance remains to be determined. Our data in C. elegans indicate that cleavage-induced structural switching is a novel mechanism to switch between the coding and non-coding outputs of a cncRNA depending on the status of the cell. In the case of xbp-1, cells can use this switch to fine-tune their response to injury and other stresses via different cellular responses.
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marc Hammarlund (marc.hammarlund@yale.edu).
Materials Availability
Plasmids and worm lines generated in this study are available upon request.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans
C. elegans were maintained on nematode growth media (NGM) plates seeded with Escherichia coli OP50 at 20 °C. Hermaphrodites were used for all experiments, and males were only used for crossing. The developmental stage of the animals used in each assay is reported in the METHOD DETAILS section below.
METHOD DETAILS
Transgenic animal generation
For transgenic animals with extrachromosomal arrays, 20 ng/μL of relevant constructs were mixed with 2.5 ng/μL of Pmyo-2::mCherry (injection marker) and DNA ladder (Promega, G5711, to adjust the total concentration of the injection mix to ~150 ng/μL), and microinjected into the gonads of young adults. Transgenic animals were first selected based on the expression of Pmyo-2::mCherry, and then confirmed by PCR genotyping of the relevant constructs.
CRISPR Mutations
The four xbp-1 alleles (wp45, wp46, wp79, wp80) were generated by CRISPR as previously described (Arribere et al., 2014; Farboud and Meyer, 2015; Paix et al., 2014). Briefly, sgRNAs were ordered from IDT and cloned into our sgRNA plasmid backbone by simple PCR. Repair templates were either directly ordered from IDT (ssODN), or PCR amplified from genomic sequence (dsDNA). An injection mix of 50 ng/μL of pDD162 (Peft-3::Cas9), 40 ng/μL of relevant sgRNA(s), 30 ng/μL per 125 nt ssODN or per 50 nt dsDNA of repair template if using, 30 ng/μL of dpy-10 sgRNA, and 26 ng/μL of ssODN AF-ZF-827 (dpy-10 repair template), was injected into the gonads of young adults. F1 progenies of the injected parents were examined for dpy-10 phenotype, and dumpy/roller worms were subject to sequencing. F2 progenies homozygous for the mutation were again confirmed by sequencing and were outcrossed >3X prior to any experiments.
Molecular Biology
Gateway recombination (Invitrogen) was used to generate Punc-25::xbp-1::unc-54 3’ UTR, Punc-25::xbp-1::xbp-1 3’ UTR, Pxbp-1::xbp-1::xbp-1 3’ UTR, Punc-25::gfp::xbp-1 3’ UTR, and Punc-25::gfp::unc-54 3’ UTR. Gibson cloning was used to generate Punc-25::CTE::xbp-1 3’UTR and Punc-25::xbp-1 3’ fragment. The CTE sequence was synthesized by IDT. Other entry pieces were amplified using Phusion polymerase (NEB, M0530) from worm lysate or previous plasmids (Kosmaczewski et al., 2015). Gibson assembly was also used to generate the various deletion constructs on the basis of the Punc-25::xbp-1 3’ fragment construct. Site-directed mutagenesis was used to introduce the C159G and G177C point mutations to the Punc-25::CTE::xbp-1 3’UTR construct. All construct and primer sequences are available upon request.
