SUMMARY
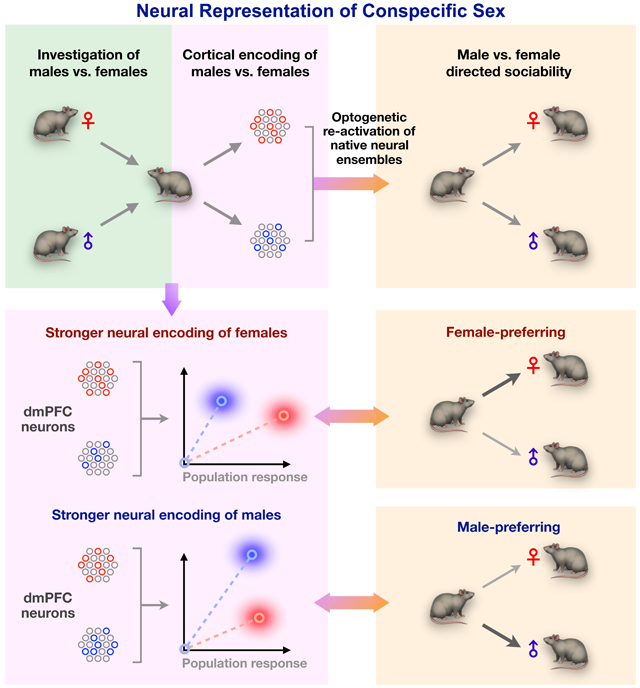
A central question related to virtually all social decisions is how animals integrate sex-specific cues from conspecifics. Using microendoscopic calcium imaging in mice, we find that sex information is represented in the dorsal medial prefrontal cortex (dmPFC) across excitatory and inhibitory neurons. These cells form a distributed code that differentiates the sex of conspecifics and is strengthened with social experience. While males and females both represent sex in the dmPFC, male mice show stronger encoding of female cues, and the relative strength of these sex representations predicts sex preference behavior. Using activity-dependent optogenetic manipulations of natively active ensembles, we further show that these specific representations modulate preference behavior toward males and females. Together, these results define a functional role for native representations of sex in shaping social behavior, and reveal a neural mechanism underlying male- vs. female-directed sociality.
Graphical Abstract
INTRODUCTION
In order to navigate the social world, animals must integrate environmental and social cues from conspecifics to make decisions that secure their survival, health, and reproductive vitality. Representation and discrimination of conspecific sex—the recognition of another as male or female—is critical for social interaction, including behavioral decisions to preferentially engage with same- vs. opposite-sex conspecifics (Chen and Hong, 2018; Li and Dulac, 2018). Previous work in mice has revealed that encoding of conspecific sex depends on chemosensory signaling and involves subcortical nodes including the amygdala and the hypothalamus (Chen and Hong, 2018; Li et al., 2017; Remedios et al., 2017), leading to a canonical view that processing of sex information depends primarily on subcortical circuits. However, how conspecific sex is represented beyond subcortical processing is poorly understood. More importantly, despite advances in identifying specific subcortical areas that encode conspecific sex, it is not clear whether representations at the level of natively active neural ensembles actually influence animal behavior. As neural representations of male and female may not precisely map onto molecularly separable cell types (Kim et al., 2019; Moffitt et al., 2018), direct manipulation of natively active neurons is necessary to understand how sex representations affect behavior. To date, this has not been explored and presents a critical gap in understanding how the brain shapes social decisions.
Recent work has illuminated the role of the medial prefrontal cortex (mPFC) in encoding general social information and in shaping social behavior such as dominance and sexual behavior (Kingsbury et al., 2019; Liang et al., 2018; Murugan et al., 2017; Nakajima et al., 2014; Wang et al., 2011). As sex recognition is deeply embedded in these behavioral processes, such findings raise the question of whether cortical circuits play a role in sex recognition and the control of sex-typical behaviors that drive social interaction. In this study, we employ in vivo microendoscopic calcium imaging in freely behaving mice to explore the involvement of dorsal mPFC (dmPFC) ensembles in the encoding of conspecific sex and control of social behavior. We find that conspecific sex is represented in the dmPFC in a distributed population code that recruits both excitatory and inhibitory subpopulations. Male but not female mice show a bias toward encoding of female cues, and this relative strength of sex representation in males predicts their sex preference behavior. Finally, activation of these same ensembles modulates their preference towards male vs. female, defining a functional role for native sex representations in shaping behavior.
RESULTS
dmPFC neurons encode conspecific sex
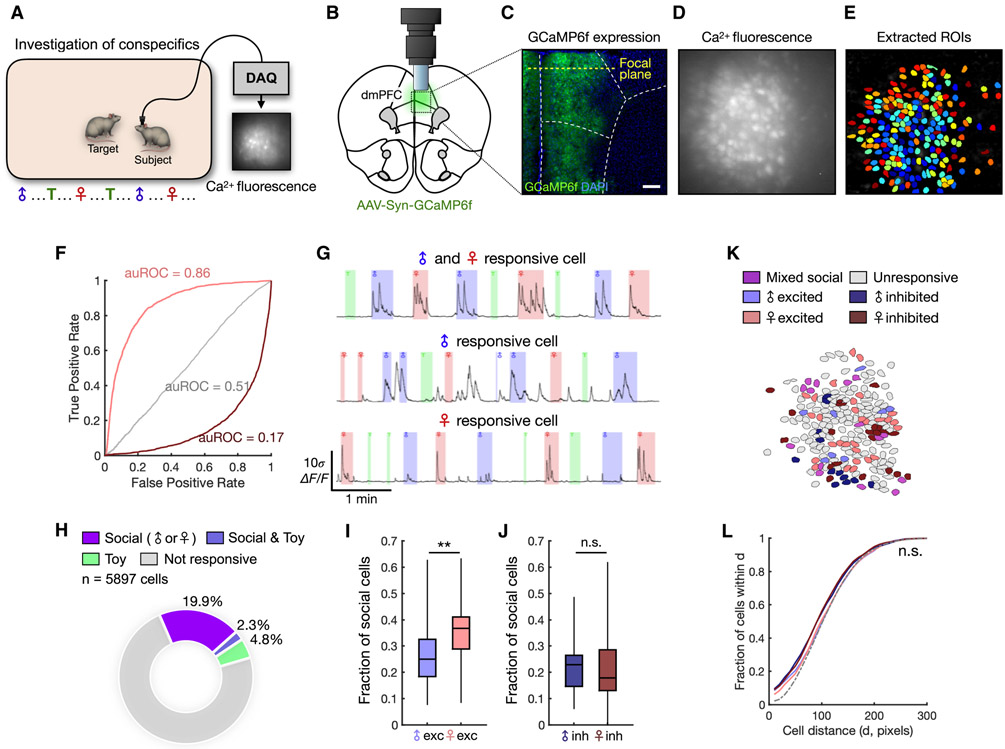
In order to first explore whether dmPFC neurons encode male- and female-specific information, we performed in vivo microendoscopic calcium imaging (Ghosh et al., 2011) of dmPFC neurons during natural investigation of male and female conspecifics (Figure 1A). To optically record from dmPFC neurons, we injected an adeno-associated virus (AAV) expressing the fluorescent calcium indicator GCaMP6f (Chen et al., 2013) and implanted a gradient refractive index (GRIN) lens above the dmPFC (Figure 1B). Expression of GCaMP6f and placement of the lens were confirmed histologically (Figure 1C). During imaging, we presented each subject animal with 8 novel males and 8 novel females interleaved with novel objects. Each stimulus animal was presented for 20-30 s, during which the subject animal was free to investigate. After imaging sessions, activity signals associated with single cells were extracted using independent component analysis as previously described (Kingsbury et al., 2019; Mukamel et al., 2009) and are reported throughout as relative change in fluorescence (ΔF/F) (Figures 1D-E). We recorded a total of 5897 dmPFC neurons across 28 animals, including both males and females.
Figure 1. dmPFC neurons encode conspecific sex during natural interaction.
(A) Schematic of social investigation assay. Eight novel male and eight novel female conspecifics are presented interleaved with a novel object, and the subject animal is allowed to freely investigate.
(B) Illustration of the microendoscope placed above the dmPFC.
(C) Example image showing expression of GCaMP6f in neuronal cell bodies. The location of the focal plane is estimated based on the estimated working distance of the lens.
(D) Imaging field of view showing raw calcium fluorescence from one animal (max projection).
(E) ROIs corresponding to single neurons extracted from the field of view in (D).
(F) Receiver operating characteristic (ROC) curves computed from three example neurons that are female-excited (auROC = 0.86), female-suppressed (auROC = 0.17), or not responsive to female (auROC = 0.51).
(G) Example calcium traces from social-mixed (top), male (middle), and female (bottom) neurons.
(H) Distribution of social cells (responding to either male or female interaction) and toy-responsive cells from all recorded animals.
(I) Fractions of male- and female-excited cells among socially responsive dmPFC neurons recorded from males (p = 0.0071). Mixed cells responding to more than one category constituted 6.29 ± 1.18% (mean ± SEM) of socially responsive cells. We also observed a higher fraction of female-excited cells when normalizing the sampling of male and female investigation (see Methods, Figure S2A-B).
(J) Fractions of male- and female-inhibited neurons recorded from male animals (p = 0.59).
(K) Example field of view showing spatial locations of male and female cells.
(L) Cumulative histogram showing the mean fraction of cells within a given pairwise distance (x-axis), compared between subsets of functionally defined neurons.
In (H) n = 28 animals (including males and females); (I-J) Mann-Whitney U test, n = 23 animals; (L) two-sample Kolmogorov-Smirnov test, n = 23 animals. **p < 0.01, n.s. = not significant. Scale bar = 200 μm.
In order to determine whether single neurons selectively encode sex-specific information, we computed a receiver operating characteristic (ROC) curve for each neuron/stimulus (male/female/toy) relationship, which quantifies its stimulus detection strength over a range of binary thresholds (Figure 1F). Using this approach, we identified a substantial fraction (22%) of dmPFC neurons that showed significant responses during investigation of social stimuli (Figure 1G, H). Many neurons were selectively tuned to either male or female investigation (Figures 1I-J, S1C-D). Interestingly, in male animals, a larger fraction of neurons showed increased activity during interaction with female compared to male conspecifics (Figure 1I), suggesting a bias toward encoding of female cues. However, when we examined this in female mice, we found that comparable fractions of neurons responded to male and female conspecifics (Figure S1C-D), suggesting that stronger encoding of female cues is specific to the male dmPFC.
This intriguing observation raises the questions of how female and male conspecifics are differentially encoded in the male brain and how this representation is linked to behavior. Although single cells encoding sex-specific information preferentially responded to male or female cues, male and female cells had overall similar response amplitude and reliability (Figure S1E-G), suggesting that their basic response properties are not significantly different. Moreover, male- and female-encoding cells were not selectively tuned to investigation of sex-specific odor cues (Figure S1I), suggesting that sex representation is unlikely solely due to olfactory inputs. Lastly, to examine whether these neurons were intermingled or separately clustered within the dmPFC, we analyzed the spatial distributions of sex-encoding cells. We found no differences in the distributions of sex-encoding compared to non-encoding cells (Figures 1K-L), indicating that sex-encoding cells are not spatially organized. Collectively, these results show that single neurons in the dmPFC encode the multisensory variable of sex, and in males, form a stronger representation of female compared to male cues.
