Abstract
Natural exosomes can express specific proteins and carbohydrate molecules on the surface and hence have demonstrated the great potentials for gene therapy of cancer. However, the use of natural exosomes is restricted by their low transfection efficiency. Here, we report a novel targeting tLyp-1 exosome by gene recombinant engineering for delivery of siRNA to cancer and cancer stem cells. To reach such a purpose, the engineered tLyp-1-lamp2b plasmids were constructed and amplified in Escherichia coli. The tLyp-1-lamp2b plasmids were further used to transfect HEK293T tool cells and the targeting tLyp-1 exosomes were isolated from secretion of the transfected HEK293T cells. Afterwards, the artificially synthesized siRNA was encapsulated into targeting tLyp-1 exosomes by electroporation technology. Finally, the targeting siRNA tLyp-1 exosomes were used to transfect cancer or cancer stem cells. Results showed that the engineered targeting tLyp-1 exosomes had a nanosized structure (approximately 100 nm) and high transfection efficiency into lung cancer and cancer stem cells. The function verifications demonstrated that the targeting siRNA tLyp-1 exosomes were able to knock-down the target gene of cancer cells and to reduce the stemness of cancer stem cells. In conclusion, the targeting tLyp-1 exosomes are successfully engineered, and can be used for gene therapy with a high transfection efficiency. Therefore, the engineered targeting tLyp-1 exosomes offer a promising gene delivery platform for future cancer therapy.
Keywords: Targeting tLyp-1exosomes, Engineering, Transfection, Gene therapy, Lung cancer
Graphical abstract
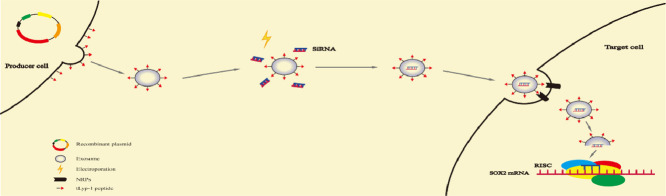
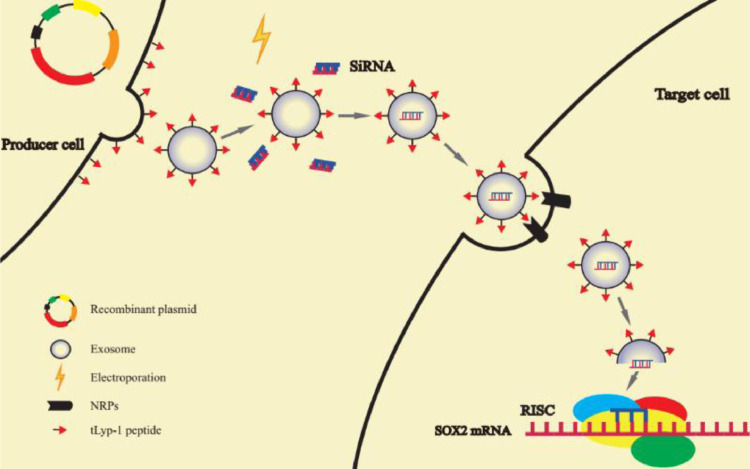
The tLyp-1-lamp2b plasmid transfected HEK293T cells can secreted tumor targeting tLyp-1 exosomes. By electroporation technology, targeting tLyp-1 exosomes were loaded with siRNA. When targeting tLyp-1 exosome ruptured in cytoplasm, siRNA was loaded into the RNA-induced silencing complex (RISC). The sense (passenger) strand was degraded while the antisense (guide) strand directs RISC to mRNA that has a complementary sequence, thereby resulting in the silence of target gene.
1. Introduction
Lung cancer remains to be the leading cause of cancer-related deaths worldwide. As one of the two main subtypes, non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers [1]. The major strategy for treatment of lung cancer is a comprehensive approach, including surgical therapy, chemotherapy and radiotherapy. For treatment of NSCLC, chemotherapy plays a critical role as the surgical treatment is only used at the early stage of the cancer. However, clinical studies shows that NSCLC has a very limited response to the chemotherapy [2].
Gene therapy has demonstrating a promising approach for treatment of NSCLC, and it has been simply defined as the genetic modification of cells to produce a therapeutic effect [3]. Gene therapy is also called human gene transfer that is the therapeutic delivery of nucleic acids (DNA or RNA) into a patient's cells as a drug to treat disease [4,5]. There are several methods by which to achieve gene therapy purpose. One strategy involves the deletion and addition of specific DNA fragments through gene editing. The other one introduces exogenous genes into the cells to cleave specific sequence in the mRNA transcript of the faulty gene, disrupting translation of the faulty mRNA, and hence correcting the expression of the faulty gene [6]. In the lasts years, a number of approaches have been developed for delivery of nucleic acids aimed at achieving therapeutic purposes, mainly including viral vectors [7], and non-viral vectors [8]. However, viral vectors are toxic and immunogenic while non-viral vectors have low transduction efficiency, and larger particle size, which are easy to induce immune response and to be rapidly cleared by reticuloendothelial system (RES). Therefore, non-immunogenicity, tissue specificity and non-toxicity play an important role in the clinical application of gene delivery vectors.
In the present study, we developed a novel kind of engineered targeting tLyp-1 exosomes for efficient delivery of siRNA into human cancer cells. In this construct, a membrane penetrating peptide tLyp-1 gene was engineered into a kind of exsomes by gene recombinant technique, 3 kinds of siRNA for silencing SOX2 gene were designed and encapsulated into the targeting tLyp-1 exosomes by electroporation technique.