xbp-1 RT-PCR assay
Worms of N2 wild-type, xbp-1(wp45) or xbp-1(wp46) were harvested from fully-populated NGM plates just before starving. Next, they were washed multiple times in M9 buffer, nutated in M9 with or without 5 μg/mL tunicamycin for 3 h, spun down, resuspended in TRIzol reagent (Invitrogen), frozen at −80 °C overnight, and then subjected to freeze/thaw in liquid nitrogen/37°C incubator 3 times. RNA was isolated from the aqueous phase following manufacturer’s instructions. First-strand cDNA was synthesized with AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent, #200436). PCR was carried out using Phusion polymerase (NEB, M0530) with intron-spanning primers listed in KEY RESOURSES TABLE. Products were digested with MseI (NEB, R0525) and then resolved on a 2% agarose gel.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tunicamycin | Sigma-Aldrich | Cat # 654380 |
| Levimisole | Santa Cruz | Cat # sc-205730 |
| TRIzol Reagent | Invitrogen | Cat # 15596026 |
| 0.05 μm microbeads for axotomy | Polysciences | Cat # 08691 |
| Critical Commercial Assays | ||
| AffinityScript Multiple Temperature cDNA Synthesis Kit | Agilent | Cat # #200436 |
| SuperScript™ III Reverse Transcriptase | Invitrogen | Cat # 18080093 |
| AmpliScribe T7-Flash transcription kit | Epicentre | Cat # ASF3257 |
| SuperScript™ II Reverse Transcriptase | Invitrogen | Cat # 18064014 |
| Deposited Data | ||
| RNA-seq comparing between RtcB mutant and control animals with and without neuronal injury | This manuscript | Table S1 |
| Experimental Models: Organisms/Strains | ||
| C.elegans: N2 wild-type | Caenorhabditis Genetics Center (CGC) | N2 (ancestral) |
| C.elegans: oxIs12[Punc-47::GFP, lin-15+] X (Note: GABA-specific GFP. The grey “Control” used in all axotomy plots) | Caenorhabditis Genetics Center (CGC) | WormBase ID: WBTransgene00001635 |
| C.elegans: rtcb-1(gk451) I / hT2[bli-4(e937), let-?(q782), qIs48] I;III; wpIs63; oxIs12[Punc-47::GFP, lin-15+] X (Note: rtcb-1 null) | Kosmaczewski et al., 2015 | XE1728 |
| C.elegans: zcIs4[Phsp-4::GFP] V (Note: UPR reporter) | Caenorhabditis Genetics Center (CGC) | SJ4005 |
| C.elegans: xbp-1(wp45) III (Note: xbp-1(uncleavable)) | This manuscript | XE2037 |
| C.elegans: xbp-1(wp45) III; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2038 |
| C.elegans: xbp-1(wp45) III; zcIs4[Phsp-4::GFP] V | This manuscript | XE2234 |
| C.elegans: xbp-1(wp46) III (Note: xbp-1(deletion)) | This manuscript | XE2048 |
| C.elegans: xbp-1(wp46) III; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2049 |
| C.elegans: xbp-1(wp46) III; zcIs4[Phsp-4::GFP] V | This manuscript | XE2235 |
| C.elegans: rtcb-1(gk451) I; xbp-1(zc12) III / hT2[bli-4(e937), let-?(q782), qIs48] I;III; wpIs63; oxIs12[Punc-47::GFP, lin-15+] X (Note: rtcb-1; xbp-1 double mutant) | This manuscript | XE1928 |
| C.elegans: rtcb-1(gk451) I; xbp-1(zc12) III / hT2[bli-4(e937), let-?(q782), qIs48] I;III; wpIs63; oxIs12[Punc-47::GFP, lin-15+] X; wpEx355[Punc-25::xbp-1::unc-54 3’ UTR + Pmyo-2::mCherry] | This manuscript | XE2236 |
| C.elegans: rtcb-1(gk451) I; xbp-1(zc12) III / hT2[bli-4(e937), let-?(q782), qIs48] I;III; wpIs63; oxIs12[Punc-47::GFP, lin-15+] X; wpEx356[Punc-25::xbp-1::xbp-1 3’ UTR + Pmyo-2::mCherry] | This manuscript | XE2237 |
| C.elegans: rtcb-1(gk451) I; xbp-1(zc12) III / hT2[bli-4(e937), let-?(q782), qIs48] I;III; wpIs63; oxIs12[Punc-47::GFP, lin-15+] X; wpEx290[Pxbp-1::xbp-1::xbp-1 3’ UTR + Pmyo-2::mCherry] | This manuscript | XE1948 |
| C.elegans: wpEx342[Punc-25::xbp-1 3’ fragment + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2181 |
| C.elegans: xbp-1(wp46) III; wpEx342[Punc-25::xbp-1 3’ fragment + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2238 |
