Abstract
Potyvirus species associated with yellow leaf stripe disease of Indian narcissus (Narcissus tazetta L.) var. Paperwhite has been studied by sequence analyses of ~ 1.5 kb genomic fragments obtained from seven RT-PCR amplifications of infected samples. Sequence analysis revealed the occurrence of three potyvirus species: cyrtanthus elatus virus-A (CEVA: KF430815, KF430816, KM066973, KM066974); narcissus yellow stripe virus (NYSV: KM066972, JQ686724) and narcissus degeneration virus (NDV: MK572806). The existence of three potyvirus species: CEVA, NYSV and NDV are being reported in Indian narcissus.
Keywords: N. tazetta var. paperwhite, Leaf yellow stripe, RT-PCR amplification, Sequence identity, Phylogenetic relationships, CEVA, NYSV, NDV
Introduction
Indian narcissus (Narcissus tazetta L.) of the family Amaryllidaceae is a bulbous ornamental plant. It is popular for its beautiful flowers in the garden beds and used as a cut-flower for bouquets, vases and also for the production of fragrant oil and perfumes in India. N. tazetta is common ornamental species in the Mediterranean region (from Portugal to Turkey), considered as its native place, though, its dissemination extended throughout the Asian countries with demand-based international trading (Harvey and Selby 1997; Hanks 2002; Kamenetsky and Okubo 2012). Now, N. tazetta are widely grown in China, Israel, India, and Japan, and the large volumes of field-grown cut-flowers are traded, along with other commercially important flowers and pot-grown plants as well as the bulbs (Hank and Chastagner 2018).
Narcissus are reported worldwide to be infected by narcissus mosaic virus (NMV), narcissus yellow stripe virus (NYSV), narcissus late season yellow virus (NLSYV), narcissus degeneration virus (NDV), narcissus latent virus (NLV), narcissus tip necrosis virus (NTNV), cyrtanthus elatus virus-A (CEVA) and ornithogalum mosaic virus (OrMV) causing streaks, yellow stripe and tip necrosis symptoms on the leaf (Brunt 1977, 2008; Wylie and Jones 2012). Other viruses such as raspberry ring spot virus (RRSV), nerine latent virus (NeLV), narcissus symptomless virus (NSV), arabis mosaic virus (ArMV), cucumber mosaic virus (CMV), tobacco rattle virus (TRV) and tomato black ring virus (TBRV) are also reported to infect narcissus though their frequency of infection is reported considerably less as compared to above said viruses (Brunt 1995). Literature survey revealed that infection of said viruses reduced the quality and productivity of narcissus bloom and bulbs (Hanks and Chastagner 2018). Among them, potyviruses are the most prevalent viruses of narcissus including NYSV, NLSYV, NDV, CEVA and OrMV (Chen et al. 2006; Yadav and Khan 2008; Kumar et al. 2015; Ohshima et al. 2016; Raj et al. 2018).
Narcissus propagates vegetatively through its bulbs and, therefore, growing plants, if infected by virus, continuously pass infection from generation to generation through propagations of infected mother stocks (Milosevic et al. 2012). Consequently, whole population of narcissus may be infected by the virus if not protected timely and hence reliable early diagnosis and identification of virus is essential for designing their efficient disease management. In the Indian scenario, previous works revealed that narcissus are infected by potyviruses based on study of symptoms, electron microscopy, molecular weight of protein subunits and virus detection by serological methods, and reverse transcription-polymerase chain reaction (RT-PCR) followed by sequence analyses of PCR amplicons (Aminuddin et al. 1999; Yadav and Khan 2008, 2015; Chandel et al. 2010; Kumar et al. 2015). In the present study, we report the existence of three potyvirus species: CEVA, NYSV and NDV in N. tazetta var. Paperwhite based on the analyses of 3′partial genome sequences of viral genome amplified by RT-PCR using potyvirus degenerate primers.
Materials and methods
For detection and molecular characterization of virus, leaf samples from eight (N. tazetta L.) var. Paperwhite plants showing yellow stripe symptoms were collected from three locations: cultivated fields, garden beds, and experimental plots of CSIR- National Botanical Research Institute (NBRI), Lucknow. The total genomic RNA was isolated from 100 mg leaf samples of infected, healthy narcissus plants and a positive control (Kumar et al. 2015) using Sigma kit Spectrum™ Plant total RNA kit (Sigma-Aldrich, Missouri, USA) and used as template for RT-PCR.
For potyvirus detection, RT-PCRs were performed in Thermal cycler (PTC 200 DNA engine of MJ Research, USA) following the protocol described elsewhere (Raj et al. 2019) using total RNA and Pot-I/Pot-II degenerate primers capable of amplifying 3′-partial genome of all potyvirus (Gibbs and Mackenzie 1997). The obtained PCR products were separated through electrophoreses in 1% agarose gel to check the presence of amplicon using standard DNA marker.
