Summary
Bacteria and their toxins are associated with significant human morbidity and mortality. While a few bacterial toxins are well characterized, the mechanism of action for most toxins has not been elucidated, thereby limiting therapeutic advances. One such example is the highly potent pore-forming toxin hemolysin BL (HBL), produced by the gram-positive pathogen Bacillus cereus. However, how HBL exerts its effects and whether it requires any host factors is unknown. Here, we describe an unbiased genome-wide CRISPR/Cas9 knockout screen that identified LPS-Induced TNF-α Factor (LITAF) as the HBL receptor. Using LITAF-deficient cells, a second, subsequent whole-genome CRISPR/Cas9 screen identified the LITAF-like protein CDIP1 as a second, alternative receptor. We generated LITAF-deficient mice, which exhibit marked resistance to lethal HBL challenges. This work outlines and validates an approach to use iterative genome-wide CRISPR/Cas9 screens to identify the complement of host factors exploited by bacterial toxins to exert their myriad of biological effects.
Keywords: Bacillus cereus, Hemolysin BL, LITAF, CDIP1, toxin receptor
Graphical Abstract

In Brief
Bacillus cereus hemolysin BL (HBL) is a potent pore-forming-toxin, which rapidly lyses nearly all mammalian host cells. However, how HBL exerts these effects is unknown. Here, using sequential, whole-genome CRISPR/Cas9 screens, Liu et al identify LPS-induced factor LITAF and its related protein CDIP1 as the heretofore unrecognized HBL toxin receptors.
Introduction
Bacillus cereus, a spore-forming, Gram-positive bacterium, is an important human pathogen commonly associated with foodborne outbreaks, hospital infections, and endophthalmitis that often results in blindness (Dierick et al., 2005; Hori et al., 2017; Saleh et al., 2012; Viel-Theriault et al., 2019; Zhang et al., 2018). With advances in rapid diagnosis, B. cereus has increasingly been identified as a pathogen linked to acute severe infections and deaths in children and immunocompromised patients (Dierick et al., 2005; Ishida et al., 2019; Saleh et al., 2012; Viel-Theriault et al., 2019). However, the molecular mechanisms underlying its pathophysiology remain elusive.
One of the B. cereus’ major virulence factors is the highly potent hemolysin BL (HBL) (Beecher et al., 2000; Beecher et al., 1995; Mathur et al., 2019; Schoeni and Wong, 2005; Stenfors Arnesen et al., 2008). HBL is a unique tripartite pore-forming toxin (PFT), consisting of three components: cellular binding B subunit (42.5 kD) and two lytic subunits, L1 (43.8 kD) and L2 (49.3 kD), which together are required to induce cytotoxicity (Sastalla et al., 2013). Although HBL toxin has been known for thirty years and is well established to be critical for pathogenesis (Beecher et al., 2000; Beecher et al., 1995; Mathur et al., 2019; Schoeni and Wong, 2005; Stenfors Arnesen et al., 2008), the molecular mechanism underlying the interaction of the toxin and target cells remains unknown. Currently, it is believed that no cellular host proteins are required for the cytolytic action of HBL, largely because the toxin has been shown to form pores in protein-free liposomes (Mathur et al., 2019). Here, we present in vitro and in vivo data demonstrating that HBL strictly requires a cellular receptor(s) for its toxicity and use sequential unbiased genome-wide CRISPR/Cas9 knockout screens to identify these critical host factors. This strategy suggests a general approach to identify the complement of host factors exploited by other bacterial toxins for their pathogenic action.
Results
Potent in vitro and in vivo toxicity of HBL
To characterize HBL, we performed comprehensive analyses on its cytotoxicity both in vitro and in vivo. We found that HBL could rapidly lyse nearly all cells tested, often in the low nanomolar range (Figures 1A and 1B), including the well-characterized 60 human cancer cell lines (NCI-60 panel) used by the National Cancer Institute to screen compounds for anticancer activity (Monks et al., 1991; Shoemaker, 2006). HBL could also rapidly kill spontaneously beating iPSC-derived human cardiomyocytes (Figure 1C). Moreover, HBL lysed these cells rapidly, as for instance in the case of CHO (Chinese hamster ovary) cells, often within minutes of toxin addition (Figure 1D and SI Video). This rapidity, along with the observation of membrane blebbing, suggests that this pore-forming toxin likely kills cells via disrupting plasma membrane integrity.
Figure 1. HBL is a major virulence factor of B. cereus.

A. HBL’s cytotoxicity on a set of human and mouse cells. ECs, primary endothelial cells from mouse lung; BMDMs, mouse bone marrow-derived macrophages. Cells were incubated with HBL for 2 h, followed by an MTT assay for assessing cell viability. Data are represented as mean ± SD.
B. Inhibitory effects of HBL on proliferation of the NCI-60 cell panel 48 h after toxin incubation. Each line represents one cell line.
C. Cytotoxicity of HBL on iPSC-induced human cardiomyocytes. Cells were incubated with 10 nM HBL for 30 min.
D. Rapid killing of CHO cells by HBL. The same cells before and 8 min after HBL (10 nM) treatment are shown. The arrows indicate the featured membrane blebbing caused by HBL-induced membrane rupture.
E. In vivo toxicity of HBL. C57BL/6 mice were challenged (I.V.) with one or two doses of HBL as indicated, and survival monitored. Of note, all mice succumbed to one dose of 50 μg/kg (bodyweight) within 2 h of injection. Mice all survived one dose of 25 μg/kg, but were all killed following a 2nd 25 μg/kg dose administered 24 h after the first injection.
F. HBL-deficient isogenic B. cereus strain was greatly attenuated in the mouse infection model. 2 ×105 colony-forming units of WT or HBL-deficient B. cereus bacteria were injected (I.P.) into C57BL/6 mice.
Patients with acute systemic B. cereus infections often have a rapid progressive course of their disease with a high mortality rate (Dierick et al., 2005; Ishida et al., 2019). To investigate the role of HBL in B. cereus pathogenesis, we challenged mice with various doses of HBL by daily I.V. administration. We found that the toxin was indeed highly toxic, such that one dose of 50 μg/kg (bodyweight) was uniformly lethal to mice within 2 h (Figure 1E). Although all mice could survive a single dose of 25 μg/kg, these animals all succumbed within an hour of a second dose of 25 μg/kg (Figure 1E). We also infected mice with the wild-type (WT) B. cereus, as well as an HBL-deficient isogenic mutant strain (ΔHBL). While most mice infected with WT bacteria succumbed to the infection within 24 h (73%, n=11), all mice (n=11) challenged with B. cereus (ΔHBL) survived the challenge without any obvious ill-effects (Figure 1F). Together, these in vitro and in vivo studies demonstrate that HBL is potent toxic to a wide range of mammalian cells and is a major virulence factor of the B. cereus pathogen.
Requirement for an HBL cellular receptor
While the structures of the two HBL lytic subunits (HBL-L1 and HBL-L2) remain unknown, the crystal structure of the cellular binding B component (HBL-B) has been solved (Madegowda et al., 2008). Although HBL-B does not have recognized sequence homologies, it is structurally highly similar to HlyE (ClyA or SheA), a single-protein cytolysin found in some Gram-negative pathogens including Salmonella enterica serovar Typhi and Shigella flexneri (del Castillo et al., 1997; Ludwig et al., 2004; Madegowda et al., 2008; Mueller et al., 2009; Oscarsson et al., 2002; Song et al., 1996; von Rhein et al., 2009; Wallace et al., 2000). To date, it is believed that no cellular component from the target cells (besides the lipid bilayer membrane) is required for the function of these toxins (Mathur et al., 2019). However, the high potency of HBL in mediating the cytolytic activity suggests that a specific cellular receptor may be exploited by HBL to concentrate on the target cell surface. To test whether a cellular factor is required for HBL’s cytotoxicity, we performed an ethyl methanesulfonate (EMS)-induced random mutagenesis in the near-haploid CHO cells (Liu and Leppla, 2003b). Remarkably, we were able to isolate HBL-resistant mutant clones from EMS-treated cells (but not from untreated cells) through HBL selection (Figures 2A and 2B). These independent HBL-resistant cells were not only resistant to HBL in a cytotoxicity assay, but also abolished their binding to HBL-B (Figure 2C). These results demonstrate that EMS exposure resulted in mutation of a CHO cell gene(s) essential for the toxin’s binding, implicating that a cellular receptor(s) is required for the toxin’s action.
