Abstract
Background/purpose
Oral squamous cell carcinoma (OSCC) is one of the most lethal malignancies which accounts for approximately 90% of all malignant oral tumours. SAMMSON is a lncRNA located on chromosome 3p13–3p14 and is known to act as an oncogene in several malignancies. However, its expression and clinical significance in oral squamous cell carcinoma (OSCC) remain mostly unclear. In this study, we investigated the expression and clinical relevance of lncRNA SAMMSON in human OSCC.
Materials and methods
Human OSCC cell lines (Tca8113, SCC9, SCC25, CAL27, HN12, HSU3, FADU) and a human normal oral keratinocyte cell line (HNOK) were used to detect the difference of SAMMSON expression. A total of 90 OSCC patients confirmed by pathological and clinical diagnoses at the Hospital of Stomatology, Department of Periodontology, Shandong University were enrolled. The mRNA expression level was analyzed by reverse transcription PCR (QRT-PCR). Statistical analyses including Student's t-test, chi-square method, Kaplan-Meier method, Univariate and, Multivariate Cox regression analysis were performed to analyse all data.
Results
This study showed that the expression of SAMMSON was significantly increased in OSCC tissues and cell lines. High SAMMSON expression was significantly associated with TMN stage, tumour differentiation, lymph node metastasis distant metastasis and neighboring tissue infiltration. Patients with high expression of SAMMSON had poor overall survival and disease-free survival compared to those with low levels. Cox regression analysis showed that SAMMSON could act as an independent prognostic factor in OSCC.
Conclusion
Serum SAMMSON expression was associated with tumour SAMMSON expression. ROC curve analysis indicated the high diagnostic sensitivity and specificity of serum SAMMSON expression in OSCC patients as compared to other traditional serum biomarker SCCA, TSGF, and CEA. These results indicated that SAMMSON might play an essential role in OSCC progression and could serve as a novel prognostic and diagnostic biomarker in OSCC.
Keywords: SAMMSON, Biomarker, Oral squamous cell carcinoma
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of oral cancer, 90% of which is originating from squamous cells.1,2 Despite an enormous amount of available reports on diagnosis and combined treatment of OSCC with surgery, radiotherapy, and chemotherapy, morbidity, and mortality rate in OSCC in recent years have not been improved significantly.1 Tobacco use and alcohol consumption have been identified as risk factors of OSCC.3 In recent years, few molecular targets have been identified in OSCC,4, 5, 6 which makes it difficult to develop targeted therapy for OSCC. Currently, standard histological evaluation is still the primary means in OSCC diagnostic procedures and therapeutic decision-making. Hence, the discovery of new potential biomarkers for prognosis prediction may provide improved treatments for OSCC patients.
Long non-coding RNAs (lncRNAs) are a novel class of RNA transcripts longer than 200 nucleotides which involve in many cellular and developmental processes, including cell proliferation, apoptosis, and differentiation, as well as cancer progression.7, 8, 9 SAMMSON is a lncRNA located on chromosome 3p13–3p14 and is known to act as an oncogene in several malignancies.10, 11, 12 It has been first identified to be expressed explicitly in the vast majority (>90%) of melanomas, but not in normal adult tissues, indicating SAMMSON as an attractive therapeutic target for melanoma.10 However, its expression in OSCC is unknown. In this study, we aimed to explore the diagnostic and prognostic value of SAMMSON in OSCC. We observed that SAMMOSON is highly expressed in OSCC tissue and further evaluated the potential diagnostic and prognostic potential of SAMMSON expression in tumor tissues and serum samples from OSCC patients.
