Dear Editor,
We previously reported in this Journal 1 that cell-surface Glucose Regulated Protein 78 (CS-GRP78), also termed heat shock protein A5 (HSPA5), could be a possible route for SARS-CoV-2 internalization. We predicted the binding site on the spike protein of SARS-CoV-2 that can recognize CS-GRP78. A recent communication by Braun and colleagues reported that the spike glycoprotein of the SARS-CoV-2 bears many conserved motifs to the previously determined human coronavirus strains such as HKU1, 229E, NL63, OC43, MERS-CoV, and SARS-CoV.2 However, we would like to emphasize that using a simple bioinformatics approach can suggest a possible role of the GRP78 in T cell cross immunization against COVID-19. Based on the findings, we can conclude that mild human coronaviruses can be useful as a vaccination against COVID-19.
For SARS-CoV-2, the binding site to CS-GRP78 was predicted to be the nine residues CNGVEGFNC (C480-C488 region of the spike glycoprotein) in the S1 C-terminal domain (the receptor-binding domain (RBD)).1 This region (C480-C488) of the spike glycoprotein represents the best fit for the 13-residue Pep42 cyclic peptide (CTVALPGGYVRVC), which was reported earlier to be selectively associated with CS-GRP78 in cancer cells.3
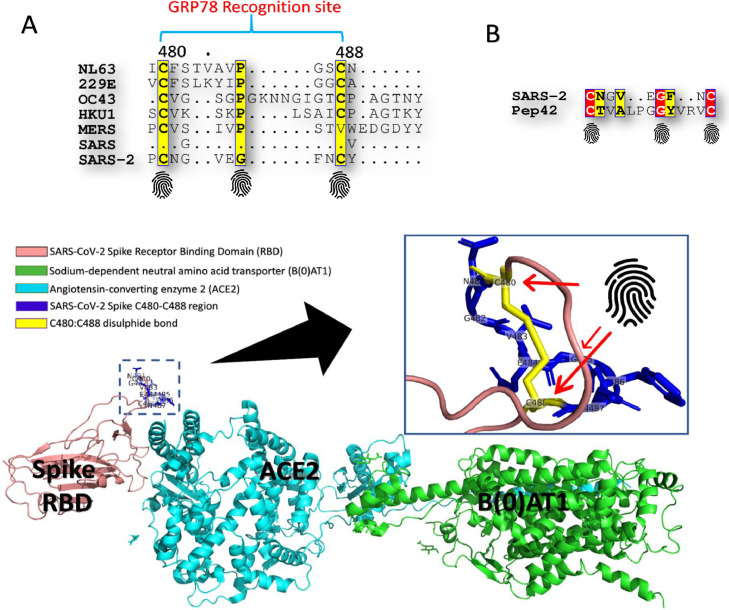
Fig. 1 A shows part of the multiple sequence alignment (MSA) of the seven human coronavirus strains (NL63, 229E, OC43, HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2). The most conserved residues in SARS-CoV-2 compared to other strains are C480, G485, and C488 (yellow columns). These three residues are fingerprints for CS-GRP78 recognition as they resemble the C1, G8, and C13 in the Pep42 cyclic peptide (red columns in Fig. 1B). These findings are in good agreement with the work of Braun and coworkers,1 where the GRP78 recognition site that we are reporting here lies in the most conserved S1 C-terminal RBD of the spike glycoprotein.2 The three fingerprint residues are found in the four mild HCoV strains (NL63, 229E, OC43, and HKU1) and the SARS-CoV-2.
Fig. 1.