Exosomes are nano-sized membrane vesicles (30–100 nm in diameter) secreted by numerous cell types such as tumor cells, T cells, B cells, dendritic cells (DC), mast cells and epithelial cells. Exosomes have natural properties because they are derived from the fusion of multivesicular bodies (MVBs) with plasma membranes [9]. Exosomes play an important role in cellular communication, especially in immune responses and cancer. When exosomes containing mRNA and microRNA are transferred to recipient cells, the mRNA and microRNA still maintain their functions and change the behavior of recipient cells [10], [11]. Despite signal-carrying capacity of exosomes, scientists have paid more attention of their potential in drug delivery for cancer therapy in the last few years. For examples, Sun et al. prepared exosomes loaded with curcumin and showed better solubility and anti-inflammatory bioactivity than the traditional curcumin [12]. Tian et al. loaded hydrophilic compounds doxorubicin into exosomes by electroporation [13]. In addition, exosomes are cell derived nano-scale membrane vesicles that are considered as siRNAs or miRNAs delivering tools to knockdown genes [14]. Current preliminary studies exhibit that the exosome-mediated nucleic acid delivery in vivo does not cause short-term innate immune activation nor obvious side effects.
tLyp-1 peptide (amino-acid sequence CGNKRTR), a newly reported hepta-peptide, is a ligand which selectively target to neuropilin1 (NRP1) and to neuropilin2 (NRP2), both which are highly expressed in tumor tissues including non-small cell lung cancer, and serve as the receptor targets in cancer drug delivery systems [15], [16], [17]. tLyp-1 is a tumor homing penetrating membrane peptide which can not only target the tumor [18], but also penetrate the tumor blood vessels and stroma to reach deep into the tumor [19]. Therefore, tLyp-1 modified exosomes can be expected to have active-targeting functions and penetrate extensively into tumor parenchyma.
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA molecules, 20–25 base pairs in length, similar to single-stranded microRNA (miRNA) [20]. Both siRNA and miRNA can play the role of RNA interference (RNAi) in live cells. siRNA interferes with the expression of specific genes with complementary nucleotide sequences by degrading mRNA after transcription, thus preventing translation [21]. siRNA is often used to treat a disease caused by abnormally gene expression, such as viral infection and cancer [22].
The objective of the present study was to develop the targeting tLyp-1 exosomes for improving the targeting ability and transfection capability of natural exosomes, and to confirm the function. During the engineering process, a plasmid vector, which encoded the fusion protein of tLyp-1-lamp2b, was firstly designed for transfecting human embryonic kidney HEK293T tool cells. The transfected HEK293T cells were able to express exosome transmembrane proteins (lamp2b) and tLyp-1 targeting peptide, and therefore, the targeting tLyp-1 exosomes were collected from the transfected HEK293T cells supernatants. The targeting tLyp-1 exosomes were used as the gene delivery vector to encapsulate siRNA by an electroporation approach. To verify the function, the targeting siRNA tLyp-1 exosomes were further applied to human non-small cells lung cancer (NSCLC) cells for observing gene silencing efficiency and action effect.
2. Materials and methods
2.1. Materials and cells
The pEGFP-C1 was from Inovogen Technology Co. (Beijing, China). Antibody against lamp2b was purchased from Abcam (Cambridge, UK) and antibody against GAPDH and SOX2 were purchased from Cell Signaling Technology (Beverly, MA, USA). siRNA was synthesized by RiboBio company (Guangzhou, China). Other reagents were from Beijing Chemical Reagents (Beijing, China).
Human cervical cancer Hela cells, human non-small cell lung cancer (NSCLC) A549 cells and human embryonic kidney HEK293T cells were purchased from Institute of Basic Medical Science, Chinese Academy of Medical Science (Beijing, China).
Hela cells and HEK293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) medium (Macgene, Beijing, China) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin (Macgene, Beijing, China). A549 cells were cultured in RPMI-1640 medium (Macgene, Beijing, China) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin (Macgene, Beijing, China). The cells were cultured in humidified atmosphere of 5% CO2 at 37 °C. The cells for all experiments were in logarithmic phase of growth.
Human NSCLC A549 stem cells were induced and cultured in DMEM-F12 medium supplemented with 2% B27 supplements (Gibco), epidermal growth factor (EGF, 20 ng/ml), basic fibroblast growth factor (bFGF, 20 ng/ml), leukemia inhibitory factor (LIF, 10 ng/ml) and insulin (181.6 ng/ml). Cells were cultured under an atmosphere of 5% CO2 at 37 °C.
Escherichia coli (i.e., DH5α competent cells) single colony was picked to 5 ml LB liquid medium (Sigma-Aldrich, St. Louis, USA), incubated at 37 °C in 100–200 r/min shaker for 12 h.
2.2. Engineering and transfection of tLyp-1-lamp2b plasmids
Total RNA was extracted from Hela cells using total RNA extraction kit, which was reversely transcribed into cDNA using reverse transcription kit (Solarbio, Beijing, China). tLyp-1 + lamp2b gene was amplified by PCR using cDNA as template, purified and recovered by using gel recovery kit (Solarbio, Beijing, China). The pEGFP-C1 and tLyp-1-lamp2b genes were digested by Bgl II and BamH I enzymes, then connected by T4 ligase. The connection products were transformed into DH5α competent cells (stored at −80 °C, Thermo Scientific, Waltham, USA) and cultured for 12 h. Single colony was culled for plasmid extraction. Lipofectamine 3000 transfection reagent (Invitrogen, USA) were used to transfect pEGFP-C1 plasmid vector expressing tLyp-1-lamp2b fusion proteins into HEK293T cells.
2.3. Identification of tLyp-1-lamp2b plasmids
A volume of 0.5 µl forward primer, 0.5 µl reverse primer, 0.2 µl primestar, 1 µl tamplate, 10 µl CG buffer, 1.6 µl dNTP and 6.2 µl H2O, were mixed together on ice and then PCR reaction was performed. The end product DNA was loaded onto 1% agarose gels. After the agarose gels were cooled to about 60 °C (not hot), and then 10 000 × Goodview dye was added in the gels and mixed evenly. The gels were poured into the tray gently. The DNA sample and loading buffer were mixed, and then the mixture was slowly added to the well using a micropipettor. The electrophoresis was performed at 100 V.
2.4. Isolation and purification of exosomes
Exosomes were isolated by differential centrifugation and micro-filtration. Firstly, the HEK293T cells were incubated for 15 h in exosome-free FBS, which had been centrifuged at 100 000 × g before use. The culture supernatants were collected from cell culture medium, centrifuged at 300 × g for 10 min and 2000 × g for 10 min to remove cells, and at 10 000 × g for 30 min to remove cell debris. After ultracentrifugation at 120 000 × g for another 70 min, exosomes were obtained. Exosomes were rinsed with phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4 and 2 mM KH2PO4, pH 7.4), and then ultra-centrifuged again. Afterwards, exosomes were re-dispersed in PBS (pH 7.4). The protein concentrations of the exosomes suspensions were determined using BCA assays (Thermo Scientific, Waltham, USA). All procedures were performed at 4 °C.
