Abstract
Mechanical loading is a potent strategy to induce bone formation, but with aging, the bone formation response to the same mechanical stimulus diminishes. Our main objectives were to (i) discover the potential transcriptional differences and (ii) compare the periosteal cell proliferation between tibias of young-adult and old mice in response to strain-matched mechanical loading. First, to discover potential age-related transcriptional differences, we performed RNA sequencing (RNA-seq) to compare the loading responses between tibias of young-adult (5-month) and old (22-month) C57BL/6N female mice following 1, 3, or 5 days of axial loading (loaded versus non-loaded). Compared to young-adult mice, old mice had less transcriptional activation following loading at each time point, as measured by the number of differentially expressed genes (DEGs) and the fold-changes of the DEGs. Old mice engaged fewer pathways and gene ontology (GO) processes, showing less activation of processes related to proliferation and differentiation. In tibias of young-adult mice, we observed prominent Wnt signaling, extracellular matrix (ECM), and neuronal responses, which were diminished with aging. Additionally, we identified several targets that may be effective in restoring the mechanoresponsiveness of aged bone, including nerve growth factor (NGF), Notum, prostaglandin signaling, Nell-1, and the AP-1 family. Second, to directly test the extent to which periosteal cell proliferation was diminished in old mice, we used bromodeoxyuridine (BrdU) in a separate cohort of mice to label cells that divided during the 5-day loading interval. Young-adult and old mice had an average of 15.5 and 16.7 BrdU+ surface cells/mm, respectively, suggesting that impaired proliferation in the first 5 days of loading does not explain the diminished bone formation response with aging. We conclude that old mice have diminished transcriptional activation following mechanical loading, but periosteal proliferation in the first 5 days of loading does not differ between tibias of young-adult and old mice.
Keywords: AGING, BONE, MECHANICAL LOADING, PROLIFERATION, RNA-SEQ/TRANSCRIPTOMICS
Introduction
Osteoporotic fractures occur in one in three women and one in five men over age 50, costing more than $22 billion annually in the United States.(1–4) Given this burden, treatment options for osteoporosis remain inadequate.(5) Antiresorptive drugs, which work by inhibiting bone resorption, are most commonly prescribed, yet they can cause negative side effects.(6–8) The two anabolic US Food and Drug Administration (FDA)-approved drugs, which work by promoting bone formation, also carry safety concerns for long-term use.(9) Thus, there remains a need for additional anabolic strategies to increase bone mass and prevent osteoporotic fractures.(10)
Mechanical loading is a potent strategy to induce bone formation,(11,12) but with aging, the bone formation response to the same mechanical strain stimulus diminishes.(13–17) Clinically, skeletal loading that increases bone mass in young-adult women is often ineffective in older women.(18–20) This age-related decline remains incompletely understood, despite loading being effectively modeled in mice.(21,22) Understanding the drivers of this diminished loading response may identify additional strategies to increase bone mass and prevent fractures.
We have previously shown that following mechanical loading, bones of old mice do not increase Wnt signaling as strongly as young-adult mice.(23) A recent microarray study that assessed transcriptomic changes in bone during the early (<24-hour) window after loading also showed age-related changes in Wnt signaling.(24) The authors suggested that aged bone retains the ability to respond acutely to loading but is unable to translate these responses into functional bone formation. Therefore, we first asked what transcriptional differences underlie the diminished loading response with aging beyond the early 24-hour window, during a time of active bone formation.
Wnt signaling plays a critical role in controlling cell proliferation,(25) and another finding of the microarray study was that expression of proliferation-related genes was higher in young-adult mice but lower in old mice.(24) This finding is compelling because loading is known to induce the proliferation of osteoblasts.(26,27) Moreover, the proliferative response of osteoblast-like cells following in vitro fluid flow was diminished in cells from old mice versus young-adult mice.(16) For these reasons, we asked if proliferation was diminished in old mice relative to young-adult mice following in vivo loading.
To discover potential age-related transcriptional differences, we performed RNA sequencing (RNA-seq) and compared the loading responses in tibias of young-adult (5-month) and old (22-month) mice following 1, 3, or 5 days of axial loading. To directly test the extent to which proliferation was diminished in old mice, we used bromodeoxyuridine (BrdU) in a separate cohort of mice to label all cells that divided during the 5-day loading interval and counted histologically the number of cells that arose via proliferation.
Materials and Methods
Mice
Female C57BL/6N mice were obtained at 5 months old and 22 months old from the aged rodent colony at the NIH National Institute on Aging (Bethesda, MD, USA), which is managed by Charles River Laboratories (Worcester, MA, USA). Relative to young-adult mice (5 months old), old mice (22 months old) are known to have diminished loading-induced bone formation (Supporting Fig. S1).(13–17) Following a 1-week to 2-week acclimation period, mice were separated into the following groups: five mice/day/age for the RNA-seq experiment and 10 mice/age for the proliferation experiment. Female mice were selected because: (i) osteoporosis is most prevalent in females(1); (ii) the diminished loading response with aging has been most consistently shown in female mice(13–17); and (iii) male mice often fight, which can confound the effects of loading.(28) Mice were housed in groups of up to five animals and kept on a 12-hour light/dark cycle under standard conditions with ad libitum access to water and chow (Purina, St. Louis, MO, USA; 5053 and 5058). Mice were loaded for either one, three, or five bouts of daily loading. Time points were selected to sample distinct phases of the loading response (Fig. 1A): early mechanosensation (day 1), activation of the bone formation cascade prior to matrix deposition (day 3), and active bone formation (day 5). All included mice were healthy throughout the experiment. Four hours after their last loading bout, mice were euthanized through CO2 asphyxiation. All animal work was approved by and in compliance with the Washington University IACUC.
Fig. 1.
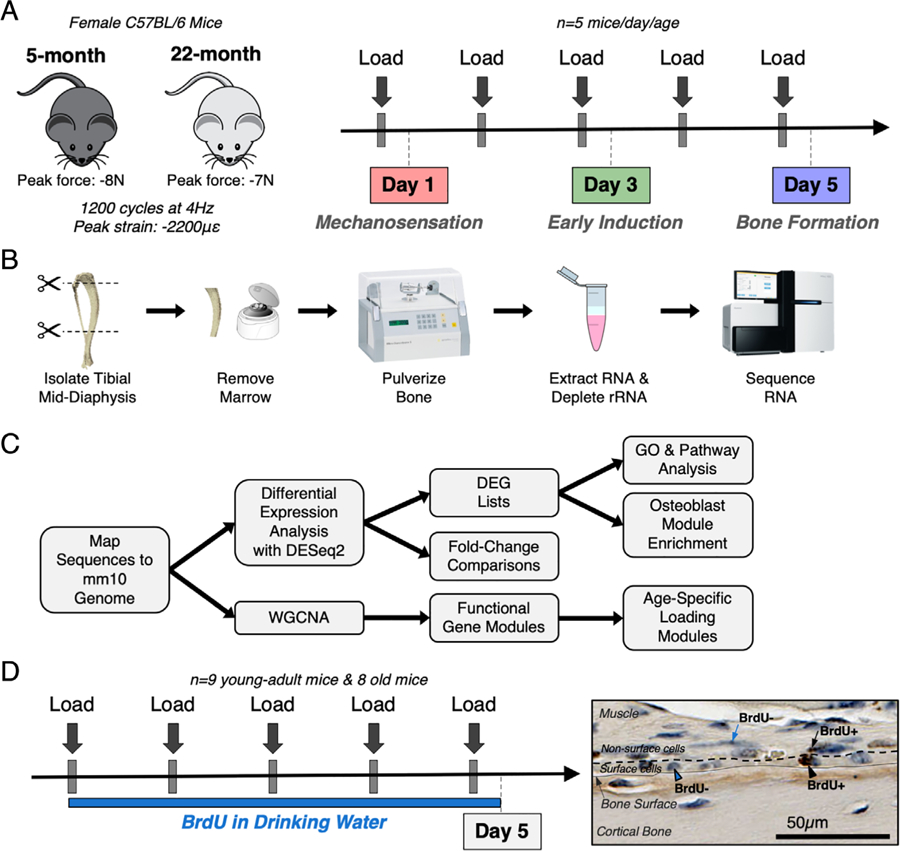
Experimental design for RNA-seq and cell proliferation studies. (A) Thirty female C57BL/6N mice (5 mice/day/age) were subjected to daily in vivo axial tibial compression for either 1 (day 1), 3 (day 3), or 5 (day 5) bouts and euthanized 4 hours after their final bout of loading. (B) The loaded (right) and non-loaded (left) tibial mid-diaphyses were isolated, removed of marrow, and pulverized. RNA was extracted and sequenced. (C) Sequencing reads were mapped to mm10 genome. First, differential expression analysis was performed using DESeq2 to obtain fold changes for each gene at every time point and lists of DEGs using a fold-change threshold of >1.5 or <0.67 and an FDR of 0.05. Pathway and GO analyses were performed using the DEG lists. Second, WGCNA was used to cluster genes into functional modules. (D) Seventeen female C57BL/6N mice (nine young-adult, eight old) were subjected to five bouts of daily in vivo axial tibial compression. Mice were given BrdU to label proliferating cells from the start of loading until euthanasia, 4 hours after their final bout. Surface and non-surface cells were counted as either BrdU+ or BrdU− using BIOQUANT. BrdU = bromodeoxyuridine; DEG = differentially expressed gene; FDR = false discovery rate; GO = gene ontology; WGCNA = weighted gene co-expression network analysis.