Material and methods
Cell culture
Human OSCC cell lines (Tca8113, SCC9, SCC25, CAL27, HN12, HSU3, FADU) and a human normal oral keratinocyte cell line (HNOK) were cultured in Dulbecco's modified Eagle medium (DMEM, Thermofisher Scientific, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Thermofisher Scientific) and 100U/ml penicillin/streptomycin. All cell lines were cultured at 37 in a humidified atmosphere with 5% CO2.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
RNAs were isolated using RNeasy Mini (Qiagen, Shanghai, China) for >50 000 cells and reverse transcribed into cDNAs using QuantiTect Reverse Transcription (Qiagen 205311). SAMMSON expression was normalized to GAPDH. SAMMSON forward, TTCCTCAACTATGCAACTCAA; reverse, TAGACTACGGGCTCATGACTT; GAPDH forward, GGAGCGAGATCCCTCCAAAAT; reverse, GGCTGTTGTCATACTTCTCATGG. Relative expression is calculated using the formula –ΔΔCt (-ddCt), where Ct stands for threshold cycles.
Clinical data collection
A total of 90 OSCC patients confirmed by pathological and clinical diagnoses at Hospital of Stomatology, Department of Periodontology, Shandong University were enrolled. All the protocols including clinical data and sample collection were approved and were under the supervising board of the Hospital of Stomatology, Shandong University. The methods were carried out following the approved guidelines. All patients met the enrolment criteria and provided signed informed consent. The collected information includes medical imaging results, surgery procedures, pathological classifications, sensitivity to chemotherapy, recurrence, tumor-free survival length, and overall survival. Tumor and adjacent normal tissues were obtained from the patients before they received any chemotherapy or radiotherapy. Table 1 presents the relevant clinicopathological characteristics. All tissue samples were fixed in 4% PFA and embedded in paraffin.
Table 1.
Clinicopathological features and SAMMON expression in OSCC patients.
| Chinicopathological characteristics | Total | high expression | low expression | X2 | P value |
|---|---|---|---|---|---|
| Gender | |||||
| male | 56 | 32 | 24 | 3.025 | 0.127 |
| female | 34 | 13 | 21 | ||
| Age | |||||
| ≤60 | 52 | 21 | 31 | 4.555 | 0.054 |
| >60 | 38 | 24 | 14 | ||
| Tumor size | |||||
| T1 | 36 | 16 | 20 | 2.352 | 0.503 |
| T2 | 26 | 15 | 11 | ||
| T3 | 15 | 6 | 9 | ||
| T4 | 13 | 8 | 5 | ||
| Distant metastasis | |||||
| Positive | 42 | 30 | 12 | 11.576 | 0.001 |
| Negative | 48 | 15 | 33 | ||
| Differentiation | |||||
| high | 17 | 12 | 5 | 14.472 | 0.001 |
| moderate | 25 | 18 | 7 | ||
| poor | 48 | 15 | 33 | ||
| Lymph node metastasis | |||||
| Positive | 46 | 31 | 15 | 11.383 | 0.001 |
| Negative | 44 | 14 | 30 | ||
| TMN stages | |||||
| I | 26 | 8 | 18 | 16.716 | 0.001 |
| II | 23 | 7 | 16 | ||
| III | 18 | 12 | 6 | ||
| IV | 23 | 18 | 5 | ||
| Cancer invasion depth | |||||
| ≤8 mm | 42 | 15 | 27 | 6.429 | 0.02 |
| >8 mm | 48 | 30 | 18 | ||
| Tumor location | |||||
| cheek | 34 | 18 | 16 | 1.487 | 0.685 |
| gangiva | 32 | 12 | 20 | ||
| Tonuge | 16 | 7 | 9 | ||
| Mouth floor | 8 | 5 | 3 | ||
Statistical analyses. Statistical analysis was performed using SPSS vX software (IBM, SPSS, Chicago, IL, USA). The Student's t-test was used to compare SAMMSON expression levels in the cell lines and tumor tissues. The data are shown as mean and standard derivation. The association between the clinical features and SAMMSON expression was analyzed by the chi-square method. Kaplan-Meier method with the log-rank test was used to analyze the overall survival and disease-free survival of OSCC patients. Univariate and Multivariate Cox regression analysis was applied to evaluate the prognostic value of SAMMON.