For cloning and sequencing, the amplicons were eluted using Wizard SV Gel and PCR Clean-Up System (Promega, CA, USA). The purified products were ligated into pGEM-T Easy Vector System and transformed into competent E. coli (DH5α) cells. The transformants were screened by restriction digestion with EcoR1 enzyme and three positive clones of each sample were sequenced. The obtained sequence data were analysed and assembled using BIOEDIT tool (https://www.mbio.ncsu.edu/bioedit/bioedit.html) to eliminate any sequence ambiguity, and consensus sequences were determined and submitted to the GenBank database.
The sequences were analyzed using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for potyvirus identification. Also, the DiAlign tool (https://www.genomatix.de/cgi-bin/dialign/dialign.pl) was used to obtain nucleotide and amino acid identities with the selected potyviruses listed in Table 1. The phylogenetic analysis of sequences was performed by Molecular Evolutionary Genetics Analysis (MEGA) v7.0 tool using the Maximum Likelihood algorithm at 1000 bootstrap value (Kumar et al. 2016).
Table 1.
Details of sequence data of samples under study and other potyvirus isolates taken for this study
| Virus | Isolate | Host | Location | Genome length (nt) | GenBank Accession | Source |
|---|---|---|---|---|---|---|
| NYSV | NAR-1 | Narcissus tazetta | India | 1559 | JQ686724 | Under study |
| NYSV | NAR-2 | Narcissus tazetta | India | 1559 | KM066972 | Under study |
| NYSV | Zhangzhou | Narcissus tazetta | China | 1669 | AJ311372 | Chen et al. (2003) |
| NYSV | NaKM1-CL7 | Narcissus tazetta | Japan | 1738 | LC158481 | Ohshima el al. (2018) |
| NYSV | NsN14-CL6 | Narcissus tazetta | Japan | 1738 | LC158491 | Ohshima el al. (2018) |
| NYSV | NaN14-CL2 | Narcissus tazetta | Japan | 1738 | LC158490 | Ohshima el al. (2018) |
| INP | Lucknow 2 | Narcissus tazetta | India | 2912 | EU888298 | Yadav and Khan (2008) |
| NLSYV | NaF1-CL4 | Narcissus tazetta | Japan | 1737 | LC158449 | Ohshima el al. (2016) |
| NLSYV | Hangzhou 2 | Narcissus tazetta | China | 1668 | AJ493579 | Chen et al. (2003) |
| NLSYV | – | Narcissus cv. Missouri | UK | 1608 | EU887015 | Monger and Nixon (2008) |
| INP | Lucknow | Narcissus spp. | India | 1558 | DQ991145 | Yadav and Khan (2008) |
| NDV | NBRI-NDV1 | Narcissus tazetta | India | 1577 | MK572806 | Under study |
| NDV | NaKM9-CL5 | Narcissus spp. | Japan | 1612 | LC158497 | Ohshima el al. (2016) |
| NDV | NaSG8-CL3 | Narcissus tazetta | Japan | 1612 | LC158500 | Ohshima el al. (2016) |
| NDV | NV-3 | Narcissus tazetta | China | 1566 | EU200456 | Wylie and Jones (2012) |
| NDV | NaN14-CL8 | Narcissus tazetta | Japan | 1615 | LC158506 | Ohshima el al. (2016) |
| NDV | NaF1-CL16 | Narcissus tazetta | Japan | 1612 | LC158495 | Ohshima el al. (2016) |
| NDV | NaKM9-CL18 | Narcissus tazetta | Japan | 1612 | LC158498 | Ohshima el al. (2016) |
| CEVA | NBRI-2 | Narcissus tazetta | India | 1601 | KF430815 | Under study |
| CEVA | NBRI-1 | Narcissus tazetta | India | 1601 | KF430816 | Under study |
| CEVA | NBRI-4 | Narcissus tazetta | India | 1601 | KM066974 | Under study |
| CEVA | NBRI-3 | Narcissus tazetta | India | 1601 | KM066973 | Under study |
| CEVA | WA-1 | Cyrtanthus elatus virus | Australia | 1056 | GU812282 | Wylie et al. (2010) |
| CEVA | NaN19-CL3 | Cyrtanthus elatus | Japan | 1825 | LC158493 | Ohshima el al. (2016) |
