Abstract
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer death worldwide. Endoplasmic reticulum stress (ERS) is generally activated in HCC and is important for the sensitivity of HCC to anticancer drugs. ERS-dependent autophagy is a crucial mechanism affecting the sensitivity of HCC to anticancer drugs, but the mechanism by which ERS regulates autophagy is not well understood. Zinc finger protein 263 (ZNF263) is a transcription factor member of the zinc finger family. However, its functional role in HCC remains to be studied. In the current study, we investigated the role of ZNF263 in regulating ERS-induced chemoresistance in HCC and its possible mechanism. We found that ZNF263 was the most significant ERS-specific super-enhancer bounding transcriptional factor and was up-regulated in HCC patients and cell lines. Further, ZNF263 expression correlated with ERS, clinical stage and shorter survival in HCC patients. ZNF263 knockdown by RNA interference results in decreased proliferation, apoptosis resistance, and chemoresistance. Further study showed that ZNF263 increased chemoresistance by activating ERS-related autophagy. In conclusion, our study highlights ZNF263 as a functional ERS-related tumor activator and indicates it as a potential target for HCC therapy.
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent type of liver cancer and is the sixth most common and aggressive malignancy, especially in East Asia such as China [1,2]. Although recent studies have yielded novel therapeutic options, chemotherapy resistance remains the major obstacle and the clinical outcome of HCC patients remains unsatisfactory [3]. Therefore, more studies are urgently needed for identifying novel therapeutic strategies to overcome chemoresistance in HCC.
Endoplasmic reticulum stress (ERS) is a disorder of the physiological function of the endoplasmic reticulum caused by oxidative stress, chemical damage and other reasons, which results in misfolded or unfolded proteins accumulate in the cellular endoplasmic reticulum [4,5]. Our previous research results show that HCC cells under endoplasmic reticulum stress state have significantly decreased sensitivity to doxorubicin [6]. Previous study from others has found that lung cancer cells treated with tunicamycin, the endoplasmic reticulum stress inducer, have reduced sensitivity to cisplatin [7].
Multiple studies have found that endoplasmic reticulum stress can induce autophagy [8,9,10]. Autophagy plays an important role in maintaining cell homeostasis in the degradation and recovery of misfolded proteins and damaged organelles [11]. The occurrence of autophagy is closely related to multidrug resistance of tumors [12]. Our previous results showed that ERS is universally activated in hepatocellular carcinoma [13] and that ERS-induced autophagy is an important cause of reduced sensitivity of hepatocellular carcinoma cells to sorafenib [14]. However, the underlying mechanism by which ER stress induces autophagic responses in HCC remains largely unknown, and the mechanism of ER stress-induced chemoresistance is also unclear.
Zinc finger protein (ZNF) 263 is a transcriptional factor member of the zinc finger family [15]. According to previous reports, ZNF263 is involved in the genesis and development of a variety of tumors and the regulation of genes. It is reported that ZNF263 may be a key transcription factor for cholangiocarcinoma (CCA) [16], gastric cancer [17], and hypothalamic hamartoma (HH) [18]. Previous study demonstrated that ZNF263 is up-regulated in the blood of HCC patients compared with the health control group, and may participate in tumorigenesis [19]. However, little is known regarding the functional role of ZNF263 in HCC. In addition, its relationship with ERS-related autophagy and chemotherapy sensitivity has not been reported.
Here, we demonstrated that ZNF263 may participate in the regulation of HCC chemotherapy sensitivity through ERS-related autophagy, providing new ideas for the treatment of hepatocellular carcinoma.
Materials and methods
Reagents
The human HCC cell lines (PLC/PRF/5, Hep3B, LM3, SK-HEP-1, HepG2 and Huh7) and normal hepatic cell line (LO2) were obtained from the Shanghai Cell Bank (Chinese Academy of Sciences, Shanghai, China). Sorafenib was purchased from Aladdin (Shanghai, China). Tunicamycin (TM) was purchased from Abcam (Cambridge, UK). Cell Counting Kit-8 (CCK-8) was purchased from BestBio (Nanjing, China). 4-phenylbutyrate (4-PBA) and 3-methyladenine (3-MA) were purchased from Sigma (USA). Antibody against GRP78 was purchased from Biogot Biotechnology (USA). Antibody against ZNF263 was purchased from Atlas antibodies (Bromma, Sweden). Antibody against Beclin 1 and LC 3 were purchased from Abcam (Cambridge, UK). Antibody against XBP-1 s was purchased from CST (USA). Antibody against GAPDH was purchased from Bioss antibodies (Beijing, China). Antibody against β-actin was purchased from ZSGB-BIO Inc. (Beijing, China). The ZNF263 small interfering RNA (siRNA) and control siRNA were purchased from GenePharma (Shanghai, China).
Human samples and datasets
Our study was approved by the Ethics Committee of Anhui Medical University (ethical approval No.20040158), and written informed consents from all patients was obtained. We studied 138 patients diagnosed with HCC. The fresh liver tissues and adjacent nontumor liver tissues were surgically collected at The First Affiliated Hospital of Anhui Medical University between 2004 and 2012. All cases were classified according to the International Union Against Cancer Tumor Node Metastasis (TNM) classification. The clinical data included age, gender, history of hepatitis, history of cirrhosis, tumor size, clinical stages and degree of differentiation were collected from medical records.
The UALCAN web (http://ualcan.path.uab.edu) [20] was utilized to analysis relative expression levels of the target proteins from The Cancer Genome Atlas (TCGA) database. Expression levels of ZNF263 were compared between HCC and normal control groups. Survival curves were generated by UALCAN.
Sequencing and analysis
For Chromatin immunoprecipitation-sequencing (CHIP-Seq) and RNA-sequencing (RNA-Seq), samples were prepared and sent to Guangzhou Eipbiotec Co., Ltd. (Guangzhou, China). CHIP-Seq was performed by Active Motif (Carlsbad, TX, USA), as described [21]. In brief, 1 × 107 HepG2 cells were first cross-linked and fixed in a 1% formaldehyde solution, followed by the addition of 125 μmol/L glycine. Cells were then washed twice in ice-cold PBS and flash frozen at −80 °C. The histone mark H3K27ac (ab4729, Abcam, Cambridge, MA, USA) was used, which distinguish active enhancer [22]. Total RNA was extracted from HepG2 cell treated with TM (2.5 mM, 24 h) or with the vehicle control, using the RNeasy Mini Kit from Qiagen (cat no;74104). To determine the top enriched DNA-Binding Motifs, the Homer findPeaks tool (http://homer.ucsd.edu/homer/ngs/peakMotifs.html) was used with default parameters. Motifs were ranked according to adjusted P-value.
Immunostaining and tissue microarray
To measure the expression of ZNF263, GRP78, Beclin1 and LC3, immunohistochemistry was applied on paraffin embedded tissue arrays contained HCC tissues and paired adjacent nontumor liver tissues. For analysis, the percentage of positive cells (0 for <10%, 1 for 10%–25%, 2 for 26%–50%, 3 for 51%–75%, and 4 for >75%) and cell staining intensity (0 for negative, 1 for weak intensity, 2 for moderate, and 3 for strong) were evaluated in five random fields in each tissue. The percentage of positive cells score multiplied by cell staining intensity score was used for ZNF263 score, as following: high, ≥6; low, <6.