| C.elegans: wpEx323[Punc-25::xbp-1 3’ fragmentΔAUG + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2063 |
| C.elegans: wpEx322[Punc-25::xbp-1 3’ fragmentfs-AUG + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2062 |
| C.elegans: wpEx329[Punc-25::GFP::xbp-1 3’UTR] | This manuscript | XE2094 |
| C.elegans: wpEx357[Punc-25::GFP::xbp-1 3’UTR + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2239 |
| C.elegans: wpEx358[Punc-25::GFP::unc-54 3’UTR] | This manuscript | XE2240 |
| C.elegans: wpEx359[Punc-25::GFP::unc-54 3’UTR + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2241 |
| C.elegans: wpEx341[Punc-25::CTE::xbp-1 3’ UTR + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2180 |
| C.elegans: wpEx327[Punc-25::xbp-1 3’ fragmentΔ1–189 + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2092 |
| C.elegans: wpEx325[Punc-25::xbp-1 3’ fragmentΔ190–378 + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2067 |
| C.elegans: wpEx353[Punc-25::xbp-1 3’ fragmentΔ379–567 + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2231 |
| C.elegans: wpEx354[Punc-25::xbp-1 3’ fragmentΔ568–702 + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2232 |
| C.elegans: wpEx326[Punc-25::xbp-1 3’ fragmentΔS134–209 + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2072 |
| C.elegans: wpEx309[Punc-25::xbp-1::xbp-1 3’ UTR + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2034 |
| C.elegans: wpEx309[Punc-25::xbp-1uncleavable::xbp-1 3’ UTR + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2036 |
| C.elegans: wpEx337[Punc-25::CTE::xbp-1 3’ UTRC159G + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2174 |
| C.elegans: wpEx338[Punc-25::CTE::xbp-1 3’ UTRC159G; G177C + Pmyo-2::mCherry]; oxIs12[Punc-47::GFP, lin-15+] X | This manuscript | XE2175 |
| C.elegans: xbp-1(wp79) III; oxIs12[Punc-47::GFP, lin-15+] X (Note: xbp-1(no-stem)) | This manuscript | XE2223 |
| C.elegans: xbp-1(wp79) III; zcIs4[Phsp-4::GFP] V | This manuscript | XE2229 |
| Oligonucleotides | ||
| Forward primer for xbp-1U and xbp-1S in RT-PCR assay: GAGACAAAAAGAAG-GAAAGATCAGC | IDT | N/A |
| Reverse primer for xbp-1U and xbp-1S in RT-PCR assay: CTCCGCTTGGGCT-CTTGAGATG | IDT | N/A |
| Forward primer for ama-1S control in RT-PCR assay: TGTCAGGATCGAAGGGATCGAAG | IDT | N/A |
| Reverse primer for ama-1S control in RT-PCR assay: CGGTGAGGTCCAT-TCTGAAATC | IDT | N/A |
| Northern probes: see Table S3 | This manuscript | N/A |
| Reverse transcription random primer used in RNA-seq library preparation: AGACGTGTGCTCTTCCGATCTNNNNNN | IDT | N/A |
| ssDNA adaptor used in RNA-seq library preparation: /5Phos/NNNNNGATCGTCGGACTGTAGAACTCTGAAC/3InvdT/ | IDT | N/A |
| In vitro transcribed RNA for SHAPE-MaP: See Table S4 | This manuscript | N/A |
| Recombinant DNA | ||
| pDD162 | Dickinson et al., 2013 | Addgene #47549 |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Micro-Manager | Edelstein et al., 2014 | https://micro-manager.org/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| CASAVA-1.8.2. | Illumina | https://bioweb.pasteur.fr/packages/pack@casava@1.8.2 |
| Bowtie2 v2.2.4 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR version 2.4.2a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Meta-chart | N/A | https://www.meta-chart.com/ |
| ShapeMapper/SuperFold | Siegfried et al., 2014; Smola et al., 2015 | www.chem.unc.edu/rna/software.html |
UPR Fluorescent Reporter Assay
UPR assay was performed as described previously (Kosmaczewski et al., 2015). Briefly, animals at the L4 stage were placed on NGM plates with or without 5 μg/mL tunicamycin for 10–24 h as indicated. Phsp-4::GFP expression was then visualized by imaging.