| CEVA | – | Vallota speciosa | New Zealand | 1602 | DQ417604 | Unpublished |
| CEVA | – | Narcissus sp. | UK | 1560 | FJ032248 | Unpublished |
| CEVA | WA-2 | Cyrtanthus elatus | Australia | 1048 | GU812283 | Wylie et al. (2010) |
| OrMV | Hangzhou | Narcissus | China | 1667 | AJ493580 | Chen et al. (2003) |
| JYMV | J2 | Japanese yam | Japan | 876 | AB027009 | Fuji et al. (2000) |
NYSV: narcissus yellow stripe virus; INP: Indian narcissus virus; NLSYV: narcissus late season yellow virus; CEVA: cyrtanthus elatus virus-A; NDV: narcissus degeneration virus; JYMV: Japanese yam mosaic virus; OrMV: ornithogalum moasic virus
Results
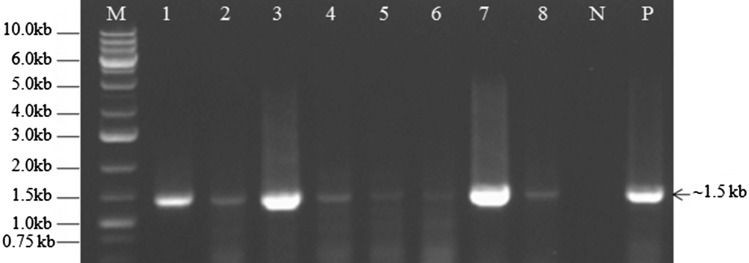
During our surveys, leaf yellow stripe symptoms on N. tazetta var. Paperwhite plants were observed in cultivated fields, garden beds and experimental plots of CSIR-NBRI, Lucknow. The RT-PCRs using potyvirus degenerate primers (Pot-I/Pot-II) revealed the presence of expected size ~ 1.5 kb amplicons in all eight leaf samples similar to as the positive control, while no such amplicon could be obtained in a healthy narcissus sample, indicated the presence of potyvirus (Fig. 1).
Fig. 1.
Gel electrophoresis of RT-PCR products obtained by Potyvirus generate primers (Gibbs and Mackenzie 1997) showing successful amplification of expected size ~ 1.5 kb band in all eight infected narcissus samples (lanes 1–8) similar to a positive control (lane P) but no such amplicon was obtained in the healthy narcissus sample (lane N) taken as negative control. M = 1.0 kb DNA ladder marker
The obtained RT-PCR amplicons of ~ 1.5 kb size were cloned and transformed into E. coli cells as described in MM. The plasmids from positive clones when digested with EcoR1 showed presence of about 1.5 kb of DNA insert. The three positive clones of each sample were sequenced and consensus sequences of each sample were submitted in GenBank under the accession numbers: KF430816 (NBRI-1), KM066974 (NBRI-2), KM066973 (NBRI-3), KF430815 (NBRI-4), JQ686724 (NAR-1), KM066972 (NAR-2) and MK572806 (NBRI-NDV1R).
The submitted NAR-1 and NAR-2 sequences shared 100% nucleotide sequence identity with each other and 75–97% with other NYSV sequences reported all over the word during the BLASTn analysis. They also revealed highest 97% nucleotide identity with NYSV-Zhangzhou (Acc. No. AJ311372) reported from China and 72–77% identities with NLSYV (the other potyvirus species) reported on the same host, N. tazetta. BLASTn analysis of NBRI-1, NBRI-2, NBRI-3 and NBRI-4 sequences revealed 88–100% nucleotide sequence identities to each other and 80–98% with CEVA. The highest 98% nucleotide sequence identity was obtained with CEVA reported from Australia (GU812283) and New Zealand (DQ417604), while 67–68% identities were with NDV reported all over the world on N. tazetta. Whereas, NBRI-NDV1R sequence showed highest 97% nucleotide sequence identity with partial polyprotein gene of Chinese narcissus potyvirus (ChNP)-Chongming Island (AJ311374), ChNP-Zhangzhou (AJ311373); narcissus degeneration virus (NDV)-Zhangzhou (AM182028) reported from Republic of China. NBRI-NDV1R also showed 96% identity with NDV-NV-3 (China, EU200456); NDV-NaN14-CL8 (Japan, LC158506); NDV-NaN9-CL12 (Japan, LC158504); NDV-Marijiniup 2 (Australia, JQ395041); NDV-NaF1-CL16 (Japan, LC158495); NDV-NaN9-CL14 (Japan, LC158505), NDV-NaN9-CL9 (Japan, LC158503) and NDV-NaF1-CL14 (Japan, LC158494) isolates.