2.5. Encapsulation of siRNA by exosomes
To encapsulate siRNA into the exosomes, 50 µg Cy3-siRNA and 100 µg exosomes were mixed together in 200 µl electroporation buffer. The exosomes were electro transfected at 400 V in 96-well plates using X-porator H1 electroporation apparatus (Yida, Soochow, China). To ensure that the plasma membranes of the exosomes were fully recovered, the mixture was incubated at 37 °C for 30 min. Free siRNA was removed and encapsulation efficiency of siRNA by exosomes was determined by fluorescence spectrophotometer (emission at 590 nm, and excitation at 550 nm).
2.6. Particle size by NTA measurement
Particle sizes and Brownian motion were measured by nanoparticle tracking analysis (NTA). Purified exosomes (100 ml; 10 ng/ml) were performed on a NanoSight LM10-HSB instrument (A&P Instrument Co., Salisbury, UK). The data of size distribution and Brownian motion were captured and analyzed with the NTA 2.2 Analytical Software Suite.
2.7. Morphology by TEM observation
A transmission electron microscope (TEM, FEI Company, OR, USA) was used to further observe the morphology of the exosomes. Purified exosomes from HEK293T cells were re-suspended in PBS (pH 7.4) and fixed with 2% paraformaldehyde for 0.5 h. A volume of 8 µl mixture was then dropped onto EM grids. After drying for 30 min, exosomes were stained with 1% uranyl acetate twice every 6 min. The dried grids were detected using a transmission electron microscope at 120 kV.
2.8. Cellular uptake by flow cytometry
To estimate cellular uptake qualitatively, cancer cells were seeded into chambered coverslips at a density of 1.5 × 105 cells/well. After incubation for 24 h, the cells were treated with 3, 3′-dioctadecyloxacarbocyanine perchlorate (DiO, 5 µM) labelled natural exosomes or targeting tLyp-1 exosomes for another 6 h. The cells were then washed with cold PBS (pH 7.4) three times, and then re-suspended in 300 µl PBS (pH 7.4). DiO fluorescence intensity was measured by FACScan flow cytometer (FACScan, Becton Dickinson, San Jose, CA, USA) with 10 000 events collected. The excitation was set at 484 nm and the emission was set at 501 nm.
2.9. mRNA expression by real-time qRT-PCR
Total RNA was extracted using trizol reagents (Invitrogen, Beijing, China) according to the manufacturer's protocol. The concentration of extracted RNA was determined with Nano-300 reader (YPH-BIO, Beijing, China). An amount of 500 ng total RNA was then reverse-transcribed into cDNA by PrimeScript RT reagent (Takara Bio. Inc., Shiga, Japan). Real time qRT-PCR analysis was performed with CFX96 Opticon (Bio-Rad, Hercules, USA) in 25 µl reaction volume by using the SYBR Prime-Script PCR (Takara Bio Inc., Shiga, Japan). Each experiment was repeated for triplicates. Each gene expression level was normalized to GAPDH mRNA content. Relative quantification was calculated with the 2−ΔΔct method [23].
2.10. Protein expression by Western blotting
Cells were seeded into 6-well plates and cultured over 24 h until the density reached 70% confluence. After exposure to different agents at fixed intervals in the presence of 10% FBS, cells were harvested in lysis buffer containing 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 10 µg/ml leupeptin, and 10 µg/ml aprotinin. Cells were then centrifuged at 4 °C for 20 min at 12 000 rpm (rpm) to remove the precipitated fragments. The protein concentration was measured by Bradford protein assay. Equal amount of total protein was loaded and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to PVDF membranes (Millipore Corporation, Temecula, USA) and then blocked with 5% BSA (w/v) for 1 h at room temperature. The membranes were incubated with primary antibodies overnight at 4 °C and then with horse radish peroxidase (HRP) linked secondary antibody at room temperature for 1 h according to the optimized dilution ratio. The blots were visualized with Clarity-ECL Western blotting detection reagents (Thermo, Beijing local agent, China) by following the instructions of the manufacturer.
2.11. Identification of NSCLC stem cells
The phenotypes of the NSCLC stem cells were identified by flow cytometry (FCM), including CD44 and CD24. Briefly, NSCLC stem cells and SOX2 gene silenced NSCLC stem cells were collected. Then, the cells were incubated at room temperature for 30 min with anti-human CD44 fluorescein isothiocyanate (FITC)-conjugated antibody (BD, 10 µl per test). The cells were washed with PBS (pH7.4) twice and re-suspended in 300 µl PBS (pH 7.4), and the fluorescence intensity was measured by FCM.
To identify CD24, anti-human CD24 phycoerythrin (PE)-conjugated antibody (BD, Beijing, China; 5 µl per test) were separately added using the procedures described above except for the perforation.
Both NSCLC stem cells and SOX2 gene silenced NSCLC stem cells were incubated with FITC-conjugated mouse IgG or PE-conjugated mouse IgG as isotype controls, respectively. The excitation wavelengths were set at 488 nm for both FITC and PE, and the emission wavelengths were set at 530 nm for both FITC and 564 nm for PE.
2.12. Statistical analyses
Data were presented as the mean ± standard deviation (SD). One-way analysis of variance was used to determine significance among groups, after which, post hoc tests with the Bonferroni correction were used for multiple comparisons between individual groups. P < 0.05 was considered significant.