Cell culture and treatment
All cells were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The final concentrations of sorafenib were 10 μmol/L, TM was 2.5 μmol/L, 3-MA was 1 mmol/L, and 4-PBA was 1 mmol/L. For knocking down ZNF263, cells were transfected with siRNA targeting ZNF263, according to the manufacturer's protocol. The sequence of si-ZNF263-1 was: 5′-CCGUAUAAAUGUACCCUUUTT-3′, si-ZNF263–2: 5′-GCAGCCAAAGAAACUCCAUTT-3′, si-ZNF263–3: 5′-GGAUAUGCAGAGAGAGCUUTT-3′, si-NC: 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were collected for further experiments at different time points after transfection.
Cell viability
CCK-8 was applied to measure cell viability, and cells (1 × 105 cells/ml) were implanted in a 96-well plate and cultured for 24 h. The optical density (OD) value was determined by a microplate reader (Infinite M1000 Pro, TECAN, Switzerland) at 450 nm.
Apoptosis analysis
Flow cytometry was used to measure the apoptotic rates of HCC cells. In brief, cells were collected and then centrifugation at 2000 rpm for 5 min. The Annexin V-FITC Apoptosis Detection Kit (BD Biosciences) was applied, according to manufacturer's instructions.
Immunofluorescence staining
The HCC cells were fixed with 4% paraformaldehyde and stained with ZNF263 rabbit antibody (1:300, Atlas Antibodies, Bromma, Sweden), LC3 mouse antibody (1:300, Santa Cruz Biotechnology, CA, USA) and GRP78 mouse antibody (1:200, Santa Cruz Biotechnology, CA, USA) overnight at 4 °C. After washed three times, the cells were then stained with Alexa Fluor 594 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-mouse IgG secondary antibodies for 2 h at room temperature. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and photos were taken randomly under a confocal laser scanning microscope (Olympus).
Transmission electron microscopy
Transmission electron microscopy (TEM), the gold standard for autophagosome measurement, was applied. In brief, cells were harvested and fixed in 2.5% glutaraldehyde (SPI Supplies) overnight at 4 °C. The cells were then post-fixed with 1% osmium tetroxide for 1 h at room temperature, dehydrated with gradient ethanol, infiltrated, and embedded in Epon 812 (SPI Supplies). Ultrathin sections (70 nm) were cut with a NOVA ultramicrotome (LKB Biotechnology). The sections were double-stained with 1% uranyl acetate and lead citrate. Images were taken with a JEM-1230 transmission electron microscope (Tokyo, Japan).
Protein extraction and Western blotting
Western blot was performed with various primary antibodies at 4 °C, and subsequently incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature, as described previously [13]. The target protein bands were visualized using an enhanced chemiluminescence detection reagent (Thermo Fisher). Image J (National Institutes of Health, Bethesda, MD, USA) was used to assess the intensity of target bands. In the current study, GAPDH or β-actin were used as internal controls.
Statistical analysis
Values are expressed as mean ± SD. ANOVA, student's t-tests, Mann-Whitney U test, Kruskal-Wallis tests, Chi-squared test or Kaplan-Meier test were applied for statistical analysis when appropriate. A two-sided P value <0.05 was considered significant.
Results
ZNF263 expression was up-regulated in HCC patients and correlated with GRP78 levels and poor prognosis
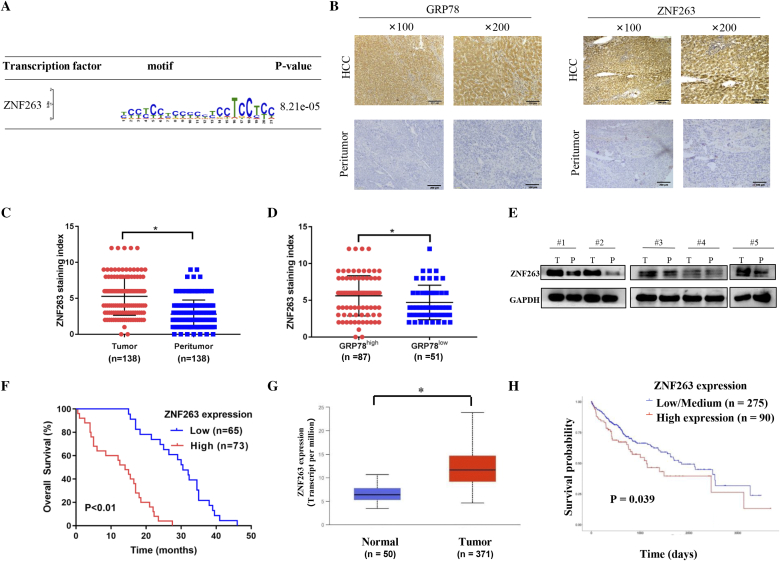
Super-enhancers (SEs), which differ from typical enhancers, are ultra-long cis-acting elements with transcription-enhancing activity that regulate key genes, and has been reported to promote oncogene activation in tumor [23,24]. Further, SEs landscape is significantly reprogrammed in HCC cells [24]. Because key transcriptional factors that bound to SEs can play important roles in cancer, we utilized H3K27ac CHIP-Seq and RNA-Seq to uncover key transcriptional factor which bound to ERS-specific SEs in HCC. We showed that, among the upregulated gene, some genes were differentially expressed by ERS and might be regulated by SEs. Further analysis showed that ZNF263 was the most significant ERS-specific transcriptional factor in HCC cell lines (HepG2 and Huh7 cells) (Fig. 1A).
Fig. 1.
ZNF263 expression is up-regulated in HCC patients and correlated with ER stress and poor prognosis. (A) Detailed information of ZNF263 binding motifs and associated P values. (B) Representative images of paired HCC tissues and adjacent nontumor liver tissues sections with GRP78 and ZNF263 immunohistochemical staining. (C) ZNF263 staining index grouped by tumor and peritumor. (D) ZNF263 staining index grouped by GRP78 level. (E) Protein expression of ZNF263 in paired HCC tissues and adjacent nontumor liver tissues. (F) Kaplan-Meier curves of overall survival in low-ZNF263 and high-ZNF263 groups. (G) ZNF263 expression in HCC tissues and normal liver tissues from TCGA data set. (H) Kaplan-Meier survival analysis in low/medium-ZNF263 and high-ZNF263 groups from TCGA data set. *P < 0.05.
Immunohistochemistry and western blot were used to investigate the expression levels of ZNF263 and GRP78 in human HCC tissues. As shown in Fig. 1B and C, ZNF263 was up-regulated in HCC tissues compared to the adjacent nontumor liver tissues (P < 0.05). Further, we found higher ZNF263 levels in group with higher GRP78 levels (P < 0.05) (Fig. 1D). Western blot also confirmed that the protein expression level of ZNF263 was up-regulated in HCC tissues compared with the adjacent nontumor liver tissues (Fig. 1E). Correlation analysis demonstrated that higher ZNF263 levels correlated with higher GRP78 levels in HCC patients (P = 0.005) (Table 1). We also investigated the correlation of ZNF263 with clinic-pathological features in HCC patients. The results showed that, there were no correlation between ZNF263 levels and parameters, including age, gender, history of hepatitis and history of cirrhosis (P > 0.05 for all) (Table 2). On the other hand, higher ZNF263 expression correlated with larger HCC tumor size (P = 0.016), advanced clinical stage (state III–IV) (P = 0.014) and loss of differentiation (P = 0.037) in HCC patients (Table 2). There was a correlation between tumor size and degree of differentiation (P < 0.05) (Table 2). As shown in Fig. 1F, patients with high ZNF263 levels showed worse overall survival compared with patients with low ZNF263 expression (P < 0.01). Analysis of the publicly available TCGA data sets confirmed that ZNF263 expression was significantly higher (P < 0.05, Fig. 1G) in HCC tissues and higher ZNF263 expression correlated with worse survival (P = 0.039, Fig. 1H).