Furthermore, the sequence analysis using Genomatix DiAlign tool employing NAR-1 sequence revealed its highest 91% and 85% identity at nucleotide (nt) and amino acid (aa) levels, respectively, with NAR2 sequence. It also showed 86–87% and 83–84% identities at nt and aa levels, respectively, with other isolates of NYSV (LC158490, LC158491, LC158481, AJ311372) reported from Japan and China (Table 2). The identities were 77% and 82% at nt and aa levels, respectively, with Indian narcissus virus (INV, EU888298). While, the identities were lesser, 54–57% at nt and 60–62% at aa level with NLSYV isolates reported from China and the United Kingdom (Table 2). The NBRI-1 sequence revealed highest 97% and 98% identity at the nt and aa levels, respectively, with CEVA isolate (DQ417604) reported from New Zealand. It also showed 94% identity at nt and 97–98% identities at the aa levels with NBRI-3 and NBRI-4 sequences under study while, identities were 80% at nt and 92% at aa with NBRI-2 sequence. It also showed identities were 75–80% at nt and 80–92% at aa with other isolates of CEVA reported from Japan (NaN19-CL3, LC158493) and Australia (WA-1, GU812282) (Table 2). The analysis of NBRI-NDV1R sequence revealed identity from 94 to 95% and 96 to 97% at nt and aa levels, respectively, with NDV isolates reported from Japan (NaKM9-CL5: LC158497, NaSG8-CL3: LC158500) and China (NV-3: EU200456) (Table 2). BLASTn and Genomatix DiAlign-based sequence analyses of NBRI-1, NBRI-2, NBRI-3, NBRI-4; NAR-1, NAR-2; and NBRI-NDV1R sequences under study with the published available sequences in GenBank putatively identified them as potyvirus isolates belonging to CEVA, NYSV and NDV, respectively.
Table 2.
Pair wise percent identities of NYSV (NAR-1: JQ686724), NDV (NDV1R: MK572806) and CEVA (NBRI1: KF430816) sequences under study at nucleotide (nt) and its amino acid (aa) with respective sequences of other potyviruses available in GenBank using DiAlign tool
| Gene Bank Accession | Virus | Isolate | Location | Percent identity nt (aa) |
|---|---|---|---|---|
| Sequence identities of NYSV NAR-1 (JQ686724) with other potyvirus isolates | ||||
| KM066972 | NYSV | NAR-2 | India | 91 (90) |
| AJ311372 | NYSV | Zhangzhou | China | 87 (89) |
| LC158481 | NYSV | NaKM1-CL7 | Japan | 86 (89) |
| LC158491 | NYSV | NsN14-CL6 | Japan | 86 (89) |
| LC158490 | NYSV | NaN14-CL2 | Japan | 86 (89) |
| LC158449 | NLSYV | NaF1-CL4 | Japan | 58 (68) |
| AJ493579 | NLSYV | Hangzhou 2 | China | 57 (69) |
| EU887015 | NLSYV | – | UK | 54 (67) |
| DQ991145 | INP | Lucknow | India | 32 (44) |
| EU888298 | INP | Lucknow 2 | India | 77 (85) |
| AJ493580 | OMV | Hangzhou | China | 34 (49) |
| AB027009 | JYMV | J2 | Japan | 51 (53) |
| Sequence identities of NDV NBRI-NDV1 (MK572806) with other potyvirus isolates | ||||