In vivo mechanical loading
Mice were anesthetized (3% isoflurane) and subjected to loading each morning for the specified number of bouts. With the mice prone, the right legs (tibias) were placed vertically in the loading fixture, with the knee positioned superiorly in a semispherical cup (10-mm-diameter) attached to the system actuator, and the foot held in a static fixture inferiorly (20 degrees of dorsiflexion). A pre-load (−0.5 N) was applied, and tibias were subjected to axial compression for 1200 cycles/day (4-Hz triangle waveform with a 0.1-s rest-insertion after each cycle) using the Electropulse 1000 materials testing system (Instron, Inc., Grove City, PA, USA). This loading protocol is anabolic for both cortical and trabecular bone in young-adult mice.(13,29–31) We used a strain-matched study design. Based on prior strain gauging analyses,(32) age-specific peak forces of −8N and −7 N were selected for 5-month-old and 22-month-old mice, respectively, to engender average peak periosteal strains of −2200 µε at the cortical mid-shaft.(15) Peak compression occurs 5 mm proximal to the distal tibiofibular junction at the posterolateral apex of the tibial cross-section. Corresponding tensile strains on the anteromedial surface were approximately 1200 µε.(32) After each loading bout, buprenorphine (0.1 mg/kg subcutaneously) was delivered to mitigate pain from loading,(23) and mice were returned to their cages to resume unrestricted activity. The left tibias served as non-loaded, contralateral controls.
RNA isolation
Left and right tibias were stripped of muscle, cut at the distal tibiofibular junction and 2 mm distal to the tibial plateau (Fig. 1B), centrifuged to remove the bone marrow,(23,33) and snap frozen in liquid nitrogen. Samples were stored at −80°C until RNA extraction. Samples were pulverized using a mikro-dismembrator (B. Braun Biotech International, Hessen, Germany), and total RNA was extracted using TRIzol (Ambion, Austin, TX, USA) with the Total RNA Purification Kit (Norgen, Thorold, ON, Canada; #17200). RNA concentrations were measured with the Nanodrop ONE (Thermo Scientific, Waltham, MA, USA), and RNA quality was evaluated with the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The median RNA integrity number (RIN) was 7.5 with a range of 6.6 to 8.7 (Supporting Fig. S2). No samples were excluded.
RNA sequencing
A total of 60 samples (30 pairs of tibias) were subjected to RNA-seq (Fig. 1B). For each of the three time points, five pairs of tibias from each age were obtained. Ribosomal RNA (rRNA) was removed using the Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA), and RNA-seq with the HiSeq 3000 (Illumina) was performed at 1 × 50 bp on 1 µg of RNA by the Washington University Genome Technology Access Center. The RNA-seq reads were aligned to the mouse genome (mm10) using Spliced Transcript Alignment to a Reference (STAR) (version 2.4.2a).(34) Gene counts were determined by the number of uniquely aligned, unambiguous reads (Subread:featureCount, version 1.4.6) and annotated (GENCODE v9; https://www.gencodegenes.org/). Only nonredundant, uniquely aligned reads were used to estimate the expression level of genes. All gene-level transcript counts were normalized for library size (R/Bioconductor DESeq2; Bioconductor Open Source Software for Bioinformatics; https://bioconductor.org/).(35) Genes that were lowly expressed (counts per million reads mapped [CPM] < 1) in all samples at any time point were filtered out. After filtering, 16,430 genes remained for downstream analysis. Differential expression analysis was performed separately for each age and time-point combination as a comparison of loaded versus non-loaded paired samples, yielding six lists of differentially expressed genes (DEGs) for each age (3 time points * 2 [up] upregulated and [down] downregulated lists). Data were further explored using principal component analysis (Supporting Fig. S3). For each sample, the reads per kilobase of transcript per million mapped reads (RPKM) values for all 16,430 genes in the data set were used as inputs.
Gene ontology and pathway analyses
For the gene ontology (GO) analysis (Fig. 1C), the 12 DEG lists were input into the Database for Annotation, Visualization and Integrated Discovery (DAVID)(36) with annotations limited to Mus musculus. Biological process GO terms were obtained for each list. For the pathway analysis, the 12 DEG lists (2 ages * 3 time points * 2 up and down lists) were input into PANTHER (version 14.1; http://pantherdb.org/)(37) to search for enriched pathways. GO terms and pathways were arranged temporally by age and then alphabetically.
Co-expression network analysis for module construction
Weighted Gene Co-expression Network Analysis (WGCNA) was used to generate two co-expression networks for data from young-adult and old mice.(38) The RPKM values for each gene across all samples for each age were used as inputs. The networks were created using a power threshold of six according to the scale-free topology criterion previously outlined.(39) To identify modules associated with loading, the eigengene of each module was calculated and correlated with loading (loaded versus non-loaded) within each network. Pathways represented in the modules associated with loading (“Loading Modules”) were characterized using PANTHER.(37) The expression of the top 20 upregulated genes (by fold change [FC]) in each module were visualized using a heat map.
In vivo proliferation labeling
Mice were subjected to daily loading as stated above (Fig. 1D). From the start of loading on day 1 until euthanasia on day 5, mice were continuously administered BrdU (Sigma-Aldrich, St. Louis, MO, USA) via their drinking water (0.8 mg/mL with 5% sucrose, prepared and changed daily). BrdU labels all cells that arise via proliferation throughout administration. This offered an advantage over relying on histological markers such as proliferating cell nuclear antigen (PCNA) or Ki-67, which only identify cells that are actively proliferating at euthanasia. Thus, our design labeled all cells that arose via proliferation during the same 5-day loading interval that we profiled transcriptionally. Three mice (one young-adult and two old) were excluded due to injury, resulting in n = 9 young-adult and n = 8 old mice for the study.
Histology to assess proliferation
Left and right tibias were collected and fixed in 10% neutral-buffered formalin (NBF). Intestines were collected as a positive, internal control for BrdU incorporation. After 48 hours in NBF, tissues were washed in PBS, and tibias were decalcified in 14% ethylenediaminetetraacetic acid (EDTA) for 2 weeks. Both tissues were processed for paraffin sectioning by the Washington University Musculoskeletal Research Center Histology Core.
Streptavidin-Biotin (SB; Invitrogen, Carlsbad, CA, USA; #93–3943) and Avidin-Biotin (AB; Invitrogen; #8800–6599-45) kits were used for BrdU staining. Manufacturer instructions were followed with minor modifications. Briefly, endogenous peroxidase activity was blocked by immersion in 3% hydrogen peroxide in methanol for 10 min. Antigen retrieval was accomplished with either 0.1% trypsin in PBS (SB) or kit reagents (AB) per instructions. A harsh denaturation step consisting of either 2 N HCl at 37°C (SB) or 4 N HCl at room temperature (AB) was included to access the BrdU antigen incorporated into dividing cells. Biotinylated anti-BrdU antibody was added to the tissue sections with either kit reagents (SB) or at a concentration of 1:500 in blocking solution (AB) at 4°C overnight. Streptavidin-peroxidase (SB) or avidin-horseradish peroxidase (HRP) (AB) was added to the samples (1:100 in blocking solution) for 1 hour at room temperature, and diaminobenzidine (DAB) (ImmPACT; Vector Laboratories, Burlingame, CA, USA) was used to stain the tissue. Last, hematoxylin was applied to the tissue sections for 3 min to counterstain the cells.
Histological analysis
Samples were imaged on a NanoZoomer 2.0-HT system (Hamamatsu Photonics K.K., Hamamatsu City, Japan) in bright-field. A 2-mm region of interest (ROI) was identified approximately 5 mm distal to the tibial plateau. We have previously determined this region to span the site 5 mm proximal to the distal tibiofibular junction, where peak compression and bone formation are observed.(32) BIOQUANT (Osteo II) (BIOQUANT, Nashville, TN, USA) was used to manually count BrdU+ cells, BrdU− cells, and bone surface length. Cells were further categorized as “surface” cells (Fig. 1D) if they were directly on the bone surface or “non-surface” cells if they were not directly adjacent to the bone surface (ie, in the periosteum more than one cell layer above the surface). Only the periosteal surface was analyzed because the BrdU+ marrow sometimes separated from or overlapped with the endocortical surface, preventing reliable analysis of this surface.
Statistical analysis
To define DEGs, we used the following criteria: an FC (loaded/non-loaded) of >1.5 or <0.67 and a false discovery rate (FDR) of 0.05. For the GO analysis, only significant terms were included (Bonferroni p < .05). For the pathway analysis, a Fisher’s exact test was used with an FDR of 0.05. For the co-expression network analysis, the significance threshold was set using a Bonferroni correction (p = .05/38 = 0.0013). For the proliferation analysis, we analyzed two independent slides for the first nine samples (four young-adult and five old) and averaged the counts. Because the counts between the two slides were not significantly different (p = 0.84 by Wilcoxon matched-pairs signed rank test, n = 9 per group), only a single slide was analyzed for the remaining eight samples. A nonparametric Wilcoxon matched-pairs signed rank test was used to compare the number of BrdU+ cells/length and BrdU+ cells/total cells between loaded and non-loaded samples within an age for both surface and non-surface counts (GraphPad Prism 8.0; GraphPad Software, Inc., La Jolla, CA, USA; n = 9 young-adult and n = 8 old). An unpaired Mann-Whitney test was used to compare the loaded counts between young-adult and old mice.