Results
SAMMSON is significantly up-regulated in OSCC tissues and cell lines
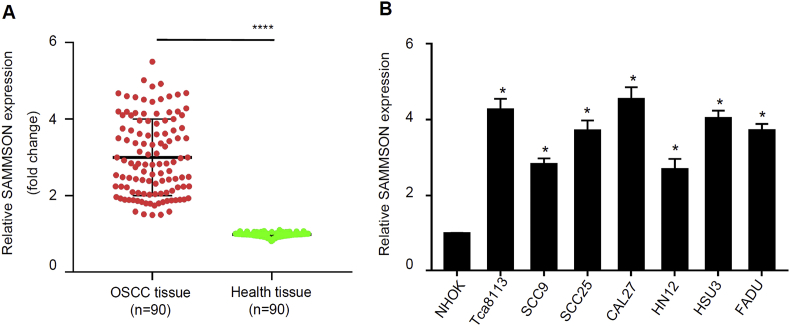
To explore its possibility as a functional biomarker in OSCC, we first analyzed SAMMSON expression in a cohort of 90 OSCC patient samples and adjacent normal healthy tissues by qRT-PCR. In the current study, tumor tissues and morphologically normal peri-cancerous tissues were obtained by a pathologist. In all 90 patients, SAMMSON expression levels were all significantly higher in the tumor tissues than the normal tissues, with an average increase of 3-fold (Fig. 1A, p < 0.001). This observation indicated that there is a possible involvement of SAMMSON in OSCC. To further investigate its role in OSCC, we characterized SAMMSON expression in OSCC cell lines using qRT-PCR. We found that SAMMSON expression levels were all significantly up-regulated in 7 OSCC cell lines compared to normal human oral keratinocyte cell line NHOK (Fig. 1B, p < 0.05). These results suggested that abnormal SAMMSON expression has a potential role in OSCC pathogenesis.
Figure 1.
SAMMSON is significantly up-regulated in OSCC tissues and cell lines. A. Relative SAMMSON expression levels in a cohort of 90 OSCC patient samples and adjacent normal healthy tissues (n = 90 per group; mean ± s.e.m.; paired Student's t-test, ∗∗∗∗P < 0.001). B. Relative SAMMSON expression levels in OSCC cell lines and NHOK cell line (n = 3 per group; mean ± s.e.m.; unpaired Student's t-test, ∗P < 0.05).
To further investigate the clinical significance of SAMMSON expression in OSCC, we then grouped 90 OSCC patients into high and low SAMMSON expression groups determined based on the median expression level. The 45 patients with the highest SAMMSON expression were categorized as the SAMMSONhigh group, and the rest 45 patients with the lower SAMMSON levels constituted the SAMMSONlow group. Patients’ clinical characteristics are summarized in Table 1. The chi-square test did not show any association between SAMMSON expression and age, gender, tumor location or size. Interestingly, we observed SAMMSON expression levels were associated with TMN stage, tumor differentiation, lymph node metastasis distant metastasis and neighboring tissue infiltration (p < 0.001, Table 1). In the SAMMSONhigh group, 30 out of 45 patients (66.7%) were in TMN stage III-IV. By contrast, stage III-IV patients only constituted 24.4% (11/34) of the SAMMSONlow group (χ2 = 16.7671, p = 0.001). Patients with distant metastasis in the SAMMSONlow group (26.7%) are significantly lower (30/45, χ2 = 11.576, p < 0.001) in the SAMMSONhigh group. Furthermore, patients with lymph node metastasis were 68.8% of SAMMSONhigh group, significantly higher than patients in SAMMSONlow group (χ2 = 11.383, p = 0.001). In agreement with this, patients with higher infiltration were 66,7% of the SAMMSONhigh group, significantly lower (30/45, χ2 = 6.429, p = 0.02) in the SAMMSONhigh group. These observations revealed a positive association between SAMMSON expression and poor clinical characteristics. SAMMSON may have an essential role in the progression of OSCC.