| LC158497 | NDV | NaKM9-CL5 | Japan | 94 (96) |
| LC158500 | NDV | NaSG8-CL3 | Japan | 94 (96) |
| EU200456 | NDV | NV-3 | China | 95 (97) |
| LC158506 | NDV | NaN14-CL8 | Japan | 94 (97) |
| LC158495 | NDV | NaF1-CL16 | Japan | 95 (96) |
| LC158498 | NDV | NaKM9-CL18 | Japan | 94 (96) |
| LC158449 | NLSYV | NaF1-CL4 | Japan | 37 (53) |
| AJ493579 | NLSYV | Hangzhou 2 | China | 38 (54) |
| EU887015 | NLSYV | – | UK | 38 (52) |
| DQ991145 | INP | Lucknow | India | 96 (97) |
| EU888298 | INP | Lucknow 2 | India | 35 (50) |
| AJ493580 | OMV | Hangzhou | China | 38 (49) |
| AB027009 | JYMV | J2 | Japan | 48 (51) |
| Sequence identities of CEVA NBRI-1 (KF430816) with other potyvirus isolates | ||||
| KF430815 | CEVA | NBRI-2 | India | 75 (80) |
| KM066973 | CEVA | NBRI-3 | India | 94 (97) |
| KM066974 | CEVA | NBRI-4 | India | 94 (98) |
| DQ417604 | CEVA | – | New Zealand | 97 (98) |
| LC158493 | CEVA | NaN19-CL3 | Japan | 78 (89) |
| GU812282 | CEVA | WA-1 | Australia | 80 (92) |
| FJ032248 | CEVA | – | UK | 77 (86) |
| GU812283 | CEVA | WA-2 | Australia | 79 (92) |
| LC158449 | NLSYV | NaF1-CL4 | Japan | 39 (54) |
| AJ493579 | NLSYV | Hangzhou 2 | China | 37 (53) |
| EU887015 | NLSYV | – | UK | 37 (52) |
| DQ991145 | INP | Lucknow | India | 57 (70) |
| EU888298 | INP | Lucknow 2 | India | 35 (51) |
| AJ493580 | OrMV | Hangzhou | China | 35 (53) |
| AB027009 | JYMV | J2 | Japan | 45 (50) |
NYSV: narcissus yellow stripe virus, NLSYV: narcissus late season yellows virus, CEVA: cyrtanthus elatus virus A, INP: Indian narcissus virus, JYMV: Japanese yam mosaic virus, NDV: narcissus degeneration virus, OrMV: ornithogalum mosaic virus
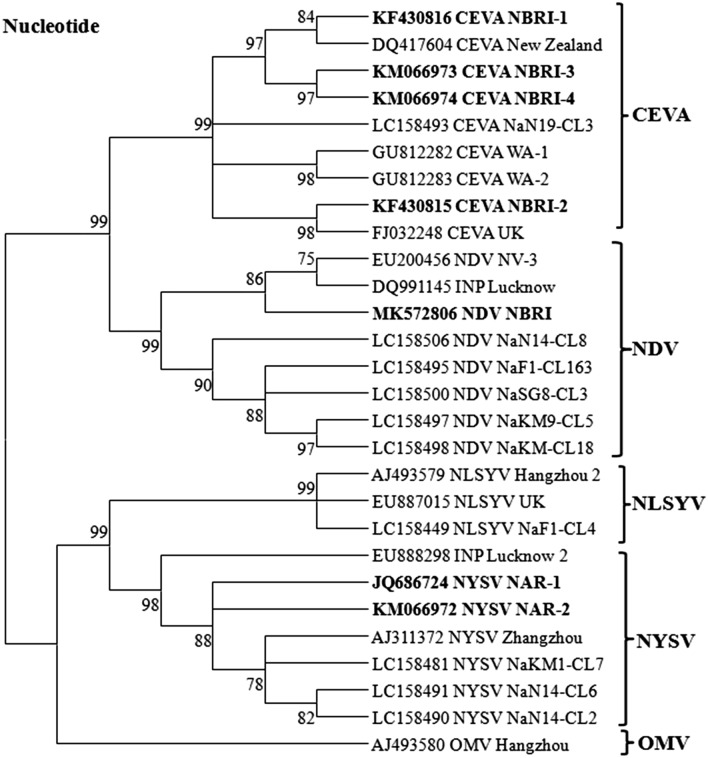
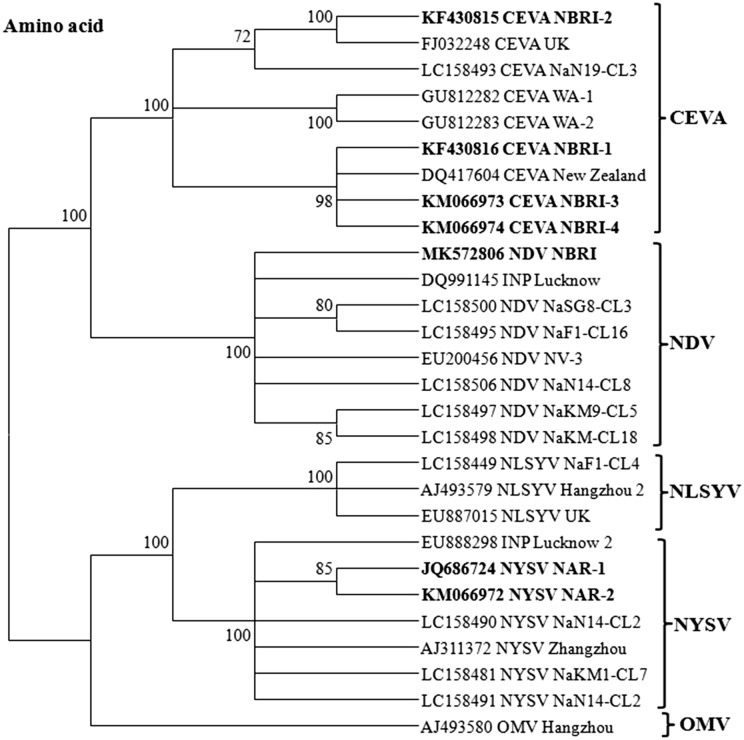
Phylogenetic analysis of all sequences under study was performed both at nucleotide (Fig. 2) and amino acid (Fig. 3) sequence levels with the selected sequences of NYSV, CEVA and NDV reported worldwide along with some other closely related potyvirus as an out group sequences by MEGA tool. Phylogeny revealed more or less similar results both at nucleotide and amino acid sequence levels for NYSV and CEVA sequences under study (Figs. 2, 3). The NAR-1 and NAR-2 sequences clustered together and showed closest phylogenetic relationships with 99% bootstrap value with NYSV-Zhangzhou reported from China (Chen et al. 2003) and other NYSV isolates (LC158481, LC158490, and LC158491) reported from Japan (Ohshima el al. 2018). They also showed a close homology with NLSYV isolates reported from China (AJ493579, Chen et al. 2003) and UK (EU887015). While, they showed distant relationships with INV (EU888298) reported from India (Yadav and Khan 2015), and other potyviruses considered in the present study (Figs. 2, 3).