Table 1.
Gene co-occurrent alteration between ZNF263 and GRP78.
| ZNF263 | GRP78 |
||
|---|---|---|---|
| Positive | Negative | P value | |
| Positive | 54 | 33 | 0.005 |
| Negative | 19 | 32 | |
Table 2.
Correlation between ZNF263 expression and clinicopathological features.
| ZNF263 expression |
P value | |||
|---|---|---|---|---|
| No. of patients | High (n, %) | Low (n, %) | ||
| Age, years | ||||
| <60 | 87 | 44, 50.5% | 43, 49.5% | 0.475 |
| ≥60 | 51 | 29, 56.9% | 22, 43.1% | |
| Gender | ||||
| Male | 104 | 53, 51.0% | 51, 49.0% | 0.425 |
| Female | 34 | 20, 58.8% | 14, 41.2% | |
| History of hepatitis | ||||
| No | 38 | 20, 52.6% | 18, 47.4% | 0.969 |
| Yes | 100 | 53, 53.0% | 47, 47.0% | |
| History of cirrhosis | ||||
| No | 73 | 37, 50.7% | 36, 49.3% | 0.612 |
| Yes | 65 | 36, 55.4% | 29, 44.6% | |
| Tumor size, cm | ||||
| <5 | 32 | 8, 25.0% | 24, 75.0% | <0.001 |
| 5–10 | 71 | 39, 54.9% | 32, 45.1% | |
| ≥10 | 35 | 26, 74.3% | 9, 25.7% | |
| Clinical stages | ||||
| I–II | 76 | 33, 43.4% | 43, 56.6% | 0.014 |
| III–IV | 62 | 40, 64.5% | 22, 35.5% | |
| Degree of differentiation | ||||
| High | 35 | 12, 34.3% | 23, 65.7% | 0.037 |
| Moderate | 77 | 45, 58.4% | 32, 41.6% | |
| Poor | 26 | 16, 61.5% | 10, 38.5% | |
ZNF263 expression in HCC cells
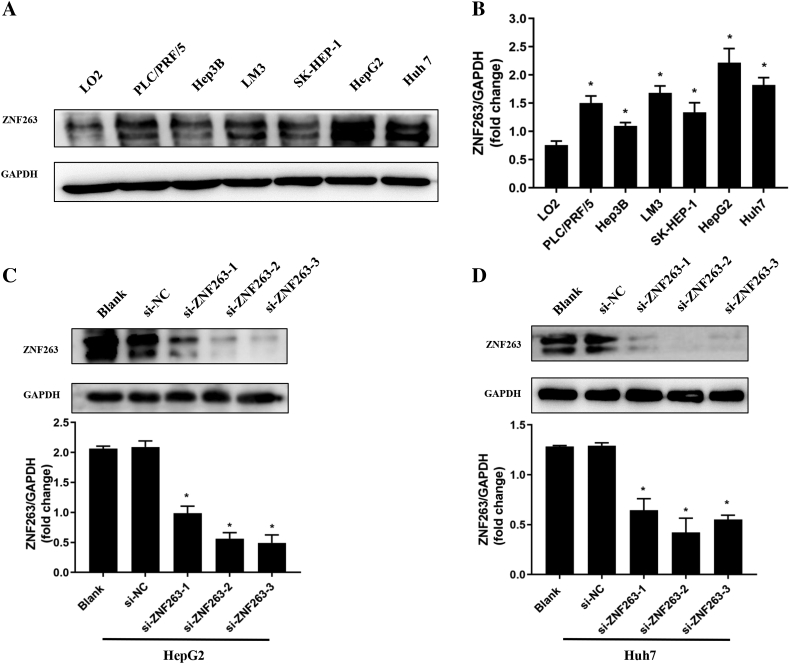
The expression level of ZNF263 in HCC cell lines was investigated by western blot. As shown in Fig. 2A and B, ZNF263 level was significantly higher in HCC cell lines including PLC/PRF/5, Hep3B, LM3, SK-HEP-1, HepG2 and Huh7, compared with the normal hepatic cell line LO2 (P < 0.05 for all). According to the expression level of ZNF263 in HCC cell lines, the HepG2 and Huh7 cells were selected for further experiments.
Fig. 2.
ZNF263 expression in HCC cell lines. (A) Protein expression of ZNF263 in HCC cell lines. (B) Quantification of the relative levels of ZNF263 in HCC cell lines by western blotting. *P < 0.05 vs LO2. (C) Representative images of western blotting and ZNF263 expression in HepG2 cell with RNA interfering. *P < 0.05 vs si-NC. (D) Representative images of western blotting and ZNF263 expression in Huh7 cell with RNA interfering. *P < 0.05 vs si-NC.
To determine the functional role of ZNF263 in HCC progression, siRNA-mediated ZNF263 knockdown in HepG2 and Huh7 cells were performed. As shown in Fig. 2C and D, the efficiency of ZNF263 knockdown with siRNA was confirmed by comparison with negative control group. Among the three siRNAs, si-ZNF263-3 in HepG2 and si-ZNF263-2 in Huh7 cells generated the most consistent knockdown results, and were therefore chosen for further experiments.
ZNF263 knockdown inhibited proliferation and promoted apoptosis in HCC cell
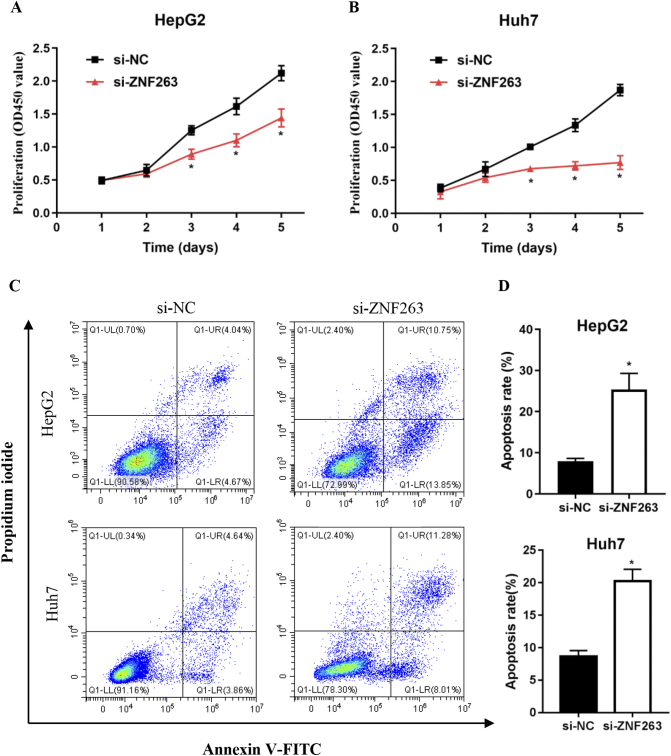
As shown in Fig. 3A and B, CCK-8 assays demonstrated that ZNF263 knockdown significantly reduced cell proliferation in HepG2 and Huh7 cells. Furthermore, flow cytometry assay showed that cell apoptosis rate was significantly higher in ZNF263 siRNA-treated groups than in negative control groups in HepG2 and Huh7 cells (Fig. 3C and D). These results indicated that ZNF263 knockdown reduced proliferation and induced apoptosis in HepG2 and Huh7 cells.