Population representations in the dmPFC uniquely encode conspecific sex
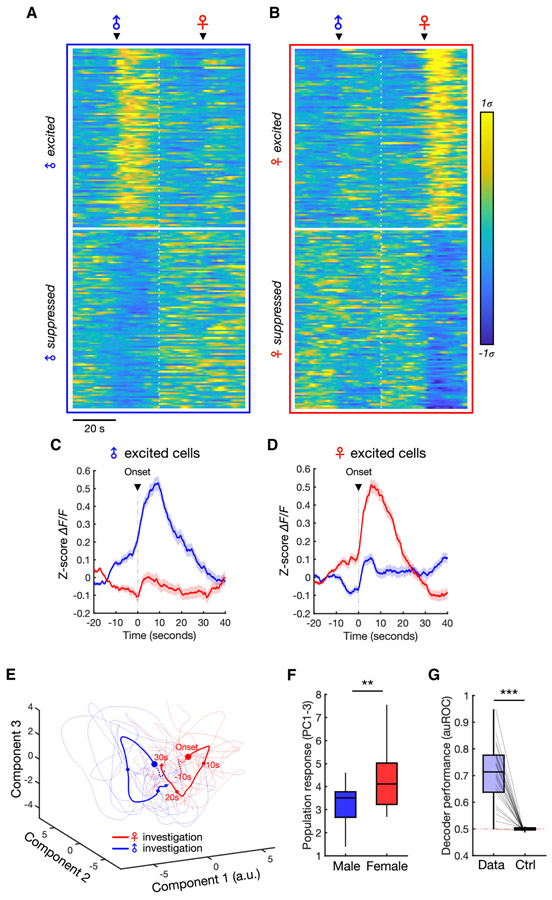
Single neurons in the dmPFC formed largely non-overlapping subpopulations that responded specifically to male or female investigation (Figures 2A-D, S1K), suggesting that male and female interaction may elicit unique and separable trajectories of population activity. In order to explore this, we projected single-trial population dynamics during investigation bouts onto principal components. This revealed a clear separation of single-trial responses associated with male vs. female interaction (Figure 2E-F), indicating that the population may discriminate sex information. Still, while this visualization suggests separable patterns of activity, sex representations may not be temporally stable or general enough to decode sex across time and across novel animals. In order to test whether sex representations are stable and general, we constructed linear discriminant decoders to predict the sex identity of novel animals using population activity. The cross-validated performance of these decoders was well above chance levels (Figure 2G), indicating that unique and stable response patterns in the dmPFC encode the sex of conspecifics. Importantly, the fact that sex could be decoded across novel animals (Figure 2G) suggests that different animals of the same sex evoked consistent activity patterns, indicating a general representation of sex itself. Consistent with male mice having more female-than male-responsive cells (Figure 1I), we also found that male animals showed higher amplitude population responses during female investigation compared to male investigation (Figure 2F), whereas females did not show this difference (Figure S1H). Together, these results suggest that dmPFC neurons encode sex stably at the population level, and that in males, this distributed representation exhibits a bias toward encoding of female cues.
Figure 2. Population representations of male and female.
(A-B) Average responses of male (A) and female (B) neurons centered around investigation onset. The top 100 neurons, sorted using rank-ordered auROC values, are shown for each cell type (showing excited or inhibited responses).
(C-D) Average responses of all male-excited (C) and female-excited (D) neurons during male and female investigation (mean ± SEM).
(E) Principal component projection of population dynamics during individual male and female investigation bouts (thin lines) and average responses (thick lines) from one example animal. Dots indicate 10-second time intervals and dotted lines indicate time before investigation onset.
(F) Comparison of the population response amplitude during male vs. female interaction in male animals. Population responses (projected to PC1-3) are measured over the first 10 seconds of interaction and averaged across bouts within each animal (Mann-Whitney U test, p = 0.0095, n = 23 animals).
(G) Performance of Fisher’s linear discriminant (FLD) decoders trained to classify male vs. female investigation using population dynamics. Cross-validated auROC is compared with performance of null models trained and tested using time-permuted calcium traces (Wilcoxon signed-rank test, p = 2.7e-5, n = 23 animals).
***p < 0.001, **p < 0.001
See also Figure S1.
Encoding of conspecific sex across distinct subpopulations
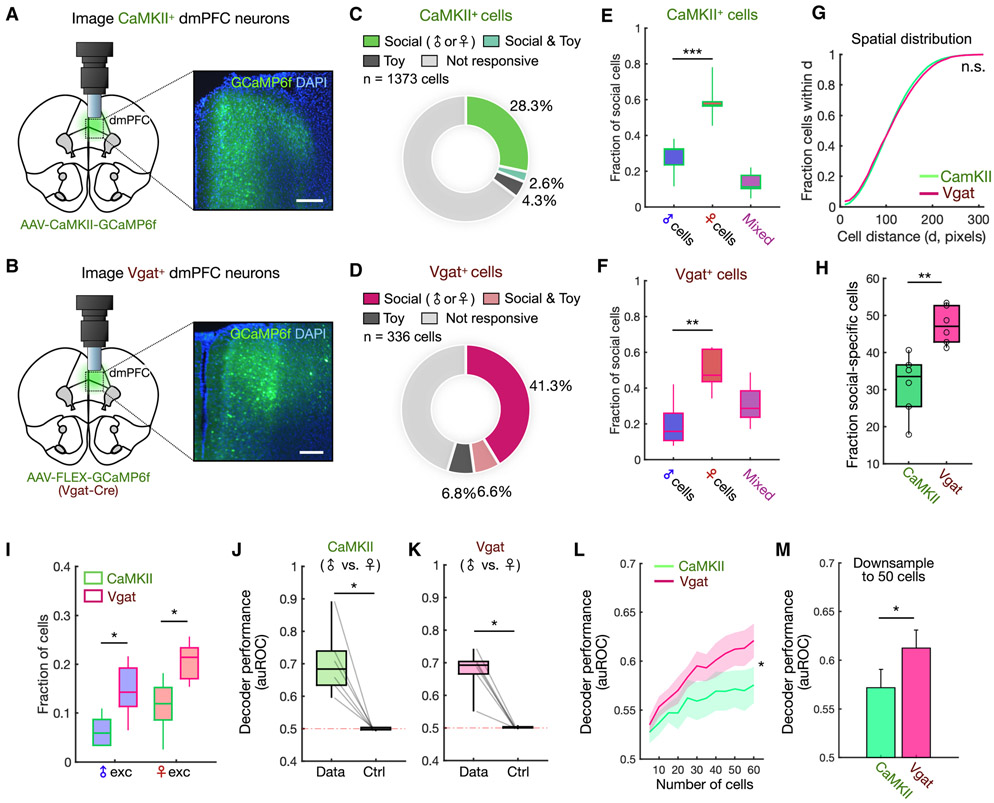
Next, we sought to gain deeper insight into how sex representations are distributed across different dmPFC neuron cell types. While encoding of conspecific sex may be distributed broadly, another possibility is that specific neuronal populations may preferentially encode sex-specific information. To this end, we explored how excitatory and inhibitory subpopulations of dmPFC neurons contribute to the encoding of conspecific sex. We injected an AAV expressing GCaMP6f in either CaMKII+ (predominantly excitatory, Figure 3A) or Vgat+ (GABAergic, Figure 3B) neurons and recorded neural activity during the social investigation assay. Overall, we recorded from a total of 1373 CaMKII+ neurons and 336 Vgat+ neurons.
Figure 3. dmPFC encoding of sex is distributed across distinct neuronal subpopulations.
(A-B) Schematic showing lens and microscope placement above dmPFC (left) and expression of GCaMP6f in CaMKII+ or Vgat+ neuron cell bodies (right). CaMKII-GCaMP labeled 25.4 ± 2.6% of dmPFC cells, and Vgat-GCaMP labeled 9.6 ± 0.3% of dmPFC cells (mean ± SEM).
(C-D) Distribution of social (male and/or female responsive) and toy-responsive neurons among CaMKII+ (n = 1373) or Vgat+ (n = 336) cells.
(E-F) Comparison of fractions of male and female cells within the CaMKII+ or Vgat+ population (E, p = 0.0001; F, p = 0.0015). We also observed a higher fraction of female cells when normalizing the sampling of male and female investigation (see Methods, Figures S2C-D).
(G) Cumulative histogram showing the mean fraction of cells within a given pairwise distance (x-axis) for CaMKII+ and Vgat+ neurons.
(H) Comparison of the percentage of social cells identified in CaMKII+ vs. Vgat+ populations (p = 0.0043).
(I) Fractions of male- and female-excited cells within the CaMKII+ and Vgat+ populations (male-excited, p = 0.031; female-excited, p = 0.012).
(J-K) Performance of Fisher’s linear discriminant (FLD) decoders trained to classify male vs. female investigation using the CaMKII+ or Vgat+ population. Cross-validated auROC is compared with performance of null models trained using time-permuted calcium traces (J, p = 0.0312; K, p = 0.0312).
(L) Decoder performance in CaMKII+ and Vgat+ cell populations as a function of ensemble size (mean ± SEM, p = 0.040 number/cell type interaction).
(M) Decoder performance of CaMKII+ vs. Vgat+ cell populations after down-sampling ensembles to 50 neurons (mean ± SEM, p = 0.0411).
In (E-F), one-way ANOVA followed by Tukey’s range test; (G) two-sample Kolmogorov-Smirnov test; (H), (M) Mann-Whitney U test; (I) Tukey’s range test; (J-K) Wilcoxon signed-rank test; (L) two-way repeated measures ANOVA. n = 6 animals for each group (CaMKII+ and Vgat+).
***p < 0.001, **p < 0.01, *p < 0.05, n.s. = not significant. Scale bar = 200 μm.
See also Figure S2.
Analysis of single neuron responses using ROC curves showed that both populations contained a substantial fraction of cells encoding social cues (Figures 3C-D). Consistent with our results above, we found that there were more female- than male-encoding cells in both subpopulations in males (Figures 3E-F, S2C-D), indicating that the bias in encoding of female cues is preserved across both excitatory and inhibitory cell types. We also found that social cells in each subpopulation were similarly spatially intermixed (Figure 3G), indicating that sex information is distributed widely and not clustered within either subtype. Interestingly however, despite similarities in spatial distributions and encoding of social information, a higher fraction of Vgat+ compared to CaMKII+ neurons were specifically responsive to conspecific sex (Figure 3H), suggesting that sex information may be more enriched in GABAergic neurons. This enrichment of sex encoding in GABAergic neurons was preserved across different response categories, and significant among male- and female-excited cells (Figures 3I, S2E).