Imaging of live animals
Animals were mounted on 3% (wt/vol) agarose pads and immobilized using 0.1% Levimisole (Santa Cruz, sc-205730). Images were taken with Olympus BX61 and processed with Micro-Manager.
Laser Axotomy
Laser axotomy was performed as described previously (Kosmaczewski et al., 2015). Briefly, worms at L4 stage were mounted on a 3% (wt/vol in M9 buffer) agarose pad, immobilized by 0.05 μm microbeads (Polysciences, #08691) with 0.02% SDS, and visualized with a Nikon Eclipse 80i Microscope. 3–4 of the 7 most posterior ventral and dorsal D-type (VD/DD) GABA motor neurons were severed using a Photonic Instruments Micropoint Laser at 10 pulses and 20 Hz. Worms were recovered to seeded NGM plates and scored for axon regeneration 24 h (unless otherwise stated) after axotomy.
Northern blots
~2,000 L4 N2 wild-type or rtcb-1(gk451) worms were picked to NGM plates with or without 50 μg/mL tunicamycin (rtcb-1 homozygotes were isolated from balanced heterozygotes), incubated at 20 °C for 3 h, and harvested from the plates with M9 buffer. RNA was isolated as in the xbp-1 RT-PCR assay. 10 μg of total RNA was separated in 1.2% agarose-37% formaldehyde gels and transferred to Zeta-Probe membranes (Bio-Rad, 1620165) by capillary transfer in 20X SSC buffer for 22h. The blots were first hybridized and visualized with the following [32P]-labeled oligonucleotide probes against the xbp-1 sequence 3’ to the IRE-1 cleavage sites (xbp-1 probes), and then stripped, and hybridized and visualized with act-1 probes as control. The hybridized signals were visualized with the Storm 860 Molecular Imager (GMI). The sequences of the probes are listed in Table S3.
RNA sequencing
RNA-seq experiments were performed to identify RtcB-dependent changes in mRNA splicing. Samples were prepared in triplicate from the following four groups: control, rtcb-1(gk451), unc-70(s1502), and rtcb-1(gk451); unc-70(s1502). For each replicate, ~2,000 L4 worms were gathered. Total RNA was extracted using TRIzol as described above. Poly-A+ RNAs were isolated using oligo d(T)25 magnetic beads (New England BioLabs) following the manufacturer’s protocol and eluted in 11 μL of water. Reverse transcription was performed in 20 μL at 25 °C for 10 min, 42 °C for 30 min, 50 °C for 10 min, 55 °C for 20 min, and 60 °C for 20 min, using Superscript III (Invitrogen) and reverse transcription random primer. The reaction was then heat inactivated at 75 °C for 15 min and RNAse H treated at 37 °C for 15 min. cDNA samples were purified using 36 μL of AMPure XP beads (Beckman Coulter Life Sciences) following the manufacturer’s protocol and eluted in 10 μL of water. Purified cDNA samples were ligated to a ssDNA adaptor at their 3’-end using CircLigase ssDNA Ligase (Epicentre) following the manufacturer’s protocol with the addition of 1M betaine and 10% PEG 6000. Ligation reactions were incubated at 60 °C for 2h, at 68 °C for 1h and heat inactivated at 80 °C for 10 min. 10 μL of water was added to each reaction and ligated products were purified using 36 μL of AMPure XP beads (Beckman Coulter Life Sciences) and dissolved in 16 μL of water. Purified ligated cDNA samples were then PCR amplified using Illumina sequencing adapters, keeping the number of cycles to the minimum needed for the detection of amplified products (8–12 cycles), and gel purified on 2% agarose gel to remove adaptor-adaptor dimers. Purified libraries were sequenced on Illumina HiSeq 2000/2500 machines producing single-end 76 nucleotide reads.