Coronavirus spike protein and GRP78 recognition site. (A) Part of the multiple sequence alignment for the spike glycoproteins of the seven reported human coronavirus strains (NL63, 229E, OC43, HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2). Yellow columns are the conserved residues among the seven HCoVs. GRP78 recognition site (C480-C488 in SARS-CoV-2) is marked at the top. (B) Pairwise sequence alignment between Pep42 and SARS-CoV-2 S (C480-C488 region). Red and yellow residues are identical and similar residues, respectively. Fingerprint residues are marked (bottom). (C) The structure of spike protein RBD (rose) bound to ACE2 (cyan) and B(0)AT1 (green) (PDB ID: 6M17) while the GRP78 recognition site (C480-C488 in spike protein RBD) is depicted in the blue sticks in the enlarged panel. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The recognition of the CS-GRP78 by the peptide Pep42 is previously reported to be restricted for the cyclic form (the first and last CYS residues form a disulfide bond).4 , 5 Interestingly, the solved structure of the SARS-CoV-2 S (PDB ID: 6M17) shows that the C480-C488 region is cyclic (a disulfide bond is formed between the residues C480 and C488). Fig. 1C shows the solved structure of the SARS-CoV-2 spike protein RBD (rose cartoon) in conjunction with the human receptor Angiotensin-Converting Enzyme 2 (ACE2) (cyan cartoon) and Sodium-dependent neutral amino acid transporter (B(0)AT1) (green cartoon). The enlarged panel shows the CS-GRP78 recognition site (C480-C488) in blue sticks with the yellow sticks representing the two CYS residues with the formed disulfide bond. The fingerprint residues (C480, G485, and C488) are marked with red arrows. As reflected in Fig. 1C, the GRP78 recognition site is surface exposed and protrude apart from the ACE2 binding site.
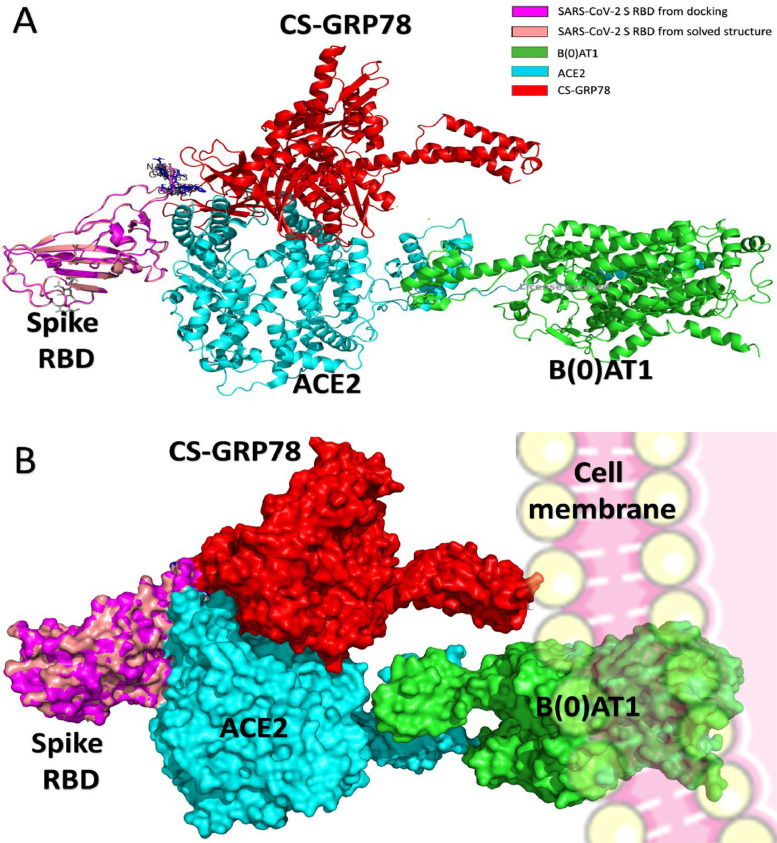
HADDOCK webserver is utilized to dock the GRP78 into spike protein RBD, and the docking complex is superimposed with the solved structure (PDB ID: 6M17) in Fig. 2 A. GRP78 can bind to the C480-C488 region of the spike protein RBD with an excellent HADDOCK score (-77 ± 3.1). The substrate-binding domain β (SBDβ) was reported to be the possible binding site to SARS-CoV-2 S RBD.1 Molecular Dynamics Simulation (MDS) (NAMD software) for the SARS-CoV-2 spike protein combined with molecular docking (HADDOCK) revealed the existence of more than four interactions (H-bonds or hydrophobic contacts) between GRP78 and C480-C488 of the SARS-CoV-2 spike protein. At least two hydrophobic contacts are formed in all the docking experiments (seven replicas are used during 100 ns MDS). This is in support of the previous reports about GRP78 recognition of the hydrophobic patches in the unfolded proteins.6 , 7
Fig. 2.