Figure 2. EMS-induced mutagenesis on CHO cells reveals that a cellular receptor is required for the HBL’s cytolytic activity.

A. Schematic representation of EMS-induced mutagenesis to isolate HBL-resistant CHO mutant cells. CHO cells (50 million) seeded in ten 10-cm dishes were treated with 0.25 mM EMS for 24 h. Then, the cells were treated with HBL (2.5 nM each component) for 3 h. After toxin removal, the toxin-treated cells were cultured for one week, allowing formation and isolation of HBL-resistant CHO mutant clones.
B. Cytotoxicity of HBL to ten independent (from different dishes) HBL-resistant clones isolated in (A). The cells were incubated with various concentrations of HBL for 2 h, followed by an MTT assay to assess cell viability. Data are represented as mean ± SD.
C. HBL-resistant CHO mutant cells cannot bind HBL. Three representative HBL-resistant clones (CHO-R1, -R2, -R3) were incubated with HBL-B for 1 h, followed by sequential staining with a mouse anti-HBL-B serum and an anti-mouse IgG conjugated with Alexa Fluor 594.
Identification of LITAF in CRISPR screen
We hypothesized that the toxin receptor could be identified by an unbiased approach, specifically a whole genome CRISPR/Cas9 knockout screen (Sanjana et al., 2014; Shalem et al., 2014). We performed such a screen on mouse macrophage RAW276.4 cells as outlined in Figure 3A, using the mouse CRISPR lentiviral pooled library-A (Sanjana et al., 2014), which covers 20,611 mouse genes, with each gene targeted by three single guided RNAs (sgRNAs). We reasoned that the HBL-resistant cells isolated after infection with a CRISPR lentiviral pooled library and subsequent toxin exposure should be enriched for cells in which the toxin receptor gene(s) had undergone sgRNA-mediated KO and its sgRNAs could be subsequently identified by Illumina deep sequencing.
Figure 3. A whole genome CRISPR screen identifies the elusive HBL cellular receptor.

A. Workflow of the CRISPR screen. RAW264.7(Cas9) cells infected with the mouse CRISPR library-A were selected for HBL-resistant cells. Representative HBL-resistant clones were isolated for further characterization. The remaining clones were pooled and subjected for Illumina deep sequencing for sgRNAs.
B. Top 10 hits from the Illumina deep sequencing were shown. All three LITAF sgRNAs were found among the top hits. LFC, Log2(fold of change).
C. HBL cytotoxicity on a set of eight representative HBL-resistant clones isolated in (A). The cells were incubated with HBL for 24 h, followed by an MTT assay assessing cell viability.
D. The sensitivity of the HBL-resistant RAW264.7 (RAW-R2) cells could be restored by reconstituting LITAF expression. The cells were incubated with HBL for 2 h, before assessing cell viability.
Data are represented as mean ± SD in C and D.
Remarkably, the lipopolysaccharide (LPS)-induced factor LPS-Induced TNF-α Factor (LITAF) appeared to be the only strong hit (Figure 3B and Table S1). All three independent LITAF sgRNAs were among the top ten hits isolated from this screen (Table S1). We also isolated a set of independent HBL-resistant clones during the screen, which were shown by cytotoxicity assay to be completely resistant to HBL (Figure 3C). Sanger sequencing demonstrated that these clones were all LITAF CRISPR edited mutants. Due to the small size deletion/insertion introduced by CRISPR editing, quantitative reverse transcriptase-PCR could not faithfully assess the mutations. Unfortunately, the antibodies available to us were not sensitive enough to detect endogenous levels of LITAF proteins. As such, we relied on genomic sequencing and functional complementation studies to characterize the isolated clones. Importantly, the sensitivity of the HBL-resistant RAW264.7 mutant cells could be restored by reconstituting LITAF expression (Figure 3D).
To test whether LITAF is a species-independent HBL receptor, we further performed two independent CRISPR screens on human cell line HT1080, using the human CRISPR lentiviral pooled library-A and –B (Sanjana et al., 2014). Strikingly, LITAF was the only gene with all six independent sgRNAs found among the top twenty-four hits from both screens (Figure S1A, Tables S2 and S3). In these CRISPR screens, we also isolated a set of independent HBL-resistant survivor clones, which were verified to be LITAF knock-out mutants (Figures S1B and S1C). However, in contrast to the results of gene editing of LITAF in RAW264.7 cells which exhibited complete HBL resistance, all human clones lacking LITAF only exhibited 5–10-fold more resistance to HBL cytotoxicity (Figure S1B). These results demonstrated the species-independent role of LITAF in HBL pathogenesis but suggested an additional receptor might operate in the human LITAF-KO HT1080 cells to mediate the residual cytotoxicity.
LITAF’s HBL receptor function
To functionally confirm the role of LITAF in mediating HBL cellular binding, we transfected a human LITAF expression vector into the HBL receptor deficient CHO-R1 cells (isolated in Figure 2). LITAF expression restored HBL sensitivity, as well as HBL binding in these cells (Figures 4A and 4B). Transfection of the LITAF cDNA into the other nine independent HBL-resistant CHO clones could also uniformly restore these cells’ sensitivity to HBL (Figure S2). We also constructed CHO cell lines complemented with the mouse or CHO versions of LITAF, which share 90% amino acid identity with human LITAF (Figure S3A). The LITAF from these species was equally active as the human LITAF in functioning as an HBL receptor (Figure S3B). We also noted that in the presence of DTSSP, a water-soluble and plasma membrane-impermeable protein crosslinker, we could detect a direct association between LITAF and HBL-B (Figure 4C).
Figure 4. Restoration of HBL sensitivity of the CHO mutant cells by LITAF expression.

A. HBL-resistant CHO-R1 cells (isolated in Fig. 2) regained HBL sensitivity by exogenous expression of LITAF. Of note, all the four LITAF-transfected CHO-R1 clones were sensitive to HBL, whereas the cells transfected with an irrelevant construct (VPS11) remained resistance to HBL. The cells were incubated with HBL for 2 h, followed by an MTT assay assessing cell viability. Data are represented as mean ± SD.
B. Exogenous expression of LITAF restores the HBL-binding ability of CHO-R1 cells. WT CHO, CHO-R1, and CHO-R1(LITAF) (transfected with LITAF) cells were incubated with HBL-B (5 nM) for 0, 3, 10, 20 min. Then cell lysates were analyzed by Western blotting using an HBL-B rat anti-serum and anti-tubulin antibody.
C. Co-immunoprecipitation of HBL-B with LITAF. CHO-R1(LITAF) cells were incubated with HBL-B (5 nM) for 2 h at 4°C. After washing, the cells were further incubated with/without the protein crosslinker DTSSP (1.2 mM) for 1 h. DTSSP is a water-soluble and plasma membrane-impermeable crosslinker (8-atom spacer arm, 12.0 Å), allowing for crosslinking cell surface proteins. Cell lysates were prepared in Triton X-100 lysis buffer, and immunoprecipitated by a goat anti-LITAF antibody or goat IgG control antibody, sequentially immunoblotted by a rat anti-HBL-B serum and anti-LITAF. Similar results were obtained in five biological repeats using a range of goat IgG controls. Purified HBL-B (2 ng) was loaded as a control.