3. Results and discussion
3.1. Engineering of targeting tLyp-1-lamp2b plasmids
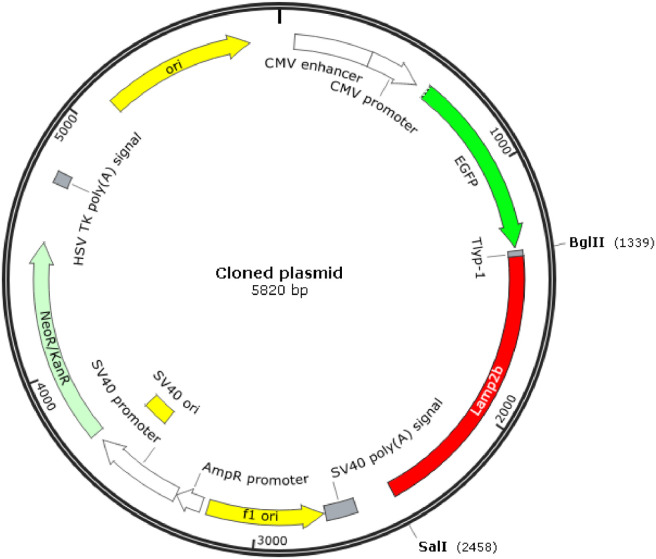
The genes of tLyp-1 and lamp2b were synthesized PCR reaction in forming a combined tLyp-1-lamp2b gene. The tLyp-1-lamp2b gene was mixed with the blank pEGFP-C1 plasmids, digested with Bgl II and BamH I enzymes, and then connected by T4 ligase, thereby producing the pEGFP-C1-tLyp-1-lamp2b plasmids, namely, the targeting tLyp-1-lamp2b plasmids (Fig. 1).
Fig. 1.
Engineering of the targeting tLyp-1-lamp2b plasmid. Artificial tLyp-1 gene and lamp2b gene were synthesized by PCR using cDNA as templates. The resultant tLyp-1-lamp2b gene was recombined into pEGFP-C1 blank plasmid through digesting with Bgl II and BamH I enzymes, and then connected by T4 ligase to form the targeting tLyp-1-lamp2b plasmid.
The engineered targeting tLyp-1-lamp2b plasmids are used for further amplifying the plasmids with DH5α competent cells, and for transfecting the tool cells (HEK293T cells). If transfecting, the tool cells will secrete the targeting tLyp-1 exosomes, in which tLyp-1 peptide are expressed on the outer membrane of the exosomes by connecting with the extra-exosomal N-terminus of lamp2b protein.
3.2. Identification of targeting tLyp-1-lamp2b plasmids
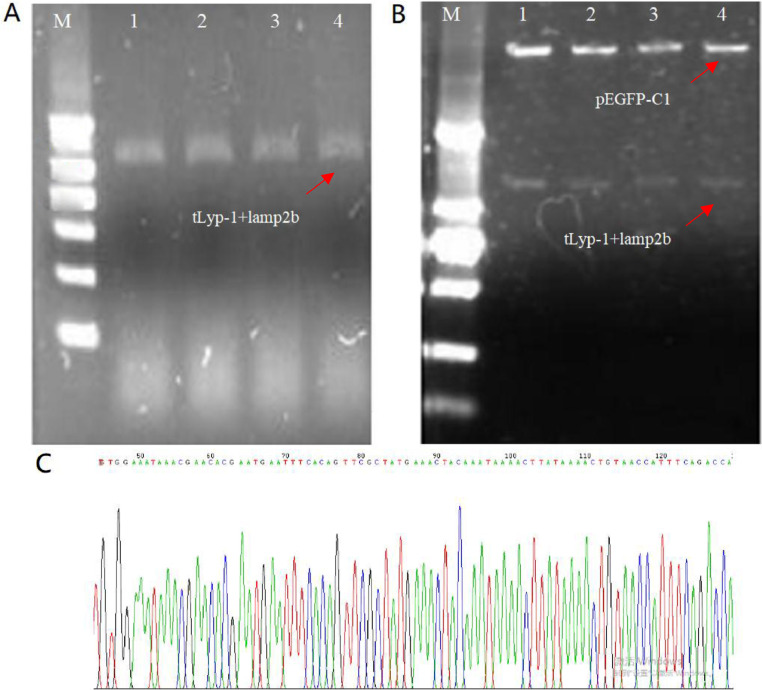
To identify the targeting tLyp-1-lamp2b plasmids, a single colony of the plasmids was selected from the DH5α competent cells, amplified in PCR, and used for colony PCR identification. The result from the bands in the electrophoresis image demonstrated that the parallel samples (1–4) produced a fragment of about 1100 bp, respectively, indicating that the tLyp-1-lamp2b genes were constructed into the plasmids (Fig. 2A). In the images, the bands were bright, without impurity band, demonstrating that there was no by-product, and the colony of plasmids was positive clone.
Fig. 2.
Identification of the targeting tLyp-1-lamp2b plasmid. (A) Colony PCR image by agarose gel electrophoresis for identification of targeting tLyp-1-lamp2b plasmids. M, 2000 bp ladder marker; 1–4, samples of tLyp-1-lamp2b plasmids. (B) PCR image by agarose gel electrophoresis for identification of the recombinant plasmid double digested with Bgl II and Sal I. M, 2000 bp ladder marker; 1–4, samples of tLyp-1-lamp2b plasmids. (C) Sequencing for tLyp-1 and part of lamp2b genes. The complete plasmid sequencing and comparison results are shown in supplementary.
Moreover, a single colony of the plasmids was selected from the DH5α competent cells, and extracted by the extraction kit for further confirming the existence of tLyp-1-lamp2b gene in the plasmids. The extracting product was then digested by Bgl II/Sal I double enzymes, and used for analysis with electrophoresis. The results from the specific bands in the electrophoresis image further demonstrated that the tLyp-1-lamp2b gene was successfully constructed into the plasmid (Fig. 2B). Besides, the gene of green fluorescence protein probe was also constructed into the plasmids.
In addition, the results of plasmid sequencing (Fig. 2C and Fig. S1) demonstrated that the homology of the sample with the original target DNA sequence reached 99.91%, and the homology of amino acid sequence reached 100%.
Through identifications from the above three aspects, the results fully confirmed that the tLyp-1+lamp2b gene was successfully inserted into the pEGFP-C1 plasmid, indicating a successful engineering of the targeting tLyp-1-lamp2b plasmid.