Results
Tibias from old mice had less transcriptional activation after loading compared to young-adult mice
At day 1, tibias from young-adult mice had 19 upregulated and 0 downregulated DEGs (Supporting Table S1) whereas old mice had no genes that met the DEG criteria (Fig. 2A). Increasing bouts of loading progressively induced more DEGs in both young-adult and old mice, but young-adult mice had more upregulated and downregulated DEGs than old mice at every time point. By day 3, young-adult mice had 430 upregulated and 122 downregulated DEGs whereas old mice had 233 and 81, respectively. At day 5, young-adult mice had 894 upregulated and 566 downregulated DEGs whereas old mice had only 353 and 50, respectively. To assess transcriptional activation beyond the number of DEGs, we compared the absolute FC inductions of the upregulated DEGs between young-adult and old mice (Fig. 2B). To compare FCs of the same genes, we restricted the comparison to the set of genes that met the DEG criteria in both ages at days 3 and 5. Young-adult mice had significantly higher FCs, indicating that even when old mice upregulate the same genes as young-adult mice, old mice upregulate them less strongly. DEGs were also compared across days to assess how the proportion of DEGs changed over time during the loading response (Fig. 2C). Young-adult and old mice shared a similar proportion of genes between days 3 and 5. In young-adult mice, eight DEGs (Wnt7b, Wnt1, Ngf, Ddn, Mt2, Gem, Rgs16, and Junb) were identified as upregulated at all three time points. To investigate if young-adult and old mice upregulated and downregulated the same genes, we compared the overlap between the two ages for upregulated and downregulated DEGs at days 3 and 5 (Fig. 2D). Although 33% to 39% of upregulated DEGs were shared between young-adult and old mice, only 3% to 4% of downregulated DEGs were shared between young-adult and old mice.
Fig. 2.
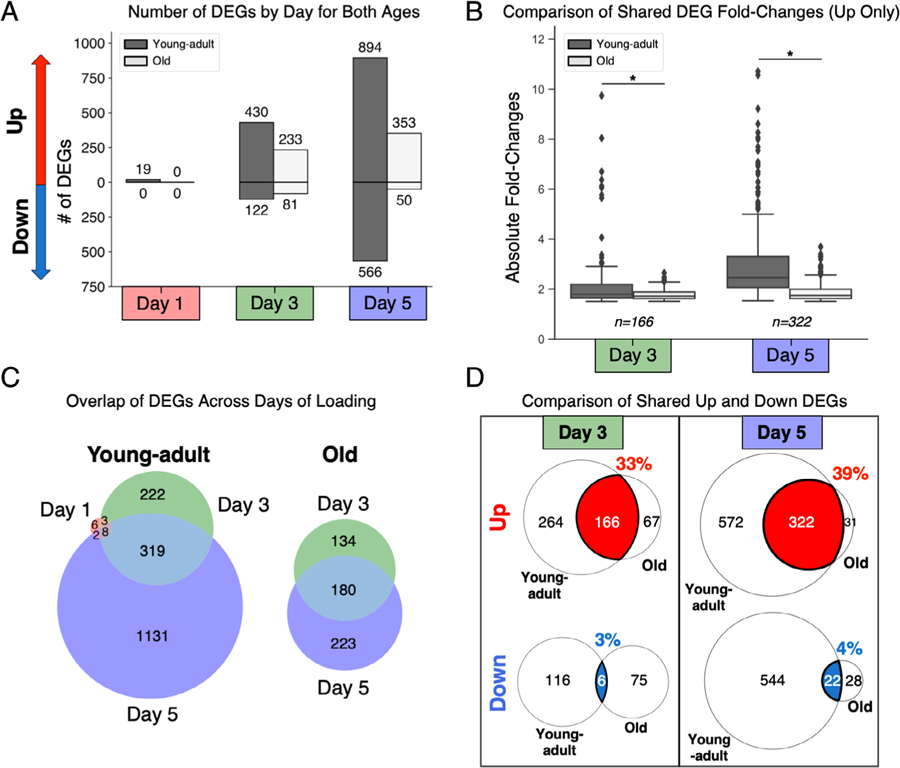
Tibias from old mice had less transcriptional induction after loading compared to young-adult mice. (A) Old mice had fewer upregulated and downregulated DEGs at every time point. (B) Old mice had lower fold-change inductions of the shared upregulated DEGs at day 3 and day 5 (*p < .0001, Wilcoxon matched-pairs test). (C) Old and young-adult mice shared a similar proportion of DEGs across days of loading. (D) Old and young-adult mice shared more upregulated DEGs than downregulated DEGs. At day 3 and day 5, 33% and 39% of upregulated DEGs were shared, respectively, whereas only 3% and 4% of downregulated DEGs were shared. DEG = differentially expressed gene.
Old mice had lower induction of Wnt-related genes at day 1 and matrix genes at day 5
The top 10 upregulated and downregulated genes by FC for each day and age were compiled (Table 1). At day 1, Wnt1 and Wnt7b were strongly induced in young-adult mice, as has been previously reported,(23,40) but Wnt1 and Wnt7b were less strongly induced in old mice, which is consistent with our previous findings.(23) Nerve growth factor (Ngf ), which is reported to act upstream of Wnt signaling in mechanically loaded bone,(41) was also strongly induced in young-adult mice but to a much lesser extent in old mice (threefold higher, young-adult versus old). In addition, dendrin (Ddn), which is expressed in brain and kidney,(42) was more strongly upregulated by loading in young-adult versus old mice (threefold higher, young-adult versus old). Fosb, which has a well-established role in the osteoblast mechanotransduction cascade,(43) was also one of the most strongly induced genes in young-adult but not old mice. Cyclooxygenase-2 (Ptgs2, commonly known as Cox-2) was more strongly induced in young-adult versus old mice (more than twofold higher). Additionally, the three genes coding for type IX collagen (Col9a1, Col9a2, Col9a3), which is found in cartilage,(44) were strongly downregulated in young-adult but not old mice at day 1, although these changes did not meet the FDR cutoff.
Table 1.
Top 10 Genes by FC for Each Day
| Young-adult | ||||||
|---|---|---|---|---|---|---|
| Day 1 |
Day 3 |
Day 5 |
||||
| Gene | FC | Gene | FC | Gene | FC | |
| Upregulated | Wnt7b | 7.78 | Serpina3m | 19.24 | Bcan | 24.93 |
| Gm42679 | 5.75 | Serpina3n | 11.83 | HapIn4 | 22.01 | |
| Wnt1 | 5.75 | CcI7 | 9.75 | Car12 | 19.17 | |
| Ngf | 4.74 | Rrad | 8.04 | Nell1 | 13.91 | |
| Ighv7-1 | 3.98 | Wnt1 | 6.70 | BCO21891 | 11.42 | |
| Ddn | 3.96 | Strati | 6.49 | Rrad | 10.73 | |
| Gm15564 | 3.64 | Wnt7b | 6.37 | Serpina3n | 10.71 | |
| Igkv15-103 | 3.15 | Ddn | 6.13 | Fam84a | 10.57 | |
| Fosb | 3.04 | Card14 | 6.06 | Slc13a5 | 9.22 | |
| Ptgs2 | 2.98 | CcI2 | 5.77 | Slc6a2 | 8.61 | |
| Downregulated | Col9a1 | 0.15 | Fgf23 | 0.20 | Perm1 | 0.10 |
| CytI1 | 0.35 | Csrp3 | 0.30 | Smco1 | 0.12 | |
| Col9a2 | 0.36 | Igkv3-4 | 0.33 | Cyp2e1 | 0.12 | |
| Col9a3 | 0.37 | Ighv11-2 | 0.35 | Gm8424 | 0.13 | |
| Igkv8-27 | 0.37 | Igkv5-39 | 0.35 | Csrp3 | 0.13 | |
| C1qtnf3 | 0.38 | Actc1 | 0.38 | Dhrs7c | 0.13 | |
| Slc6a1 | 0.43 | Pamr1 | 0.39 | Ckmt2 | 0.14 | |
| Ighv11-2 | 0.47 | Igkv14-126 | 0.40 | Fgf23 | 0.15 | |
| Hapln1 | 0.47 | Prdm8 | 0.40 | Scg3 | 0.15 | |
| Ighv4-1 | 0.48 | Perm1 | 0.41 | 9430073C21Rik | 0.16 | |
| Old | ||||||
|---|---|---|---|---|---|---|
| Day 1 |
Day 3 |
Day 5 |
||||
| Gene | FC | Gene | FC | Gene | FC | |
| Upregulated | Ngf | 1.50 | HapIn4 | 2.64 | Prss35 | 3.69 |
| Rrad | 1.48 | Slc6a2 | 2.48 | Bglap | 3.40 | |
| Gm42679 | 1.44 | Cthrcl | 2.45 | Slc13a5 | 3.32 | |
| Wnt7b | 1.44 | Fkbp11 | 2.36 | Col11a1 | 3.29 | |
| Ddn | 1.41 | II11 | 2.34 | Cthrcl | 3.20 | |
| Wnt1 | 1.39 | Wnt1 | 2.33 | Col1a1 | 2.95 | |
| Dusp6 | 1.36 | Gm10629 | 2.30 | Bglap2 | 2.89 | |
| Gem | 1.35 | Ddn | 2.28 | Smpd3 | 2.88 | |
| Ptgs2 | 1.32 | Tubb3 | 2.25 | Bcan | 2.88 | |
| Akr1 b8 | 1.30 | Lurap1I | 2.24 | Cgrefl | 2.86 | |
| Downregulated | Tigit | 0.74 | Gm28653 | 0.45 | Mpz | 0.45 |
| Tox2 | 0.74 | Abra | 0.48 | Hepacam2 | 0.50 | |
| 1110001J03Rik | 0.76 | Gbp10 | 0.51 | Cldn19 | 0.52 | |
| Gm10451 | 0.76 | Atp1b4 | 0.52 | Ncmap | 0.52 | |
| Mcee | 0.77 | 2310020H05Rik | 0.52 | Mal | 0.53 | |
| Phpt1 | 0.77 | Islr2 | 0.53 | Igkj5 | 0.55 | |
| Gm26613 | 0.77 | Fgf23 | 0.53 | Ly6d | 0.55 | |
| Igkv5-48 | 0.78 | Klf15 | 0.55 | Gm15675 | 0.56 | |
| RP23-385F8.1 | 0.78 | Snai3 | 0.55 | Gjc3 | 0.56 | |
| Gm43618 | 0.80 | Gm8424 | 0.55 | Ugt8a | 0.56 | |
Bold denotes DEG (FC>1.5 or <0.67 & FDR <0.05)
At day 3, Wnt1 and Wnt7b continued to be highly upregulated by loading in young-adult mice, with FC inductions approximately threefold higher than old mice. Only young-adult mice strongly upregulated the serine protease inhibitors Serpina3m and Serpina3n, which to our knowledge have no documented role in cortical bone or the mechanical loading response. Notably, these genes had the highest FCs at day 3. Chemokine (C-C motif) ligand 7 (Ccl7), a Wnt-activating cytokine that is secreted by osteocytes in response to in vitro mechanical stimulation and protects against cell death,(45) was more highly induced in young-adult than old mice (approximately fivefold, young-adult versus old) at day 3. A related cytokine, Ccl2, which also protects against cell death,(45) was more strongly induced in young-adult mice (approximately threefold, young-adult versus old). Ras-related glycolysis inhibitor and calcium channel regulator (Rrad) was strongly upregulated at days 3 and 5 in young-adult but not old mice. Rrad is critical for bone homeostasis, and its deletion results in lower bone mineral density (BMD) and higher bone marrow adipose tissue.(46) Last, at day 3, fibroblast growth factor 23 (Fgf23), which is critical for phosphate homeostasis,(47) is more strongly downregulated in young-adult mice (2.5-fold, young-adult versus old).