SHAPE-MaP
Four RNA species (xbp-1 3’ fragment, gfp::xbp-1 3’ UTR (GFP-UTR), CTE::xbp-1 3’ UTR (CTE-UTR), and xbp-1::xbp-1 3’ UTR (full-length xbp-1)) were transcribed in vitro from PCR products using the AmpliScribe T7-Flash transcription kit (Epicentre), and purified using RNeasy columns (QIAGEN). The primers used and the sequences of the RNAs generated are listed in Table S4. For each transcript, 1 μg of RNA in 14 μl final volume was heated to 95 °C for 2 min, placed on ice for 2 min, folded at 22 °C for 20 min with the addition of 4 μl of 5X folding buffer (500 mM tris HCl pH 7.5, 500 mM KCl and 50 mM MgCl2), probed at 22 °C for 5 min with the addition of 2 μL NAI (2 M, Sigma-Aldrich) or DMSO, and quenched by the addition of 90 μL of stop solution (3 M β-mercaptoethanol, 508 mM sodium acetate and 15 μg glycoblue). Following a 5 min incubation at room temperature, samples were ethanol precipitated, washed with 70% ethanol, resuspended in water, and subjected to MaP reverse transcription (requiring Superscript II and the addition of Mn2+ to the RT buffer(Smola et al., 2015)) using random nonamer primers. Double-stranded cDNA was synthesized in Second Strand Synthesis Reaction Buffer (NEB) with Second Strand Synthesis Enzyme mix (NEB), purified using PureLink PCR micro spin column (Thermo Fisher Scientific), eluted in water, and sent to the Yale Center for Genome Analysis to be fragmented using the Nextera DNA Flex Library Prep Kit (Illumina) and sequenced on an Illumina NovaSeq sequencer producing paired-end 150 nucleotide reads.
Life span assay
For each genotype, ~100 L4 animals were picked to NGM plates seeded with OP50 bacteria. The animals were kept at 20°C and fed with OP50 during the assay. During the first 5 days, animals were transferred to a fresh seeded plate every other day to separate them from their off springs. Viability was scored every other day. Death was scored by failure to respond to touching.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of axotomy data
For all axotomy experiments, the relative lengths (Figure 1D) of all successfully cut axons (indicated by the presence of a severed distal stump) were measured with ImageJ and plotted with GraphPad Prism. All scoring processes were carried out blindly. No data were excluded. Measurements were taken from distinct samples for each plot shown. n represents the number of axons cut in each group (i.e. number of dots plotted in each violin), and the value of n can be found in figure legends. The black bar in each figure shows the median; median was used instead of mean because the distribution of relative length of cut axons is often not normal. Two-tailed P values were calculated using the Kolmogorov-Smirnov test (2-tailed K-S test). The K-S test calculates empirical P value comparing the continuous probability distribution of a sample with a reference probability distribution, and does so by quantifying the distance between the two distribution functions. The K-S test does not assume or require normal distribution of the data. This is appropriate in our case because in many cases our data are not normally distributed. P values are reported as asterisks in figures: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Analysis of life span data
When conducting the life span assay, worms were counted dead only when the corpse could be found. Disappeared worms (which might have crawled out of plate or into cracks) were excluded. n represents the number of worms tested in each group, and the value of n can be found in figure legends. Survival curves were plotted with GraphPad Prism. Significance was calculated using the log rank Mantel-Cox test and is reported as asterisks in figures.