3.3. Transfection efficiency of targeting tLyp-1-lamp2b plasmids into HEK293T cells
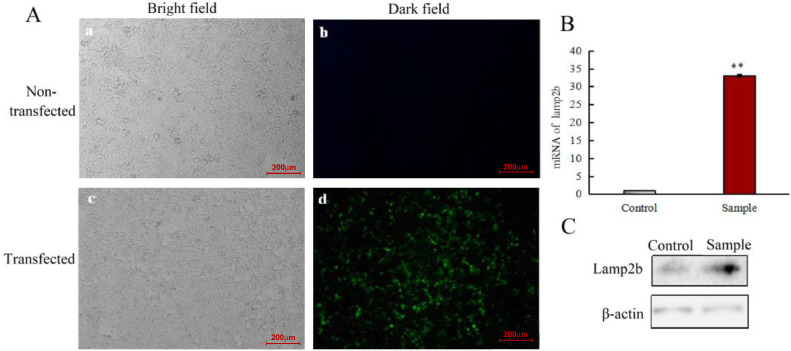
To obtain the targeting tLyp-1 exosomes, the targeting tLyp-1-lamp2b plasmids were transfecting into tool cells (HEK293T cells). The expression of green fluorescent protein was used to validate whether tLyp-1-lamp2b plasmids were expressed in HEK293T cells successfully. The results showed that the transfection efficiency was above 70% (Fig. 3A).
Fig. 3.
Transfection efficiency of the targeting tLyp-1-lamp2b plasmids in HEK293T cells. (A) Microscopy images of human embryonic kidney HEK293T cells. (a):image of non-transfected HEK293T cells under fluorescent microscope in bright field; (b): image of non-transfected HEK293T cells under fluorescent microscope in dark field; (c): image of transfected HEK293T cells under fluorescent microscope in bright field; (d): image of transfected HEK293T cells under fluorescent microscope in dark field. The results indicate that the green fluorescent protein is detected as a marker of the transfection, demonstrating a successful transfection in HEK293T cells using the targeting tLyp-1-lamp2b plasmids. (B) mRNA expression of lamp2b in HEK293T cells after transfection with targeting tLyp-1-lamp2b plasmids. Real-time qRT-PCR analysis was performed on HEK293T cells that were either non-transfected (control) or transfected (sample) with a plasmid encoding tLyp-1+Lamp2b. (C) Protein expression of lamp2b in HEK293T cells after transfection with targeting tLyp-1-lamp2b plasmids. Western blot analysis was performed on HEK293T cells that were either non-transfected (control) or transfected (sample) with a plasmid encoding tLyp-1+lamp2b. *P < 0.01 vs. control. Data are presented as mean ± SD (n = 3).
Furthermore, the mRNA and protein levels of lamp2b after transfection at 48 h were assessed using real time qRT-PCR and Western blotting, respectively. In contrast to non-transfected HEK293T cells, the results showed that both the mRNA and protein expressions of lamp2b were evidently up-regulated in the transfected HEK293T cells (Fig. 3B and 3C).
The above results demonstrated that targeting tLyp-1-lamp2b plasmids were successfully transfected into HEK293T tool cells, and the tLyp-1-lamp2b protein was successfully expressed in the tool cells.
In this study, tLyp-1 gene is transfecting with lamp2b gene into the HEK293T cells because lamp2b gene is a specific gene, which expresses on the membrane protein of exosome [24]. Transfection using tLyp-1-lamp2b fusion plasmid can lead to the expression of targeting peptide tLyp-1 on the outer membrane of exosomes, while targeting peptide tLyp-1 modified exosomes can be secreted and collected from the transfected HEK293T cells.
Actually, tLyp-1 peptide is a cell penetration-peptide, which can selectively target both NRP1 and NRP2 receptors that highly express in neoplastic vessels and tumor cells [25]. Accordingly, tLyp-1 modified exosomes can be used for targeting cancer cells, and for increasing the penetration of cancer cells. Such functions are especially useful for delivery of gene because the un-modified natural exosomes often have low transfection efficiency when being used as gene delivery vectors [26].
3.4. Isolation and characterization of natural and targeting tLyp-1 exosomes
The isolation of targeting tLyp-1 exosomes was obtained from the culture supernatants of tLyp-1-lamp2b plasmids transfected or non-transfected HEK293T cells by ultra-centrifugation. After 5 times of centrifugations, the purified targeting tLyp-1 exosomes and natural exosomes were isolated from the transfected and non-transfected HEK293T cells, respectively.
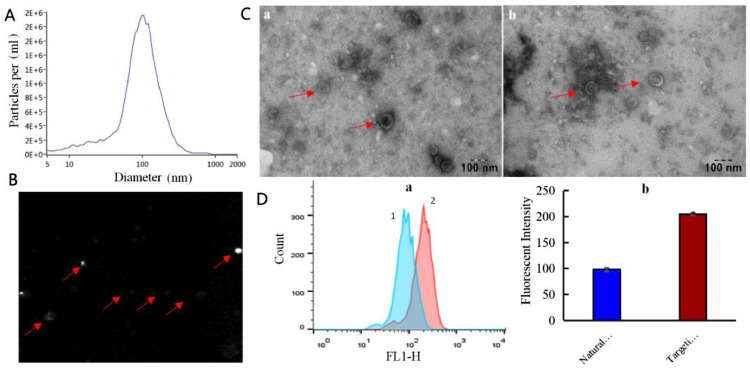
The results from NTA demonstrated that the targeting tLyp-1 exosomes were approximately 100 nm in diameter (Fig. 4A). In the diluted solution, targeting tLyp-1 exosomes exhibited an evidently Brownian motion, suggesting that the nano-sized exosomes were not aggregated but well single distributed (Fig. 4B).
Fig. 4.
Characterization of the engineered targeting tLyp-1 exosomes. (A) Size distribution and movement of targeting tLyp-1 exosomes. The targeting tLyp-1 exosomes were measured by nanoparticle tracking analysis (NTA) with NanoSight LM10-HSB instrument. (B) Image of Brownian motion of the tLyp-1 exosomes in PBS (pH7.4) solution. The targeting tLyp-1 exosomes were photographed by NTA system. (C) Representative TEM images of natural and engineered exosomes. (a): natural exosomes; (b): targeting tLyp-1 exosomes. (D) Uptake of the engineered targeting tLyp-1exosomes into A549 cells. Both natural exosomes and engineered exosomes were labeled with DiO dye (green fluorescence). (a): mean fluorescent intensity was measured by FACS. 1: natural exosomes; 2: targeting tLyp-1 exosomes. (b): the quantitative results of fluorescent intensity. Data are presented as mean ± SD (n = 3). *P < 0.05 vs natural exosomes.