Fig. 3.
ZNF263 knockdown inhibited proliferation and promoted apoptosis in HCC cell. (A) and (B) CCK-8 assay was used to examine the proliferation of HepG2 and Huh7 cells. (C) Representative images of flow cytometry in HepG2 and Huh7 cells. (D) Quantification of cell apoptosis rate in HepG2 and Huh7 cells. *P < 0.05.
ZNF263 knockdown increases drug sensitivity of HCC cells
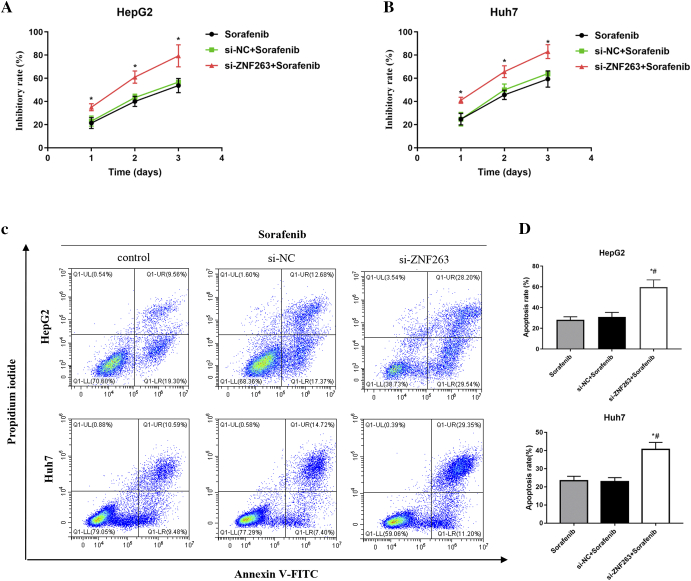
As shown in Fig. 4A and B, CCK-8 assays demonstrated that ZNF263 knockdown significantly increased cell inhibitory rate in sorafenib-treated HepG2 and Huh7 cells. As shown in Fig. 4C and D, cell apoptosis rate was significantly higher by ZNF263 knockdown in sorafenib-treated HepG2 and Huh7 cells. These results indicated that ZNF263 knockdown reduced chemoresistance in HepG2 and Huh7 cells.
Fig. 4.
ZNF263 knockdown increases drug sensitivity of HCC cells. (A) and (B) CCK-8 assay was used to examine the inhibitory rate of sorafenib-treated HepG2 and Huh7 cells. *P < 0.05 vs si-NC + sorafenib. (C) Representative images of flow cytometry in sorafenib-treated HepG2 and Huh7 cells. (D) Quantification of cell apoptosis rate in sorafenib-treated HepG2 and Huh7 cells. *P < 0.05 vs sorafenib, #P < 0.05 vs si-NC + sorafenib.
Taken together, these results indicated that ZNF263 might play an important role in cancer progression in HCC.
ZNF263 activates autophagy in HCC
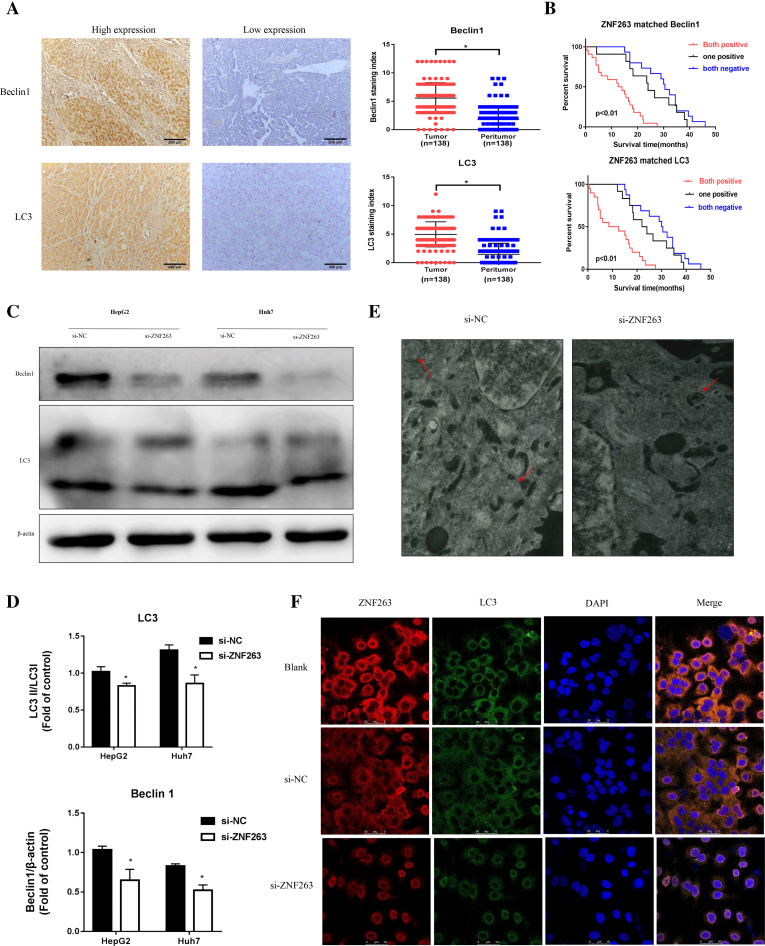
As shown in Fig. 5A, Beclin1 and LC3 staining density were significantly higher in HCC tissues compared to the adjacent nontumor liver tissues (P < 0.05 for both, respectively). As shown in Table 3, the positive expression of ZNF263 was significantly correlated with the positive expression of Beclin1 and LC3 in HCC tissues (P < 0.05 for both). Importantly, further survival analysis showed that ZNF263+/Beclin1+ double-positive group and ZNF263+/LC3+ double-positive group had worse overall survival than other groups (P < 0.01 for both, respectively) (Fig. 5B).
Fig. 5.
ZNF263 activates autophagy in HCC. (A) Representative images of HCC tissues and adjacent nontumor liver tissues and quantification of Beclin1 and LC3 staining index. * P < 0.05. (B) Kaplan-Meier curves of overall survival in ZNF263-/Beclin1- double-negative, ZNF263+ or Beclin1+ one positive and ZNF263+/Beclin1+ double-positive groups; ZNF263-/LC3- double-negative, ZNF263+ or LC3+ one positive and ZNF263+/LC3+ double-positive groups. (C) Protein expression of Beclin1 and LC3 in HepG2 and Huh7 cells. (D) Quantification of the relative levels of Beclin1 and LC3 in HepG2 and Huh7 cells by western blotting. * P < 0.05 vs si-NC. (E) Representative images of transmission electron microscopy in HepG2 cell. (F) Representative images of ZNF263 (red) and LC3 (green) double immunostaining in Huh7 cell. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Gene co-occurrent alteration between ZNF263, Beclin1 and LC3.
| ZNF263 | Beclin1 |
LC3 |
||||
|---|---|---|---|---|---|---|
| Positive | Negative | P value | Positive | Negative | P value | |
| Positive | 49 | 24 | 0.021 | 48 | 25 | 0.013 |
| Negative | 31 | 34 | 29 | 36 | ||
Western blot analysis showed that, ZNF263 knockdown significantly reduced Beclin1 (P < 0.05 for both) and LC3 II/I ratio (P < 0.05 for both) in HepG2 and Huh7 cells (Fig. 5C and D). The results of TEM showed that, there was decreased autophagic vacuoles accumulation in ZNF263 knockdown group of HepG2 cell (Fig. 5E). Moreover, immunofluorescence also verified that LC3 levels was lower in ZNF263 knockdown Huh7 cell (Fig. 5F).