At day 5, in young-adult mice, extracellular matrix (ECM) components, including brevican (Bcan) and hyaluronan and proteoglycan link protein 4 (Hapln4), were the most highly upregulated. Wnt1 continued to be more strongly induced in young-adult mice (threefold-young-adult versus old), but Wnt7b was only modestly upregulated at day 5, and this upregulation was similar in both ages. In old mice, Bglap, Col11a1, and Col1a1 were the most highly upregulated matrix components, yet the FCs of these three genes in young-adult mice were still approximately twofold higher. Carbonic anhydrase 12 (Car12), a gene induced after fracture,(48) was more highly induced in young-adult than old mice (approximately sevenfold, young-adult versus old). Nell1, which is osteoanabolic,(49) was strongly induced in young-adult but not old mice at day 5.
GO analyses show less process activation in old mice
We assessed GO process enrichment using separate upregulated and downregulated DEG lists (Fig. 3A) and identified 33 total processes (Fig. 3B). Most of these processes (82%, 27/33) were identified in young-adult mice, whereas fewer (58%, 19/33) were identified in old mice at any time point. At day 1, no processes were enriched in either age, likely due to the low number of DEGs at that time point. At day 3, cell-cell signaling, which was driven by expression changes in gap junctions and signaling molecules such as Wnt ligands, was up in young-adult but not old mice. At days 3 and 5, in young-adult mice, processes involved in bone formation were enriched, including collagen fibril organization and osteoblast differentiation, yet most of these processes were not identified until day 5 in old mice. Further, at day 5, young-adult mice showed enrichment for proliferative processes including positive regulation of mesenchymal cell proliferation, but this signal was absent in old mice. At day 5, in young-adult mice, brown fat cell differentiation, gluconeogenesis, and metabolic processes were down, suggesting a role for metabolic changes during the bone formation phase of the loading response. At day 5, pathways exclusively upregulated in old mice included endochondral ossification and negative regulation of canonical Wnt signaling pathway. The negative regulation of canonical Wnt signaling pathway was driven by a set of genes that included NKD inhibitor of WNT signaling pathway 2 (Nkd2) and SRY-box 9 (Sox9), two genes that have been shown to inhibit Wnt signaling in bone.(50,51) Myelination, identified at day 5, was the only downregulated pathway in old mice.
Fig. 3.
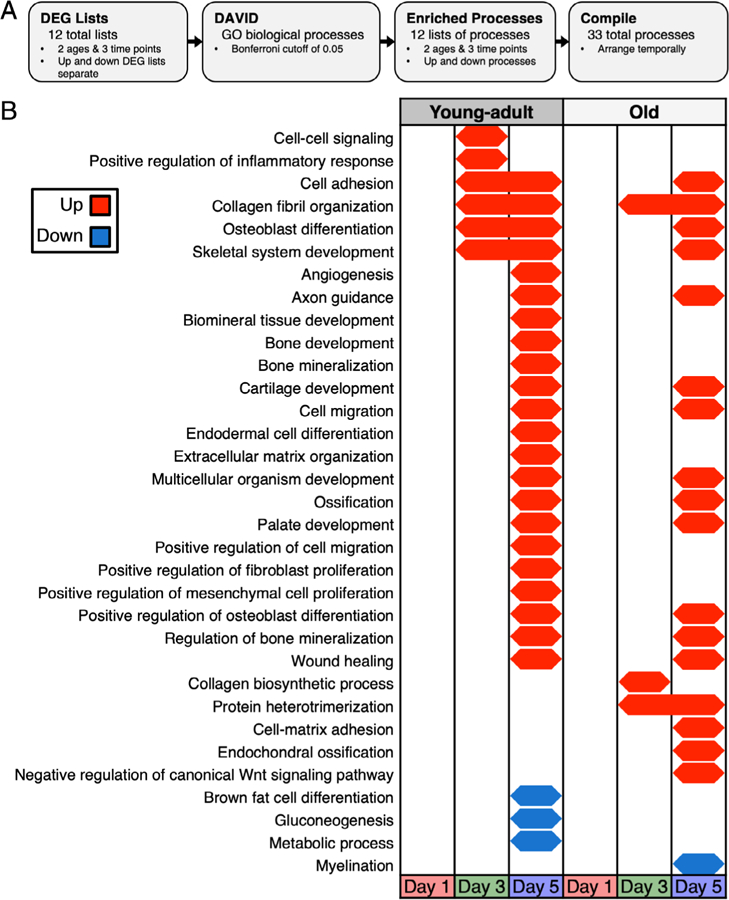
Temporal map of GO biological processes showed less activation in old mice. (A) DEG lists for each age and day combination (upregulated and downregulated DEGs separate) were input to DAVID to identify enriched GO biological processes. (B) Old mice had fewer enriched processes, and some processes were enriched later relative to young-adult mice. Young, but not old mice, downregulated metabolic processes at day 5. DEG = differentially expressed gene; GO = gene ontology.
Pathway analyses show less pathway activation in old mice
We also assessed pathway enrichment in separate upregulated and downregulated DEG lists for each day (Fig. 4A) and identified 14 total pathways. At all days, in young-adult mice, angiogenesis was enriched in upregulated DEGs (Fig. 4B). At day 1, gonadotropin-releasing hormone receptor, which affects BMD,(52) was up exclusively in young-adult mice. At days 3 and 5, integrin signaling was up in both young-adult and old mice whereas Wnt signaling and cadherin signaling were exclusively up in young-adult mice. The Alzheimer disease–presenilin pathway was also up in young-adult mice at these days but was not identified until day 5 in old mice. At day 5, axon guidance mediated by semaphorins, Huntington disease, ionotropic glutamate receptor, and plasminogen activating cascade pathways were up exclusively in young-adult mice. The enrichment of axon guidance mediated by semaphorins pathway supports the idea that a neurogenic response may play a role in the loading response.(41) This enrichment was not detected in old mice. At day 5, enkephalin release was exclusively up in old mice. Enkephalins are endogenous opioid peptides that regulate nociception(53) and have been shown to inhibit osteoblast alkaline phosphatase.(54) The only pathway that was enriched in downregulated DEGs was acetate utilization in young-adult mice at day 5. Enrichment FCs were calculated (Supporting Table S2). We repeated this analysis using combined up and down DEG lists, and the results were similar to those described here for separate up and down DEG lists (Supporting Fig. S4).
Fig. 4.