Human and mouse LITAF have three protein coding isoforms generated by differential splicing resulting in LITAF proteins of 191 amino acids (191aa), 161aa, and 152aa. We performed reverse transcriptase-PCR analysis on human HT1080, HT29, SK-MEL28, Colo205, C32, MDA-MB-231, and A549 cell lines, and found that LITAF-161 is the major isoform, with this cDNA being amplified from all these cell lines. The 152-aa isoform could only be amplified from HT29, Colo205, and A549 cells, whereas the 191-aa isoform could not be amplified from any of these cells. LITAF-161 was the isoform we demonstrated as a functional HBL receptor in our earlier analyses (Figures 3 and 4). Lacking a signal peptide, LITAF-161 has been described as a C-tail-anchored protein, attaching on the plasma membrane via its putative transmembrane domain (TMD) (residues 114–135, Figure S2A) located near its C-terminus (Lee et al., 2011). This leaves a short C-terminal 26 amino acids (residues 136–161, Figures S3A and S4) segment exposed on the cell surface. While LITAF-161 could completely restore the toxin-sensitivity of the CHO-R1 cells, LITAF-152, that lacks an intact transmembrane domain and has a completely different C-terminal sequence, could not (Figure S4A). Moreover, an N-terminal truncated mutant only consisting of the C-terminal half of LITAF (LITAF 84–161) could function as an HBL receptor (Figure S4B). In contrast, a mutant lacking only the exposed C-terminus (LITAF 1–137aa) was not functional, attesting to the importance of the C-terminal residues in mediating HBL binding (Figure S4B).
Alternative HBL receptor
While LITAF is likely to be the only HBL-receptor in RAW264.7 cells (Figure 3), our data supports the existence of an alternative receptor in other cells, such as human HT1080 cells (Figure S1B). Moreover, knockout of LITAF in mouse B16F10 cells rendered this cell only 4-fold more resistant to HBL (Figure 5). We reasoned that this alternative receptor would become the major factor regulating virulence when LITAF is absent, and thus could be identified using an additional whole genome CRISPR screen on LITAF-deficient cells. To test this, we generated B16F10 LITAF-KO cells, performed an additional whole genome CRISPR screen and then challenged these cells with high-dose HBL (10 nM) (Figures S5A and S5B). The sgRNA containing DNA fragments from these clones were sequenced, revealing that sixteen of them contained CDIP1 (cell death involved p53 target 1) sgRNAs. Of note, CDIP1, also termed LITAF-like, is the only known homolog of LITAF in mammals, with its C-terminal 80 residues highly homologous to those of LITAF (44% identical in amino acids) (Figures 5A and S5C). This portion of LITAF appears to be sufficient in functioning as an HBL receptor (LITAF 84–161) (Figure S4B).
Figure 5. CDIP1 is an alternative HBL receptor.
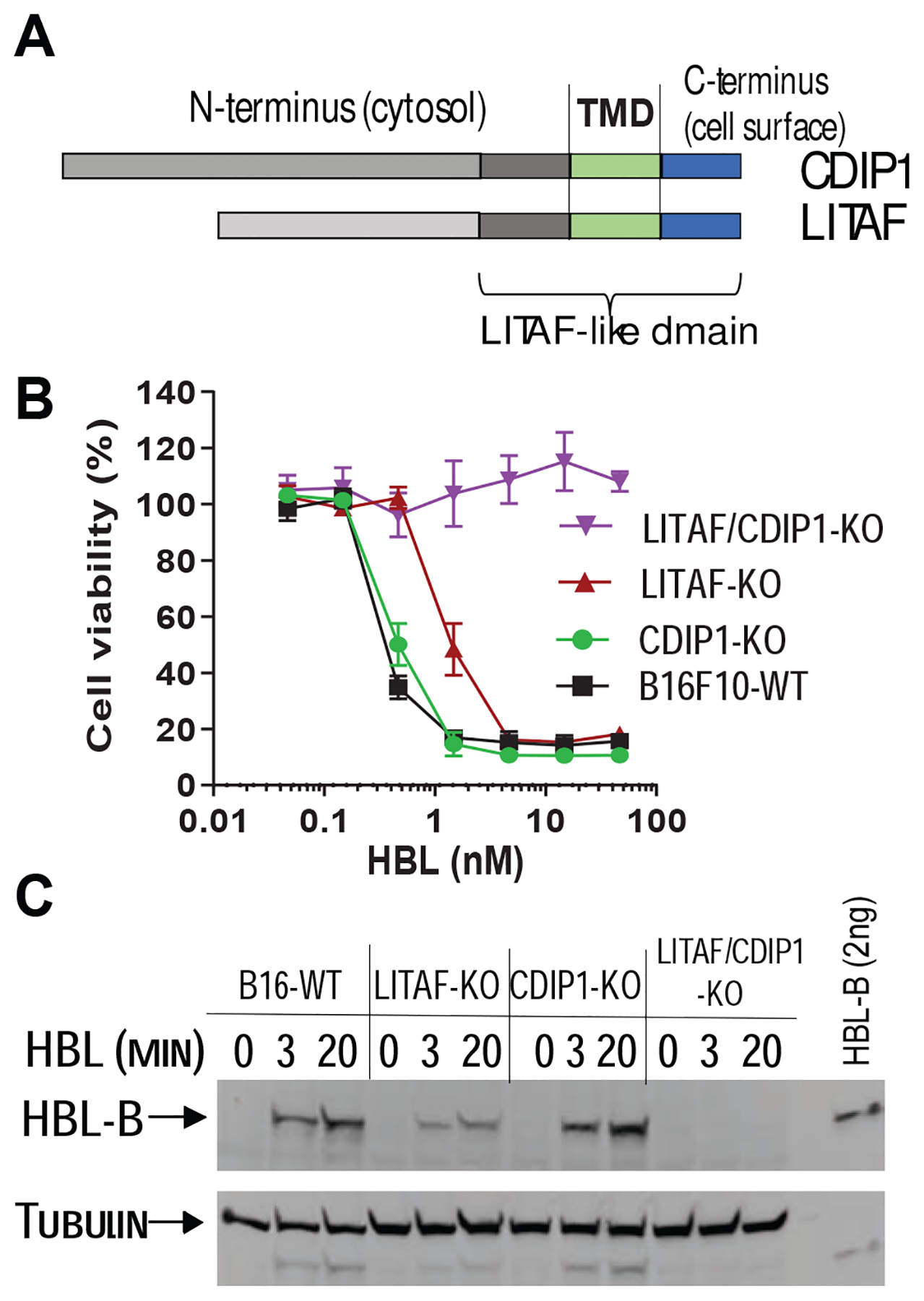
A. CDIP1 is a LITAF-like family member protein sharing 44% amino acid identity with LITAF across the last 80 aa (LITAF-like domain).
B. Cytotoxicity of HBL on LITAF-, CDIP1-, and LITAF/CDIP1-KO B16F10 cells. B16F10 cells with LITAF and/or CDIP1 knocked-out by CRISPR editing were incubated with various concentrations of HBL for 2 h, prior to measuring cell viability. Data are represented as mean ± SD.
C. HBL cell binding assay. The indicated B16F10 cells with LITAF- and/or CDIP1-KO were incubated with 2.5 nM HBL for 0, 3, 20 min. Then cell lysates were analyzed by Western blotting using a rat HBL-B anti-serum and anti-tubulin antibody.
To verify the role of CDIP1 in HBL pathogenesis, we regenerated LITAF and CDIP1 single, as well as double knockout B16F10 cells using CRISPR gene editing. As expected, LITAF-KO B16F10 cells were 4-fold more resistant to HBL (Figure 5B). While CDIP1 knockout did not affect the cells’ HBL sensitivity, the LITAF/CDIP1 double knockout cells were completely resistant to HBL and lost their capacity to bind the toxin (Figures 5B and 5C), demonstrating that CDIP1 acts as alternative receptor when LITAF is absent. Together, these results demonstrate that B. cereus HBL specifically uses these two LITAF-like proteins as cellular receptors; with LITAF serving as the major receptor, while CDIP1 functions as an alternative receptor when LITAF is absent.