Inhibition of ER stress reduced autophagy, HCC cell chemoresistance and the expression of ZNF263
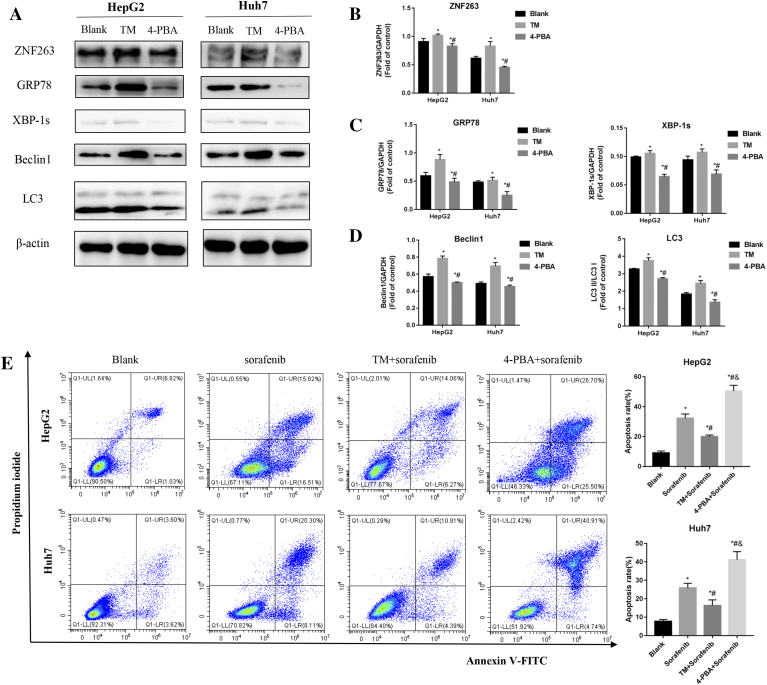
As shown in Fig. 6A–D, treatment with an ER stress activator (TM) increased protein levels of ZNF263, ER stress marker proteins (GRP78 and XBP-1 s) and autophagy marker proteins (Beclin1 and LC3) in HepG2 and Huh7 cells (P < 0.05 for both). On the other hand, treatment with an ER stress inhibitor (4-PBA) significantly reduced the expression levels of the above mentioned target proteins in HepG2 and Huh7 cells (P < 0.05 for both), as compared with the blank and TM groups (Fig. 6A–D).
Fig. 6.
Inhibition of ER stress reduced autophagy, HCC cell chemoresistance and the expression of ZNF263. (A) Protein expression of ZNF263, GRP78, XBP-1s, Beclin1 and LC3 in HepG2 and Huh7 cells. (B) Quantification of the relative levels of ZNF263 in HepG2 and Huh7 cells by western blotting. *P < 0.05 vs Blank, # P < 0.05 vs TM. (C) Quantification of the relative levels of GRP78 and XBP-1s in HepG2 and Huh7 cells by western blotting. *P < 0.05 vs Blank, #P < 0.05 vs TM. (D) Quantification of the relative levels of Beclin1 and LC3 in HepG2 and Huh7 cells by western blotting. *P < 0.05 vs Blank, #P < 0.05 vs TM. (E) Representative images of flow cytometry and quantification of cell apoptosis rate in HepG2 and Huh7 cells. *P < 0.05 vs Blank, #P < 0.05 vs sorafenib, &P < 0.05 vs TM + sorafenib.
As shown in Fig. 6E, compared with sorafenib group, cell apoptosis rate was significantly lower in TM combined with sorafenib group while significantly higher in 4-PBA combined with sorafenib group, in both HepG2 (P < 0.05) and Huh7 (P < 0.05) cells. Compared with TM combined with sorafenib group, cell apoptosis rate was significantly higher in 4-PBA combined with sorafenib group in both HepG2 and Huh7 cells (P < 0.05 for both) (Fig. 6E).
Inhibition of autophagy reduced cell chemoresistance without influence ER stress
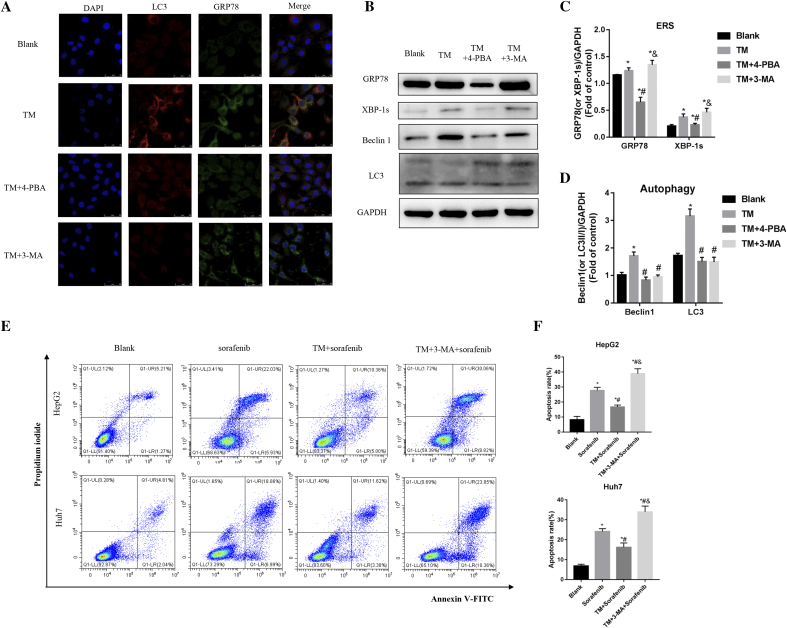
As shown in Fig. 7A, TM treatment increased ER stress marker protein (GRP78) and autophagy marker protein (LC3) in Huh7 cell, which was attenuated by co-treatment with 4-PBA. Combined TM and 3-MA treatment decreased autophagy marker protein (LC3) but not ER stress marker protein (GRP78). As shown in Fig. 7B–D, western blot results also confirmed that co-treatment with 4-PBA significantly reduced ER stress marker proteins (GRP78 and XBP-1 s) and autophagy marker protein (Beclin1 and LC3). Co-treatment with 3-MA significantly reduced autophagy marker protein (Beclin1 and LC3) but not ER stress marker proteins (GRP78 and XBP-1s).
Fig. 7.