High SAMMSON expression predicts poor prognosis in OSCC
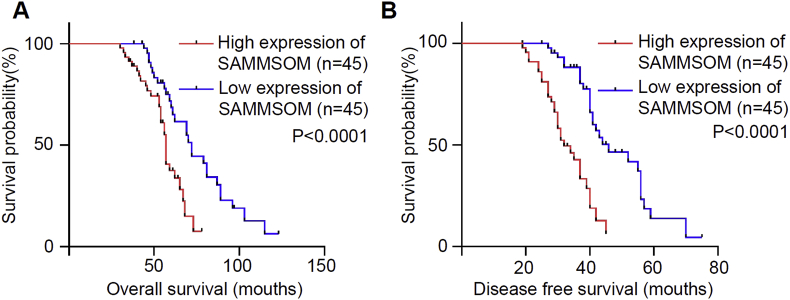
In the study of the effect of SAMMSON expression on patient survival, the Kaplan-Meier survival analysis showed poorer overall survival in the 45 SAMMSONhigh patients than in the 45 SAMMSONlow patients (Fig. 2A, p < 0.0001). The relationship between SAMMSON levels and disease-free survival rates was also analyzed by Kaplan-Meier analysis in all 90 OSCC cases, which showed high SAMMSON expression predicts poor disease-free survival of OSCC (Fig. 2B, p < 0.0001).
Figure. 2.
High SAMMSON expression predicts poor prognosis in OSCC. A:Kaplan–Meier curves for the overall survival in patients with OSCC. Patients with high SAMMON expression had significantly poorer overall survival than those with low SAMMON expression (p < 0.001, log-rank test). B:Kaplan–Meier curves for disease-free survival in patients with OSCC which are grouped based on SAMMON expression level (p < 0.001, log-rank test).
Cox regression analysis was used further determine the prognostic value of SAMMSON in OSCC. Univariate analysis of the potential overall survival prognosis factors of OSCC patients showed that SAMMSON expression levels were significantly associated with poor prognosis in OSCC patients (p < 0.05). Multivariant Cox analysis showed that SAMMSON was an independent factor for OSCC prognosis (p < 0.05). These results were summarized in Table 2. Univariate analysis on the potential disease-free survival prognosis factors of OSCC patients showed similar results, which summarized in Table 3. That SAMMSON expression levels, as well as tumor differentiation, tumor stage, lymphatic metastasis, and neighboring infiltration, were significantly associated with poor prognosis in OSCC patients (p < 0.05). Multivariant cox analysis on potential disease-free survival prognosis factors of OSCC patients further confirmed that SAMMSON was an independent factor for OSCC prognosis (p < 0.05).
Table 2.
Cox regression analysis for prognosis in OSCC patients.
| Overall survival | B | SE | Wald | df | P value | Exp(B) | 95%Exp(B) |
|
|---|---|---|---|---|---|---|---|---|
| upper limit | Lower limit | |||||||
| SAMMSON | 1.277 | 0.629 | 4.117 | 1 | 0.042 | 3.586 | 1.044 | 12.315 |
| Tumor differentiation | 0.21 | 0.412 | 0.259 | 1 | 0.0351 | 1.254 | 0.55 | 2.854 |
| TMN stage | −1.473 | 0.489 | 9.08 | 1 | 0.003 | 0.229 | 0.088 | 0.598 |
| Lymphatic metastasis | −1.237 | 0.459 | 8.72 | 1 | 0.035 | 1.984 | 0.605 | 0.998 |
| Distant metastasis | −0.797 | 0.392 | 4.133 | 1 | 0.042 | 0.451 | 0.209 | 0.972 |
| Cancer invasion depth (8 mm) | −0.998 | 0.542 | 7.33 | 1 | 0.027 | 0.179 | 0.025 | 0.993 |
| Tumor location | 0.11 | 0.17 | 0.42 | 1 | 0.517 | 1.116 | 0.8 | 1.557 |
| Tumor size | −0.115 | 0.182 | 0.398 | 1 | 0.528 | 0.897 | 0.624 | 1.274 |
| Sex | −0.002 | 0.016 | 0.017 | 1 | 0.895 | 0.998 | 0.967 | 1.03 |
| Age | −0.275 | 0.51 | 0.292 | 1 | 0.589 | 0.759 | 0.28 | 2.062 |
Table 3.