Lastly, we analyzed how information at the single neuron level in CaMKII+ and Vgat+ ensembles could be read out at the population level to discriminate conspecific sex. To explore this, we constructed linear decoders and found that male vs. female investigation could be decoded from both CaMKII+ and Vgat+ ensembles with above chance accuracy (Figures 3J-K). However, when we examined the dependence of decoder performance on population size, we observed that GABAergic neurons, when compared using the same number of neurons, were consistently better at discriminating conspecific sex (Figures 3L-M). This suggests again that sex information is more strongly encoded within inhibitory neurons. Taken together, these data demonstrate that while conspecific sex is represented more strongly in inhibitory neurons, population-level encoding of conspecific sex recruits both subpopulations of cells, which both exhibit a stronger representation of female compared to male cues.
dmPFC neurons encode conspecific sex across contexts
In order to guide behavior in an adaptive way, one may expect representations of sex to generalize across different environments as well as animal identities. To determine whether dmPFC encoding of conspecific sex is general across different environments, we followed the home cage social investigation assay with a two-chamber assay in which animals freely explored novel male and female conspecifics placed in opposing corners in a two-chamber environment (Figure 4A). As mPFC neurons may also encode spatial information (Murugan et al., 2017), we performed two sessions alternating the locations of male and female stimuli in the arena to dissociate encoding of conspecific sex from location (Figure 4A). We used DeepLabCut (Mathis et al., 2018) to automatically track the location and pose of subject animals—this was then used to identify social investigation events in an unbiased manner (see Methods). We first confirmed that single dmPFC neurons also encode sex information in the two-chamber context. Individual cells responded to male or female stimuli across sessions with alternating locations, indicating that they encode conspecific sex irrespective of spatial conjunction (Figure 4B).
Figure 4. Encoding of conspecific sex across contexts and experience-dependent changes.
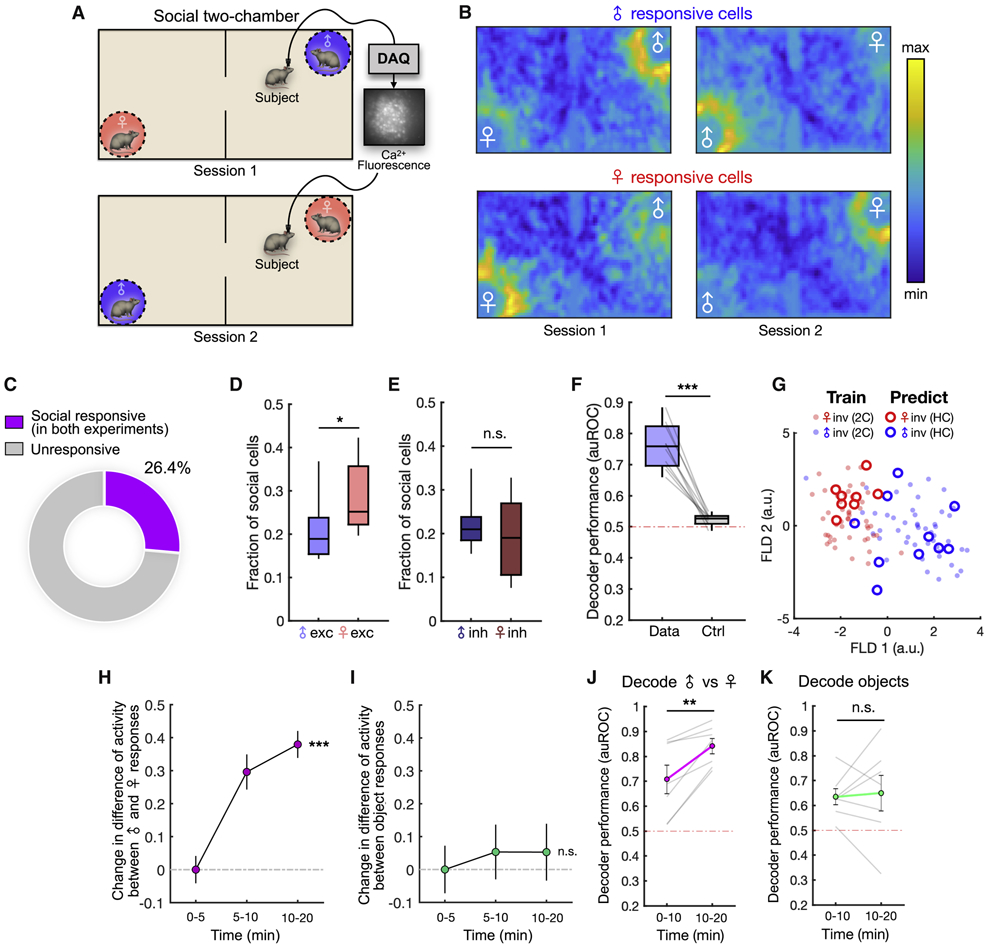
(A) Illustration of the two-chamber social exploration task. Male and female locations are switched across two sequential sessions.
(B) Neural activity heat maps showing mean responses of male-excited (top) and female-excited (bottom) neurons during the two-chamber assay.
(C) Distribution of social (male or female responsive) dmPFC neurons that are significantly responsive across both the home cage social investigation and two-chamber assays.
(D) Fractions of male- and female-excited cells that consistently encode sex information across both the home cage and two-chamber sessions (p = 0.031).
(E) Fractions of male- and female-suppressed cells that encode sex information across sessions (p = 0.450).
(F) Performance of Fisher’s linear discriminant (FLD) decoders trained using data from both the social investigation and two-chamber assays and tested on data from both. Cross-validated auROC is compared to control decoders constructed using time-permuted calcium traces (p = 0.002).
(G) Projection of mean population responses associated with male and female investigation bouts from the home cage (circles) and two-chamber (solid dots) onto FLD components computed from two-chamber data. The discrimination axis (FLD 1) generalizes to events in the home cage. Performance of cross session decoders using sex-selective cells are shown in Figures S3A-B.
(H) Change in activity difference of male and female cells evoked by male vs. female investigation during different time epochs in the two-chamber (mean ± SEM, p = 0.0001). Stimulus-evoked activity in each epoch is normalized to overall population activity.
(I) Change in activity difference of object-excited cells during investigation of one object vs. the other object (mean ± SEM, p = 0.10). Stimulus-evoked activity in each epoch is normalized to overall population activity.
(J) Performance of decoders to discriminate male vs. female investigation using population dynamics during the first and second half of the two-chamber session (animal locations not switched, p = 0.016).
(K) Performance of decoders to discriminate investigation of different objects using population dynamics during the first and second half of non-social two-chamber experiments (p = 0.69).
In (D-E), Mann-Whitney U test, n = 10 animals; (F) Wilcoxon signed-rank test, n = 10 animals; (H-I) One-way ANOVA, n = 291 cells (H), n = 115 cells (I); (J-K) Wilcoxon signed-rank test, n = 7 animals.
***p < 0.001, **p < 0.01, *p < 0.05, n.s. = not significant.
See also Figure S3.
By precisely mapping neuronal cell bodies across the home cage and two-chamber assays (Figure S2F, see Methods), we found that 26% of the recorded cells responded selectively across both assays (Figures 4C-E), suggesting that single cells in the dmPFC are able to encode conspecific sex consistently across different contexts. To explore whether the population representation within each animal could generalize across contexts, we trained linear decoders to discriminate male vs. female investigation across both sessions. Consistent with our observations of shared encoding at single neuron levels, population decoders could predict sex identity across both contexts (Figure 4F). Further, male and female investigation events were generally well separated by a discrimination axis defined in only one context (Figure 4G, S3A-B). Together, these data suggest that dmPFC representations of male and female cues are general across distinct contexts.
Representation of conspecific sex is strengthened by short-term social experience
Previous work has suggested that in subcortical regions, neural representations of conspecific sex are sharpened over the course of days or weeks (Li et al., 2017; Remedios et al., 2017). Even within the course of one interaction episode, accumulation of social cues may dynamically shape subsequent interaction. We therefore explored whether and how neural representations of sex may change over a faster time scale during a single exploration episode in the two-chamber. Interestingly, we found that the mean activity of sex-encoding cells during investigation of male and female conspecifics changed over the course of the session (Figures S3C-D) such that the difference in response strength between the two stimuli grew larger (Figure 4H). This was not true for cells that responded to non-social objects (Figure 4I), suggesting that single cell discriminability of sex is specifically sharpened over time. Indeed, population decoders trained and tested during the first and second halves of the session showed significantly higher performance in the second half (Figure 4J), but not for decoding of two objects (Figure 4K), suggesting an increase in population-level sex discriminability that depends on social experience. These data suggest that although representations of sex are stable and general, they can also be sharpened by social experience.
In order to further analyze how changes in responses to male and female cues affect neural tuning, we constructed generalized linear models (GLM) to model single neuron activity as a function of behavioral variables, including social investigation (Figure S3F). Analysis of the coefficients fit to single neuron GLMs showed a significant increase in tuning to male and female investigation for male and female cells, respectively, over time (Figures S3G-I). In contrast, cells that responded to objects did not show an increase in tuning (Figures S3J-K), further indicating that encoding of conspecific sex, but not non-social stimuli, is sharpened with social experience. While previous work has found that subcortical encoding of social information can be strengthened over days (Li et al., 2017; Remedios et al., 2017), these results suggest that cortical sex representations may be more amenable to short-term sharpening.
Activity of sex-encoding cells predicts behavioral sex preference
Internal representations of sex-specific social cues are thought to play an important role in driving behavior directed toward male or female conspecifics (Chen and Hong, 2018; Li and Dulac, 2018), raising the possibility that sex representations in the dmPFC may play a role in controlling male- or female-directed sociality. Based on this, we next explored the relationship between sex encoding and sex preference behavior in individual animals.
At the behavioral level, we observed a modest group-level bias in males toward female investigation (Figure S4A), consistent with previous literature (Beny-Shefer et al., 2017; Yao et al., 2017). Interestingly, we found earlier that dmPFC neurons encoded female more strongly in distinct cell types (Figures 1I, 2F, 3E, and 3F), and across different contexts (Figure 4D), suggesting that this sex encoding bias may be linked to female preference behavior in males. To rule out the possibility that bias toward female encoding may be due to unequal sampling of investigation events and resulting differences in statistical power, we controlled this analysis by normalizing the representation of male and female investigation (see Methods). We confirmed that a larger fraction of dmPFC neurons encoded females even when behavioral events were precisely equalized (Figure S2A-D).
However, beyond this group-level female preference, individual males displayed a wide range of preference scores, with some displaying a preference for males (Figure S4A). When social stimuli were absent, the variability in behavioral preference in the two-chamber assay was significantly reduced (Figure S4C), indicating that individual preferences for male or female are driven by the presence of conspecifics and not simply by random movement. Moreover, this individual behavioral preference is relatively stable—sex preference of individual animals was significantly correlated within sessions and across consecutive days (Figures S4D-E), indicating that sex preference in individuals reflects a consistent behavioral state.