The results from TEM observations showed that both natural exosomes and targeting tLyp-1 exosomes (Fig. 4C) were round in shape, and their membrane structures could be clearly observed on the exosomes vesicles. Besides, there were no significant differences in the particle size and in the shape between natural and targeting tLyp-1 exosomes.
To identify cellular uptake, flow cytometry was used for quantitative purpose. The evaluation by flow cytometry revealed the uptake rate of targeting tLyp-1 exosomes was higher than that of natural exosomes in A549 cancer cells (Fig. 4D).
As natural exosomes express specific proteins and carbohydrate molecules on the surface, they have been used as drug delivery vectors, which can avoid the phagocytosis of mononuclear phagocyte system in blood circulation after administration [27]. Moreover, nano-sized exosomes can enhance the permeability and retention (EPR) effect in solid tumor tissue [28]. However, natural exosomes often have low transfection efficiency into eukaryocytes like cancer cells [29]. The increasing uptake efficiency of engineered exosomes was associated with the tLyp-1 peptide which was a cell membrane penetrating peptide [30]. Consequently, this study will provide a high transfection efficiency of targeting tLyp-1 exosomes when they are used as drug or gene delivery vectors for cancer therapy.
3.5. Function verification of targeting siRNA tLyp-1 exosomes in lung cancer cells
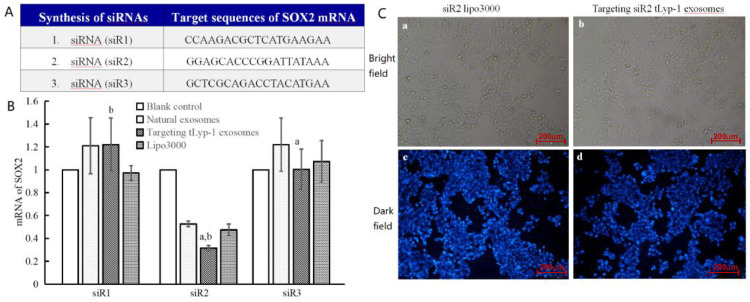
To verify the function for gene delivery of targeting tLyp-1 exosomes, three siRNAs (siR1: CCAAGACGCTCATGAAGAA; siR2: GGAGCACCCGGATTATAAA; siR3: GCTCGCAGACCTACATGAA) were designed based on the sequence of SOX2 gene, synthesized (Fig. 5A) and encapsulated into the exosomes by electroporation technology. The siRNA encapsulation efficiency of targeting exosomes was 61.53% ± 0.32%, and the siRNA encapsulation efficiency of natural exosomes was 46.87% ± 0.24% (Fig. S2).
Fig. 5.
Functional verification of the targeting siRNA tLyp-1 exosomes in silencing SOX2 gene of human NSCLC cells. (A) Designed 3 kinds of sequences for siRNA according to human SOX2 siRNAs. (B) mRNA expression of SOX2 in A549 cells after treated with lipo3000 carrying siRNA, natural exosomes carrying siRNA or targeting tLyp-1 exosomes carrying siRNA. Real-time qRT-PCR analysis was performed on A549 cells that were treated with lipo3000, natural exosomes or targeting tLyp-1 exosomes which carried 3 kinds of siRNAs, respectively. The results demonstrate that siR2 has the highest silence efficiency. P < 0.05; (a): vs natural exosomes; (b): vs. lipo3000. Data are presented as mean ± SD (n = 3). (C) Images of human NSCLC A549 cells under fluorescence microscope. (a): image of transfected A549 cells with siR2 lipo3000 under fluorescent microscope in bright field; (b): image of transfected A549 cells with the targeting siR2 tLyp-1 exosomes under fluorescent microscope in bright field; (c): image of transfected A549 cells with siR2 lipo3000 under fluorescent microscope in dark field; d, image of transfected A549 cells with targeting tLyp-1 exosomes under fluorescent microscope in dark field. siRNA (siR2) was labeled with Cy3 and blue was set as an indicator of transfection efficiency by fluorescence microscopy. The results demonstrate a successful transfection in A549 cells using the targeting siR2tLyp-1 exosomes.
The resultant targeting siRNA tLyp-1 exosomes were used to transfect NSCLC A549 cells for knocking-down SOX2 gene. In addition, a commercially available gene delivery vector (lipo3000) was used as a positive control vector.
To confirm the efficacy of designed siRNAs in knocking-down SOX2 gene, three kinds of siRNAs were separately encapsulated into natural exosomes, targeting tLyp-1 exosomes or lipo3000, and used for transfecting A549 cells. The mRNA levels of SOX2 gene in the A549 cells after transfection were assessed using real time qRT-PCR. The results showed that targeting siRNA (siR2) tLyp-1 exosomes had the most significant knock-down effect on SOX2 gene (Fig. 5B), and hence, targeting siRNA (siR2) tLyp-1 exosomes were selected for further study.
To directly observe the transfection efficiency in A549 cells, targeting Cy3 labeled siRNA (siR2) tLyp-1 exosomes were included for comparing with Cy3 labeled siRNA (siR2) lipo3000, and were evaluated under fluorescent microscope. The results showed that the targeting tLyp-1 exosomes reached the same transfection efficiency as that of lipo3000 by observing Cy3 fluorescent distribution and intensity (Fig. 5C).
The results demonstrated that transfection efficiency of targeting tLyp-1 exosomes was similar to that of commercially available transfection reagents. More importantly, targeting tLyp-1 exosomes have less immunogenic and toxic features [31], [32], which assign the favorable characteristics of the engineered targeting tLyp-1 exosomes when they are used as the gene delivery vectors for cancer therapy.