Cox regression analysis for disease-free survival in OSCC patients.
| Free diseases survival | B | SE | Wald | df | P value | Exp(B) | 95%Exp(B) |
|
|---|---|---|---|---|---|---|---|---|
| upper limit | Lower limit | |||||||
| SAMMSON | 1.15 | 0.578 | 3.955 | 1 | 0.047 | 3.157 | 1.017 | 9.804 |
| Tumor differentiation | −0.895 | 0.435 | 0.3893 | 1 | 0.048 | 0.423 | 0.18 | 0.994 |
| TMN stages | −1.308 | 0.471 | 7.716 | 1 | 0.005 | 0.27 | 0.107 | 0.68 |
| Lymphatic metastasis | −1.114 | 0.422 | 0.119 | 1 | 0.021 | 1.723 | 0.622 | 0.784 |
| Distant metastasis | −1.13 | 0.392 | 0.693 | 1 | 0.048 | 0.638 | 0.783 | 0.998 |
| Cancer invasion depth (8 mm) | −1.193 | 0.369 | 0.272 | 1 | 0.602 | 0.825 | 0.4 | 1.701 |
| Tumor location | −0.011 | 0.169 | 0.004 | 1 | 0.948 | 0.989 | 0.771 | 1.367 |
| Tumor size | −0.003 | 0.188 | 0 | 1 | 0.988 | 0.997 | 0.96 | 1.44 |
| Sex | 0.004 | 0.016 | 0.074 | 1 | 0.785 | 1.004 | 0.973 | 1.0307 |
| Age | 0.455 | 0.382 | 1.415 | 1 | 0.234 | 1.576 | 0.745 | 3.332 |
Upregulated serum SAMMSON is tumor-derived and acts as a potential diagnostic biomarker for OSCC
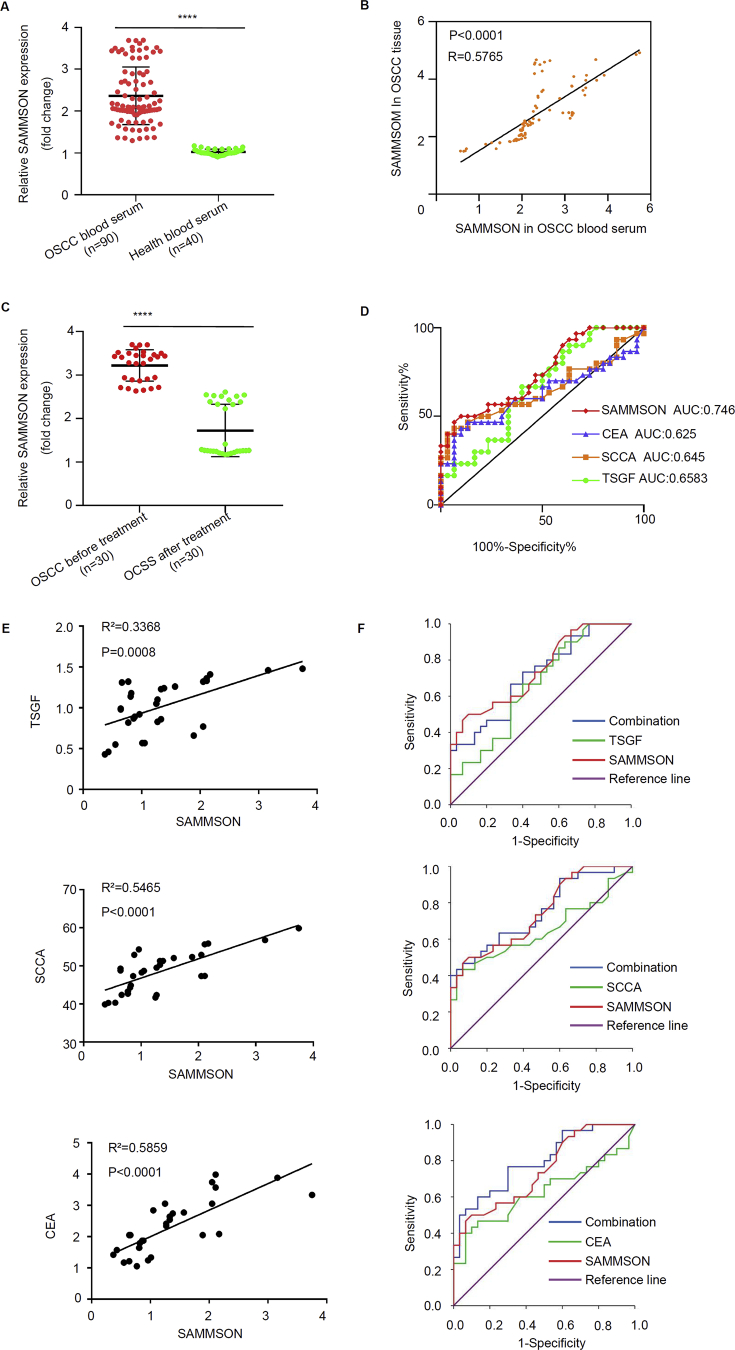
To explore the possible therapeutic application of SAMMSON as a biomarker, we first analyzed SAMMSON expression level in 90 OSCC patient serum and 40 normal healthy controls by qRT-PCR. Significantly higher SAMMSON expressions in the serum from OSCC patients were observed compared to normal healthy serum (Fig. 3A, p < 0.001). Next, we compared SAMMSON expression in the serum from OSCC patients with their tumor tissues. SAMMSON expression levels in the patient serum were positively correlated with levels in the tumor tissues using Pearson's correlation coefficient analysis (Fig. 3B, p < 0.001). These data indicated that serum SAMMSON is possibly tumor-derived. To confirm this