Interestingly, when we compared animals that displayed a stronger preference for female vs. for male interaction, female-preferring animals had significantly more female-responsive neurons than male-preferring animals (Figure S4G-H), suggesting that the relative strength of male vs. female representations is linked to behavioral preference. In light of this, we next explored the relationship between population-level encoding of sex and preference behavior. To analyze the strength of male and female representations, we measured the population response amplitude during each male/female investigation event as the Mahalanobis distance between the stimulus-evoked population vector and baseline activity. While female-preferring animals showed a stronger population response during female interaction (Figures 5A-B), male-preferring animals showed a stronger response during male interaction (Figures 5C-D). Taken together, these data suggest that a preference state is encoded in the relative activity of neural sex representations that predicts sex preference behavior (Figure 5E).
Figure 5. dmPFC neurons encode a sex preference state.
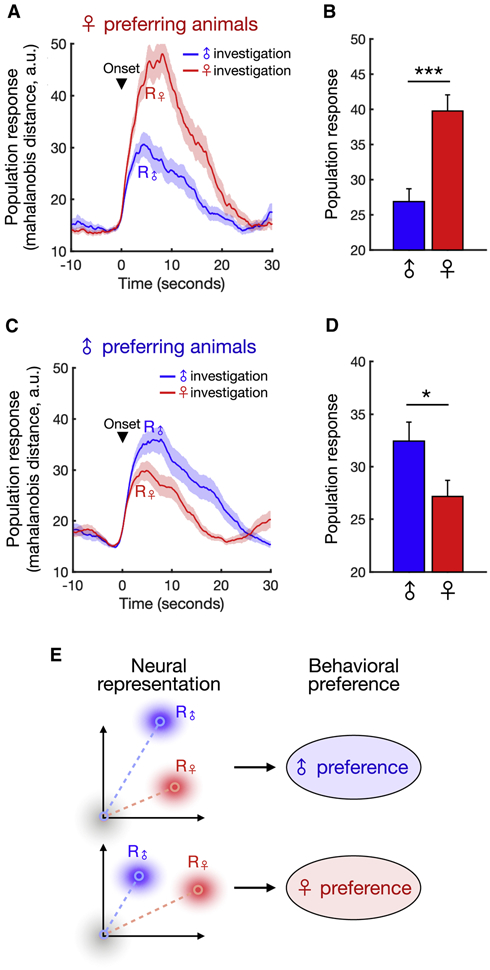
(A, C) Average population response, measured as the Mahalanobis distance between trial response vectors and baseline (see Methods) for male- and female-preferring animals evoked during male or female interaction (mean ± SEM).
(B, D) Average responses in (A) and (C) quantified over the first 10 seconds of interaction (mean ± SEM, Mann-Whitney U test, p = 1.26e-4, n = 46 male bouts and n = 42 female bouts (b), p = 0.0195, n = 60 male bouts and n = 66 female bouts (D)).
(E) Schematic showing how the relative strength of sex-specific responses predicts individual sex preference.
***p<0.001, *p < 0.05.
See also Figure S4.
Male and female neural representations modulate sex preference behavior
The strong association between the neural representation of conspecific sex and behavioral preference raises the possibility that ensemble representations may causally influence preference behavior. A typical way to examine causal influence of neural activity is to activate an anatomically or genetically defined subset of neurons (Lerner et al., 2016; Luo et al., 2008, 2018). However, accumulating evidence points to an increasingly common picture in which one neuronal subpopulation may participate in multiple representations, and one representation may recruit multiple subpopulations (Kim et al., 2019; Moffitt et al., 2018; Wu et al., 2017). Indeed, we found that sex representations in the dmPFC are distributed across excitatory and inhibitory subpopulations (Figure 3). This underscores the necessity of directly controlling neuronal populations defined by natural activity rather than by specific molecular markers.
Therefore, to test the functional role of sex representations in sex preference behavior, we employed an activity-dependent labeling strategy to express ChR2 in specific subsets of dmPFC neurons that were activated by male or female conspecifics (Figure 6A). We virally injected an AAV carrying E-SARE-CreER to express a Tamoxifen (4-TM)-inducible CreER driven by E-SARE, an activity-induced synthetic promoter that was previously shown to display greater activity-dependent induction compared to immediate early genes such as Fos or Arc (Kawashima et al., 2013; Kim et al., 2017). We also co-injected an AAV expressing Cre-dependent ChR2. This combination of AAVs allowed us to restrict ChR2 expression to the select neural population that was active at the time of 4-TM injection (Kim et al., 2017). Specific dmPFC neurons were labeled following natural exposure to either male or female conspecifics, and then optogenetically re-activated several weeks later in the two-chamber assay to measure sex preference. Histological analysis showed robust expression of ChR2 in animals exposed to social stimuli at the time of 4-TM induction, but not in animals lacking 4-TM injection (Figures S5A-B), confirming specific labeling of neurons that were active during social interaction.
Figure 6. Male and female cells modulate male vs. female interaction.
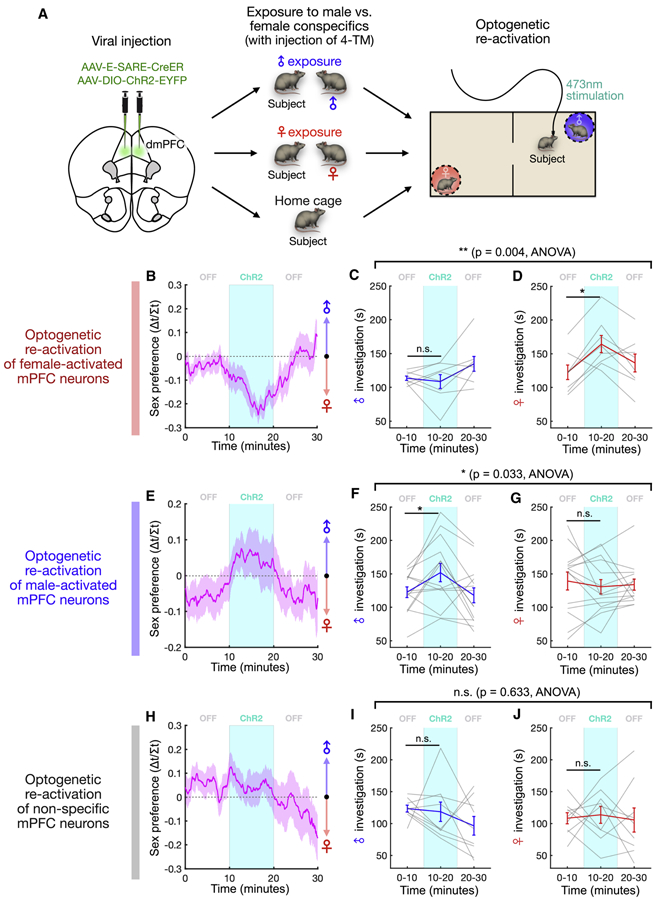
(A) Experimental paradigm used to test the causal role of male and female representations in sex preference behavior. After viral injection of E-SARE-CreER and Cre-dependent ChR2 constructs, animals interact with either male or female conspecifics followed by tamoxifen injection, and activity-defined cells are optically stimulated in the social two-chamber (see Methods).
(B, E, H) Time course of sex preference in the male/female two-chamber session (mean ± SEM). Optical stimulation of female cells induces a bias toward female interaction (B), stimulation of male cells induces a bias toward male interaction (E), and stimulation of non-specific cells induces no bias (H).
(C-D) Male or female interaction time (%) before, during, and after stimulation of female-activated neurons (mean ± SEM, p = 0.004, light/sex interaction in two-way repeated measures ANOVA; C, p = 0.84, D, p = 0.016, Wilcoxon signed-rank test; n = 8 animals).
(F-G) Male or female interaction time (%) before, during, and after stimulation of male-activated neurons (mean ± SEM, p = 0.033, light/sex interaction in two-way repeated measures ANOVA; F, p = 0.042, G, p = 0.50, Wilcoxon signed-rank test; n = 14 animals).
(I-J) Male or female interaction time (%) before, during, and after stimulation of non-specific neurons (mean ± SEM, p = 0.63, light/sex interaction in two-way repeated measures ANOVA; I, p = 0.36, J, p = 0.32, Wilcoxon signed-rank test; n = 9 animals).
***p<0.001, *p < 0.05, n.s. - not significant.
See also Figures S5 and S6.
In order to further validate the efficacy and specificity of this strategy, we examined the overlap between EYFP+ cells labeled by E-SARE following social or non-social exposure and cells expressing the immediate early genes Fos and Arc following a second exposure to males or females (Figure S5C). We found that male-induced E-SARE EYFP+ cells showed a substantially higher overlap with male-induced Fos/Arc+ cells, compared to that of EYFP+ cells induced by females or in home cage controls (Figure S5D). Conversely, female-induced E-SARE EYFP+ cells showed a substantially higher overlap with female-induced Fos/Arc+ cells, compared to that of EYFP+ cells induced by males or in home cage controls (Figure S5E). This suggests that the E-SARE labeling strategy captures specific ensembles of neurons that encode sex-specific cues across distinct social experiences.
We then optogenetically re-activated neurons that were naturally activated by exposure to females (Figure S5F-H). Stimulation of female cells resulted in an acute change in sex preference behavior toward female-directed interaction (Figure 6B, S5I). This was driven by a specific increase in the time spent investigating the female conspecific, as male investigation time remained unchanged (Figures 6C-D). In striking contrast, stimulation of male cells had the opposite effect on sex preference behavior (Figure 6E, S5J), resulting in an acute bias toward male interaction driven by a specific increase in male-directed investigation (Figures 6F-G). This suggests that sex preference behavior is modulated by specific subpopulations of neurons that are defined by their native activation in response to male or female stimuli. Indeed, optogenetic re-activation of neurons that were activated during a home cage exposure without male or female stimuli had no effect on preference behavior (Figures 6H-J, S5K).
Finally, we analyzed the axonal projection patterns of male- and female-activated dmPFC neurons (Figure S6). Both male and female cells showed broad projection patterns to several subcortical structures involved in social behavior and motivation. Although we observed a trend toward stronger nucleus accumbens and dorsal striatum projections in female cells (Figure S6B), there was no overall difference between the projection patterns of male and female cells. This suggests that their opposing effects on sex preference behavior are unlikely due to gross differences in their axonal projection patterns.
Together, these data demonstrate that two subsets of dmPFC neurons—which are separable subpopulations that are natively activated in response to sex-specific social cues—modulate interaction with male vs. female conspecifics. These results firmly establish a functional role for cortical representations of conspecific sex in sex preference behavior.
DISCUSSION
Using microendocsopic calcium imaging, we found that the dmPFC uses a distributed neural code, which recruits both excitatory and inhibitory subpopulations, to represent conspecific sex. While both male and female dmPFC encodes conspecific sex, an overall stronger encoding of female conspecifics is specifically observed in males but not females, and the relative strength of male vs. female representations predicts sex preference behavior. Further, using activity-dependent optogenetic manipulations of natively active ensembles, we found that cortical representations of conspecific sex modulate sex preference behavior in males. Together, these results demonstrate a functional role for neural representations of sex in shaping behavior and present a neural mechanism for cortical control of male- and female-directed sociality.