In vivo role of LITAF in HBL pathogenesis
To determine the in vivo role of LITAF in HBL pathogenesis, we generated two LITAF mutant mouse lines using the CRISPR/Cas9 method (Wang et al., 2013) (Figure S6): one bearing a 5.6-Kb LITAF gene fragment deletion (missing the majority of the coding sequence), termed LITAF KO allele (LITAF−); the other with a 7-bp deletion in exon 3 that leads to a frameshift mutation causing a replacement of LITAF C-terminal 22 amino acids with 41 unrelated residues (thus likely affecting HBL binding), termed C-terminal frameshift mutant (LITAFCFS). The LITAFCFS was generated by the LITAF sgRNA that was frequently found in HBL-resistant RAW264.7 cells from the CRISPR screen (Figures 3C and 3D), cutting upstream of the stop codon in the last coding exon (Figure S6A). This altered transcript did not appear to be subject to nonsense mediated decay, since it could readily be amplified and cloned into an expressing plasmid by reverse transcriptase-PCR from tissues of LITAFCSF/CSF mice. The complementation experiment showed that this C-terminal frameshift mutant could not function as an HBL receptor (Figure S6C). LITAFCSF/CSF and LITAF−/− mice did not exhibit obvious phenotypes. To verify that LITAFCSF and LITAF− alleles are null-mutations for HBL binding, we isolated bone-marrow-derived macrophages (BMDMs) from LITAFCSF/CSF, LITAF−/−, and their littermate control WT mice. While WT BMDMs were highly sensitive to HBL, BMDMs from both LITAFCSF/CSF and LITAF−/− mice were completely resistant to HBL (Figures 6A and 6B), demonstrating that both the LITAF deletion and the C-terminal frameshift are null-mutations for HBL binding and that LITAF is the only HBL receptor in primary mouse macrophages.
Figure 6. In vivo role of LITAF in HBL pathogenesis.

A. Cytotoxicity of HBL on primary BMDMs and ECs derived from LITAF−/− mice and WT control mice. Cells were incubated with HBL for 2 h, followed by an MTT assay assessing cell viability. Note, the BMDMs from LITAF−/− mice were completely resistant to HBL, while the ECs were 15- fold more resistant to HBL, compared to the WT control cells.
B. Primary BMDMs and ECs from LITAFCFS/CFS mice were analyzed as in (A).
C. LITAF/CDIP1 double KO ECs were completely resistant to high concentrations of HBL. LITAF/CDIP1 double KO ECs were generated by further knocking out of CDIP1 in LITAF−/− ECs using CRISPR editing. Cells were treated with 50 nM HBL for 2 h.
D. RBCs from LITAFCFS/CFS mice were completely resistant to HBL-induced hemolysis. RBCs from LITAFCFS/CFS and WT control mice were incubated with HBL for 30 min. RBCs lysed by 0.5% SDS was set as 100% of hemolysis.
E, F. LITAFCFS/CFS and LITAF−/− mice are more resistant than WT control mice to HBL challenge. While one dose of 50 μg/kg HBL (I.V.) killed all WT mice, the LITAFCFS/CFS (D) and LITAF−/− mice (E) mice could uniformly survive following two doses of 50 μg/kg HBL administered as indicated by the red arrows.
G. LITAF−/− mice were treated with three doses of 50 μg/kg HBL (I.V.) as indicated by the red arrows.
Data are represented as mean ± SD in A-D.
We further isolated primary endothelial cells (ECs) from lungs of LITAFCSF/CSF and LITAF−/− mice. The ECs from the LITAF-null mice were 15-fold more resistant to HBL, compared to WT ECs (Figures 6A and 6B). These results demonstrate that LITAF is the major HBL receptor in vivo. The residual HBL cytotoxicity in LITAF-null ECs is likely mediated by the alternative receptor CDIP1, because further knockout of CDIP1 in the LITAF-null ECs by CRISPR editing rendered the cells completely resistant to HBL (Figure 6C).
HBL (as it is termed a hemolysin) can rapidly lyse red blood cells (RBCs) (Sastalla et al., 2013). To examine the role of LITAF in HBL’s hemolytic activity, RBCs from LITAF−/− mice and their littermate control mice were isolated. Notably, while WT RBCs could be efficiently lysed, the RBCs from LITAF−/− mice were completely resistant to the toxin (Figure 6D), demonstrating that LITAF is also the only HBL receptor in RBCs and is required for the observed hemolytic activity.
Lastly, we challenged the LITAFCSF/CSF and LITAF−/− mice and their littermate control mice with lethal doses of HBL. Both the LITAF−/− and LITAFCSF/CSF mice were markedly more resistant to HBL than their WT littermate control mice. While all WT mice succumbed to only one dose of 50 μg/kg HBL (I.V.), all the LITAFCSF/CSF and LITAF−/− mice uniformly survived two doses of 50 μg/kg HBL administration (Figures 6E and 6F). To evaluate the in vivo toxicity mediated by the alternative minor receptor, we challenged the LITAF−/− mice with multiple HBL doses and did observe that the third injection could kill ~50% of the LITAF−/− mice (Figure 6G). Together, while demonstrating the major role of LITAF in mediating the HBL toxicity in vivo, these results also suggest the alternative HBL receptor (CDIP1) may mediate lethality both in vitro and in vivo when much higher concentrations of HBL are reached.
Discussion
As noted, LITAF was named as LPS-Induced TNF-α Factor, and is among a set of host proteins upregulated in response to microbial infections (Myokai et al., 1999). Following bacterial pathogen infections, LITAF expression rapidly increases and is believed to play an important role in inducing many cytokines/chemokines including TNF-α (Bushell et al., 2011; Merrill et al., 2011; Tang et al., 2006). As such, B. cereus appears to have evolved to co-opt this upregulated host protein to potentiate HBL’s cytolytic action. It should also be noted that PFTs are the largest class of virulence factors of human bacterial pathogens, damaging target cells through formation of sophisticated membrane-crossing pore structures (Dal Peraro and van der Goot, 2016). Because PFTs are often found to form ion conductive pores on artificial lipid bilayers, PTFs, like HBL, are not generally believed to require a cellular receptor for their actions (Dal Peraro and van der Goot, 2016). Here, however, we provide evidence that disputes this commonly held assumption and have identified the two alternative surface receptors required for HBL cytotoxicity. Furthermore, this work outlines and validates the use of iterative CRISPR screens to identify the full complement of host factors exploited by a wide range of bacterial toxins that impact human health.
The Human Protein Atlas (www.proteinatlas.org) data base suggests that both LITAF and CDIP1 are expressed ubiquitously. Since knockout of both LITAF and CDIP1 is required for the cells originally express both receptors to reach completely resistance to HBL, these two LITAF-like proteins appear to function as alternative, independent receptors, with LITAF serving as the major receptor, while CDIP1 as the minor one (Figure 5). This is reminiscence of the fact that anthrax toxin also uses two alternative receptors, the major receptor CMG2 (Capillary morphogenesis protein-2) and the minor receptor TEM8 (tumor endothelium marker-8), to mediate the toxin’s cytotoxic action (Bradley et al., 2001; Liu et al., 2009; Liu et al., 2012; Scobie et al., 2003). While CMG2 has ten-fold higher affinity than TEM8 for anthrax toxin, whether LITAF has a higher affinity than CDIP1 for HBL toxin requires further investigation.
LITAF was reported as a C-tail-anchored membrane protein regulating the endosomal trafficking and signal attenuation of ErbB receptors via recruiting the ESCRT (endosomal sorting complex required for transport) machinery (Lee et al., 2012). However, this function requires an N-terminal PSAP tetrapeptide motif (17PSAP20, Figure S3), which we have shown is not required in mediating cell surface HBL binding. Recent evidence suggests that LITAF is a zinc-binding monotopic membrane protein with both its termini located in the cytosol (Ho et al., 2016; Qin et al., 2016). This conformation likely results from the coordination of a zinc ion (Zn2+) in the cytosol by the four cysteine residues from the two TMD-flanked CxxC motifs (96CPSC99 and 148CPNC151, Figure S3). If this is the case, our results support the notion that an equilibrium may exist between the two conformations, so that a fraction of LITAF escapes Zn2+ coordination and displays its short C-terminal fragment on the cell surface, where it is exploited by HBL as a toxin receptor. Our work suggests that synthetic peptides containing the short C-terminal fragments of LITAF (combined with antibiotics) may serve as efficient anti-HBL decoy receptors used for targeted therapy in B. cereus infections.