3.6. Function verification of targeting siRNA tLyp-1 exosomes in lung cancer stem cells
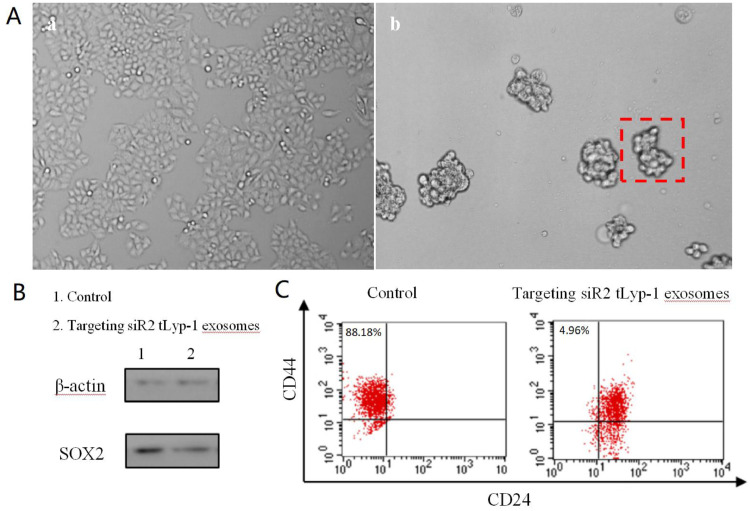
To further extend the function of targeting tLyp-1 exosomes for potential future clinical uses, targeting siRNA (siR2) tLyp-1 exosomes were used for transfecting lung cancer stem cells. Accordingly, the lung cancer stem cells were firstly induced by serum-free culture method. The result showed that the lung cancer stem cells could be obtained from the culturing at the 6th day, exhibiting cancer stem cell spheres (Fig. 6A) and up-regulation of stemness gene (CD44+/CD24-).
Fig. 6.
Functional verification of the targeting siRNA tLyp-1 exosomes in silencing SOX2 gene of human NSCLC cells and in decreasing the stemness of NSCLC stem cells. (A) Microscopic image for the appearance of human non-small lung cell (NSCLC) A549 stem cell spheres. (a): NSCLC A549 cells; (b): NSCLC A549 stem cells. (B) Knocked-down SOX protein of NSCLC A549 stem cells by targeting siR2 tLyp-1 exosomes. The study was performed by Western blotting assay. The results demonstrate that the targeting siR2 tLyp-1 exosomes enables to silence SOX2 protein in A549 stem cells as compared to untreated A549 stem cells. (C) The decreased stemness of NSCLC A549 stem cells by targeting siR2 tLyp-1 exosomes. NSCLC A549 stem cells were treated by targeting siR2 tLyp-1 exosomes, stained with anti-CD44-FITC, anti-CD24-PE, and analyzed with FACScan flow cytometer. The identification of phenotypes demonstrate that the population of CD44+/CD24- cells are reduced by knocking-down SOX2 gene.
To verify the efficacy of targeting tLyp-1 exosomes as gene delivery vectors, the lung cancer stem cells were further transfected with targeting siR2 tLyp-1 exosomes. After transfection, the protein expression of SOX2 was evaluated by Western blotting. The results showed that the protein expression of SOX2 in the lung cancer stem cells was significantly knocked-down after 48 h transfection (Fig. 6B).
To reveal the changes in stemness surface makers, CD44 and CD24 markers on the lung cancer stem cells were evaluated by flow cytometry after transfecting with targeting siR2 tLyp-1 exosomes. The results showed that the transfection of targeting siR2 tLyp-1 exosomes resulted in an evident decrease in the population of CD44+/CD24- cells (Fig. 6C).
The results indicate the targeting tLyp-1 exosomes can be used as the gene delivery vectors for effectively transfecting cancer stem cells. As cancer stem cells are characterized as the more resistant malignant cells than cancer cells [33,34], the effective transfection suggests that the targeting tLyp-1 exosomes could be the potential drug delivery carriers for a broad spectrum of anticancer drugs as well.
3.7. Illustration for engineering and mechanism of targeting siRNA tLyp-1 exosomes
For gene therapy, a high efficient delivery vector plays an important role in killing cancer and cancer stem cells. In particular, an effective transfection vector could provide a high efficacy in killing these malignant cells. In this study, the engineering and action mechanism of targeting siRNA tLyp-1 exosomes can be illustrated as the following steps: (i) the engineered tLyp-1-lamp2b plasmids are constructed and amplified in Escherichia coli; (ii) the tLyp-1-lamp2b plasmids are used to transfect HEK293T cells, and the fused protein containing targeting molecule tLyp-1 peptide is successfully expressed in the cells; (iii) the targeting tLyp-1 exosomes are secreted from transfected HEK293T cells; (iv) the artificially synthesized siRNA is encapsulated into targeting tLyp-1 exosomes by electroporation technology; (v) the targeting siRNA tLyp-1 exosomes are used to transfect cancer or cancer stem cells, and when the exosomes rupture in cytoplasm, siRNA is released from the exosomes and attracted into the RNA-induced silencing complex (RISC); and (vi) consequently, the sense (passenger) strand of siRNA is degraded in cytoplasm while the antisense (guide) strand was combined with a complementary sequence of mRNA within RISC [35], thereby resulting in the knock-down of target gene (such as SOX2 gene in this case) (Fig. 7).
Fig. 7.
Mechanism illustration of the engineered targeting exosomes for efficient delivery of siRNA into human cancer cells. The tLyp-1-lamp2b plasmid transfected HEK293T cells can secreted tumor targeting tLyp-1 exosomes. By electroporation technology, targeting tLyp-1 exosomes were loaded with siRNA. When targeting tLyp-1 exosome ruptured in cytoplasm, siRNA was loaded into the RNA-induced silencing complex (RISC). The sense (passenger) strand was degraded while the antisense (guide) strand directs RISC to mRNA that has a complementary sequence, thereby resulting in the silence of target gene. The targeting tLyp-1 exosomes demonstrate highly transfection efficiency in NSCLC cells and highly SOX2 gene silencing ability in NSCLC stem cells.