Cortical representation of conspecific sex
To navigate the social world, animals must be able to recognize conspecific sex, an essential feature that defines social relationships and shapes interaction between conspecifics. Previous studies have contributed to a classic view that processing of sex-specific social cues depends almost exclusively on subcortical circuits (Chen and Hong, 2018; Li et al., 2017; Remedios et al., 2017). In contrast, growing appreciation for the role of prefrontal circuits in social behavior has spurred an effort to explore native mPFC activity during social interaction (Kingsbury et al., 2019; Liang et al., 2018; Murugan et al., 2017). As mPFC circuits have been implicated in the control of social motivational states (Franklin et al., 2017), social dominance (Kingsbury et al., 2019; Wang et al., 2011), and sexual behavior (Nakajima et al., 2014), complex social information must be integrated by mPFC neurons for animals to make adaptive decisions. Still, the basic question of whether cortical neurons play a role in sex recognition has not been addressed. Here, we found that a substantial fraction of neurons in the dmPFC of both male and female mice respond preferentially to male vs. female interaction.
Recent work has shown that neurons in the mPFC respond similarly to both male and female odor cues (Levy et al., 2019). Our study shows that, during natural interaction with conspecifics, males and females are encoded in largely non-overlapping sets of neurons that form unique population representations. This suggests that natural social stimuli may drive neural responses differently, and perhaps more strongly, than simple odor cues. Furthermore, we found that in the brains of male but not female mice, prefrontal neurons exhibit a bias toward stronger encoding of female cues which is maintained at the population level. This representational dimorphism may reflect distinct mechanisms underlying sex encoding and preference behavior in males vs. females. Indeed, subcortical sex representations, which also show a bias toward encoding of female cues in males, are selectively regulated by oxytocin signaling in males but not females (Li et al., 2017; Yao et al., 2017). More generally, whether and how cortical sex representations differ from sex encoding in subcortical regions such as the amygdala and hypothalamus (Li et al., 2017; Remedios et al., 2017) also deserve future attention. While subcortical representations are shaped over the course of days with social experience, we found that neurons in the dmPFC sharpen their sex-specific tuning on a faster timescale of 10-20 minutes. This may suggest that cortical representations of sex are more flexible, and may be more amenable to conjunctive coding schemes that link social stimuli with contextual or spatial cues (Murugan et al., 2017).
We also leveraged the optical recording approach to investigate how different populations of dmPFC neurons contribute to sex representations. By imaging both excitatory (CaMKII+) and inhibitory (Vgat+) neurons, we found that both subpopulations encode male and female cues and discriminate conspecific sex. Interestingly, while sex information was distributed across both subpopulations, male and female cues were represented more strongly by GABAergic neurons, suggesting that sex-specific social information is enriched in inhibitory neurons. These results illustrate the neural heterogeneity of native sex representations, highlighting the limitations of defining functional circuitry based molecular or anatomical profiles.
Neural representation of behavioral sex preference
Despite our current understanding of how neurons encode sex cues in the brain, how native neural representations are related to social behavior, such as opposite-sex preference, remains poorly understood. Previous studies have looked into the group-level bias that animals display toward interaction with opposite-sex conspecifics (Beny-Shefer et al., 2017; McHenry et al., 2017; Yao et al., 2017). However, this has left open the question of how male- vs. female-directed social interaction is actually controlled in individual animals. Here, we show that besides the group-level preference for females, individual males naturally exhibit distinct preferences for either male or female interaction. At the neural level, we observed a stronger representation of female in male animals across multiple contexts, and across both excitatory and inhibitory subpopulations, linking neural representations of sex to a group-level behavioral preference toward females. Yet beyond this overall enrichment of female cells in males, dmPFC ensembles also encode individuals’ sex preference states in the relative activity of male vs. female population responses. While female-preferring animals showed a stronger neural response to female conspecifics, male-preferring animals in fact showed a stronger response to males. Collectively, these data support the idea that behavioral sex preference is closely linked to a particular representation of conspecific sex in the prefrontal cortex, which separately controls male- vs. female-directed sociability.
Control of sex preference by native ensemble representations
Although some molecularly defined neural populations have been linked to social behaviors, it has been challenging to establish causal roles for native ensemble representations of sex information. In part, this is because neural representations of social features such as conspecific sex are often distributed across distinct neuronal cell types that defy precise molecular or anatomical distinction (Kim et al., 2019; Moffitt et al., 2018; Wu et al., 2017). Indeed, we show here that cortical representations of conspecific sex are distributed across both excitatory and inhibitory neural subpopulations. In the context of early sensory and valence processing, activity-dependent labeling has been used to re-activate native neural representations, yielding insights into circuit function that may be missed by more traditional approaches (Jennings et al., 2019; Marshel et al., 2019; Ramirez et al., 2015; Ye et al., 2016). Using activity-dependent labeling coupled with optogenetic stimulation, we show that neurons encoding male and female are directly involved in social behavior, providing a critical missing link between native sex representations and control of behavior. When optogenetically re-activated weeks later, neural populations that were natively activated during interaction with male or female conspecifics acutely and specifically increased male or female interaction, respectively. The fact that each subpopulation only increased social interaction with a specific sex demonstrates that male- and female-directed sociality is regulated by separable prefrontal representations. Together, the activity of these two representations effectively control a sex preference state that may be adjusted to shape behavior. As immediate early gene-based labeling methods cannot capture neural ensembles that show reduced activity during stimulus presentation, how socially-inhibited neurons play a role in sex preference behavior remains an open question for future studies.
Collectively, these results establish a novel role for the prefrontal cortex in encoding of conspecific sex and provide direct evidence of a functional role for native sex representations in shaping male- and female-directed social behavior. These findings provide new insights into how the brain transforms social information into adaptive behavior.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Weizhe Hong (whong@ucla.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets and the code that support the findings of this study are available from the corresponding author upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments were carried out in accordance with the NIH guidelines and approved by the UCLA institutional animal care and use committee (IACUC). Subject mice were male and female C57BL6/J and male Vgat-Cre mice ordered from Jackson Laboratories at 8-12 weeks of age and 24-30 g of weight. Mice were maintained in a 12 h:12h light/dark cycle (lighted hours: 10:00 pm – 10:00 am) with food and water ad libitum. All mice used for calcium imaging were individually housed for three weeks prior to experiments. Mice used for behavior experiments were individually housed for at least one week prior to experiments. All experiments were performed during the dark cycle of the animals.
METHOD DETAILS
Surgical procedures
Viral injections for imaging and GRIN lens implantations
Mice were anaesthetized with 1.0 to 2.0% isoflurane. Viral injections and lens implantation in the dorsomedial prefrontal cortex (dmPFC; also prelimbic cortex, PL) were done as previously described (Kingsbury et al., 2019). Specifically, we bilaterally injected 300 nl (on each side) of virus (AAV1.Syn.GCaMP6f.WPRE.SV40 for non-specific neuron imaging, AAV1-CaMKII-GCaMP6f-WPRE for CaMKII+ cells, or 500 nl (on each side) of AAV1-Syn-FLEX-GCaMP6f for Vgat+ cells) (Penn Vector Core) at 30 nl min−1 into the dmPFC using the stereotactic coordinates (AP: +2.0 mm, ML: ± 0.3mm, DV: −1.8mm to bregma skull surface). 1 week after injection, a 1.9mm diameter circular craniotomy was centered at the coordinates (AP: +2.0 mm, ML: 0.0 mm), and the GRIN lens (Edmund Optics; 1.8mm) was implanted above the injection site at a depth of −1.6mm ventral to the bregma skull surface and secured to the skull using super glue and dental cement. Mice were given one subcutaneous injection of Ketoprofen (4mg/kg) on the same day of surgery and Ibuprofen in drinking water (30mg/kg) starting on surgery day for 4 days. Mice were individually housed after surgery for three weeks. Then, the microscope together with a plastic baseplate were placed on top of the lens. We adjusted the position of the microscope until the cells and blood vessels appeared sharp in the focal plane and secured this position using dental cement.
The typical distance between the GRIN lens implant and the imaging focal plane is ~200 um. Although some tissue above the dmPFC must be removed in order to implant the lens, we have established in our previous work that the implant in the dmPFC does not disrupt normal social interaction (Kingsbury et al., 2019). We have also compared sex preference behavior in implanted animals with that of un-implanted animals (Figure S4B) and found that implanted and un-implanted animals do not show significant differences in sex preference behavior.
For non-specific and CaMKII+ cell imaging, we used wild type mice, and for GABAergic cell imaging, we used Vgat-Cre mice. All mice were handled and habituated to wearing the head-mounted microscope for at least 4 days before imaging experiments. While CaMKII promoter in AAV labels ~20% of inhibitory neurons in somatosensory cortex (Nathanson et al., 2009), 1-2% of CaMKII+ cells express GABAergic cell markers in the mPFC (Zhang et al., 2017). This suggests that, while CaMKII+ cells are predominantly excitatory in the prefrontal cortex, a very small fraction could be inhibitory.
Viral injections for activity-dependent labeling experiments
For activity-dependent optogenetics experiments, mice were bilaterally injected with 650 nL (each side) of a mixture of two viruses (AAV5-E-SARE-CreER (Kim et al., 2017) and AAV2-EF1a-DIO-hChR2-EYFP; ratio 1:5) at 40 nl min−1 into dmPFC using the stereotactic coordinates (AP: +2.0 mm, ML: ± 0.3mm, DV: −1.8mm to bregma skull surface). Optic fiber ferrules were implanted 4mm above the injection site (DV: −1.4mm to bregma skull surface). In order to create enough space for attachment of both optic fibers, the left ferrule was implanted at an angle 20 degrees from the midline DV axis, to the same depth of −1.4mm to bregma skull surface. Implants were secured to the skull using super glue and dental cement. For recovery, mice were given one subcutaneous injection of Ketoprofen (4mg/kg) on the same day of surgery and Ibuprofen in drinking water (30mg/kg) starting on surgery day for 4 days. Mice were individually housed after surgery. The same procedure was followed for the E-SARE validation experiment, except that 450 nL of a mixture of AAV5-E-SARE-CreER and AAV2-EF1a-DIO-EYFP (ratio 1:5) was used and that no fiber ferrules were implanted.
Histological analysis of dmPFC neuron projections and E-SARE validation
After calcium imaging and optogenetics experiments were completed, mice were transcardially perfused with 4% paraformaldehyde (PFA). Brains were post-fixed in 4% PFA overnight at 4 °C and cryo-protected for 48-72 h in 30% sucrose at 4°C before freezing in OCT on dry ice. 50-μm coronal sections were obtained using a cryostat, and sections were stained with DAPI (1:5,000 dilution) and mounted on slides. Images were acquired using a fluorescence microscope (Invitrogen EVOS FL Auto 2 Imaging System or Leica DM6 automated microscope) to confirm the position of lens implantation and expression of GCaMP6f (Figure 1C) or E-SARE-CreER-driven ChR2-EYFP (Figures S5-S6).