Fig. 2.
Phylogenetic tree showing relationships of sequences under study: NYSV (KM066972 and JQ686724); CEVA (KF430816, KM066974, KM066973, and KF430815) and NDV (NBRI-NDV1R: MK572806) at nucleotide sequence level with the respective sequences of other selected potyvirus isolates reported worldwide (listed in Table 1). The tree was constructed employing the MEGA v7.0 tool and the Maximum Likelihood method with 1000 bootstrap replicates. The tree is drawn to scale with branch lengths measured in the number of substitutions per site and the percentage of trees in which the associated taxa clustered together shown next to the branches
Fig. 3.
Phylogenetic tree showing relationships of sequences under study: NYSV (KM066972 and JQ686724); CEVA (KF430816, KM066974, KM066973, and KF430815) and NDV (NBRI-NDV1R: MK572806) at amino acid sequence level with the respective sequences of other selected potyvirus isolates reported worldwide (listed in Table 1). The tree was constructed employing the MEGA v7.0 tool and the Maximum Likelihood method with 1000 bootstrap replicates
During analysis of CEVA sequences under study both at nucleotide and amino acid levels with the sequences of other potyvirus isolates considered for study, the NBRI-1, NBRI-3 and NBRI-4 showed close relationships and clustered with CEVA-New Zealand isolate (DQ417604). Contrary to this, NBRI-2 showed closest relationship and clustered with CEVA-WA-1 isolate (GU812282) reported from Australia (Wylie et al. 2010) and CEVA-NaSG8-CL3 isolate (LC158493) from Japan (Ohshima el al. 2016). These results of phylogeny analyses both at the level of nucleotide and amino acid sequences clearly indicate sequence diversity among sequences under study (Figs. 2, 3).
The NBRI-NDV1R sequence clustered with reported NDV isolates and revealed closest relationships with NDV-NV-3 isolate (EU200456) from China (Wylie and Jones 2012) and Indian narcissus virus (DQ991145) from India which was subsequently identified as lycoris virus (Yadav and Khan 2008). Our sequence also showed close relationships with other isolates of NDV: NaSG8-CL3 (LC158500), NaKM9-CL5 (LC158497) and NaKM9-CL18 (LC158498) reported from Japan (Ohshima el al. 2016), whereas distant relationships with other potyviruses considered in the present study at nucleotide and amino acid levels (Figs. 2, 3). Based on pair-wise sequence identity and phylogenetic analyses, the NBRI-1, NBRI-2, NBRI-3 and NBRI-4; NAR-1 and NAR-2 and NBRI-V1R sequences from narcissus were identified as isolates of CEVA, NYSV and NDV, respectively.
Discussion
The yellow stripe symptoms on leaves were observed in several N. tazetta var. Paperwhite plants growing in cultivated field, garden beds and experimental plots at CSIR-NBRI, Lucknow. The exhibited disease symptoms in narcissus were found similar to those described earlier for potyvirus in India (Aminuddin et al. 1999; Khan and Yadav 2008; Kumar et al. 2015) and from abroad (Wylie et al. 2014; Ohshima et al. 2018) hence infection of potyvirus was suspected. Therefore, detection of potyvirus was attempted by RT-PCR using Pot I and Pot II degenerate potyvirus primers capable of amplifying conserved 3′-UTR to partial Nlb region and as used for detection of a variety of potyviruses (Gibbs and Mackenzie 1997).
The analyses of nucleotide and amino acid sequence data of cloned PCR products resulted in identification of three potyvirus species: NYSV, CEVA and NDV in accordance to the ICTV species demarcation criteria suggested for potyvirus speciation that coat protein (CP) coding region sequences were considered usually to be < 80% amino acid and < 76% nucleotide sequence identities (Adams et al. 2005; Zerbini et al. 2012). The pair-wise sequence identity and phylogenetic analysis of NYSV and CEVA isolates revealed that sequences under study were genetically diverse and showed approximately 33% nucleotide sequence diversity among them. NYSV and CEVA were reported early in 1908 for the first time in narcissus in the UK (Darlington 1908) and India (Kumar et al. 2015), however, NDV has been detected in narcissus for the first time in India.