Inhibition of autophagy reduced cell chemoresistance without influence ER stress. (A) Representative images of LC3 (red) and GRP78 (green) double immunostaining in Huh7 cell. (B) Protein expression of GRP78, XBP-1 s, Beclin1 and LC3 in Huh7 cell. (C) Quantification of the relative levels of GRP78 and XBP-1s in Huh7 cell by western blotting. *P < 0.05 vs Blank, #P < 0.05 vs TM, & P < 0.05 vs TM + 4-PBA. (D) Quantification of the relative levels of Beclin1 and LC3 in Huh7 cell by western blotting. *P < 0.05 vs Blank, #P < 0.05 vs TM, &P < 0.05 vs TM + 4-PBA. (E) Representative images of flow cytometry in HepG2 and Huh7 cells. (F) Quantification of cell apoptosis rate in HepG2 and Huh7 cells. *P < 0.05 vs Blank, #P < 0.05 vs sorafenib, &P < 0.05 vs TM + sorafenib. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As shown in Fig. 7E and F, compared with sorafenib group, cell apoptosis rate was significantly lower in TM combined with sorafenib group while significantly higher in 3-MA combined with TM and sorafenib group, in both HepG2 (P < 0.05) and Huh7 (P < 0.05) cells. Compared with TM combined with sorafenib group, cell apoptosis rate was significantly higher in 3-MA combined with TM and sorafenib group in both HepG2 and Huh7 cells (P < 0.05 for both) (Fig. 7E and F).
ZNF263 knockdown inhibited HCC cell chemoresistance via ER stress- dependent autophagy
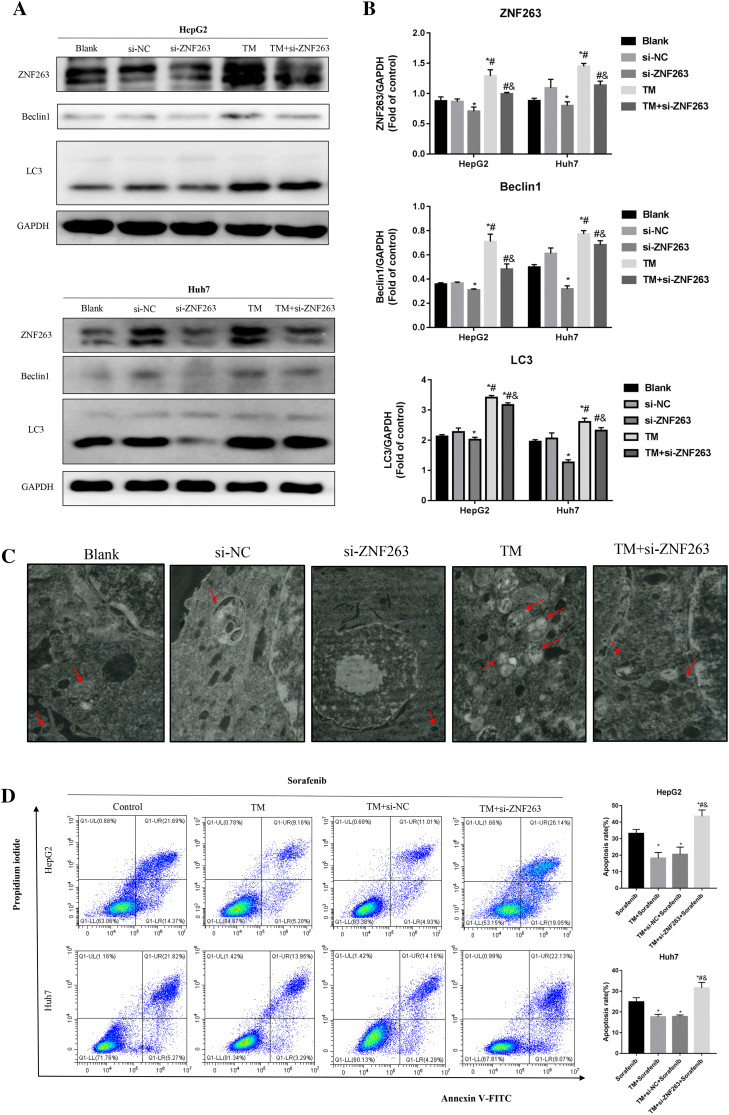
As shown in Fig. 8A and B, TM treatment significantly increased the protein levels of ZNF263, Beclin1 and LC3 in HepG2 and Huh7 cells. As shown in Fig. 8C, TM treatment increased the number of autophagic vacuoles in HepG2 cell. Further knockdown of ZNF263 significantly attenuated the above mentioned enhanced autophagic flux effect of TM (Fig. 8A, B and C).
Fig. 8.
ZNF263 knockdown inhibited HCC cell chemoresistance via ER stress-dependent autophagy. (A) Protein expression of ZNF263, Beclin1 and LC3 in HepG2 and Huh7 cells. (B) Quantification of the relative levels of ZNF263, Beclin1 and LC3 in HepG2 and Huh7 cells by western blotting. *P < 0.05 vs si-NC, #P < 0.05 vs si-ZNF263, &P < 0.05 vs TM. (C) Representative images of transmission electron microscopy in HepG2 cell. (D) Representative images of flow cytometry and quantification of cell apoptosis rate in HepG2 and Huh7 cells. *P < 0.05 vs sorafenib, #P < 0.05 vs TM + sorafenib, &P < 0.05 vs TM + si-NC + sorafenib.
As shown in Fig. 8D, cell apoptosis rate was significantly lower by TM treatment in sorafenib-treated HepG2 and Huh7 cells. ZNF263 knockdown combined with TM treatment significantly increased cell apoptosis rate compared with sorafenib treatment group and TM treatment group in HepG2 and Huh7 cells (Fig. 8D).
Discussion
At present, resistance to a combination of treatments including chemotherapy, radiotherapy and immunotherapy has troubled oncologists and clinicians around the world. How to overcome this obstacle is an issue of widespread concern. Elucidating potential mechanisms could greatly increase the efficiency of cancer treatment. In the current study, we investigated the potential role of the ERS-specific transcription factor ZNF263 in ERS mediated resistance to apoptosis in HCC. The key findings of this study are that: (1) ZNF263 level was up-regulated in HCC tumor tissues compared with its expression in the adjacent nontumor tissues, and higher ZNF263 level was associated with ER stress activation and worse clinical outcome in HCC patients; (2) Reduced ZNF263 expression attenuated HCC cell viability, chemoresistance, and increased cell apoptosis; (3) ZNF263 participated in ER stress and autophagy regulation in HCC cells; (4) ZNF263 knockdown inhibited HCC cell chemoresistance via ER stress-dependent autophagy. These data suggested that ZNF263 might represent a potential diagnostic biomarker and therapeutic target for HCC.
ERS represents a defense mechanism for cell survival [25,26,27]. Previous studies indicated that the ERS pathway is activated in HCC [13,28]. Importantly, it has been demonstrated that ERS activities affect chemotherapeutic drug resistance in HCC [6,29,30]. Furthermore, evidence from us and others indicated that ERS can prevent chemotherapy-induced apoptosis by inducing autophagy activation, while autophagy deficiency leads to increased apoptosis during ERS in HCC [31]. Tian and other studies found that ERS may up-regulate cellular autophagy through mammalian target of rapamycin (mTOR) and Beclin-1, leading to reduced sensitivity of ovarian cancer cells SKOV3 to cisplatin. Inhibition of cell autophagy can effectively enhance the sensitivity of ovarian cancer cells to cisplatin [32]. Similarly, we also found in previous work that ERS can induce autophagy in HepG2 liver cancer cells. Autophagy can protect liver cancer cells and inhibit apoptosis but does not protect normal liver cells. The use of autophagy inhibitor 3−MA can increase the oxaliplatin-induced Apoptosis of liver cancer cells [14,33]. The different response between normal cells and tumor cells to chemoresistance is important to understand the mechanism of autophagy regulation in the endoplasmic reticulum stress of tumor cells. Understanding the role of ERS-induced autophagy flux accompanying chemoresistance in HCC would lead to potential effective therapeutic options.