For analysis of overlap between male/female-induced E-SARE-CreER and male/female-induced Fos/Arc (Figures S5C-E), coronal sections were obtained at 40 μm using a cryostat (Leica) at −20°C. Sections containing the dmPFC were washed in 1X PBS, blocked with 10% Normal Donkey Serum for 1 h at room temperature, and incubated with rabbit anti-Arc at 1:500 and rabbit anti-Fos at 1:500. The following day, sections were incubated with donkey anti-rabbit Alexa 568 antibody at 1:500. Images were acquired using a confocal microscope (Zeiss LSM 880). Overlap between EYFP+ cells and Fos/Arc+ cells was measured and quantified using CellProfiler 3.0 (McQuin et al., 2018). For each condition, ≥10 sections from four independently injected hemispheres were analyzed and quantified.
For analysis of neural projections in E-SARE-CreER labeled cells (Figure S6), coronal sections were obtained at 35 μm using a cryostat (Leica) at −20°C. Sections were washed in 1X PBS, blocked with 10% normal donkey serum for 1 h at room temperature, and incubated in chicken anti-GFP antibody at 1:500 overnight. The following day, sections were incubated with donkey anti-chicken Alexa 488 antibody at 1:500. Images were acquired using a fluorescence microscope (Leica DM6 automated microscope). Using FIJI (ImageJ), fluorescence intensity was measured for dmPFC injection sites, as well as the following axon terminal sites: nucleus accumbens, dorsomedial striatum, dorsolateral striatum, basolateral amygdala, lateral hypothalamus, and ventral tegmental area. For each region of interest, area (square microns) and raw intensity were measured for six to eight hemisections. The total area and total raw intensity were computed as a sum of each value for all hemisections, and total raw intensity was normalized to total area. Finally, each axon terminal region was normalized to the dmPFC injection site.
Behavior Assays
Home cage social investigation assay
For imaging during home cage social investigation, animals were outfitted with the head-mounted microscope and briefly habituated for 2-3 minutes in their home cage. Subject animals were then presented manually with 8 different male and female conspecifics, as well as a novel object (a toy mouse), for 20-30 second periods in which they were permitted to freely investigate the stimuli. For each session, there were a total of 8 presentations of male conspecifics (8 unique and novel animals), 8 presentations of female conspecifics (8 unique and novel animals), and 8 presentations of the toy. Conspecifics and the toy were presented in a pseudorandomized order such that no type of stimulus was presented twice in a row. Sessions typically lasted 12-15 minutes. In order to preserve naturalistic behavior of subject animals as much as possible, each bout of social interactions is allowed for around 20-30 s, as opposed to a strictly fixed window. In each bout, animals were permitted to initiate investigate events at will (they were not cued or forced to investigate stimulus animals) and ongoing investigation events were, as much as possible, not artificially ended by experimenters.
As each stimulus animal is presented for a much shorter period of time (20-30 s) compared to a typical resident-intruder assay, subject animals rarely display aggressive or mating behavior toward presented conspecifics; it usually takes a longer interaction time for these behaviors to occur. Because of these conditions, we did not observe aggressive or mating behavior during the 20-30 s presentations of stimulus animals. Therefore, while aggressive and mating behaviors are part of the natural behaviors directed towards different conspecific sexes, the encoding of male or female conspecifics is not due to the presence or absence of aggressive or mating behavior.
Neural responses to odor cues and juvenile animals
In order to examine how sex-encoding dmPFC neurons respond to sex-specific odor cues, we performed additional sessions of the social investigation assay (described above) that were immediately followed by presentation of soiled bedding material gathered from cages with novel male or female conspecifics (Figure S1I). Soiled bedding was presented to subject animals for 20-30 s intervals during which subjects were permitted to freely investigate and sniff bedding material. Male and female bedding were alternately presented four separate times. All bedding investigation events were manually annotated.
We also explored how sex-encoding neurons respond during investigation of male and female juveniles (Figure S1J). For these experiments, we performed the social investigation assay as described above, and immediately followed this with alternating presentations of 4 novel male juveniles and 4 novel female juveniles (~8 weeks of age). Juvenile investigation events were manually annotated using the same criteria as adult investigation events.
Two-chamber social exploration assay
For imaging in the two-chamber assay, animals were outfitted with the head-mounted microscope, briefly habituated in their home cage for 2-3 minutes, and then placed in a 24” x 47” two-chamber arena with novel male and female conspecifics placed in opposing corners underneath barred pencil cups (Figure 4A). The subject animals were then allowed to freely move about the environment and investigate social stimuli at will for the duration of the session.
For experiments described in Figures 4A-B, two 20-minute sessions were performed in immediate succession and the positions of the male and female conspecifics were swapped in each one. The bottoms of pencil cups were fitted with petri dishes in order prevent excrement and odor cues from contaminating the floor of the arena.
For experiments described in Figures 4C-H, two-chamber sessions were 30 minutes long and stimulus animals were not altered in any way throughout the experiment. These experiments immediately followed home cage social investigation sessions, and the microscope, LED power and fluorescence collection settings were not changed across the two sessions. For experiments presented in Figure 4H, datapoints from the first 20 mins were presented in Figure 4H (so that comparison between conditions is consistent with other analyses in Figure 4I-K). Datapoints from the full 30 mins session was presented in Figure S3E.
For experiments described in Figures 4I-K, two-chamber sessions were 20 minutes long. Each animal underwent one recording session in which male and female conspecifics were placed beneath the cups, and one recording session in which non-social novel objects were placed beneath the cups, on separate days.
For behavior experiments using non-implanted animals (Figures S4C-F), two-chamber sessions were 30 minutes long using male/female, male/empty cup, female/empty cup, or 2 empty cups. For experiments used to analyze the stability of sex preference over time, animals were first habituated to the social chamber on the day prior to experiments, and then underwent two consecutive days of two-chamber experiments using novel conspecifics on both days. In each session, animals were first habituated to the environment using empty cups for 10 minutes. Social stimuli were then introduced under the cups, and animals explored for 20 minutes.
Analysis of animal behavior
For the home cage social investigation assay, behavior videos were recorded with a video camera at 20 frames per second (fps) and manually annotated frame by frame to identify onset and offset times for individual investigation bouts. Investigation bouts were considered to be epochs of at least one second where the subject animal displayed directed interaction with the stimulus and its head was facing and in physical contact with the stimulus. Animals displayed similar frequencies of social vs. toy investigation but spent significantly more time investigating social stimuli per-bout than the toy (Figures S1A-B).
For two-chamber experiments, behavior videos were recorded at 20 fps and the location and pose of the animal were automatically tracked using DeepLabCut (DLC) (Mathis et al., 2018), a neural network framework that we trained using manually annotated video frames. For each frame, we extracted DLC output for the animal’s nose, left ear, right ear, and tail coordinates. We considered social investigation events to be periods lasting at least 1 second where the animal’s head was within 3 inches of the center of the cup, and the angle between its head and the vector between its head and the cup was < 90 degrees. Behavior annotations were converted into binary vectors that denote precisely when animals are engaged in social investigation for downstream analysis.
In all behavior experiments, we measured the bias between male vs. female interaction using a sex preference score, computed as the fractional difference in male vs. female investigation time (T♂−T♀) / (T♂+T♀). In the home cage social investigation assay, male and female investigation time were each normalized by the total amount of time male and female conspecifics were presented in the session. While we observed a modest female preference in our behavior experiments, consistent with previous reports, there was large variability in preference scores in individual animals that was greater than expected just from random movement (Figure S4C). Sex preference scores of individual animals were also consistent across two days of testing (Figure S4E) with different stimulus animals, indicating stability of the behavioral state. We observed no difference in sex preference behavior in animals wearing the microendoscope and animals without any implant (Figure S4B).
In order to examine whether sex preference behavior in males was altered by sexual experience, we measured sex preference in the two-chamber assay on two consecutive days, between which subjects had two 30-minute exposures to novel female conspecifics in their home cage. During these female exposure sessions, mice were freely permitted to engage in mating behavior with females. We found that sex preference scores were not significantly different between sessions, suggesting that sexual experience does not alter preference behavior (Figure S4F).
For Figure 1H, both male and female animals were used. For Figures S1C-D and S1H, female animals were used. For all other figures, male animals were used.
Activity-dependent labeling and stimulation of male and female cells
For optogenetics experiments, animals were first virally injected with Tamoxifen-inducible E-SARE-CreER and Cre-dependent ChR2 constructs (described above). 3 weeks after injection, animals underwent a social exposure paradigm in which they interacted with either male or female conspecifics, or they remained in their home cage with no social stimulus. On each of two consecutive days, animals were presented sequentially with 6 novel male or female intruders in their home cage and allowed 10 minutes of interaction time with each one over a total period of 1 hour. For home cage controls, animals were left in their home cage for one hour, and an experimenter briefly placed their hand in the cage once every 10 minutes to control for effects related to human stimulus. At the end of the social exposure session on each day, animals were injected with 20mg/kg of 4OH-tamoxifen (4-TM). Prior to social exposure and E-SARE::ChR2 induction, animals were habituated to the induction room for 1 week, handled for 5 days, and habituated to IP injection with daily saline injections for 4 days.
In order to validate the efficacy and specificity of the activity-dependent labeling strategy, we followed a previously established procedure (Kim et al., 2017) by measuring the overlap between E-SARE labeled cells and cells that express immediate early genes Fos and Arc following a second exposure (Figures S5C-E). Animals first underwent the E-SARE-CreER induction as described above on two consecutive days. On each day, animals were exposed to either male conspecifics, female conspecifics, or a home cage control over a period of one hour, immediately followed by 4-TM injection (20mg/kg). This was repeated on two consecutive days (one exposure and one injection on each day). Cre-dependent EYFP was used in place of ChR2 to visualize individual cell bodies. Three weeks later, each animal underwent a second exposure with novel male conspecifics or female conspecifics. For the second exposure, four novel animals were presented for 15 min each for a total of one hour. One hour following this second exposure, animals were anaesthetized with isoflurane and were transcardially perfused with 4% paraformaldehyde (PFA). Brains were sectioned to perform histological analysis of E-SARE::EYFP and expression of Fos and Arc.
In order to test whether neural populations active during interaction with male or female conspecifics control the sex preference of animals, we optically stimulated male and female cells during the social two-chamber assay. After habituation to the two-chamber environment for at least one day with empty cups, animals were placed in the same arena with novel male and female conspecifics in either corner, as described above for imaging experiments. After a 10-minute baseline period, dmPFC neurons were stimulated for 10 minutes using a 473 nm laser (4 mW/mm2). The stimulation applied a protocol of repeated sequences of 3 seconds light-on and 2 seconds light-off, at 20hz with 20 ms light pulses. This stimulation epoch was then followed by another 10-minute baseline period. These experiments were performed 4-5 weeks following social exposure and 4-TM injection to allow time for sufficient ChR2 expression.