In the present study, Indian NYSV sequences showed highest sequence identities at nucleotide and amino acid levels, and close phylogenetic relationships with NYSV isolates reported from China (Chen et al. 2003) and from Japan (Ohshima et al. 2016). This may be due to the migration of virus through some human activity, through transport of any planting material and import of the bulbs from Japan and China to India at initial stages of establishment of narcissus cultivation in Indian continent. However, we could not trace any evidence if infected narcissus bulbs were imported to India from China or Japan.
The occurrence of lycoris virus (Yadav and Khan 2008), Indian narcissus virus (Yadav and Khan 2015), CEVA (Kumar et al. 2015) and a distinct potyvirus (Chandel et al. 2010) having close resemblance with NLSYV and NYSV are already reported on narcissus from India based on analyses of their partial genome sequences. The complete genome sequences of CEVA and NYSV infecting narcissus have been published recently by us (Raj et al. 2018, 2019). We report here the existence of three potyvirus species: CEVA, NYSV and NDV in N. tazetta var. Paperwhite. The probable reasons for occurrence of three species at NBRI, Lucknow may be because of mixing and pooling of bulbs from infected/symptomatic narcissus plants collected from all the plots during their harvest and storage.
Conclusion
In this study, the existence of CEVA, NYSV and NDV has been investigated in Indian N. tazetta var. Paperwhite exhibiting leaf yellow stripe symptoms based on sequence analyses of about 1.5 kb genomic fragments obtained from seven infected samples by RT-PCR amplification using potyvirus degenerate primers. The study suggests prevalence of diverse potyvirus species present in the vicinity and probability of emergence of new recombinant species is high. Therefore, information may be useful for understanding virus epidemiology and designing the disease management strategies in interest of the narcissus growers.
Acknowledgements
Thanks are due to the Director, CSIR-NBRI, Lucknow, India for necessary laboratory facilities. R. Raj is thankful to the University Grant Commission for Rajiv Gandhi National Fellowships and AcSIR for Ph.D. Thanks are also due to Dr. R. K. Roy, the Head, Floriculture Department, CSIR-NBRI, Lucknow for allowing narcissus leaf sample collection. This study is funded by the In-house project OLP105.
Compliance with ethical standards
Conflict of interest
There is no conflict of interest.
References
- Adams MJ, Antoniw JF, Fauquet CM. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch Virol. 2005;150:459–479. doi: 10.1007/s00705-004-0440-6. [DOI] [PubMed] [Google Scholar]
- Aminuddin KJA, Raj SK. Association of an unknown potyvirus isolate with a severe mosaic disease of Narcissus tazetta L. Ind J Exp Biol. 1999;37:1034–1036. [Google Scholar]
- Brunt AA. Some hosts and properties of Narcissus latent virus, a carlavirus commonly infecting narcissus and bulbous iris. Ann Appl Biol. 1977;87:355–364. doi: 10.1111/j.1744-7348.1977.tb01900.x. [DOI] [Google Scholar]
- Brunt AA. Bulb and corm crops. Narcissus. In: Loebenstein G, Lawson RH, Brunt AA, editors. Virus and virus-like diseases of bulb and flower crops. Chichester: John Wiley and Sons; 1995. pp. 322–334. [Google Scholar]
- Brunt AA. Narcissus mosaic virus. Ann Appl Biol. 2008;58:13–23. doi: 10.1111/j.1744-7348.1966.tb05066.x. [DOI] [Google Scholar]
- Chandel V, Singh MK, Hallan V, Zaidi AA. Evidence for the occurrence of a distinct potyvirus on naturally growing Narcissus tazetta. Arch Phytopathol Plant Protect. 2010;43:209–214. doi: 10.1080/03235400701722145. [DOI] [Google Scholar]
- Chen J, Chen JP, Langeveld SA, Derks AFLM, Adams MJ. Molecular characterization of carla- and potyviruses from Narcissus in China. J Phytopathol. 2003;151:26–29. doi: 10.1046/j.1439-0434.2003.00674.x. [DOI] [Google Scholar]
- Chen J, Lu YW, Shi YH, Adams MJ, Chen JP. Complete nucleotide sequence of the genomic RNA of Narcissus yellow stripe virus from Chinese narcissus in Zhangzhou city, China. Arch Virol. 2006;15:1673–1677. doi: 10.1007/s00705-006-0788-x. [DOI] [PubMed] [Google Scholar]