possibility, we analyzed 30 matched pre- and post-surgery serum samples from 30 OSCC patients. A significant decrease of SAMMSON expression in post-surgery serum samples was observed compared to their matched pre-surgery expression levels (p < 0.001, Fig. 3C). We then evaluated serum SAMMSON levels as a diagnostic biomarker using receiver operating characteristic (ROC) curve analysis. Comparing to traditional serum biomarker SCCA, TSGF, and CEA, the AUC of SAMMSON was higher, which indicated the high diagnostic sensitivity and specificity of serum SAMMSON expression in OSCC patients. The SAMMSON expression levels were positively correlated with SCCA, TSGF, and CEA expression levels respectively (Fig. 3E). The sensitivity of using SAMMSON and CEA was higher than using SAMMSON single through ROC curve analysis for synergistic diagnosis, but there are no significant effect when combinedly using SAMMSON and SCCA or TSGF (Fig. 3F).
Figure 3.
Upregulated serum SAMMSON is tumor derived and acts as a potential diagnostic biomarker for OSCC. A. Relative SAMMSON expression levels in OSCC patient serum samples and healthy serum controls (p < 0.001). B. Pearson correlation coefficient analysis on SAMMSON expression level in tumor and serum samples from OSCC patients (p < 0.001). C. Relative SAMMSON expression in serum samples from 30 matched pre- and post-surgery serum samples (n = 30 per group; mean ± s.e.m.; paired Student's t-test, ∗p < 0.001). D. The ROC curve analysis of SAMMSON, CEA, SCCA, and TSGF (n = 30 per group; bivariate regression analysis, p < 0.001). E. The correlation among seral SAMMSON, SCCA, CEA and TSGF levels (n = 30 per group; mean ± s.e.m.; paired Student's t-test, ∗p < 0.001). F. The ROC curve analysis of synergistic diagnosis (n = 30 per group; bivariate regression analysis, p < 0.001).