Binary vectors denoting male and female investigation were automatically extracted using DeepLabCut (Mathis et al., 2018) (described above). These were then smoothed using a 10-minute moving average window, and (T♂−T♀) / (T♂+T♀) was computed to obtain time courses for sex-preference behavior throughout the experiment. Male and female investigation time was computed and compared across 10-minute windows corresponding to the baseline and stimulation epochs. For each cohort, we performed sham control sessions in which animals were attached to the optic fiber, but no light was delivered (Figures S5I-K). Two sham sessions were performed on different days and behavior for each animal was averaged across sham sessions.
While optogenetic stimulation may have an effect over a timescale of seconds, this temporal resolution is constrained by the experimental assays used to measure this effect. Also, optogenetic stimulation may or may not result in acute changes in behavior (e.g. activating the neurons when the animals are facing away from stimulus animals may not elicit a strong effect). Thus, due to the nature of the behavioral assay and its natural variability, we have to measure this over a period of time. This has been the typical way to operationalize and analyze sex preference behavior in the established literature (Beny-Shefer et al., 2017; Yao et al., 2017).
For experiments using male-exposed animals (stimulating male cells), n = 14 animals, for experiments using female-exposed animals (stimulating female cells), n = 8 animals, and for home cage non-social control experiments, n = 9 animals.
Extraction of Calcium Signals
Motion-correction and preprocessing
During behavior experiments, calcium fluorescence videos were recorded through customized miniature microscopes (UCLA miniscope) at 30Hz using custom-written data acquisition software as previously described (Kingsbury et al., 2019). Raw videos from each imaging session were processed using a MATLAB implementation of the NoRMCorre algorithm to correct for motion-induced artifacts across frames (Pnevmatikakis and Giovannucci, 2017). In order to normalize image frames prior to cell sorting, (F-F0)/F0 (ΔF/F) was applied to each frame, where F0 was the de-trended mean frame. ΔF/F normalized videos were de-noised using an FFT spatial band-pass filter in ImageJ (v1.52a, U.S. National Institutes of Health), and spatially down sampled by a factor of 2 prior to ROI identification and cell sorting.
For comparison of cell encoding across the home cage and two-chamber assays (Figures 4C-H), we performed these two experiments in immediate succession without removing the head-mounted microscope or changing the recording settings (Figure S2F), such that identical cells in the same imaging view were recorded across two assays. Calcium fluorescence videos from these two sessions were concatenated and preprocessed together, and single cell ROIs were segmented (described below) across both experiments. These procedures ensured that cells in both sessions were precisely aligned, allowing analysis of the exact same cells across sessions.
Segmentation and ROI Identification
We identified putative cell bodies for extraction of neural signals using an established pipeline as previously described and validated (Kingsbury et al., 2019). Specifically, we employed an automated ROI detection algorithm that uses principal (PCA) and independent component analysis (ICA) to extract spatial filters based on spatiotemporal correlations among pixels (Mukamel et al., 2009). Independent components were manually inspected to remove components that did not represent cell bodies, and binary thresholding was applied to remove contributions from pixels outside the bounds of putative neurons. Spatial filters were then applied to the ΔF/F movie to extract the calcium traces. All traces from recorded cells were manually inspected to ensure quality signals. Specifically, putative neurons that had abnormally shaped cell bodies (abnormally large or small), or that had calcium transients with low signal-to-noise were excluded from further analysis (<5% of all putative neurons were excluded in this manner).
For home cage social investigation imaging experiments in Figure 1, a total of 5829 (mean ± SEM = 211 ± 9) single neurons were analyzed (n = 23 males and 5 females). For imaging of GABAergic (Vgat+) neurons, a total of 366 (mean ± SEM = 61 ± 4) neurons were analyzed (9.6% ± 0.3 of all cells, n = 6 males). For imaging of glutamatergic (CaMKII+) neurons, a total of 1373 (mean ± SEM = 229 ± 7) neurons were analyzed (25.4% ± 2.6 of all cells, n = 6 males).
For two-chamber imaging experiments, a total of 6686 (mean ± SEM = 216 ± 6) neurons were analyzed (n = 14 males). For Figures 4A-B, n = 7 males. For figures 4C-H, n = 10 males. For Figures 4I-K, n = 7 males.
For all experiments, a single neuron refers to one calcium trace extracted from an ROI, identified as described above, from one recording session.
Analysis of single cell responses during social interaction
Prior to downstream analysis, all ΔF/F calcium traces were z-scored and are presented throughout in units of standard deviation (σ). Responses of single neurons during social interaction events were quantified using ROC (receiver operating characteristic) analysis as previously described (Kingsbury et al., 2019; Li et al., 2017). Upon application of a binary threshold to the ΔF/F signal and comparison with a binary event vector denoting behavior, event detection based on neural activity can be measured using the true positive rate (TPR) and the false positive rate (FPR) over all time-points. Plotting the TPR against the FPR over a range of binary thresholds, spanning the minimum and maximum values of the neural signal, yields an ROC curve that describes how well the neural signal detects behavior events at each threshold (Figure 1F). We used the area under the ROC curve (auROC) as a metric for how strongly neurons were modulated during social interaction. For each neuron/behavior category, the observed auROC was compared to a null distribution of 1,000 auROC values generated from constructing ROC curves over randomly permuted calcium signals (traces that were circularly permuted using a random time shift). A neuron was considered significantly responsive (α = 0.05) if its auROC value exceeded the 95th percentile of the random distribution (auROC < 2.5th percentile for inhibited responses, auROC > 97.5th percentile for excited responses).
We analyzed the difference in stimulus-evoked activity in dmPFC neurons over the course of the two-chamber session (Figure 4H, I, S3E). We normalized differential activity in each epoch by the average activity of that cell. This normalization ensures that changes in differential activity are not due to overall changes in spontaneous activity, but rather reflect changes in the relative responsiveness of neurons to male and female stimuli.
Because some behavior events occur at different frequencies, there may be differences in the effective statistical power for the bootstrap ROC analysis between different event types (e.g. male and female investigation). Although this is unlikely to substantially affect measures of significantly responsive neurons, we performed additional control analyses to confirm that differences in fractions of responsive neurons were not due to unequal class sampling (Figure S2A-D). To this end, we normalized male/female representation by uniformly rescaling each male or female investigation bout for each session (using nearest-neighbor interpolation) by scaling factors s♂ = mean(T♂, T♀)/T♂ and s♀ = mean(T♂, T♀)/T♀ where T♂ and T♀ are the fractions of total time spent investigating male and female conspecifics. Corresponding epochs of the neural traces were equivalently rescaled. Following this, the total number of frames within a session that correspond to male and female investigation are equalized. The ROC bootstrap analysis was then performed again using these rescaled bouts and calcium traces, and fractions of significant cells for each category were computed. The differences in male- and female-responsive cells observed across all animals and in female-preferring animals were robust in this control analysis (Figures S2A-D, S4G-H).
In order to examine the coding consistency of sex-specific neurons across social investigation and two-chamber assays, we compared their stimulus-evoked activity in response to male vs. female stimuli in both experiments. We found that 72.2% of male-excited cells, 66.5% of female-cells, 64.7% of male-inhibited cells, 72.0% of female-inhibited cells (p < 0.001, bootstrap test) showed consistent increased or decreased activity in response to social stimulus investigation.
Cell tuning analysis using models of single neuron activity
In order to analyze how single dmPFC neurons change their tuning to male and female interaction over time, we used gaussian generalized linear models (GLM) to model calcium activity in individual neurons as a function of several factors (Figure S3F-K). This approach discounts contributions to neural activity that may be explained by other variables such as approach behavior or speed, so that weights fit to social interaction provide a measure of individual neural tuning to social stimuli. We used five variables to model cell activity: male and female interaction events, social approach, the animal’s speed, and the decay of overall neural population activity over time. Binary-valued vectors denoting interaction and approach events were first smoothed using an exponential filter (tau = 3 sec), and the animal’s speed was smoothed using a 3 sec moving average. For experiments with objects instead of conspecifics in the corners, behavior vectors denoted interaction with the right or left object, defined using the same criteria as described above for male and female interaction. GLM weights were fit using MATLAB (glmfit), and weights corresponding to male/female or object investigation were averaged across stimulus excited cells (defined using ROC analysis). Separate models were constructed for different epochs of the two-chamber session.
Analysis of population dynamics during behavior
Principal Component Analysis
To visualize population responses during social interaction (Figure 2E), we applied principal component analysis (PCA) to obtain components that capture the covariance of the neural population during interaction events (Cunningham and Yu, 2014). Calcium traces were first smoothed using a 5-second moving window average. Trial-averaged responses were computed over a time window of 20 seconds after event onset for male and female investigation events and concatenated, and responses for each neuron were formed into a matrix to perform PCA. Population trajectories for individual investigation bouts were then projected onto the first 3 principal components for visualization (Figure 2E, one example animal), and trial-averaged responses were also projected in thick lines.
Strength of population responses
In order to measure population responses associated with male and female investigation (Figure 5A-D), we used the Mahalanobis distance, which provides a measure of the separation between two population vectors while accounting for the covariance structure of the underlying distribution. The Mahalanobis distance between two population vectors was computed as:
where x is a population response vector at some timepoint, b is the average population vector over all baseline frames (where the animal does not exhibit social investigation behavior), and S is the covariance matrix computed over all baseline frames. For population response time-courses (Figures 5A, C), the Mahalanobis distance for individual investigation bouts was computed over a window of 10 seconds prior to 30 seconds after event onset. Here, we compared the average population responses evoked by male and female investigation and compared these across the top 25% male-preferring and top 25% female-preferring animals (Figures 5B, D). For each animal, a population preference score was defined as the fractional difference in average male vs. female response vectors, (R♂−R♀) / (R♂+R♀).
Male vs. female population decoder analysis
In order to measure population-level encoding of conspecific sex by dmPFC neurons, we constructed statistical models to predict the sex identity (male vs. female) of events based on population activity. For this, we used binary Fisher’s linear discriminant (FLD) classifiers.
For all classifiers used to quantify sex discrimination within the home cage and two-chamber sessions, training sets and test sets were constructed using population vectors evoked during male and female investigation events. Classifiers were constructed separately for each animal and used neurons only from that animal. We used 10-fold cross validation to measure classifier performance. For each cross-validation fold, the test set was a continuous 10% epoch of the data, and the remaining 90% training set was used to construct the model. For each fold, partial least-squares regression (PLS) was used to reduce the dimensionality of the training data, and the top 15 PLS components were retained for FLD analysis. Model performance was measured using the area under the ROC curve (auROC) for test data projected onto the Fisher discriminant. Overall model performance for each animal was calculated as the average over 50 folds where the training and test sets were randomly redrawn, and folds where both male and female events were not represented in the training and test set were dropped. Models were compared with null models constructed using data with randomly permuted calcium traces.