The zinc finger family is one of the largest family of transcription factors in the human genome. Recent studies revealed that the zinc finger family includes both oncogenes and tumor suppressor genes [34,35]. Furthermore, ZNF proteins play key role in cancer progression. However, very little is known about the function of ZNF263 in HCC progression, except that it is predicated to involve in tumorigenesis. Herein, we found that ZNF263 was overexpressed in human HCC tissues and correlated with ER stress activation, advanced tumor stages and poor clinical survival. Our finding is consistent with previous study, which reported the upregulation of ZNF263 in the blood of HCC patients [19]. ZNFs have been reported to induce cancer progression by activating cell proliferation, migration, invasion, and regulating cell apoptosis [34]. The present study showed that, knockdown of ZNF263 in HCC cell lines was associated with decreased proliferation, chemoresistance and increased apoptosis. These findings suggest that ZNF263 can participate in tumor progression in HCC, especially chemoresistance, which might represent a potential therapy target.
Autophagy is a conserved intracellular process and plays an important role in preserving cellular homeostasis by degrading and recycling misfolded proteins and damaged organelles [11]. Autophagy is strongly induced upon environmental stresses and in most contexts promotes tumorigenesis [11,33,36,37]. Previous studies by us [14,33,38] and other groups [39,40,41] have demonstrated that cell autophagy is involved in HCC progression, which results in drug resistance and poor prognosis. Recently, it has been revealed that ZNFs may play important role in autophagy regulation in cancer. For example, ZNF306 has been reported to regulate autophagy genes expression in cancer cells [42]. Overexpression of Tristetraprolin, the most well-known ZNF protein, decreases autophagy in lung adenocarcinoma cells [43]. In addition, ZNF32 knockdown significantly promotes autophagy in breast cancer cells [44]. However, the role of ZNF263 in autophagy regulation in HCC cell remains largely unknown. Considering the important role of autophagy in HCC, we investigated the effect of ZNF263 on autophagy regulation. Herein, we found that ZNF263 knockdown markedly suppressed HCC cell autophagy formation capability. Furthermore, ZNF263 knockdown also attenuated the increased autophagy formation by TM treatment. Our previous studies have demonstrated the important role of ERS in autophagy accumulation, and the potential role of ERS-related autophagy in chemoresistance and apoptosis resistance in HCC [14,33]. Therefore, we further investigated the effect of ZNF263 knockdown on ERS-induced autophagy accumulation and ERS-induced chemoresistance in HCC cells. Our results showed that, ZNF263 knockdown effectively improved the chemoresistance effect induced by ERS-related autophagy in HCC cells. Hence, these results suggest that ZNF263 can participate in upregulating cell ERS and autophagy in HCC, which might be an important mechanism for tumor progression.
The mechanism for ERS-induced autophagy remains largely unknown. It has been suggested that the IRE1-JNK pathway and PERK-eIF2α pathway are important for ERS-induced autophagy activation [45,46]. Our previous research also showed that miR-663, as an endoplasmic reticulum stress factor-specific microRNA, can be activated by autophagy in hepatocellular carcinoma [47]. And the unfolded protein response (UPR) PERK-ATF4-Beclin1 pathway may be involved in ERS-induced autophagy activation [14]. In this study, it was demonstrated that knockdown of ZNF263 can inhibit autophagy and inhibit the expression of Beclin1 and LC3. Moreover, the expression of ZNF263 was positively correlated with the expression of Beclin1 and LC3 in the tissue chip. We thus speculate that ZNF263 activating autophagy may also be related to the proven Beclin1 related pathway. Recently, autophagy has been shown to be related to another transcription factor, ZNF32, of the zinc finger protein family. ZNF32 can activate the AKT/mTOR pathway, thereby reducing the severe autophagy activity of breast cancer cells [44]. According to bioinformatics data, it is speculated that ZNF263 may pass the same path, while pushes autophagy in the opposite direction. A study published this year demonstrated that the ZNF family is related to the methylation stability of the target gene promoter [48]. We speculate that ZNF263 activation induced by ERS may cause a series of methylation of autophagy inhibitory proteins' promoters, which in turn inhibits the transcription of these genes, and leads to the activation of autophagy in the environment of endoplasmic reticulum stress. In addition, our previous data suggest that ZNF263 may affect mitochondrial function. Changes in mitochondrial function may be one of the factors affecting autophagy. Perhaps ZNF263 also regulates autophagy in organelles such as mitochondria or endoplasmic reticulum. Although we identified ZNF263 as an ER stress specific transcriptional factor for autophagy activation in the current study, further studies are warranted to determine the potential mechanism for ER stress related autophagy regulation.
In summary, this study demonstrated that the transcription factor ZNF263, one of the ERS-specific transcription factors, may be involved in the process of drug resistance of HCC caused by ERS-related autophagy. Targeting the transcription factor ZNF263 and targeting the corresponding ERS-specific transcription factors can bring new ideas to the clinical treatment and medication of hepatocellular carcinoma with apoptosis resistance in the future.
Author's contributions
Cui Jie: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing. Liu Jiatao: Conceptualization, Investigation, Validation, Methodology, Review & Editing. Fan Lulu: Conceptualization. Zhu Yue: Formal analysis. Zhou Bei: Investigation. Wang Yu: Formal analysis. Hua Wei: Investigation. Wei Wei: Conceptualization & Methodology. Sun Guoping: Supervision, Funding Acquisition, Review & Editing.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81872047).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
No additional acknowledgements.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Llovet J.M., Burroughs A., Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Asghar U., Meyer T. Are there opportunities for chemotherapy in the treatment of hepatocellular cancer? J Hepatol. 2012;56:686–695. doi: 10.1016/j.jhep.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Kaneko M., Imaizumi K., Saito A., Kanemoto S., Asada R., Matsuhisa K. ER stress and disease: toward prevention and treatment. Biol Pharm Bull. 2017;40:1337–1343. doi: 10.1248/bpb.b17-00342. [DOI] [PubMed] [Google Scholar]
- 5.Corazzari M., Gagliardi M., Fimia G.M., Piacentini M. Endoplasmic reticulum stress, unfolded protein response, and cancer cell fate. Front Oncol. 2017;7:78. doi: 10.3389/fonc.2017.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan L., Sun G., Ma T., Zhong F., Lei Y., Li X. Melatonin reverses tunicamycin-induced endoplasmic reticulum stress in human hepatocellular carcinoma cells and improves cytotoxic response to doxorubicin by increasing CHOP and decreasing survivin. J Pineal Res. 2013;55:184–194. doi: 10.1111/jpi.12061. [DOI] [PubMed] [Google Scholar]