Splice junction analysis of RNA-seq data
Following the library preparation protocol, raw reads contained the following features: NNNNN-insert-adapter, where the 5N sequence composes the Unique Molecular Identifier (UMI), and adapter is the 3’-Illumina adapter that can occasionally be present within the read (mainly in adapter-adapter dimers). The UMI was used to discard PCR duplicates and count single ligation events. Base calling was performed using CASAVA-1.8.2. The Illumina TruSeq index adapter sequence was then trimmed when present by aligning its sequence, requiring a 100% match of the first five base pairs and a minimum global alignment score of 60 (Matches: 5, Mismatches: -4, Gap opening: -7, Gap extension: -7, Cost-free ends gaps). The UMI was clipped from the 5’-end and kept within the read name, for marking PCR duplicates. Reads were then depleted of rRNA, tRNA, snRNA, snoRNA and miscRNA, using Ensembl 80 annotations, as well as from RepeatMasker annotations, using strand-specific alignment with Bowtie2 v2.2.4 (Langmead and Salzberg, 2012). Next, reads were aligned to the C. elegans WBcel235 genome assembly using STAR version 2.4.2a (Dobin et al., 2013) with the following non-default parameters: --alignEndsType EndToEnd --outFilterMultimapNmax 100 --seedSearchStartLmax 15 --sjdbScore 10 --outSAMattributes All -limitBAMsortRAM 3221225472. Genomic sequence indices for STAR were built including exon-junction coordinates from Ensembl 80. Finally, the STAR SJ.out.tab output files were parsed and the coverage of individual splice sites was compared between control and rtcb-1(gk451) (Table S1, “no neuronal injury” tab), and between unc-70(s1502) and rtcb-1(gk451); unc-70(s1502) (Table S1, “neuronal injury” tab). For each comparison, we considered only splice sites i) were detected in at least two out three replicates of genotype 1, ii) were supported by at least three uniquely mapped reads in genotype 1 and iii) had a splice site coverage difference of at least 2-fold between the two genotypes after normalizing for changes in RNA abundance (equation 1). A value of 0.01 was added to splice site coverages equal to 0 in genotype 2. Only uniquely mapped reads were considered in the calculation of splice site coverages and RPKM (Read Per Kilobase Million) values. Non-canonical splice site with >10-fold difference in either of the two comparisons were annotated based on WormBase annotation (Table S2). Venn diagrams were drawn with Meta-chart.
| Equation 1 |
where ss coverage 1 is the splice site coverage in genotype 1, RPKM 1 is the mRNA’s RPKM value in genotype 1, ss coverage 2 is the splice site coverage in genotype 2, and RPKM 2 is the mRNA’s RPKM value in genotype 2.
Analysis of SHAPE-MaP data
Raw sequencing data were processed using the ShapeMapper pipeline (version 2.1.3)(Siegfried et al., 2014; Smola et al., 2015) with default parameters. SHAPE reactivities were only computed for nucleotides possessing sequencing depths above 1,000 in both modified and untreated samples. Nucleotides not passing this filter were treated as “no data” and excluded from downstream analysis. Secondary structure predictions and base pairing probabilities were generated using the SuperFold algorithm and SHAPE reactivities as restraints(Smola et al., 2015) with the following parameters: SHAPEintercept = −0.6, SHAPEslope = 1.8, trimInterior = 50, partitionWindowSize = 1200, PartitionStepSize = 100, foldWindowSize = 3000, foldStepSize = 300, maxPairingDist = 600.
Supplementary Material
Table S1. Junction analysis of the RNA-seq data comparing RtcB mutant and control animals.
(Tab 1) Comparison between RtcB mutant and control animals without neuronal injury. (Tab 2) Comparison between RtcB mutant and control animals with neuronal injury, using a mutation in β-spectrin (unc-70) to trigger spontaneous axon breaks. (Tab 3) Definitions of the column names used in Tabs 1 and 2.
Related to: Figure 1B; Results section “Blocking xbp-1 cleavage versus ligation results in opposite phenotype in axon regeneration”.
Table S2. Annotations of the genes identified in the junction analysis.
Related to: Figure 1B; Results section “Blocking xbp-1 cleavage versus ligation results in opposite phenotype in axon regeneration”.
HIGHLIGHTS.
Cytoplasmic cleavage of the xbp-1 mRNA produces a biologically active ncRNA.
The xbp-1 ncRNA impacts neuronal regeneration and animal lifespan in vivo.
The function of the xbp-1 ncRNA requires the formation of an RNA stem.
The xbp-1 ncRNA represents an XBP-UPR-independent output of the xbp-1 locus.