4. Conclusions
In sum, we report a novel targeting tLyp-1 exosome by gene recombinant engineering for delivery of siRNA to cancer and cancer stem cells. The targeting tLyp-1 exosomes have a nanosized structure (approximately 100 nm) and can be produced in large scale by amplification using cell culture technology. The present study reveals the full engineering process of the targeting tLyp-1 exosomes, demonstrates their high transfection efficiency into lung cancer and cancer stem cells, and confirm the functions of targeting siRNA tLyp-1 exosomes in knocking-down the target gene of cancer cells and in decreasing stemness of the cancer stem cells. The results indicate that the targeting tLyp-1 exosomes can be used a powerful gene delivery platform for cancer therapy, and hence are promising for broad potentials in the biomedical fields.
Conflict of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Acknowledgments
The research leading to these results has received funding from National Natural Science Foundation of China (Grant nos. 81673367 and 81874303) and Beijing Natural Science Foundation (Key Grant no. 7181004).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2019.04.002.
Appendix. Supplementary materials
References
- 1.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y. International association for the study of lung cancer/American thoracic society/European respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan T.S., Hsu C.C., Pai V.C., Liao W.Y., Huang S.S., Tan K.T. Metronomic chemotherapy prevents therapy-induced stromal activation and induction of tumor-initiating cells. J Exp Med. 2016;213(13):2967–2988. doi: 10.1084/jem.20151665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hutson T.H., Foster E., Moon L.D., Yáñez-Muñoz R.J. Lentiviral vector-mediated RNA silencing in the central nervous system. Hum Gene Ther Methods. 2014;25(1):14–32. doi: 10.1089/hgtb.2013.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mulligan R.C. The basic science of gene therapy. Science. 1993;260(5110):926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 5.Eugene H., Kaji E.H., Leiden J.M. Gene and stem cell therapies. JAMA. 2001;285(5):545–550. doi: 10.1001/jama.285.5.545. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Naldini L. Ex vivo gene transfer and correction for cell based therapies. Nat Rev Genet. 2011;12(5):301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 8.Kim C.S., Duncan B., Creran B., Rotello V.M. Triggered nanoparticles as therapeutics. Nano Today. 2013;8(4):439–447. doi: 10.1016/j.nantod.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Théry C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569e79. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 10.Théry C., Ostrowski M., Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581e93. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 11.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosomemediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654e9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 12.Sun D., Zhuang X., Xiang X., Liu Y., Zhang S., Liu C. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18(9):1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y., Li S., Song J., Ji T., Zhu M., Anderson G.J. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 14.Wahlgren J., De L Karlson T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40(17):e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1 dependent cell, vascular and tissue penetration. Proc Natl Acad Sci USA. 2009;106(38):16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami T., Tokunaga T., Hatanaka H., Kijima H., Yamazaki H., Abe Y. Neuropilin 1 and neuropilin 2 co-expression is significantly correlated with increased vascularity and poor prognosis in nonsmall cell lung carcinoma. Cancer. 2002;95(10):2196–2201. doi: 10.1002/cncr.10936. [DOI] [PubMed] [Google Scholar]
- 17.Roth L., Agemy L., Kotamraju V.R., Braun G., Teesalu T., Sugahara K.N. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene. 2012;31(33):3754–3763. doi: 10.1038/onc.2011.537. [DOI] [PubMed] [Google Scholar]
- 18.Jubb A.M., Strickland L.A., Liu S.D., Mak J., Schmidt M., Koeppen H. Neuropilin-1 expression in cancer and development. J Pathol. 2012;226(1):50–60. doi: 10.1002/path.2989. [DOI] [PubMed] [Google Scholar]
- 19.Roth L., Agemy L., Kotamraju V.R., Braun G., Teesalu T., Sugahara K.N. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene. 2012;31(33):3754–3763. doi: 10.1038/onc.2011.537. [DOI] [PubMed] [Google Scholar]
- 20.Melnikova I. RNA-based therapies. Nat Rev Drug Discov. 2007;6(11):863–864. doi: 10.1038/nrd2314. [DOI] [PubMed] [Google Scholar]
- 21.Sarett S.M., Werfel T.A., Lee L., Jackson M.A., Kilchrist K.V., Brantley-Sieders D. Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. Proc Natl Acad Sci USA. 2017;114(32):E6490–E6497. doi: 10.1073/pnas.1621240114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Caby M.P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. Exosomallike vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 25.Bielenberg D.R., Pettaway C.A., Takashima S., Klagsbrun M. Neuropilins in neoplasms: expression, regulation, and function. Exp Cell Res. 2006;312(5):584–593. doi: 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 26.Syn N.L., Wang L., Chow E.K., Lim C.T., Goh B.C. Exosomes in cancer nanomedicine and immunotherapy: prospects and challenges. Trends Biotechnol. 2017;35(7):665–676. doi: 10.1016/j.tibtech.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Hadla M., Palazzolo S., Corona G., Caligiuri I., Canzonieri V., Toffoli G. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine. 2016;11(18):2431–2441. doi: 10.2217/nnm-2016-0154. [DOI] [PubMed] [Google Scholar]
- 28.Biswas S., Torchilin V.P. Nanopreparations for organelle-specific delivery in cancer. Adv Drug Deliv Rev. 2014;66:26–41. doi: 10.1016/j.addr.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciobanasu C., Dragomir I., Apetrei A. The penetrating properties of the tumor homing peptide LyP-1 in model lipid membranes. J Pept Sci. 2019;25(3):e3145. doi: 10.1002/psc.3145. [DOI] [PubMed] [Google Scholar]
- 30.H Rashed M., Bayraktar E., K Helal G., Abd-Ellah M.F., Amero P., Chavez-Reyes A. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. 2017;18(3):E538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao W., Du Y., Zhang C., Pan F., Yao Y., Zhang T. Exosomes: the next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2018;28(18):S1742–S7061. doi: 10.1016/j.actbio.2018.12.045. [DOI] [PubMed] [Google Scholar]
- 32.van der Meel R., Fens M.H., Vader P., van Solinge W.W., Eniola-Adefeso O., Schiffelers R.M. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release. 2014;195:72–85. doi: 10.1016/j.jconrel.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 33.Batlle E., Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 34.Jin X., Jin X., Kim H. Cancer stem cells and differentiation therapy. Tumour Biol. 2017;39(10):1–11. doi: 10.1177/1010428317729933. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.