- Darlington HR. Yellow-stripe of daffodils. J R Hort Soc Lond. 1908;34:161. [Google Scholar]
- Fuji S, Iida T, Nakamae H. Selection of an attenuated strain of Japanese yam mosaic virus and its use for protecting yam plants against severe strains. Nippon Shokubutsu Byori Gakkaiho. 2000;66:35–39. [Google Scholar]
- Gibbs A, Mackenzie A. A primer pair for amplifying part of the genome of all potyvirids by RT-PCR. J Virol Methods. 1997;63:9–16. doi: 10.1016/S0166-0934(96)02103-9. [DOI] [PubMed] [Google Scholar]
- Hanks GR (2002) Narcissus and daffodil, In: Hanks GR (Eds.), The genus narcissus. Horticulture Research International, Kirton, UK, London: Taylor and Francis. ISBN 978–0–415–27344–2
- Hanks GR, Chastagner GA (2018) Diseases of Daffodil (Narcissus), In: McGovern R, Elmer W (Eds.), Handbook of Florists' Crops Diseases. ISBN: 978–3–319–39668–2
- Harvey BMR, Selby C. Micropropagation of Narcissus (Daffodils) In: Bajaj YPS, editor. Biotechnology in agriculture and forestry, high-tech and micropropagation. Berlin Heidelberg: Springer-Verlag; 1997. pp. 225–251. [Google Scholar]
- Kamenetsky Rina and Okubo Hiroshi, eds. (2012) Ornamental Geophytes: From Basic Science to Sustainable Production. CRC Press. ISBN 978–1–4398–4924–8
- Kumar S, Raj R, Kaur C, Raj SK, Roy RK. First report of Cyrtanthus elatus virus A in Narcissus tazetta in India. Plant Dis. 2015;99:1655. doi: 10.1094/PDIS-02-15-0211-PDN. [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic S, Cingel A, Jevremović S, Stanković I, Bulajić A, Krstić B, Subotić A. Virus elimination from ornamental plants using in vitro culture techniques. J Pest Phytomed. 2012;27:203–211. doi: 10.2298/PIF1203203M. [DOI] [Google Scholar]
- Monger WA, Nixon TP (2008) Potyviruses found in UK Narcissus. Accession number EU887015. https://www.ncbi.nlm.nih.gov/nuccore/EU887015
- Ohshima K, Mitoma S, Gibbs AJ. The genetic diversity of narcissus viruses related to Turnip mosaic virus blur arbitrary boundaries used to discriminate potyvirus species. PLoS ONE. 2018;13:e0190511. doi: 10.1371/journal.pone.0190511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Nomiyama R, Mitoma S, Honda Y, Yasaka R, Tomimura K. Evolutionary rates and genetic diversities of mixed potyviruses in Narcissus Infection. Genet Evol. 2016;45:213–223. doi: 10.1016/j.meegid.2016.08.036. [DOI] [PubMed] [Google Scholar]
- Raj R, Ct K, Agrawal L, Chauhan PS, Kumar S, Raj SK. 3-Biotech. 2018;8:168–173. doi: 10.1007/s13205-018-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj R, Susheel K, Lalit A, Chauhan PS, Raj SK. Complete genome sequence analysis of Narcissus yellow strip virus infecting Narcissus tazetta in India. 3-Biotech. 2019;9:409. doi: 10.1007/s13205-019-1939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SJ, Jones MGK. Complete genome sequences of seven carlavirus and potyvirus isolates from Narcissus and Hippeastrum plants in Australia, and proposals to clarify their naming. Arch Virol. 2012;157:1471–1480. doi: 10.1007/s00705-012-1319-6. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Li H, Sivasithamparam K, Jones MG. Complete genome analysis of three isolates of Narcissus late season yellows virus and two of Narcissus yellow stripe virus: three species or one? Arch Virol. 2014;159:1521–1525. doi: 10.1007/s00705-013-1969-z. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Nouri S, Coutts BA, Jones MGK. Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch Virol. 2010;155:1171–11744. doi: 10.1007/s00705-010-0682-4. [DOI] [PubMed] [Google Scholar]
- Yadav N, Khan JA. Identification of a potyvirus associated with mosaic disease of Narcissus sp. in India. Plant Pathol. 2008;57:394. doi: 10.1111/j.1365-3059.2007.01655.x. [DOI] [Google Scholar]
- Yadav N, Khan JA. Molecular identification of a new strain Narcissus yellow stripe virus associated with severe mosaic disease of narcissus from India. Ind Phytopathol. 2015;68:444–448. [Google Scholar]
- Zerbini FM, French R, Rabenstein F, Stenger DC Valkonen JPT (2012) Family Potyviridae. In: Adams MJ, Lefkowitz EJ, Carstens EB, King AMQ (Eds), Virus taxonomy: 9th report of the International Committee on Taxonomy of Viruses Elsevier, Amsterdam. pp 1069–1089