In summary, these data demonstrate that serum SAMMSON can act as a potential diagnostic biomarker in OSCC.
Discussion
Despite an enormous amount of available reports and significant progress on diagnosis and combined treatment of OSCC with surgery, radiotherapy, and chemotherapy, the 5-year survival rate of OSCC and the prognosis of patients with OSCC remains unsatisfactory.13 Thus, it is critical to have a better and boarder understanding of the molecular events associated with OSCC development and progression to improve the clinical strategies and therapeutics. lncRNAs are shown to be involved in many cellular and developmental processes, including cell proliferation, apoptosis, and differentiation, as well as cancer progression.14,15 Recently, various IncRNAs, such as HOTAIR, TUG1, MALAT1, and ANDRIL, have been identified to be associated with OSCC progression.16, 17, 18, 19, 20, 21
SAMMSON is a lncRNA which was shown to be up-regulated and predicted poor prognosis of various cancers.10, 11, 12 However, to our knowledge, the role of lncRNA SAMMSON in the carcinogenesis of OSCC remains unclear.
In our study, we found that SAMMSON expression was significantly increased in OSCC tumor tissues and cell lines in comparison to adjacent healthy tissues and human normal oral keratinocyte cell line HNOK respectively. The relative expression level of SAMMSON in tumors of OSCC patients was associated with their TNM stage, tumor differentiation, lymph node metastasis, distant metastasis, and neighboring tissue infiltration. Kaplan-Meier and Cox regression analyses showed the prognostic value of SAMMSON. Patients with high SAMMSON expression had poor overall survival and disease-free survival. These results were similar to the presented researches. Yang et al. find that SAMMSON overexpression in hepatocellular carcinoma cells and it could promote migration and invasion of cancer cells by inhibiting the expression miR-9-3p as a sponge role.22 It has been demonstrated SAMMSON was up-regulated and increased the proliferation rate of glioblastoma cells as well as SAMMSON overexpression caused the down-regulated expression of miR-622.23 In papillary thyroid carcinoma, SAMMSON has been demonstrated a novel diagnostic and prognostic biomarker, which contributing progression to cancer by p30/Sp1 axis.24 Furthermore, our study indicated that serum SAMMSON is tumor-derived as Pearson's correlation coefficient analysis showed a positive correlation between serum SAMMSON levels and tumor SAMMSON levels. Serum SAMMSON expression as a potential diagnostic biomarker was evaluated using ROC curve analysis, which indicated the high diagnostic sensitivity and specificity of serum SAMMSON expression in OSCC patients.
This study firstly demonstrated the effect of lncRNA SAMMSON in OSCC. These findings highlight the clinical relevance of SAMMSON in OSCC patients and imply that SAMMSON may play a vital role in predicting OSCC progression. The multivariate analysis implied that high SAMMSON expression was a potential independent prognostic factor for overall survival and disease-free survival of OSCC patients. ROC curve analysis suggested SAMMSON has higher diagnostic sensitivity than traditional biomarkers such as SCCA, TSGF, and CEA. Therefore this study provides a new prospects for clinical diagnose and SAMMOSON can be used as a potential biomarker and therapeutic target in OSCC. However there are some limitations in this study, the possible target gene and signal pathway have been researched, and in our further study, we will explore the impact on biological phenotype and which genes would be regulated.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgements
Not Applicable.