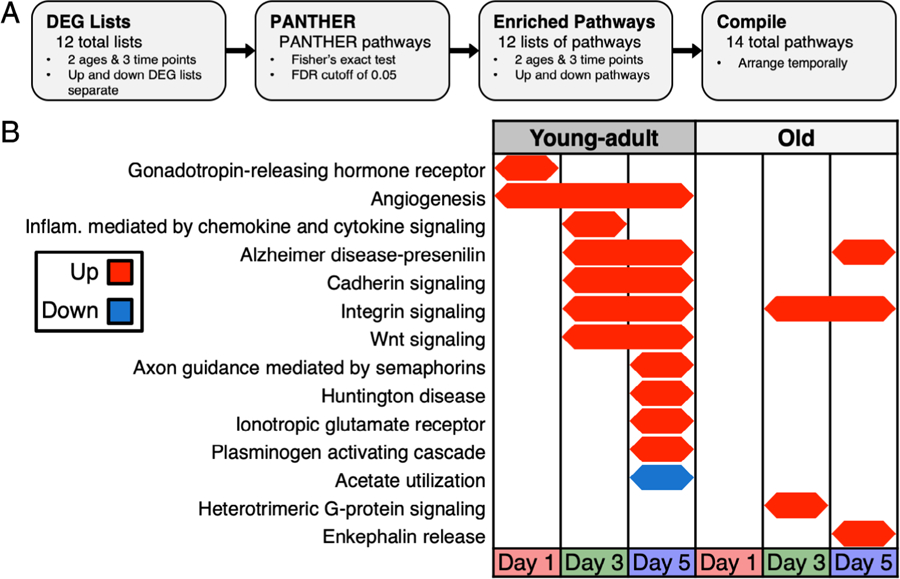
Temporal map of PANTHER pathways showed fewer enriched pathways in old mice. (A) DEG lists for each age and day combination (up and down DEGs separate) were input to PANTHER to identify enriched pathways. (B) Old mice lacked enrichment of several pathways including cadherin signaling and Wnt signaling yet had activation of other pathways including heterotrimeric G-protein signaling and enkephalin release that were absent in young-adult mice. DEG = differentially expressed gene.
Co-expression network analysis identified a loading module in each age
Separate co-expression networks for young-adult and old mice were constructed using WGCNA. Both networks consisted of 38 co-expression modules (Fig. 5A). To identify modules associated with loading, we correlated each module’s eigengene with loading in both the young-adult and old networks. In each age, only a single module had a significant association with loading, and these two modules were termed the Young-adult Loading Module and the Old Loading Module. Only 29% of the genes were shared between the two modules. However, the top 10 GO hits for each age showed a nearly identical osteoanabolic signal, driven by terms including collagen fibril organization, collagen metabolic process, and osteoblast differentiation (Fig. 5B). Visualization of the top 20 genes (by FC at any day) in the two loading modules revealed that even the genes most highly expressed in old mice were more highly expressed in young-adult mice (Fig. 5C). The 956 genes unique to the Young-Adult Loading Module were enriched for Wnt signaling and neuronal projection (Supporting Table S3). The 1201 genes unique to the Old Loading Module were enriched for negative regulation of Wnt signaling, including Notum, which inactivates Wnt ligands and negatively regulates bone mass.(55)
Fig. 5.
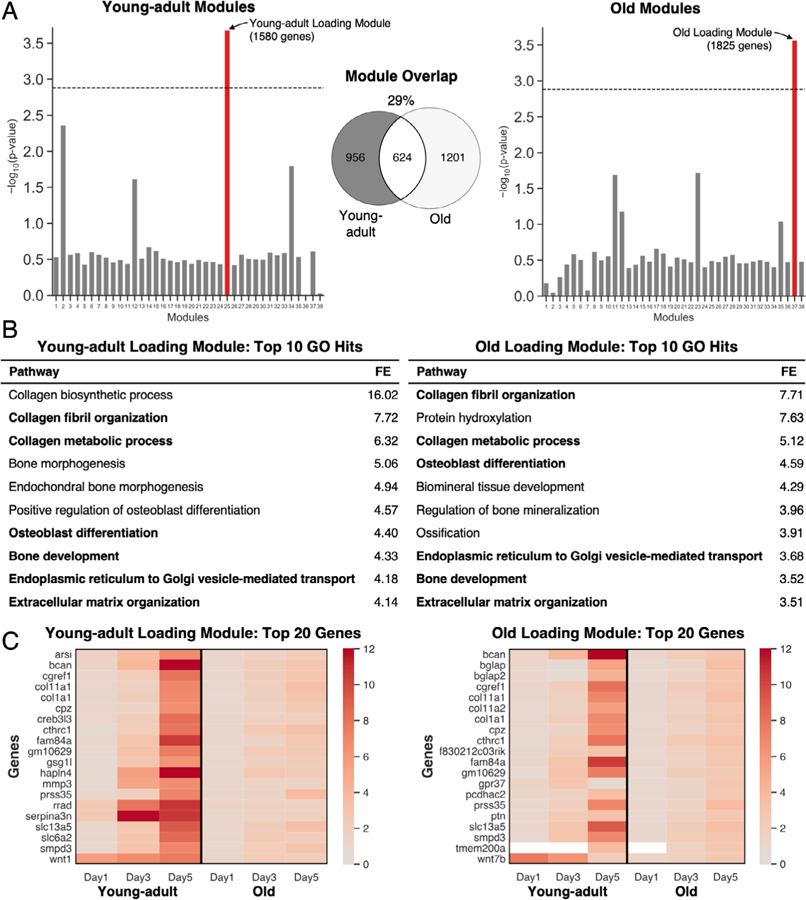
A distinct loading-responsive gene module was identified for each age using WGCNA. (A) For each age, RPKM values were used to cluster genes into 38 functional modules. Each age had one module that was significantly enriched for loading (red). The significance threshold (dashed line) was set using a Bonferroni correction (p = .05/38 = .0013); 29% of the genes in these modules were shared between young-adult and old. (B) GO analysis of the loading modules showed high enrichment of collagen-related and osteoblast-related processes (bold: shared between young-adult and old). (C) The top 20 upregulated genes (by fold-change) in each module were induced in both ages but to a lesser magnitude in old mice (white: expression not detected). FE = fold-enrichment; GO = gene ontology; WGCNA = weighted gene co-expression network analysis.
Old mice showed a similar periosteal proliferative response compared to young-adult mice
BrdU+ cells were observed in both the intestinal samples and the marrow of all bone samples, confirming that BrdU was taken up by every animal and that the BrdU staining was successful (Supporting Fig. S5A). In both young-adult and old mice, the non-loaded periosteal (Ps) surface displayed few BrdU+ cells, whereas the loaded periosteal surface displayed numerous BrdU+ cells (Fig. 6A). In young-adult and old mice, the non-loaded surface had an average of 1.6 and 1.7 BrdU+ surface cells/mm, respectively, whereas the loaded surface had an average of 15.5 and 16.7 BrdU+ surface cells/mm, respectively (p < .01, Fig. 6B). We repeated the analysis after normalizing the number of BrdU+ cells to the total number of cells, and the same trends were observed (Supporting Fig. S5B). Both ages showed that ~30% to 40% of cells on the loaded surface had arisen via proliferation, which is consistent with our recent data assessing the periosteal lamellar bone formation responses in 5-month-old and 12-month-old mice.(27) We defined “non-surface” cells as those that were not directly adjacent to the bone surface and assessed the number of these cells that arose via proliferation. Both young-adult and old mice showed a statistically significant increase in the number of BrdU+ non-surface cells with loading (p < .02, Fig. 6C). Although there was no significant difference in loaded counts between the young-adult and old mice, the median number of 30 in young-adult mice compared to 12 cells in old mice may indicate more nonresponders in old mice. We observed no difference in the total number of cells between the loaded and non-loaded surfaces in either age (Supporting Fig. S5C). Nor did we observe any differences in apoptosis by TUNEL staining between ages or with loading (Supporting Fig. S6A,B). Overall, young-adult and old mice showed a similar proliferative response to the same loading stimulus.
Fig. 6.
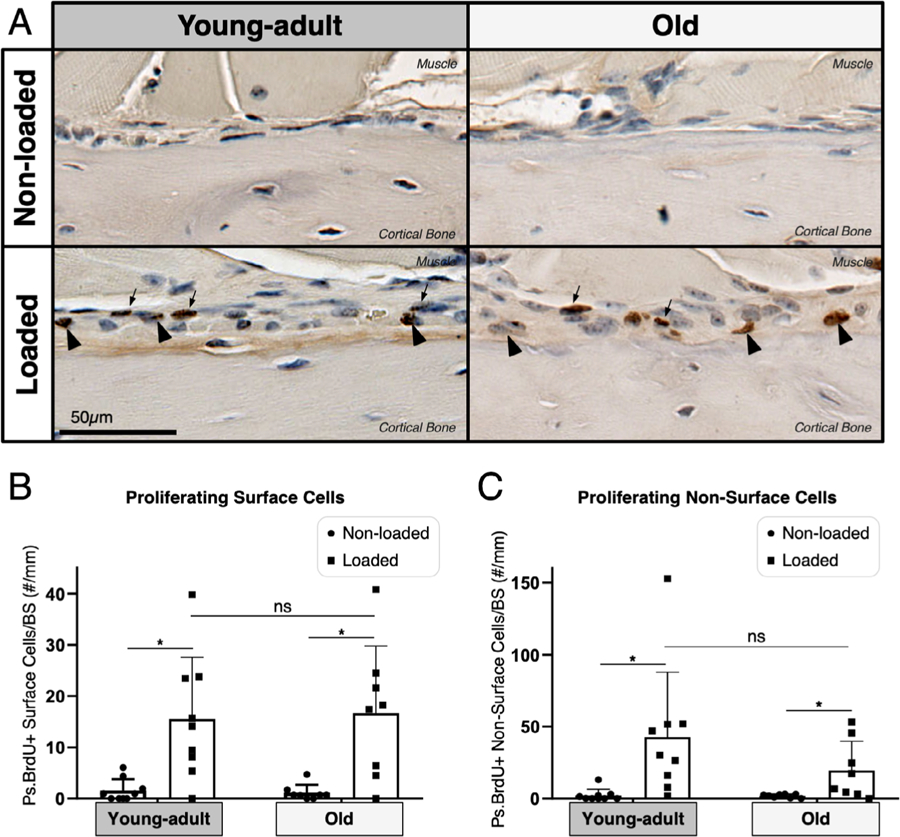
Old mice showed a similar periosteal proliferative response compared to young-adult mice. (A) BIOQUANT was used to count BrdU+ and BrdU− cells 2-mm region of interest (ROI), shown at magnification ×40. (B) Both young-adult (n = 9) and old (n = 8) mice showed a significant increase in proliferating cells on the periosteal (Ps) surface (arrowheads) following loading (*p < .02, Wilcoxon matched-pairs signed rank test). (C) Both young-adult and old mice showed a significant increase in proliferating non-surface cells (arrows) following loading (*p < .02, Wilcoxon matched-pairs signed rank test).