Missense mutations in LITAF cause Charcot-Marie-Tooth disease (CMT) (Guimaraes-Costa et al., 2017; Street et al., 2003), the most common hereditary peripheral neuropathy having no effective treatment. Identification of the toxin receptor function of LITAF and CDIP1 in this work will open new avenues in understanding their structure/function relationship, elucidating their elusive pathophysiological roles in diseases.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shihui Liu (SHL176@pitt.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all data generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse studies
All animal studies were carried out in a pathogen-free facility maintaining full accreditation by the American Association for the Accreditation of Laboratory Animal Care (AAALAC) and in accordance with protocols approved by the University of Pittsburgh Institutional Animal Care and Use Committee (Protocol #:19054269). Mice are housed in cages with 24-hour free access to food and water, with each cage holds a maximum of 4 male or 5 female mice at constant temperature (23°C) and humidity (55% ± 10%) with a 12-hour light/dark cycle (lights on 7:00 am). Mice with indicated phenotypes in C57BL/6 background, 8–12-week-old, both male and female, were used. To avoid the potential degradation of the toxin by gastric acid and enzymes, we chose I.V. route rather than the oral gavage for the toxin administration. Therefore, in HBL challenge experiments, 8–10 week old male and female mice with various genotypes were injected (I.V.) with one to three doses of HBL in 0.2 ml PBS. B. cereus ATCC 10876 was obtained from ATCC, and its HBL-deficient isogenic mutant strain was generated by a Cre-LoxP based mutagenesis as described previously (Sastalla et al., 2013). B. cereus bacteria were grown in Luria-Bertani medium at 37°C overnight, and subcultured (1:50) for 2–3 h before use. For infection studies, mice were injected (I.P.) with 2 × 105 (colony-forming units) B. cereus strains as described (Mathur et al., 2019). Our previous studies demonstrate that 5–10 mice per treatment group are sufficient for statistical analyses in toxin challenge experiments (Liu et al., 2013). Thus, we use 5–10 or more mice per group to ensure statistical power. The mice were grouped based on genotypes. When the mice in same genotype received different treatments, they were randomly assigned to treatment groups. All toxin-challenged or infected mice were monitored twice daily for one week post-challenge for signs of malaise or mortality.
LITAF mutant mice
LITAF mutant mouse lines were generated using the CRISPR/Cas9 method (Wang et al., 2013; Yang et al., 2013). Two CRISPR sgRNAs were designed and made using T7 in vitro transcription to target its exons. The sgRNA-a (CCTGTGGCTGGTCCCGGCAT) cuts shortly after translation initiation in the first coding exon and sgRNA-b (GTTCTGCGTAGACGCCCTAC) (the one frequently found in HBL-resistant RAW264.7 cells in CRISPR screen) cuts upstream of the stop codon in the last coding exon (Figure S6A). These sgRNAs were co-microinjected (20 ng/μl each sgRNA) with 50 ng/μl Cas9 mRNA (Trilink BioTechnologies) into fertilized eggs collected from C57BL/6N mice (Charles River). The injected embryos were cultured overnight in M16 medium (Millipore) and then implanted into the oviducts of pseudopregnant foster mothers. Mice born to the foster mothers were genotyped by PCR and DNA sequencing to identify founders with the desired mutations. Genotyping primers are listed in Table S4. We identified three different mutations among them: (1) a 21-bp deletion (in-frame) in exon 1 guided by sgRNA-a; (2) a 5.6-Kb deletion mediated by the two sgRNAs’ double cuts (Figure S5A), termed LITAF+/−; (3) a 7-bp deletion (frameshift) in exon 3 guided by sgRNA-b (Figure S5B), named LITAF+/CFS. The last two mutant mice were bred further and used in this work.
Cell culture
CHO cells were grown in AMEM (α-minimal essential medium) supplemented with 10% FBS. Other cell lines were grown in DMEM supplemented with 10% FBS.
BMDMs were isolated as described previously (Liu et al., 2010; Newman et al., 2010). BMDMs were cultured in L929 conditioned DMEM with 10% fetal bovine serum (FBS). ECs were isolated and cultured as described previously (Reynolds and Hodivala-Dilke, 2006). Briefly, three mouse lungs were digested with type I collagenase and plated on gelatin, fibronectin and collagen-coated flasks. The cells were then subjected to sequential negative sorting by magnetic beads coated with a sheep anti-rat antibody using a Fc Blocker (rat anti-mouse CD16/CD32, BD Pharmingen Cat. No. 553142) to remove macrophages and positive sorting by magnetic beads using an anti-intermolecular adhesion molecule 2 (ICAM2 or CD102) antibody (BD Pharmingen Cat. No. 553326) to isolate ECs (ICAM2 positive cells). The cells other than ECs (non-ECs, ICAM2 negative cells) were also isolated simultaneously as controls. These primary cells were used within 5 passages after isolation. iPSC-derived human myocardiocytes were generated using a well-characterized in vitro differentiation protocol (Burridge et al., 2014).
METHOD DETAILS
HBL cytotoxicity assay
Cells grown in 96-well plates (50–80% confluence) were incubated with various concentrations of HBL (1 nM HBL defined as 1 nM each of B, L1, and L2 components) for 2 h or 24 h. Cell viabilities were then assayed by MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, Sigma Cat. No. M5655) as described previously (Liu and Leppla, 2003a), expressed as % of signals of untreated cells. The NCI-60 panel evaluation was done through the NCI-60 DTP Human Tumor Cell Line Screening Services (Monks et al., 1991; Shoemaker, 2006). In brief, the NCI-60 panel cells (60 human cancer cell lines) grown in 96-well plates were incubated with various concentrations of HBL for 48 h. Cell proliferation inhibition was assessed by the relative total protein after the toxin incubation to the total protein before the toxin addition. The HBL toxin components, HBL-B, HBL-L1, and HBL-L2, were purified from a non-virulent B. anthracis strain as previously described (Sastalla et al., 2013).
Cellular binding assay
For HBL cell binding assay, cells grown to 80% confluence in 12-well plates were incubated with 10 nM HBL-B for up to 20 min. Cell lysates were prepared and separated on SDS-PAGE and analyzed by Western blotting using a rat anti-HBL-B serum prepared in our laboratory.
Hemolysis
RBCs from WT and LITAF mutant mice were washed with PBS twice, resuspended in 10 volumes of PBS, and incubated with various concentrations of HBL for 30 min at 37°C. After incubation, unlysed erythrocytes were removed by centrifugation. The extent of hemoglobin release was quantified by measuring the A540 nm of the supernatant using a plate reader. The hemolytic activity of HBL was expressed as a percentage of the hemolytic activity of 0.5% of SDS.
EMS-induced random mutagenesis
The near-haploid CHO cells were used for the EMS-induced random mutagenesis. 50 million CHO cells seeded in ten 10-cm dishes were treated with 0.25 mM EMS (Sigma Cat. No. M0880) for 24 h. 25 million CHO cells grown in five 10-cm dishes were used as no EMS treatment controls. Then the cells were treated with HBL (2.5 nM each component) for 3 h. The treated cells were changed with fresh medium, cultured for one week, allowing formation of HBL-resistant CHO mutant colonies. While no HBL-resistant colonies formed in the five dishes without EMS treatment, we were able to isolate 10 independent HBL-resistant clones from each of the ten dishes. These clones were defective in HBL binding.