We found that decoders performed well above chance levels when constructed using a mixture of training data from both experiments (continuous 90% epochs) and tested on events from both (Figure 4F). For cross-session decoders (Figures S3A-B), the analysis was performed as described above, except the training set comprised male and female investigation events from the entire home cage or two-chamber session, and the test set those from the entire other session. For each animal, the neural populations for both cross-session decoders were down-sampled to contain only the population of cells that were significantly responsive to social stimuli based on ROC analysis. Performance may be slightly higher in decoders trained on two-chamber data and tested using home cage data because more training data is available from the longer two-chamber session. Figure 4G shows the projection of mean population vectors, from one example animal, associated with all investigation bouts from both home cage and two-chamber experiments onto FLD components that are defined using data from only the two-chamber session.
For the analyses in Figures 3L-M, where the performance of sex discrimination decoders using CaMKII+ neurons is compared with that of GABAergic neurons, neural populations were randomly down-sampled on each cross-validation fold to a subset of N neurons (on the x-axis in Figure 3L). 500 iterations of down sampling and cross validation were performed for each ensemble size. Partial least-squares regression for dimensionality reduction was not used for this analysis, as some cell subsets smaller than 15 are sampled (the number of PLS components used in other decoder analyses).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses for this study were conducted using GraphPad Prism (v8.4.0) or custom routines in MATLAB (Mathworks) and are described in the respective figure legends and Methods. All bar plots with error bars represent mean ± SEM; all box and whisker plots represent the median, interquartile range (box), and min and max (whiskers) of the underlying distribution. Statistical significance was defined with α < 0.05 using two-tailed tests unless otherwise specified. Full statistics for all ANOVA analyses are reported in Table S1. For comparisons of cell pairwise distance distributions, two-sample Kolmogorov-Smirnov tests were used. Resampling methods based on temporally permuting calcium traces (described in Methods) were used to assess significance of auROC values for social modulation of neural signals and performance of FLD decoders.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Arc antibody (rabbit) | Synaptic Systems | Cat# 156003 |
| Fos antibody (rabbit) | Synaptic Systems | Cat# 226003 |
| GFP antibody (chicken) | Aves Labs | Cat# 1020 |
| Donkey anti-chicken Alexa 488 antibody | Jackson Immunoresearch | Cat# AB_2340375 |
| Donkey anti-rabbit Alexa 568 antibody | ThermoFisher | Cat# A10042 |
| Bacterial and Virus Strains | ||
| AAV1-Syn-GCaMP6f-WPRE-SV40 | Penn vector core | Cat# 37087 |
| AAV1-CaMKII-GCaMP6f-WPRE-SV40 | Penn vector core | Cat# 100834 |
| AAV1-Syn-FLEX-GCaMP6f-WPRE-SV40 | Penn vector core | Cat# 100833 |
| AAV5-ESARE-ERT2-CreERT2-PEST | Kim et al., 2017 | N/A |
| AAV2-EF1a-DIO-hChR2-EYPP | UNC vector core | N/A |
| AAV2-EF1a-DIO-EYPP | UNC vector core | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratories | Stock 000664, RRID: IMSR_JAX:000664 |
| Mouse: Slc32a1tm2(cre)Lowl/J (Vgat-ires-cre) | Jackson Laboratories | Stock 016962, RRID: IMSR_JAX:016962 |
| Software and Algorithms | ||
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| ImageJ | NIH | https://imagej.nih.gov/ij/index.html; RRID: SCR_003070 |
| DeepLabCut | Mathis et al., 2018 | mousemotorlab.org/deeplabcut |
| Miniscope Controller | UCLA Miniscope | https://github.com/daharoni/Miniscope_DAQ_Software |
| NoRMCorre | Pnevmatikakis and Giovannucci, 2017 | https://github.com/flatironinstitute/NoRMCorre |
| CellSort | Mukamel et al., 2009 | https://github.com/mukamel-lab/CellSort |
| CellProfiler | McQuin et al., 2018 | https://cellprofiler.org |
| Other | ||
| Microendoscope | UCLA Miniscope | http://miniscope.org |
| Nanoinjector | World Precision Instruments | Cat# Nanoliter 2000 |
| Superfrost Plus slides | Fisher Scientific | Cat# 22-037-246 |
Neurons in dmPFC respond selectively to male vs. female social interaction
Male mice exhibit a bias toward stronger encoding of female vs. male cues
Sex encoding in dmPFC predicts behavioral sex preference in individuals
Activation of sex-encoding cells controls male vs. female preference behavior
Kingsbury et al. find that prefrontal cortex neurons represent the sex of conspecifics during social interaction. Male animals show stronger encoding of female than male cues, which predicts their behavioral preference toward interaction with females. Activation of sex-encoding cells shapes preference behavior by controlling male- vs. female-directed sociality.
ACKNOWLEDGEMENTS
We thank P. Golshani, D. Aharoni, and P. Zhao (UCLA) for technical support on miniature microendoscopic imaging, and H. Bito (University of Tokyo, Japan) for the E-SARE construct. This work was supported in part by NIH grants F31-MH117966, T32-NS058280, T32-NS048004, R01-NS113124, an ARCS Fellowship, a Marion Bowen Postdoctoral Grant, a NARSAD Young Investigator grant, a Brain Research Foundation grant, a Searle Scholars Award, a Klingenstein-Simons Fellowship, a Packard Foundation Fellowship, and a McKnight Scholar Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Beny-Shefer Y, Zilkha N, Lavi-Avnon Y, Bezalel N, Rogachev I, Brandis A, Dayan M, and Kimchi T (2017). Nucleus Accumbens Dopamine Signaling Regulates Sexual Preference for Females in Male Mice. Cell Rep. 21, 3079–3088. [DOI] [PubMed] [Google Scholar]
- Chen P, and Hong W (2018). Neural Circuit Mechanisms of Social Behavior. Neuron 98, 16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JP, and Yu BM (2014). Dimensionality reduction for large-scale neural recordings. Nat. Neurosci 17, 1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Silva BA, Perova Z, Marrone L, Masferrer ME, Zhan Y, Kaplan A, Greetham L, Verrechia V, Halman A, et al. (2017). Prefrontal cortical control of a brainstem social behavior circuit. Nat. Neurosci. 20, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal A El, and Schnitzer MJ (2011). Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings JH, Kim CK, Marshel JH, Raffiee M, Ye L, Quirin S, Pak S, Ramakrishnan C, and Deisseroth K (2019). Interacting neural ensembles in orbitofrontal cortex for social and feeding behaviour. Nature 565, 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, and Bito H (2013). Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods 10, 889–895. [DOI] [PubMed] [Google Scholar]
- Kim CK, Ye L, Jennings JH, Pichamoorthy N, Tang DD, Yoo A-CW, Ramakrishnan C, and Deisseroth K (2017). Molecular and Circuit-Dynamical Identification of Top-Down Neural Mechanisms for Restraint of Reward Seeking. Cell 170, 1013–1027.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Yao Z, Graybuck LT, Kim TK, Nguyen TN, Smith KA, Fong O, Yi L, Koulena N, Pierson N, et al. (2019). Multimodal Analysis of Cell Types in a Hypothalamic Node Controlling Social Behavior. Cell 179, 713–728.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury L, Huang S, Wang J, Gu K, Golshani P, Wu YE, and Hong W (2019). Correlated Neural Activity and Encoding of Behavior across Brains of Socially Interacting Animals. Cell 178, 429–446.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Ye L, and Deisseroth K (2016). Communication in Neural Circuits: Tools, Opportunities, and Challenges. Cell 164, 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DR, Tamir T, Kaufman M, Parabucki A, Weissbrod A, Schneidman E, and Yizhar O (2019). Dynamics of social representation in the mouse prefrontal cortex. Nat. Neurosci [DOI] [PubMed] [Google Scholar]
- Li Y, and Dulac C (2018). Neural coding of sex-specific social information in the mouse brain. Curr. Opin. Neurobiol 53, 120–130. [DOI] [PubMed] [Google Scholar]
- Li Y, Mathis A, Grewe BF, Osterhout JA, Ahanonu B, Schnitzer MJ, Murthy VN, and Dulac C (2017). Neuronal Representation of Social Information in the Medial Amygdala of Awake Behaving Mice. Cell 171, 1176–1190.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Zhang L, Barbera G, Fang W, Zhang J, Chen X, Chen R, Li Y, and Lin D-T (2018). Distinct and Dynamic ON and OFF Neural Ensembles in the Prefrontal Cortex Code Social Exploration. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, and Svoboda K (2008). Genetic dissection of neural circuits. Neuron 57, 634–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, and Svoboda K (2018). Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Kim YS, Machado TA, Quirin S, Benson B, Kadmon J, Raja C, Chibukhchyan A, Ramakrishnan C, Inoue M, et al. (2019). Cortical layer-specific critical dynamics triggering perception. Science (80-. ). 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, and Bethge M (2018). DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci 21, 1281–1289. [DOI] [PubMed] [Google Scholar]
- McHenry JA, Otis JM, Rossi MA, Robinson JE, Kosyk O, Miller NW, McElligott ZA, Budygin EA, Rubinow DR, and Stuber GD (2017). Hormonal gain control of a medial preoptic area social reward circuit. Nat. Neurosci 20, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac C, et al. (2018). Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science (80-. ). 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, and Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan M, Jang HJ, Park M, Miller EM, Cox J, Taliaferro JP, Parker NF, Bhave V, Hur H, Liang Y, et al. (2017). Combined Social and Spatial Coding in a Descending Projection from the Prefrontal Cortex. Cell 171, 1663–1677.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Görlich A, and Heintz N (2014). Oxytocin modulates female sociosexual behavior through a specific class of prefrontal cortical interneurons. Cell 159, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, and Callaway EM (2009). Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. Neuroscience 161, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, and Giovannucci A (2017). NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Liu X, MacDonald CJ, Moffa A, Zhou J, Redondo RL, and Tonegawa S (2015). Activating positive memory engrams suppresses depression-like behaviour. Nature 522, 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remedios R, Kennedy A, Zelikowsky M, Grewe BF, Schnitzer MJ, and Anderson DJ (2017). Social behaviour shapes hypothalamic neural ensemble representations of conspecific sex. Nature 550, 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu J, Zhu H, Zhang Q, Lin Z, and Hu H (2011). Bidirectional Control of Social Hierarchy by Synaptic Efficacy in Medial Prefrontal Cortex. Science (80-. ). 334. [DOI] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329.e6. [DOI] [PubMed] [Google Scholar]
- Yao S, Bergan J, Lanjuin A, and Dulac C (2017). Oxytocin signaling in the medial amygdala is required for sex discrimination of social cues. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, et al. (2016). Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences. Cell 165, 1776–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Daly KM, Liang B, Zhang L, Li X, Li Y, and Lin D-T (2017). BDNF rescues prefrontal dysfunction elicited by pyramidal neuron-specific DTNBP1 deletion in vivo. J. Mol. Cell Biol 9, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and the code that support the findings of this study are available from the corresponding author upon reasonable request.