Discussion
Our main objectives were to (i) discover the potential transcriptional differences and (ii) compare the periosteal cell proliferation between tibias of young-adult and old mice following strain-matched mechanical loading. First, we used RNA-seq to profile the transcriptomes in the interval spanning from the early response (day 1) through active bone formation (days 3 and 5). Compared to young-adult mice, old mice had less transcriptional activation at each time point, as measured by both the number of DEGs induced and their FCs. Old mice engaged fewer GO processes and pathways and had less activation of processes involving proliferation and differentiation, consistent with previous findings.(24) We observed prominent Wnt signaling, ECM, and neuronal responses, which were all diminished to some extent with aging. We also identified targets that had diminished upregulation in old mice, which may inform therapeutic approaches to restore the aged loading response. Next, we built on previous work(16,27,56) analyzing the aged proliferative response to mechanical loading. Specifically, we used a strain-matched study design to facilitate young-adult versus old comparisons in vivo and BrdU incorporation to assess proliferation. We found that following 5 days of loading, a similar number of surface cells arose via proliferation in young-adult and old mice, implying that the proliferative capacity of old mice in the early interval is not impaired.
Previous studies suggested that old mice are “equally able” to sense loading stimuli yet are unable to coordinate the same bone formation response.(24) In our study, however, the lack of DEGs in old mice at day 1 suggests that early mechanosensing is blunted in old mice, which is consistent with the age-related degeneration of the mechanosensing osteocyte network.(57,58)
Our RNA-seq results support previous studies that implicate increased Wnt signaling as a fundamental feature of the loading response.(23,40) Wnt1 and Wnt7b were strongly induced at all days in young-adult mice but to a lesser extent in old mice. Despite their low absolute expression levels in bone (based on RPKMs), both Wnt1 and Wnt7b are known to be important. Specifically, gain-of-function studies of Wnt1 and Wnt7b in osteoblast-lineage cells have shown that both ligands are osteoanabolic.(59–61) Further, human WNT1 mutations have been associated with early-onset osteoporosis and osteogenesis imperfecta.(62) The cellular sources and targets of these ligands in bone remain incompletely understood. Osteocytes are thought to be the predominant source of Wnt1, which likely targets both adjacent osteocytes and osteoblasts.(60) Even less is known about the sources and targets of Wnt7b. Future studies should elicit the functional roles of these ligands in the loading response. In our pathway analysis, Wnt signaling was exclusively up in young-adult mice. Wnt-related genes Wnt1, Wnt7b, Wnt10b, and Fzd1 drove the activation of the Alzheimer disease–presenilin pathway at days 3 and 5 in young-adult mice, but only day 5 in old mice. Together, these pathway analysis results indicate less and slower activation of Wnt-related processes with aging. In old mice, the GO finding of negative regulation of canonical Wnt signaling pathway and the enrichment of negative regulation of Wnt signaling in the Old Loading Module suggest that Wnt signaling may be actively dampened in old mice (rather than simply not stimulated as strongly). This finding supports the strategy of disinhibiting Wnt signaling to restore the aged loading response (eg, anti-sclerostin antibodies). Finally, although Wnt signaling was robustly enriched, we did not observe consistent indication of noncanonical versus canonical signaling with loading or aging, likely because many of these downstream signaling targets are post-transcriptionally regulated.
In addition, our RNA-seq findings point to a number of targets that might be explored to restore the mechanical loading response in aged bone. First, like the Wnt ligands, Ngf was mechanoresponsive in young-adult mice at each time point but showed blunted upregulation in old mice. Notably, Ngf has been reported to act upstream of Wnt signaling, and inhibiting NGF signaling in young-adult mice mimicked aging by reducing both loading-induced bone formation and Wnt signaling.(41) Thus, the extent to which (re-)activating NGF signaling in old mice could restore loading-induced Wnt signaling and bone formation merits further exploration. Second, one of the drivers of the enriched negative regulation of Wnt signaling in the Old Loading Module was Notum, a secreted enzyme that inactivates Wnt ligands and is expressed by osteoblast-lineage cells.(63) Global Notum knockout led to a striking increase in cortical thickness,(55) and oral inhibitors of Notum increased cortical bone thickness in a dose-dependent manner.(64) Thus, Notum inhibition may offset the impaired Wnt ligand upregulation in aging and rescue the loading response. Third, the higher induction of Ptgs2 (Cox-2) in young-adult versus old mice suggested that prostaglandin signaling is attenuated in the aged loading response. Past work on the role of Cox-2 in loading-induced bone formation has been conflicting,(65–67) but enhancing prostaglandin signaling protected against age-related bone loss.(68) Because many FDA-approved drugs effectively modulate prostaglandin signaling, targeting this pathway should be considered. Fourth, Nell1 upregulation was also attenuated in old mice. Notably, Nell1 plays a role in osteoporotic bone loss, and delivering Nell-1 to ovariectomized mice increased bone formation by enhancing Wnt signaling.(49) Thus, Nell-1 may also be an attractive target to restore the loading response in aging, and the first human trial for Nell-1 was recently approved.(69) Last, both Fosb and Junb, which are members of the mechanoresponsive AP-1 transcription factor family, are upregulated at day 1 in young-adult but not old mice.(43) Thus, although the AP-1 family plays a role in a range of contexts in humans,(70) targeting AP-1 in bone is a possible strategy to restore the loading response in aging.
We also consistently identified changes in ECM-related genes. At day 5, when histological bone formation actively occurs, the upregulation of ECM proteins represented the largest signal. In young-adult mice, a total of 23 collagen transcripts (including Col1a1 and Col1a2) were among the upregulated DEGs at day 5 (Supporting Table S4). Most of these genes (16/23, 70%) were also upregulated DEGs in old mice, but their FCs were lower. Thus, in bones of old mice, the diminished induction of these collagen transcripts is consistent with the diminished bone formation (Supporting Fig. S1).(15) In the GO analysis, collagen fibril organization was up in both ages at days 3 and 5, but ECM organization was exclusively enriched in young-adult mice. These findings suggest that some aspects of the ECM response remain intact with aging while others do not. A number of noncollagenous proteins were also upregulated at day 5. In particular, Bcan and Hapln4 were up >20-fold (loaded/non-loaded). Although these two proteins have not been described in bone’s loading response, they play a role in synaptic stabilization in the brain.(71) Given the nerve-like nature of the osteocytic network, we speculate that these proteins may play a role in the bone response to loading by facilitating osteocytogenesis or osteocyte maintenance.(72)
Consistent with the nerve-like nature of the osteocytic network, we identified a consistent neuronal signal in the loading response. In fact, of the eight DEGs common to every time point in young-adult mice, two—Ddn and Ngf—are classically associated with the nervous system. First, dendrin (Ddn) was originally identified in dendrites in the brain.(73) More recently, in kidney podocytes, dendrin was shown to interact with Yes-associated protein (YAP), a key player in mechanotransduction, to induce apoptosis.(74) Either role in bone—whether functioning in osteocyte dendrites or modulating apoptosis following mechanical loading—remains plausible but should be explored further. Second, in addition to a role for NGF noted above, NGF is a potent growth factor for both sympathetic and sensory neurons.(75) Because osteocytes and osteoblasts express low levels of the high-affinity NGF receptor (TrkA), NGF may target surrounding peripheral nerves to promote nerve sprouting into bone following loading.(41) The axon guidance mediated by semaphorins pathway was exclusively enriched in young-adult mice at day 5. Whether this signal represents a cue for nerve sprouting following loading or is evidence of osteocyte-driven repurposing of neuron-associated pathways remains to be established.(76,77) The latter explanation may suggest that osteocytes induce classical “nerve genes” to remodel their dendritic processes. Either way, this neuronal response does not seem to be fully preserved with aging.