ACKNOWLEDGMENTS
This research was supported by the Gruber Science Fellowship to X.L. and C.A.D.; K99 award (HD093873) from NIH, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) to J.-D.B.; NIH (R35 GM122580) to A.J.G.; and NIH (grants R01 NS098817, and R01 NS094219) to M.H.. We thank WormBase and the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We thank the Steitz lab at Yale University, especially Dr. Joan A. Steitz and Dr. Johanna Withers for sharing reagents, protocols, and space for our northern blots.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, and Ron D (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Crerar H, Scott-Solomon E, Bodkin-Clarke C, Andreassi C, Hazbon M, Logie E, Cano-Jaimez M, Gaspari M, Kuruvilla R, and Riccio A (2019). Regulation of NGF Signaling by an Axonal Untranslated mRNA. Neuron 102, 553–563.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. (2011). Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Cheng CL, Bingman CA, Phillips GN, and Raines RT (2014). A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 42, 3931–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KK, Beltrame AL, and Raines RT (2015). Coevolution of RtcB and Archease created a multiple-turnover RNA ligase. RNA 21, 1866–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, and Soll D (2011). Archaeal 3’-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci 108, 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Bray M, Rekosh D, and Hammarskjöld ML (1997). A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol 17, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, and Meyer BJ (2015). Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D (2014). Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev 1, 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dörfler S, Walter P, and Silverman RH (1999). Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J 18, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, and Izaurralde E (1998). TAP, the Human Homolog of Mex67p, Mediates CTE-Dependent RNA Export from the Nucleus. Mol. Cell 1, 649–659. [DOI] [PubMed] [Google Scholar]
- Hammarlund M, Jorgensen EM, and Bastiani MJ (2007). Axons break in animals lacking β-spectrin. J. Cell Biol 176, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M, Nix P, Hauth L, Jorgensen EM, and Bastiani M (2009). Axon regeneration requires a conserved MAP kinase pathway. Science 323, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. (2013). CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, and Mollereau B (2014). Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci 15, 233–249. [DOI] [PubMed] [Google Scholar]
- Hollien J, and Weissman JS (2006). Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science (80-.). 313, 104–107. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, and Weissman JS (2009). Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Park KK, Yang L, Wei X, Yang Q, Cho K-S, Thielen P, Lee A-H, Cartoni R, Glimcher LH, et al. (2012). Differential Effects of Unfolded Protein Response Pathways on Axon Injury-Induced Death of Retinal Ganglion Cells. Neuron 73, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, and Struhl K (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Niwa M, and Koong AC (2015). Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin. Cancer Biol 33, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, and Martinez J (2014). The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 33, 2922–2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, and Hirokawa N (2004). Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, and Mori K (1998). Unconventional Splicing of HAC1/ERN4 mRNA Required for the Unfolded Protein Response. J. Biol. Chem 273, 1802–1807. [DOI] [PubMed] [Google Scholar]
- Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, and Hammarlund M (2014). The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep 15, 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaczewski SG, Han SM, Han B, Irving Meyer B, Baig HS, Athar W, Lin-Moore AT, Koelle MR, and Hammarlund M (2015). RNA ligation in neurons by RtcB inhibits axon regeneration. Proc. Natl. Acad. Sci. U. S. A 112, 8451–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P (2015). cncRNAs: Bi-functional RNAs with protein coding and non-coding functions. Semin. Cell Dev. Biol 47–48, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang F-X, and Wang X (2014). A Synthetic Biology Approach Identifies the Mammalian UPR RNA Ligase RtcB. Mol. Cell 55, 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, and Reed R (2002). An extensive network of coupling among gene expression machines. Nature 416, 499–506. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Nix P, Hammarlund M, Hauth L, Lachnit M, Jorgensen EM, and Bastiani M (2014). Axon regeneration genes identified by RNAi screening in C. elegans. J. Neurosci 34, 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Matsuhisa K, Kaneko M, Kanemoto S, Asada R, Imaizumi K, and Saito A (2018). Axonal Activation of the Unfolded Protein Response Promotes Axonal Regeneration Following Peripheral Nerve Injury. Neuroscience 375, 34–48. [DOI] [PubMed] [Google Scholar]