The docked complex of GRP78 (red) and S RBD (magenta) superimposed to the solved structure (PDB ID: 6M17) containing spike protein RBD (rose), ACE2 (cyan), and B(0)AT1 (green). (A) shows the cartoon representation, while (B) indicates the surface representation. The C480-C488 are labeled with its one-letter code and shown in blue. The membrane is depicted in (B) to show how the CS-GRP78 would look like when binding the spike protein RBD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As shown in the surface representation of the complex of spike protein RBD (rose), GRP78 (red), ACE2 (cyan), and B(0)AT1 (green) in Fig. 2B, the CS-GRP78 to be associate with spike protein RDB and ACE2, should also be associated with a membrane-bound protein to carry it. It was reported that CS-GRP78 was associated with murine tumor cell DnaJ-like protein 1 (MTJ-1) and that this association was essential for GRP78 membrane localization and its binding to the activated α2-macroglobulin.8 , 9 MTJ-1 may be the GRP78 carrying transmembrane protein that could be of therapeutic potential against COVID-19 and other viral infections.
Finally, the human coronaviruses NL63, 229E, OC43, and HKU1, had mildly impacted human beings (characterized by mild flu-like symptoms). People previously infected with these strains of human coronaviruses may develop immunity against SARS-CoV-2. The milder human coronaviruses could be used as a vaccine against COVID-19 as they share the same CS-GRP78 recognition region on their spikes.
Author Contributions
A.E. drafted the manuscript and draw figures, I.I. Did the MDS calculations, A.I. drafted the document, and the hypothesis, W.E. revised the manuscript and supervision.
Declaration of Competing Interest
The authors declare that there is no competing interest in this work.
References
- 1.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020 doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 3.Elfiky A.A., Baghdady A.M., Ali S.A., Ahmed M.I. GRP78 targeting: hitting two birds with a stone. Life Sci. 2020 doi: 10.1016/j.lfs.2020.118317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quinones Q.J., Ridder G.G.d., Pizzo S.V. GRP78, a chaperone with diverse roles beyond the endoplasmic reticulum. Histol Histopathol. 2008 doi: 10.14670/HH-23.1409. [DOI] [PubMed] [Google Scholar]
- 5.Kim Y., Lillo A.M., Steiniger S.C., Liu Y., Ballatore C., Anichini A. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45(31):9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 6.Tsai Y.-.L., Lee A.S. Elsevier; 2018. Cell surface GRP78: anchoring and translocation mechanisms and therapeutic potential in cancer. cell surface GRP78, a new paradigm in signal transduction biology; pp. 41–62. [Google Scholar]
- 7.Li J., Lee A.S. Stress induction of GRP78/BiP and its role in cancer. Curr Mol Med. 2006;6(1):45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- 8.Bastida-Ruiz D., Wuillemin C., Pederencino A., Yaron M., Martinez de Tejada B., Pizzo S.V. Activated α2-macroglobulin binding to cell surface GRP78 induces trophoblastic cell fusion. Sci Rep. 2020;10(1):9666. doi: 10.1038/s41598-020-66554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misra U.K., Gonzalez-Gronow M., Gawdi G., Pizzo S.V. The role of MTJ-1 in cell surface translocation of GRP78, a receptor for alpha 2-macroglobulin-dependent signaling. J Immunol (Baltimore, Md: 1950) 2005;174(4):2092–2097. doi: 10.4049/jimmunol.174.4.2092. [DOI] [PubMed] [Google Scholar]