References
- 1.Markopoulos A.K. Current aspects on oral squamous cell carcinoma. Open Dent J. 2012;6:126–130. doi: 10.2174/1874210601206010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosebush M.S., Rao S.K., Samant S. Oral cancer: enduring characteristics and emerging trends. J Mich Dent Assoc. 2012;94:64–68. [PubMed] [Google Scholar]
- 3.Moore S.R., Johnson N.W., Pierce A.M., Wilson D.F. The epidemiology of mouth cancer: a review of global incidence. Oral Dis. 2000;6:65–74. doi: 10.1111/j.1601-0825.2000.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 4.Choi S., Myers J.N. Molecular pathogenesis of oral squamous cell carcinoma: implications for therapy. J Dent Res. 2008;87:14–32. doi: 10.1177/154405910808700104. [DOI] [PubMed] [Google Scholar]
- 5.Gomes C.C., de Sousa S.F., Gomez R.S. MicroRNAs: small molecules with a potentially role in oral squamous cell carcinoma. Curr Pharmaceut Des. 2013;19:1285–1291. doi: 10.2174/138161213804805694. [DOI] [PubMed] [Google Scholar]
- 6.Momen-Heravi F., Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Bhan A., Soleimani M., Mandal S.S. Long noncoding RNA and cancer: a new paradigm. Canc Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez-Dominguez J.R., Lodish H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Tian H., Yang J., Gong Z. Long noncoding RNAs regulate cell growth, proliferation, and apoptosis. DNA Cell Biol. 2016;35:459–470. doi: 10.1089/dna.2015.3187. [DOI] [PubMed] [Google Scholar]
- 10.Leucci E., Vendramin R., Spinazzi M. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 11.Li X., Li M., Chen J. SAMMSON drives the self-renewal of liver tumor initiating cells through EZH2-dependent Wnt/beta-catenin activation. Oncotarget. 2017;8:103785–103796. doi: 10.18632/oncotarget.21792. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Vendramin R., Verheyden Y., Ishikawa H. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat Struct Mol Biol. 2018;25:1035–1046. doi: 10.1038/s41594-018-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinevici N., O'Sullivan J. Oral cancer: deregulated molecular events and their use as biomarkers. Oral Oncol. 2016;61:12–18. doi: 10.1016/j.oraloncology.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 15.Castro-Oropeza R., Melendez-Zajgla J., Maldonado V., Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. 2018;41:585–603. doi: 10.1007/s13402-018-0406-4. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y., Zhang L., Zhang L. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Li Z., Wang C. Expression of long non-coding RNA-HOTAIR in oral squamous cell carcinoma Tca8113 cells and its associated biological behavior. Am J Transl Res. 2016;8:4726–4734. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X., Liu W., Wang P., Li S. RNA interference of long noncoding RNA HOTAIR suppresses autophagy and promotes apoptosis and sensitivity to cisplatin in oral squamous cell carcinoma. J Oral Pathol Med. 2018;47:930–937. doi: 10.1111/jop.12769. [DOI] [PubMed] [Google Scholar]
- 19.Liang S., Zhang S., Wang P. LncRNA, TUG1 regulates the oral squamous cell carcinoma progression possibly via interacting with Wnt/beta-catenin signaling. Gene. 2017;608:49–57. doi: 10.1016/j.gene.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 20.Chai L., Yuan Y., Chen C., Zhou J., Wu Y. The role of long non-coding RNA ANRIL in the carcinogenesis of oral cancer by targeting miR-125a. Biomed Pharmacother. 2018;103:38–45. doi: 10.1016/j.biopha.2018.01.105. [DOI] [PubMed] [Google Scholar]
- 21.Zhang D., Sun G., Zhang H., Tian J., Li Y. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511–516. doi: 10.1016/j.biopha.2016.11.058. [DOI] [PubMed] [Google Scholar]
- 22.Yang S., Cai H., Hu B., Tu JJBr. vol. 39. 2019. LncRNA SAMMSON negatively regulates miR-9-3p in hepatocellular carcinoma cells and has prognostic values. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie J., Wang X., Liu S. LncRNA SAMMSON overexpression distinguished glioblastoma patients from patients with diffuse neurosarcoidosis. 2019;30:817–821. doi: 10.1097/WNR.0000000000001278. [DOI] [PubMed] [Google Scholar]
- 24.Shao L., Sun W., Wang Z., Dong W., Qin YJIl. 2019. Long noncoding RNA SAMMSON promotes papillary thyroid carcinoma progression through p300/Sp1 axis and serves as a novel diagnostic and prognostic biomarker. [DOI] [PubMed] [Google Scholar]