In our proliferation study, the few BrdU+ cells on the periosteal surface in non-loaded tibias indicate negligible cell proliferation in the absence of mechanical loading. With loading, the number of BrdU+ surface cells significantly increased in both ages. These surface cells—because of their location—are presumably the cells responsible for active bone formation, so old mice did not display an impaired recruitment of bone-forming surface cells via proliferation during a 5-day loading interval. Because the BrdU labeling was conducted for 5 days, we cannot rule out the possibility that old mice do not sustain this proliferative response for loading regimens exceeding 5 days. Overall, we found that ~30% to 40% of surface cells arose via proliferation following loading, which is consistent with our previous work in 5-month-old and 12-month-old reporter mice.(27) To explore the origins of these proliferating cells, we considered markers of osteoprogenitors, including Sca-1 (Ly6a)(78) and Ctsk.(79) Sca-1 was unchanged with loading in both age groups, and Ctsk was downregulated in young-adult mice at day 3. This suggests that, at these early time points, there may not be enrichment of osteoprogenitors, which is consistent with our current understanding that the initial bone formation response to mechanical loading is based on the activation and proliferation of existing osterix-lineage cells rather than the differentiation of progenitors.(27)
Our study had several limitations. First, small amounts of contaminant nonbone tissue (including muscle and bone marrow) may significantly impact RNA-seq data.(80) Therefore, we carefully removed other tissues from the tibias and used a previously published 893-gene contaminant tissue list(80) to confirm that few of our DEGs were contaminants (Supporting Fig. S7). Second, because of the minimal trabecular bone in tibias of old mice, no effort was made to separate cortical and trabecular bone, which have differential expression following loading.(40) Third, our study focused on female mice for the reasons stated above. Although we cannot be certain that our findings would be the same in male mice, both female and male C57BL/6 mice exhibit similar age-related cortical bone loss,(16) suggesting that these findings will translate to male mice. Fourth, although a sample size of 5 mice/age/day enabled the identification of many DEGs at later days, a larger sample size would have increased statistical power and allowed genes with more variable expression to be identified as DEGs. Fifth, bulk sequencing cannot distinguish whether transcriptional differences were due to changes in cellular composition (eg, a new osteoblast expressing Col1a1 after arising from cell division) or due to altered expression (eg, the same osteoblast upregulating Col1a1). Sixth, in contrast to other reports, Sost was not strongly downregulated with loading in young-adult mice at day 1 (FC of 0.99, loaded/control), day 3 (0.71), or day 5 (0.81). Similar nonsignificant changes were detected in old mice. We observed a tendency for lower overall Sost expression in old bones. Last, in our proliferation study, we administered BrdU for the entire 5-day loading interval, which allowed us to assess the cumulative number of cells that proliferated in the 5-day loading interval rather than evaluating a single time point.
In summary, old mice had less transcriptional activation following tibial loading compared to young-adult mice. Old mice engaged fewer GO processes and pathways and had less activation of processes involving proliferation and differentiation. Prominent features of the young-adult loading response were Wnt signaling, ECM, and neuronal responses, and these were diminished with aging. We identified a number of targets that may be effective in restoring the mechanoresponsiveness of aged bone, including NGF, Notum, prostaglandin signaling, Nell-1, and the AP-1 family. We also showed that young-adult and old mice had a similar periosteal cell proliferative response following five bouts of daily loading. We conclude that old mice have diminished transcriptional activation following mechanical loading, but periosteal proliferation in the first 5 days of loading does not differ between tibias of young-adult and old mice.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01-AR047867 (to MJS), T32-GM007200 (to CCS), and F30-AG066310 (to CCS). We thank the Washington University Musculoskeletal Research Center (supported by P30-AR074992) for their assistance with histology. We thank the Genome Technology Access Center (supported by P30-CA91842 and UL1TR002345) for help with sequencing and genomic analysis. Finally, we thank Brandon Coates, Jennifer Mckenzie, Nicole Migotsky, and Heather Zannit for advice and critical comments on the manuscript.
Footnotes
Disclosures
All authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Melton LJ, Chrischilles EA, Cooper C, Lane AW, Riggs BL. Perspective. How many women have osteoporosis? J Bone Miner Res 1992;7(9): 1005–10. [DOI] [PubMed] [Google Scholar]
- 2.Melton LJ, Atkinson EJ, O’Connor MK, O’Fallon WM, Riggs BL. Bone density and fracture risk in men. J Bone Miner Res 1998;13(12): 1915–23. [DOI] [PubMed] [Google Scholar]
- 3.Kanis JA, Johnell O, Oden A, et al. Long-term risk of osteoporotic fracture in Malmö. Osteoporos Int 2000;11(8):669–74. [DOI] [PubMed] [Google Scholar]
- 4.Blume SW, Curtis JR. Medical costs of osteoporosis in the elderly Medicare population. Osteoporos Int 2011;22(6):1835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khosla S, Hofbauer LC. Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol 2017;5(11): 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinerman S, Usera GL. Antiresorptive therapies for osteoporosis. Oral Maxillofac Surg Clin North Am 2015;27(4):555–60. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd AA, Gludovatz B, Riedel C, et al. Atypical fracture with long-term bisphosphonate therapy is associated with altered cortical composition and reduced fracture resistance. Proc Natl Acad Sci U S A 2017; 114(33):8722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramchand SK, Seeman E. Advances and unmet needs in the therapeutics of bone fragility. Front Endocrinol (Lausanne). 2018;9:505 10.3389/fendo.2018.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitlak BH, Burr DB, Allen MR. Pharmaceutical treatments of osteoporosis In: Burr DB, Allen MR, editors. Basic and applied bone biology. London, UK: Elsevier; 2014. p. 345–63. Available at: http://linkinghub.elsevier.com/retrieve/pii/B9780124160156000174. Accessed April 21, 2020. [Google Scholar]
- 10.Foundation for the National Institutes of Health (FNIH) Biomarkers Consortium. Foundation for the NIH Launches Bone Quality Project (Internet). Bethesda, MD: FNIH; 2013. [cited 2020 Apr 21]. Available at: https://fnih.org/news/press-releases/fnih-launches-bone-quality-project. [Google Scholar]
- 11.Wolff J [The law of bone transformation] Das gesetz der transformation der knochen. Berlin: Hirschwald; 1892. 152 p. German [Google Scholar]
- 12.Frost HM. The laws of bone structure. Springfield, IL: Thomas; 1964. [Google Scholar]
- 13.Lynch ME, Main RP, Xu Q, et al. Tibial compression is anabolic in the adult mouse skeleton despite reduced responsiveness with aging. Bone. 2011;49(3):439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willie BM, Birkhold AI, Razi H, et al. Diminished response to in vivo mechanical loading in trabecular and not cortical bone in adulthood of female C57Bl/6 mice coincides with a reduction in deformation to load. Bone. 2013;55(2):335–46. [DOI] [PubMed] [Google Scholar]
- 15.Holguin N, Brodt MD, Sanchez ME, Silva MJ. Aging diminishes lamellar and woven bone formation induced by tibial compression in adult C57BL/6. Bone. 2014;65:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meakin LB, Galea GL, Sugiyama T, Lanyon LE, Price JS. Age-related impairment of Bones’ adaptive response to loading in mice is associated with sex-related deficiencies in osteoblasts but no change in osteocytes. J Bone Miner Res 2014;29(8):1859–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birkhold AI, Razi H, Duda GN, Weinkamer R, Checa S, Willie BM. The influence of age on adaptive bone formation and bone resorption. Biomaterials. 2014;35(34):9290–301. [DOI] [PubMed] [Google Scholar]
- 18.Korpelainen R, Keinänen-Kiukaanniemi S, Heikkinen J, Väänänen K, Korpelainen J. Effect of impact exercise on bone mineral density in elderly women with low BMD: a population-based randomized controlled 30-month intervention. Osteoporos Int 2006;17(1):109–18. [DOI] [PubMed] [Google Scholar]
- 19.Howe TE, Shea B, Dawson LJ, et al. Exercise for preventing and treating osteoporosis in postmenopausal women. Cochrane Database of Syst Rev 2011;(7):CD000333 http://doi.wiley.com/10.1002/14651858.CD000333.pub2. Accessed April 21, 2020. [DOI] [PubMed] [Google Scholar]
- 20.Marques EA, Mota J, Carvalho J. Exercise effects on bone mineral density in older adults: a meta-analysis of randomized controlled trials. Age (Dordr) 2012;34(6):1493–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Souza RL, Matsuura M, Eckstein F, Rawlinson SCF, Lanyon LE, Pitsillides AA. Non-invasive axial loading of mouse tibiae increases cortical bone formation and modifies trabecular organization: a new model to study cortical and cancellous compartments in a single loaded element. Bone. 2005;37(6):810–8. [DOI] [PubMed] [Google Scholar]
- 22.Fritton JC, Myers ER, Wright TM, van der Meulen MCH. Loading induces site-specific increases in mineral content assessed by micro-computed tomography of the mouse tibia. Bone. 2005;36(6):1030–8. [DOI] [PubMed] [Google Scholar]
- 23.Holguin N, Brodt MD, Silva MJ. Activation of Wnt signaling by mechanical loading is impaired in the bone of old mice: load-induced activation of Wnt signaling impaired in bone of old mice. J Bone Miner Res 2016;31(12):2215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galea GL, Meakin LB, Harris MA, et al. Old age and the associated impairment of bones’ adaptation to loading are associated with transcriptomic changes in cellular metabolism, cell-matrix interactions and the cell cycle. Gene 2017;599:36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodine PVN, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord 2006;7(1):33–9. [DOI] [PubMed] [Google Scholar]
- 26.Turner CH, Owan I, Alvey T, Hulman J, Hock JM. Recruitment and proliferative responses of osteoblasts after mechanical loading in vivo determined using sustained-release bromodeoxyuridine. Bone. 1998;22(5):463–9. [DOI] [PubMed] [Google Scholar]