- 7.Lin Y., Wang Z., Liu L., Chen L. Akt is the downstream target of GRP78 in mediating cisplatin resistance in ER stress-tolerant human lung cancer cells. Lung Cancer. 2011;71:291–297. doi: 10.1016/j.lungcan.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Bhat T.A., Chaudhary A.K., Kumar S., O'Malley J., Inigo J.R., Kumar R. Endoplasmic reticulum-mediated unfolded protein response and mitochondrial apoptosis in cancer. Biochim Biophys Acta Rev Cancer. 2017;1867:58–66. doi: 10.1016/j.bbcan.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Sisinni, M. Pietrafesa, S. Lepore, F. Maddalena, V. Condelli, F. Esposito, et al., Endoplasmic Reticulum Stress and Unfolded Protein Response in Breast Cancer: The Balance between Apoptosis and Autophagy and Its Role in Drug Resistance. Int J Mol Sci 20 (2019). [DOI] [PMC free article] [PubMed]
- 10.Jeon Y.J., Khelifa S., Ratnikov B., Scott D.A., Feng Y., Parisi F. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell. 2015;27:354–369. doi: 10.1016/j.ccell.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y.J., Lei Y.H., Yao N., Wang C.R., Hu N., Ye W.C. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J., Fan L., Yu H., Zhang J., He Y., Feng D. Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology. 2019;70:241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou B., Lu Q., Liu J., Fan L., Wang Y., Wei W. Melatonin increases the sensitivity of hepatocellular carcinoma to sorafenib through the PERK-ATF4-beclin1 pathway. Int J Biol Sci. 2019;15:1905–1920. doi: 10.7150/ijbs.32550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frietze S., Lan X., Jin V.X., Farnham P.J. Genomic targets of the KRAB and SCAN domain-containing zinc finger protein 263. J Biol Chem. 2010;285:1393–1403. doi: 10.1074/jbc.M109.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Feng S., Yang Y. Identification of transcription factors (TFs) and targets involved in the cholangiocarcinoma (CCA) by integrated analysis. Cancer Gene Ther. 2016;23:439–445. doi: 10.1038/cgt.2016.64. [DOI] [PubMed] [Google Scholar]
- 17.Xu G., Li K., Zhang N., Zhu B., Feng G. Screening driving transcription factors in the processing of gastric cancer. Gastroenterol Res Pract. 2016;2016:8431480. doi: 10.1155/2016/8431480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitsu H., Sonoda M., Higashijima T., Shirozu H., Masuda H., Tohyama J. Somatic mutations in GLI3 and OFD1 involved in sonic hedgehog signaling cause hypothalamic hamartoma. Ann Clin Transl Neurol. 2016;3:356–365. doi: 10.1002/acn3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Qian Z., Li F., Li J., Lu Y. Integrative analysis of microarray data to reveal regulation patterns in the pathogenesis of hepatocellular carcinoma. Gut Liver. 2017;11:112–120. doi: 10.5009/gnl16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar D.S., Bashel B., Balasubramanya S., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itkonen H.M., Urbanucci A., Martin S.E., Khan A., Mathelier A., Thiede B. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics. 2019;9:2183–2197. doi: 10.7150/thno.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsang F.H., Law C.T., Tang T.C., Cheng C.L., Chin D.W., Tam W.V. Aberrant super-enhancer landscape in human hepatocellular carcinoma. Hepatology. 2019;69:2502–2517. doi: 10.1002/hep.30544. [DOI] [PubMed] [Google Scholar]
- 25.Kim I., Xu W., Reed J.C. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H. ER stress and diseases. Febs J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 27.Madden E., Logue S.E., Healy S.J., Manie S., Samali A. The role of the unfolded protein response in cancer progression: from oncogenesis to chemoresistance. Biol Cell. 2019;111:1–17. doi: 10.1111/boc.201800050. [DOI] [PubMed] [Google Scholar]
- 28.Shuda M., Kondoh N., Imazeki N., Tanaka K., Okada T., Mori K. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 29.Al-Rawashdeh F.Y., Scriven P., Cameron I.C., Vergani P.V., Wyld L. Unfolded protein response activation contributes to chemoresistance in hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2010;22:1099–1105. doi: 10.1097/MEG.0b013e3283378405. [DOI] [PubMed] [Google Scholar]
- 30.Jiang X., Kanda T., Nakamoto S., Miyamura T., Wu S., Yokosuka O. Involvement of androgen receptor and glucose-regulated protein 78 kDa in human hepatocarcinogenesis. Exp Cell Res. 2014;323:326–336. doi: 10.1016/j.yexcr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Wang N., Tan H.Y., Li S., Feng Y. Atg9b deficiency suppresses autophagy and potentiates endoplasmic reticulum stress-associated hepatocyte apoptosis in hepatocarcinogenesis. Theranostics. 2017;7:2325–2338. doi: 10.7150/thno.18225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian J., Liu R., Qu Q. Role of endoplasmic reticulum stress on cisplatin resistance in ovarian carcinoma. Oncol Lett. 2017;13:1437–1443. doi: 10.3892/ol.2017.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Ma, Y.Y. Li, J. Zhu, L.L. Fan, Du WD, C.H. Wu, et al., Enhanced autophagic flux by endoplasmic reticulum stress in human hepatocellular carcinoma cells contributes to the maintenance of cell viability. Oncol Rep 30 (2013) 433-40. [DOI] [PubMed]
- 34.Jen J., Wang Y.C. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cassandri M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3:17071. doi: 10.1038/cddiscovery.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Fazio P., Matrood S. Targeting autophagy in liver cancer. Transl Gastroenterol Hepatol. 2018;3:39. doi: 10.21037/tgh.2018.06.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Fan L., Wang H., Sun G. Autophagy, a double-edged sword in anti-angiogenesis therapy. Med Oncol. 2016;33:10. doi: 10.1007/s12032-015-0721-9. [DOI] [PubMed] [Google Scholar]
- 38.Liu J.T., Li W.C., Gao S., Wang F., Li X.Q., Yu H.Q. Autophagy inhibition overcomes the antagonistic effect between gefitinib and cisplatin in epidermal growth factor receptor mutant non—small-cell lung cancer cells. Clin Lung Cancer. 2015;16:e55–e66. doi: 10.1016/j.cllc.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 39.W.P. Xu, J.P. Liu, J.F. Feng, C.P. Zhu, Y. Yang, W.P. Zhou, et al., miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut (2019). [DOI] [PubMed]
- 40.Song J., Qu Z., Guo X., Zhao Q., Zhao X., Gao L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y.H., Ding Z.B., Zhou J., Hui B., Shi G.M., Ke A.W. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–1172. doi: 10.4161/auto.7.10.16818. [DOI] [PubMed] [Google Scholar]
- 42.Chauhan S., Goodwin J.G., Chauhan S., Manyam G., Wang J., Kamat A.M. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50:16–28. doi: 10.1016/j.molcel.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong F., Li C., Wang P., Deng X., Luo Q., Tang X. The RNA binding protein tristetraprolin down-regulates autophagy in lung adenocarcinoma cells. Exp Cell Res. 2018;367:89–96. doi: 10.1016/j.yexcr.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Zhang L., Li K., Li J., Xiang R., Zhang J. ZNF32 inhibits autophagy through the mTOR pathway and protects MCF-7 cells from stimulus-induced cell death. Sci Rep. 2015;5:9288. doi: 10.1038/srep09288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 47.Y. Huang, J. Liu, L. Fan, F. Wang, H. Yu, W. Wei, et al., miR-663 overexpression induced by endoplasmic reticulum stress modulates hepatocellular carcinoma cell apoptosis via transforming growth factor beta 1. Onco Targets Ther 9 (2016) 1623-33. [DOI] [PMC free article] [PubMed]
- 48.Yu Z., Feng J., Wang W., Deng Z., Zhang Y., Xiao L. The EGFR-ZNF263 signaling axis silences SIX3 in glioblastoma epigenetically. Oncogene. 2020;39:3163–3178. doi: 10.1038/s41388-020-1206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]