CRISPR gene editing and CRISPR pooled lentiviral library screen
The genome-wide CRISPR knockout lentiviral pooled libraries developed by Feng Zhang’s MIT laboratory (Sanjana et al., 2014) were used for our unbiased screens for host genes required for toxin activity. There are both human and mouse versions of the CRISPR libraries (Addgene Cat. # 1000000049 for human version and Cat. #1000000053 for mouse version). The libraries are divided into two half-libraries (A and B), each containing three independent sgRNAs for each of the 19,050 human genes or 20,611 mouse genes. Each half library usually provides sufficient power as an independent unbiased genome-wide screen. Because the libraries are in the format of a two vector system, meaning Sp Cas9 is encoded by a separate vector, we first transfected lentiCas9-Blast plasmid (Addgene#52962, (Ran et al., 2013)) into the cells used for the screen (RAW264.7 and HT1080 cells). We followed the protocols provided by Addgene and in reference (Sanjana et al., 2014) for the pooled lentiviral library preparations and infections. Briefly, the library DNA was packaged to form pooled lentiviral sgRNA libraries and titrated. For the mouse library A screen, 60 million RAW264.7 (Cas9-expressing) cells (1000 × coverage of the library) were infected with the lentiviral pooled library A at a M.O.I. (virus/cell ratio) of 0.3. Infected cells were divided into twelve 15-cm (diameter) culture dishes and selected with puromycin (5 μg/ml) for three days. Cells were passed by trypsinization to maintain 50% confluence for one week allowing the completion of the gene editing process. Then for each cell plate, half of the cells were frozen for the later genomic DNA isolation as the non-selected control, and half of the cells were cultured and subjected to HBL selection. In brief, cells were incubated with 2.5 nM HBL for 2 h. Dead cells were removed by replacing with fresh medium. Two days later, the survivors were treated again with 2.5 nM HBL for 2 h. Surviving cells were cultured to allow formation of cell colonies. A set of independent colonies were isolated for Sanger DNA sequencing and HBL cytotoxicity assay. The remaining cells (majority) were collected into four different pools for genomic DNA isolation and sgRNA DNA amplification by two rounds of PCR using the primers listed in Table S4. The PCR products were sequenced by Illumina deep sequencing and analyzed. The raw sequencing data trimmed off the adaptor sequence were matched to the guide sequences from the filtered library files using the Modelbased Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) algorithm (v0.5.6) count function (Li et al., 2014). Normalized sgRNA counts from HBL-resistant cells over non-toxin selected cells were calculated, and expressed as Log2[fold of change] (LFC). We also performed both human CRISPR library-A and-B screens on HT1080 (cas9) cells.
To generate B16F10 LITAF-KO cells for further CRISPR screens, we cloned the mouse LITAF sgRNA sequence (GTTCTGCGTAGACGCCCTAC), which was most frequently found in HBL-resistant cells in our initial screen (Figure 3), into the pSpCas9–2A-GFP vector (Addgene, PX458). This vector does not have a puromycin resistance gene but allows us to select the sgRNA transfected cells by sorting for GFP positive cells. We transfected the resulting LITAF sgRNA construct into B16F10 (Cas9) cells and sorted the GFP-positive cells 48 h after transfection. The GFP-positive cells were first cultured for 7 days, and then selected with 2.5 nM HBL for 2 h. As expected, all the GFP-positive cells that survived the HBL selection were LITAF KO cells, and were 4-fold more resistant to HBL, yet could still be killed by higher dose of HBL (7.5 nM). These LITAF-deficient B16F10 cells were used for the next round of whole genome CRISPR screen following the procedure as described above.
Cloning of sgRNAs into pSpCas9–2A-GFP (Addgene, #48138), pSpCas9–2A-Puro (Addgene, #48139), and LentiCRISPRv2 (Addgene, #52961) was done by following the protocol described by Feng Zhang’s MIT laboratory (Sanjana et al., 2014). X-tremeGENE™ 9 DNA Transfection Reagent was used for transfection of the plasmids into the indicated cells following the manufacturer’s manual (Roche, Cat. No.: 06366236001)
Co-immunoprecipitation
CHO cells grown in 15-cm dishes were incubated with HBL-B (10 nM) for 30 min at 4°C. After washing with PBS, the cells were incubated with/without the protein crosslinker DTSSP (3,3’dithiobis (sulfosuccinimidyl propionate), Sigma Cat. No. 803200) (1.2 mM) in PBS for 1 h. DTSSP is a popular water-soluble and plasma membrane-impermeable crosslinker, allowing for crosslinking cell surface proteins. DTSSP contains amine-reactive NHS-ester ends around an 8-atom spacer arm (12.0 Å), whose central disulfide bond can be cleaved with reducing agents (such as DTT-containing SDS-PAGE sample loading buffer), allowing separation of crosslinked products. Cells were washed, cell lysates prepared in Triton X-100 lysis buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with EDTA-free Protease Inhibitor Cocktail (Sigma Cat. No. 4693132001), and immunoprecipitated by a goat anti-LITAF antibody (R&D Systems, Cat. No. AF4695) or goat IgG control antibodies attached to SureBeads™ Protein G Magnetic Beads (BIO-RAD, Cat. No. 1614023) and sequentially immunoblotted by an anti-HBL-B serum and anti-LITAF antibody. Five biological replicates were performed under various goat IgG controls with similar results demonstrating the association of LITAF and HBL. Anti-HBL-B anti-sera were prepared by immunization of rats and mice by subcutaneous injections of HBL-B. Co-immunoprecipitation of HBL and LITAF could not be detected when incubation was done at 37°C, possibly because the association is of low affinity and easily reversed.
Reverse transcriptase PCR and cell transfection
Total RNA was prepared from the indicated human, mouse and CHO cells using TRIzol reagent (Invitrogen, Carlsbad, CA), and was used to synthesize single-strand cDNA using the SuperScript IV First-Strand Synthesis System following the manufacturer’s manual (Invitrogen, Cat. No.: 18091050). Full-length LITAF cDNA from human, mouse, and CHO cells, and the truncated variants of human LITAF cDNA were amplified by PCR and cloned into pIREShyg-2 mammalian expression vector. The primers used for cloning are listed in Table S4. X-tremeGENE™ 9 DNA Transfection Reagent was used for transfection of the plasmids into the indicated cells following the manufacturer’s manual (Roche, Cat. No.: 06366236001).
QUANTIFICATION AND STATISTICAL ANALYSIS
Survival curves were compared via Log-rank Mantel-Cox test using GraphPad Prism. P < 0.05 was considered as a significant difference. Other data represent mean values ± SD from at least three independent experiments in triplicate. In comparisons between two groups with equal variance, unpaired two-tailed Student’s t-tests was used to identify significant (P < 0.05) differences.
Supplementary Material
SI Video. Rapidly killing of CHO cells by HBL. CHO cells were incubated with HBL (10 nM) for up to 8 min. Related to Figure 1.