- Oñate M, Catenaccio A, Martínez G, Armentano D, Parsons G, Kerr B, Hetz C, and Court FA (2016). Activation of the unfolded protein response promotes axonal regeneration after peripheral nerve injury. Sci. Rep 6, 21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Wang Y, Smith HE, Lee C-YS, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, and Seydoux G (2014). Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazo A, Pérez-González A, Oliveros JC, Huarte M, Chavez JP, and Nieto A (2019). hCLE/RTRAF-HSPC117-DDX1-FAM98B: A New Cap-Binding Complex That Activates mRNA Translation. Front. Physiol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González A, Pazo A, Navajas R, Ciordia S, Rodriguez-Frandsen A, and Nieto A (2014). hCLE/C14orf166 Associates with DDX1-HSPC117-FAM98B in a Novel Transcription-Dependent Shuttling RNA-Transporting Complex. PLoS One 9, e90957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, and Walter P (2019). tRNA ligase structure reveals kinetic competition between non-conventional mRNA splicing and mRNA decay. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, and Reik W (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Luhrmann R, Soll D, et al. (2011). HSPC117 Is the Essential Subunit of a Human tRNA Splicing Ligase Complex. Science (80-.). 331, 760–764. [DOI] [PubMed] [Google Scholar]
- Popow J, Jurkin J, Schleiffer A, and Martinez J (2014). Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511, 104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath K, and Ephrussi A (2016). CncRNAs: RNAs with both coding and non-coding roles in development. Development 143, 1234–1241. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, and Walter P (1997). The Transmembrane Kinase Ire1p Is a Site-Specific Endonuclease That Initiates mRNA Splicing in the Unfolded Protein Response. Cell 90, 1031–1039. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Cox JS, and Walter P (1996). tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 87, 405–413. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JAE, and Weeks KM (2014). RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, and Weeks KM (2015). Selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala A, and Hutvagner G (2011). Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2, 853–862. [DOI] [PubMed] [Google Scholar]
- Song Y, Sretavan D, Salegio EA, Berg J, Huang X, Cheng T, Xiong X, Meltzer S, Han C, Nguyen T-T, et al. (2015). Regulation of axon regeneration by the RNA repair and splicing pathway. Nat. Neurosci 18, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, and Shuman S (2011). RtcB Is the RNA Ligase Component of an Escherichia coli RNA Repair Operon. J. Biol. Chem 286, 7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, and Parker R (2009). Stressing Out over tRNA Cleavage. Cell 138, 215–219. [DOI] [PubMed] [Google Scholar]
- Unlu I, Lu Y, and Wang X (2018). The cyclic phosphodiesterase CNP and RNA cyclase RtcA fine-tune noncanonical XBP1 splicing during ER stress. J. Biol. Chem 293, 19365–19376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, and Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Yan D, Wu Z, Chisholm AD, and Jin Y (2009). The DLK-1 Kinase Promotes mRNA Stability and Local Translation in C. elegans Synapses and Axon Regeneration. Cell 138, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Zhai R, McLean NA, Johnston JM, Misra V, and Verge VMK (2015). The Unfolded Protein Response and Cholesterol Biosynthesis Link Luman/CREB3 to Regenerative Axon Growth in Sensory Neurons. J. Neurosci 35, 14557–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, and Mori K (2001). XBP1 mRNA Is Induced by ATF6 and Spliced by IRE1 in Response to ER Stress to Produce a Highly Active Transcription Factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Junction analysis of the RNA-seq data comparing RtcB mutant and control animals.
(Tab 1) Comparison between RtcB mutant and control animals without neuronal injury. (Tab 2) Comparison between RtcB mutant and control animals with neuronal injury, using a mutation in β-spectrin (unc-70) to trigger spontaneous axon breaks. (Tab 3) Definitions of the column names used in Tabs 1 and 2.
Related to: Figure 1B; Results section “Blocking xbp-1 cleavage versus ligation results in opposite phenotype in axon regeneration”.
Table S2. Annotations of the genes identified in the junction analysis.
Related to: Figure 1B; Results section “Blocking xbp-1 cleavage versus ligation results in opposite phenotype in axon regeneration”.
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.