- 27.Zannit HM, Silva MJ. Proliferation and activation of Osterix-lineage cells contribute to loading-induced periosteal bone formation in mice. JBMR Plus. 2019;3(11):e10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meakin LB, Sugiyama T, Galea GL, Browne WJ, Lanyon LE, Price JS. Male mice housed in groups engage in frequent fighting and show a lower response to additional bone loading than females or individually housed males that do not fight. Bone. 2013;54(1):113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch ME, Main RP, Xu Q, et al. Cancellous bone adaptation to tibial compression is not sex dependent in growing mice. J Appl Physiol (1985). 2010;109(3):685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D, Brodt MD, Zannit HM, Holguin N, Silva MJ. Evaluation of loading parameters for murine axial tibial loading: stimulating cortical bone formation while reducing loading duration. J Orthop Res 2018;36:682–91. http://doi.wiley.com/10.1002/jor.23727. Accessed April 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holguin N, Brodt MD, Sanchez ME, Kotiya AA, Silva MJ. Adaptation of tibial structure and strength to axial compression depends on loading history in both C57BL/6 and BALB/c mice. Calcif Tissue Int 2013;93(3):211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel TK, Brodt MD, Silva MJ. Experimental and finite element analysis of strains induced by axial tibial compression in young-adult and old female C57Bl/6 mice. J Biomech 2014;47(2):451–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly NH, Schimenti JC, Patrick Ross F, van der Meulen MCH. A method for isolating high quality RNA from mouse cortical and cancellous bone. Bone. 2014;68:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4(9):R60 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- 37.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003;13(9):2129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformat 2008;9(1):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4(1): Article17. [DOI] [PubMed] [Google Scholar]
- 40.Kelly NH, Schimenti JC, Ross FP, van der Meulen MCH. Transcriptional profiling of cortical versus cancellous bone from mechanically-loaded murine tibiae reveals differential gene expression. Bone. 2016;86:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlinson RE, Li Z, Li Z, et al. NGF-TrkA signaling in sensory nerves is required for skeletal adaptation to mechanical loads in mice. Proc Natl Acad Sci U S A 2017;114(18):E3632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunér F, Patrakka J, Xiao Z, et al. Dendrin expression in glomerulo-genesis and in human minimal change nephrotic syndrome. Nephrol Dial Transplant. 2008;23(8):2504–11. [DOI] [PubMed] [Google Scholar]
- 43.Inoue D, Kido S, Matsumoto T. Transcriptional induction of FosB/ΔFosB gene by mechanical stress in osteoblasts. J Biol Chem 2004;279(48):49795–803. [DOI] [PubMed] [Google Scholar]
- 44.van der Rest M, Mayne R, Ninomiya Y, Seidah NG, Chretien M, Olsen BR. The structure of type IX collagen. J Biol Chem 1985;260 (1):220–5. [PubMed] [Google Scholar]
- 45.Kitase Y, Lee S, Gluhak-Heinrich J, Johnson ML, Harris SE, Bonewald LF. CCL7 is a protective factor secreted by mechanically loaded osteocytes. J Dent Res 2014;93(11):1108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Withers CN, Brown DM, Byiringiro I, et al. Rad GTPase is essential for the regulation of bone density and bone marrow adipose tissue in mice. Bone. 2017;103:270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimada T, Kakitani M, Yamazaki Y, et al. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J Clin Invest 2004;113(4):561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnsdorf EJ, Tummala P, Jacobs CR. Non-canonical Wnt signaling and N-cadherin related β-catenin signaling play a role in mechanically induced osteogenic cell fate. PLoS One. 2009;4(4):e5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.James AW, Shen J, Zhang X, et al. NELL-1 in the treatment of osteoporotic bone loss. Nat Commun 2015;6:7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting β-catenin phosphorylation in the nucleus. J Biol Chem 2009;284(5):3323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S, Kurenbekova L, Gao Y, et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene 2015;34(39):5069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uemura T, Mohri J, Osada H, Suzuki N, Katagiri N, Minaguchi H. Effect of gonadotropin-releasing hormone agonist on the bone mineral density of patients with endometriosis. Fertil Steril. 1994;62(2): 246–50. [DOI] [PubMed] [Google Scholar]
- 53.Raffa RB, Ossipov MH, Porreca F. Opioid analgesics and antagonists In: Dowd FJ, Johnson BS, Mariotti AJ. Pharmacology and therapeutics for dentistry. 7th ed. St. Louis, MO: Mosby; 2017. p. 241–56. http://www.sciencedirect.com/science/article/pii/B9780323393072000163. Accessed April 21, 2020. [Google Scholar]
- 54.Rosen H, Bar-Shavit Z. Dual role of osteoblastic proenkephalin derived peptides in skeletal tissues. J Cell Biochem. 1994;55(3):334–9. [DOI] [PubMed] [Google Scholar]
- 55.Brommage R, Liu J, Vogel P, et al. NOTUM inhibition increases endocortical bone formation and bone strength. Bone Res. Nature Publishing Group. 2019;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabahug-Zuckerman P, Liu C, Cai C, et al. Site-specific load-induced expansion of Sca-1+Prrx1 + and Sca-1 −Prrx1 + cells in adult mouse long bone is attenuated with age. JBMR Plus. 2019;3(9):e10199 10.1002/jbm4.10199. Accessed April 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tiede-Lewis LM, Xie Y, Hulbert MA, et al. Degeneration of the osteocyte network in the C57BL/6 mouse model of aging. Aging (Albany, NY). 2017;9:2190–208. 10.18632/aging.101308. Accessed April 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrell AE, Robinson ST, Silva MJ, Guo XE. Mechanosensitive Ca2+ signaling and coordination is diminished in osteocytes of aged mice during ex vivo tibial loading. Connect Tissue Res 2020;61:389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J, Tu X, Esen E, et al. WNT7B promotes bone formation in part through mTORC1. PLoS Genet. 2014;10(1):e1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joeng KS, Lee Y-C, Lim J, et al. Osteocyte-specific WNT1 regulates osteoblast function during bone homeostasis. J Clin Invest 2017; 127(7):2678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luther J, Yorgan TA, Rolvien T, et al. Wnt1 is an Lrp5-independent bone-anabolic Wnt ligand. Sci Translat Med 2018;10(466). https://stm.sciencemag.org/content/10/466/eaau7137. Accessed January 15, 2020. [DOI] [PubMed] [Google Scholar]
- 62.Laine CM, Joeng KS, Campeau PM, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med 2013;368(19):1809–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Movérare-Skrtic S, Nilsson KH, Henning P, et al. Osteoblast-derived NOTUM reduces cortical bone mass in mice and the NOTUM locus is associated with bone mineral density in humans. FASEB J 2019; 33(10):11163–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brommage R, Liu J, Vogel P, et al. NOTUM inhibition increases endocortical bone formation and bone strength. Bone Res 2019;7(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forwood MR. Inducible cyclo-oxygenase (COX-2) mediates the induction of bone formation by mechanical loading in vivo. J Bone Miner Res 1996;11(11):1688–93. [DOI] [PubMed] [Google Scholar]
- 66.Alam I, Warden SJ, Robling AG, Turner CH. Mechanotransduction in bone does not require a functional cyclooxygenase-2 (COX-2) gene. J Bone Miner Res 2005;20(3):438–46. [DOI] [PubMed] [Google Scholar]
- 67.Sugiyama T, Meakin LB, Galea GL, Lanyon LE, Price JS. The cyclooxygenase-2 selective inhibitor NS-398 does not influence trabecular or cortical bone gain resulting from repeated mechanical loading in female mice. Osteoporos Int 2013;24(1):383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang M, Feigenson M, Sheu T, et al. Loss of the PGE2 receptor EP1 enhances bone acquisition, which protects against age and ovariectomy-induced impairments in bone strength. Bone. 2015;72: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bone Biologics Receives Human Research Ethics Committee (HREC). Approval for the First Center of a Multicenter Pilot Clinical Trial to Evaluate NB1 (NELL-1/DBX®) in Australia [Internet]. AP NEWS; 2019. March 25 Available from: https://apnews.com/Business%20Wire/dca8fa880907483192342298e0cff151. Accessed April 21, 2020. [Google Scholar]
- 70.Trop-Steinberg S, Azar Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am J Med Sci 2017;353(5):474–83. [DOI] [PubMed] [Google Scholar]
- 71.Bekku Y, Su W-D, Hirakawa S, et al. Molecular cloning of Bral2, a novel brain-specific link protein, and immunohistochemical colocalization with brevican in perineuronal nets?. Mol Cell Neurosci 2003;24(1): 148–59. [DOI] [PubMed] [Google Scholar]
- 72.Qing H, Bonewald LF. Osteocyte remodeling of the perilacunar and pericanalicular matrix. Int J Oral Sci 2009;1(2):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neuner-Jehle M, Denizot J-P, Borbély AA, Mallet J. Characterization and sleep deprivation-induced expression modulation of dendrin, a novel dendritic protein in rat brain neurons. J Neurosci Res 1996;46 (2):138–51. [DOI] [PubMed] [Google Scholar]
- 74.Campbell KN, Wong JS, Gupta R, et al. Yes-associated protein (YAP) promotes cell survival by inhibiting Proapoptotic Dendrin signaling. J Biol Chem 2013;288(24):17057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen S, Levi-Montalcini R. Purification and properties of a nerve growth-promoting factor isolated from mouse sarcoma 180. Cancer Res 1957;17(1):15–20. [PubMed] [Google Scholar]
- 76.Paic F, Igwe JC, Nori R, et al. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45(4): 682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell … and more. Endocr Rev 2013;34(5):658–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Vlasselaer P, Falla N, Snoeck H, Mathieu E. Characterization and purification of osteogenic cells from murine bone marrow by two-color cell sorting using anti-Sca-1 monoclonal antibody and wheat germ agglutinin. Blood. 1994;84(3):753–63. [PubMed] [Google Scholar]
- 79.Debnath S, Yallowitz AR, McCormick J, et al. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018; 562(7725):133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ayturk UM, Jacobsen CM, Christodoulou DC, et al. An RNA-seq protocol to identify mRNA expression changes in mouse diaphyseal bone: applications in mice with bone property altering Lrp5 mutations. J Bone Miner Res 2013;28(10):2081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