Table S1. Illumina deep sequencing counts of sgRNAs in RAW264.7(Cas9) cells infected with the CRISPR lentiviral pooled library after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
Table S2. Illumina deep sequencing counts of sgRNAs in human HT1080(Cas9) cells infected with the human CRISPR lentiviral pooled library-A after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
Table S3. Illumina deep sequencing counts of sgRNAs in human HT1080(Cas9) cells infected with the human CRISPR lentiviral pooled library-B after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-LITAF antibody | R&D Systems | Cat# AF4695; RRID:AB_2135711 |
| Mouse monoclonal anti-LITAF antibody | Santa Cruz | Cat# sc-166719; RRID:AB_2135842 |
| Rabbit polyclonal anti-CDIP1 antibody | Invitrogen | Cat# PA5–20697; RRID:AB_11152803 |
| Mouse monoclonal anti-tubulin antibody | Santa Cruz | Cat# sc-8035; RRID:AB_628408 |
| Rat anti-mouse CD16/CD32 antibody | BD Pharmingen | Cat# 553142; RRID:AB_394657 |
| Rat anti-mouse ICAM2/CD102 antibody | BD Pharmingen | Cat# 553326; RRID:AB_394784 |
| Rat anti-HBL B component antiserum | This paper | N/A |
| Bacterial and Virus Strains | ||
| B. cereus ATCC 10876 | ATCC | 10876D-5 |
| Isogenic HBL-deficient B. cereus ATCC 10876 strain | (Sastalla et al., 2013) | N/A |
| DH5α chemical competent cells | New England BioLabs | Cat# C29871 |
| Endura ElectroCompetent Cells | Lucigen | Cat# 60242 |
| Pooled sgRNA library lentiviruses | This paper; (Sanjana et al., 2014) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) | Sigma | Cat# M5655 |
| Ethyl methanesulfonate (EMS) | Sigma | Cat# M0880 |
| X-tremeGENE™ 9 DNA Transfection Reagent | Roche | Cat# 06366236001 |
| cComplete, EDTA-free Protease inhibitor Cocktail | Sigma | Cat# 4693132001 |
| Type I collagenase | Life technologies | Cat# 17100–017 |
| Gelatin (2% solution type B: from bovine skin) | Sigma | Cat# G1393 |
| Collagen (ultrapure bovine) | Sigma | Cat# C4243–20ML |
| Bovine plasma fibronectin | Sigma | Cat# F4759–2MG |
| Dynabeads Sheep anti-Rat IgG | Invitrogen | Cat# 11035, |
| SureBeads™ Protein G Magnetic Beads | BIO-RAD | Cat# 1614023 |
| Endothelial cell growth supplement from bovine neural tissue | Sigma | Cat# E2759–15 mg |
| Heparin sodium salt, Grade I-A (from porcine intestine mucosa) | Sigma | Cat# H3149–100 KU |
| TRIzol reagent | Invitrogen | Cat# 15596026 |
| HBL-B protein | This paper | N/A |
| HBL-L1 protein | This paper | N/A |
| HBL-L2 protein | This paper | N/A |
| Critical Commercial Assays | ||
| SuperScript IV First-Strand Synthesis System | Invitrogen | Cat# 18091050 |
| Experimental Models: Cell Lines | ||
| Human: HT1080 | ATCC | CCL-121; RRID:CVCL_0317 |
| Hamster: CHO (CHO WTP4) | (Liu and Leppla, 2003b) | N/A |
| Mouse: B16F10 | ATCC | CRL-6475; RRID:CVCL_0159 |
| Mouse: RAW264.7 | ATCC | TIB-71; RRID:CVCL_0493 |
| Mouse: L929 | ATCC | CCL-1; RRID:CVCL_0462 |
| Human: HT-29 | ATCC | HTB-38; RRID:CVCL_0320 |
| CHO HBL-resistant mutant cells | This paper | N/A |
| HT1080 CRISPR edited mutant cells | This paper | N/A |
| RAW264.7 CRISPR edited mutant cells | This paper | N/A |
| B16F10 CRISPR edited mutant cells | This paper | N/A |
| Mouse primary endothelial cells | This paper | N/A |
| Mouse bone-marrow derived macrophages | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson laboratory | Cat# 000664 |
| Mouse: C57BL/6N | Charles River | C57BL/6NCrl |
| Mouse: LITAF+/+, LITAF+/−, and LITAF−/− mice | This paper | N/A |
| Mouse: LITAF+/+, LITAF+/CSF, and LITAFCFS/CSF mice | This paper | N/A |
| Oligonucleotides | ||
| See Table S4 for the full list of primers and oligonucleotides | Sigma | N/A |
| Recombinant DNA | ||
| Plasmid: pIRESHyg-LITAF (human) | This paper | N/A |
| Plasmid: pIRESHyg-LITAF (mouse) | This paper | N/A |
| Plasmid: pIRESHyg-LITAF (CHO) | This paper | N/A |
| Plasmid: pIRESHyg-LITAF-152 | This paper | N/A |
| Plasmid: pIRESHyg-LITAF 84–161 | This paper | N/A |
| Plasmid: pIRESHyg-LITAF 1–137 | This paper | N/A |
| Plasmid: pIRESHyg-CDIP1 | This paper | N/A |
| Plasmid: pSpCas9–2A-Puro | (Ran et al., 2013) | Addgene#48139 |
| Plasmid: lentiCas9-Blast | (Sanjana et al., 2014) | Addgene#52962 |
| Plasmid: LentiCRISPRv2 | (Sanjana et al., 2014) | Addgene#52961 |
| Mouse gRNA pooled library in lentiGuide-Puro vector | (Sanjana et al., 2014) | Addgene#1000000053 |
| Human gRNA pooled library in lentiGuide-Puro vector | (Sanjana et al., 2014) | Addgene#1000000049 |
| Software and Algorithms | ||
| Prism 7 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
Highlights.
Hemolysin BL (HBL) is the major virulence factor of human pathogen Bacillus cereus
HBL is a potent pore-forming toxin that can lyse a wide range of mammalian cells
Sequential CRISPR-based screens identify LITAF and CDIP1 as the toxin receptors for HBL
Mice deficient of LITAF are highly resistant to lethal HBL challenge
Acknowledgements
This research was supported by the institutional seed fund (S.L) and the grant R01AI145879 (S.L.) from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), and in part by the Intramural Programs of the NIAID, NIH, and the National Heart Lung and Blood Institute, NIH. GeCKOv2 CRISPR knockout pooled libraries were gifts from Feng Zhang (Addgene # 1000000049 for human version and #1000000053 for mouse version). The NCI-60 panel evaluation was done through the NCI-60 DTP Human Tumor Cell Line Screening Services. We thank Mahtab Moayeri (NIAID) for making anti-sera against HBL toxin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- Beecher DJ, Olsen TW, Somers EB, and Wong AC (2000). Evidence for contribution of tripartite hemolysin BL, phosphatidylcholine-preferring phospholipase C, and collagenase to virulence of Bacillus cereus endophthalmitis. Infection and immunity 68, 5269–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecher DJ, Pulido JS, Barney NP, and Wong AC (1995). Extracellular virulence factors in Bacillus cereus endophthalmitis: methods and implication of involvement of hemolysin BL. Infection and immunity 63, 632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, and Young JA (2001). Identification of the cellular receptor for anthrax toxin. Nature 414, 225–229. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. (2014). Chemically defined generation of human cardiomyocytes. Nature methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell KN, Leeman SE, Gillespie E, Gower AC, Reed KL, Stucchi AF, Becker JM, and Amar S (2011). LITAF mediation of increased TNF-alpha secretion from inflamed colonic lamina propria macrophages. PloS one 6, e25849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Peraro M, and van der Goot FG (2016). Pore-forming toxins: ancient, but never really out of fashion. Nat Rev Microbiol 14, 77–92. [DOI] [PubMed] [Google Scholar]
- del Castillo FJ, Leal SC, Moreno F, and del Castillo I (1997). The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Molecular microbiology 25, 107–115. [DOI] [PubMed] [Google Scholar]
- Dierick K, Van Coillie E, Swiecicka I, Meyfroidt G, Devlieger H, Meulemans A, Hoedemaekers G, Fourie L, Heyndrickx M, and Mahillon J (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J Clin Microbiol 43, 4277–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes-Costa R, Iancu Ferfoglia R, Leonard-Louis S, Ziegler F, Magy L, Fournier E, Dubourg O, Bouche P, Maisonobe T, Lacour A, et al. (2017). Phenotypic spectrum of Charcot-Marie-Tooth disease due to LITAF/SIMPLE mutations: a study of 18 patients. Eur J Neurol 24, 530–538. [DOI] [PubMed] [Google Scholar]
- Ho AK, Wagstaff JL, Manna PT, Wartosch L, Qamar S, Garman EF, Freund SM, and Roberts RC (2016). The topology, structure and PE interaction of LITAF underpin a Charcot-Marie-Tooth disease type 1C. BMC Biol 14, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori YS, Kodera S, Nagai Y, and Suzuki Y (2017). Fulminant Bacillus cereus septicaemia with multiple organ ischaemic/haemorrhagic complications in a patient undergoing chemotherapy for acute myelogenous leukaemia. BMJ Case Rep 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida R, Ueda K, Kitano T, Yamamoto T, Mizutani Y, Tsutsumi Y, Imoto K, and Yamamori Y (2019). Fatal community-acquired Bacillus cereus pneumonia in an immunocompetent adult man: a case report. BMC Infect Dis 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Chin LS, and Li L (2012). Charcot-Marie-Tooth disease-linked protein SIMPLE functions with the ESCRT machinery in endosomal trafficking. The Journal of cell biology 199, 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Olzmann JA, Chin LS, and Li L (2011). Mutations associated with CharcotMarie-Tooth disease cause SIMPLE protein mislocalization and degradation by the proteasome and aggresome-autophagy pathways. J Cell Sci 124, 3319–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Crown D, Miller-Randolph S, Moayeri M, Wang H, Hu H, Morley T, and Leppla SH (2009). Capillary morphogenesis protein-2 is the major receptor mediating lethality of anthrax toxin in vivo. Proc Natl Acad Sci U S A 106, 12424–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, and Leppla SH (2003a). Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J Biol Chem 278, 5227–5234. [DOI] [PubMed] [Google Scholar]
- Liu S, and Leppla SH (2003b). Retroviral insertional mutagenesis identifies a small protein required for synthesis of diphthamide, the target of bacterial ADP-ribosylating toxins. Mol Cell 12, 603–613. [DOI] [PubMed] [Google Scholar]
- Liu S, Miller-Randolph S, Crown D, Moayeri M, Sastalla I, Okugawa S, and Leppla SH (2010). Anthrax toxin targeting of myeloid cells through the CMG2 receptor Is essential for establishment of Bacillus anthracis infections in mice. Cell Host Microbe 8, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang Y, Hoover B, and Leppla SH (2012). The receptors that mediate the direct lethality of anthrax toxin. Toxins (Basel) 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang Y, Moayeri M, Liu J, Crown D, Fattah RJ, Wein AN, Yu ZX, Finkel T, and Leppla SH (2013). Key tissue targets responsible for anthrax-toxin-induced lethality. Nature 501, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, von Rhein C, Bauer S, Huttinger C, and Goebel W (2004). Molecular analysis of cytolysin A (ClyA) in pathogenic Escherichia coli strains. J Bacteriol 186, 5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madegowda M, Eswaramoorthy S, Burley SK, and Swaminathan S (2008). X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus. Proteins 71, 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur A, Feng S, Hayward JA, Ngo C, Fox D, Atmosukarto II, Price JD, Schauer K, Martlbauer E, Robertson AAB, et al. (2019). A multicomponent toxin from Bacillus cereus incites inflammation and shapes host outcome via the NLRP3 inflammasome. Nat Microbiol 4, 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill JC, You J, Constable C, Leeman SE, and Amar S (2011). Whole-body deletion of LPS-induced TNF-alpha factor (LITAF) markedly improves experimental endotoxic shock and inflammatory arthritis. Proceedings of the National Academy of Sciences of the United States of America 108, 21247–21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A, et al. (1991). Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 83, 757–766. [DOI] [PubMed] [Google Scholar]
- Mueller M, Grauschopf U, Maier T, Glockshuber R, and Ban N (2009). The structure of a cytolytic alpha-helical toxin pore reveals its assembly mechanism. Nature 459, 726–730. [DOI] [PubMed] [Google Scholar]
- Myokai F, Takashiba S, Lebo R, and Amar S (1999). A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: molecular cloning, sequencing, characterization, and chromosomal assignment. Proceedings of the National Academy of Sciences of the United States of America 96, 4518–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman ZL, Crown D, Leppla SH, and Moayeri M (2010). Anthrax lethal toxin activates the inflammasome in sensitive rat macrophages. BiochemBiophysResCommun 398, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscarsson J, Westermark M, Beutin L, and Uhlin BE (2002). The bacteriophage-associated ehly1 and ehly2 determinants from Escherichia coli O26:H- strains do not encode enterohemolysins per se but cause release of the ClyA cytolysin. Int J Med Microbiol 291, 625–631. [DOI] [PubMed] [Google Scholar]
- Qin W, Wunderley L, Barrett AL, High S, and Woodman PG (2016). The Charcot Marie Tooth disease protein LITAF is a zinc-binding monotopic membrane protein. Biochem J 473, 3965–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LE, and Hodivala-Dilke KM (2006). Primary mouse endothelial cell culture for assays of angiogenesis. Methods Mol Med 120, 503–509. [DOI] [PubMed] [Google Scholar]
- Saleh M, Al Nakib M, Doloy A, Jacqmin S, Ghiglione S, Verroust N, Poyart C, and Ozier Y (2012). Bacillus cereus, an unusual cause of fulminant liver failure: diagnosis may prevent liver transplantation. J Med Microbiol 61, 743–745. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastalla I, Fattah R, Coppage N, Nandy P, Crown D, Pomerantsev AP, and Leppla SH (2013). The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One 8, e76955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeni JL, and Wong AC (2005). Bacillus cereus food poisoning and its toxins. J Food Prot 68, 636–648. [DOI] [PubMed] [Google Scholar]
- Scobie HM, Rainey GJ, Bradley KA, and Young JA (2003). Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A 100, 5170–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH (2006). The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 6, 813–823. [DOI] [PubMed] [Google Scholar]
- Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, and Gouaux JE (1996). Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866. [DOI] [PubMed] [Google Scholar]
- Stenfors Arnesen LP, Fagerlund A, and Granum PE (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS MicrobiolRev 32, 579–606. [DOI] [PubMed] [Google Scholar]
- Street VA, Bennett CL, Goldy JD, Shirk AJ, Kleopa KA, Tempel BL, Lipe HP, Scherer SS, Bird TD, and Chance PF (2003). Mutation of a putative protein degradation gene LITAF/SIMPLE in Charcot-Marie-Tooth disease 1C. Neurology 60, 22–26. [DOI] [PubMed] [Google Scholar]
- Tang X, Metzger D, Leeman S, and Amar S (2006). LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: Evidence for LITAF-dependent LPS signaling pathways. Proceedings of the National Academy of Sciences of the United States of America 103, 13777–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viel-Theriault I, Saban J, Lewis A, Bariciak E, and Grynspan D (2019). A Case of Fulminant Bacillus cereus Lung Necrosis in a Preterm Neonate. Pediatr Dev Pathol, 1093526619825895. [DOI] [PubMed] [Google Scholar]
- von Rhein C, Bauer S, Lopez Sanjurjo EJ, Benz R, Goebel W, and Ludwig A (2009). ClyA cytolysin from Salmonella: distribution within the genus, regulation of expression by SlyA, and pore-forming characteristics. Int J Med Microbiol 299, 21–35. [DOI] [PubMed] [Google Scholar]
- Wallace AJ, Stillman TJ, Atkins A, Jamieson SJ, Bullough PA, Green J, and Artymiuk PJ (2000). E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100, 265–276. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, and Jaenisch R (2013). One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Zeng YX, Liang JY, Zhang L, Zhao XB, Pan JH, and Xiao RZ (2018). Rapidly Progressive, Fata Infection Caused by Bacillus Cereus in a Patient with Acute Lymphoblastic Leukemia. Clin Lab 64, 1761–1764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SI Video. Rapidly killing of CHO cells by HBL. CHO cells were incubated with HBL (10 nM) for up to 8 min. Related to Figure 1.
Table S1. Illumina deep sequencing counts of sgRNAs in RAW264.7(Cas9) cells infected with the CRISPR lentiviral pooled library after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
Table S2. Illumina deep sequencing counts of sgRNAs in human HT1080(Cas9) cells infected with the human CRISPR lentiviral pooled library-A after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
Table S3. Illumina deep sequencing counts of sgRNAs in human HT1080(Cas9) cells infected with the human CRISPR lentiviral pooled library-B after HBL selection (only sgRNAs with LFC ≥ 3 listed). Related to Figure 3.
Data Availability Statement
The published article includes all data generated or analyzed during this study.


