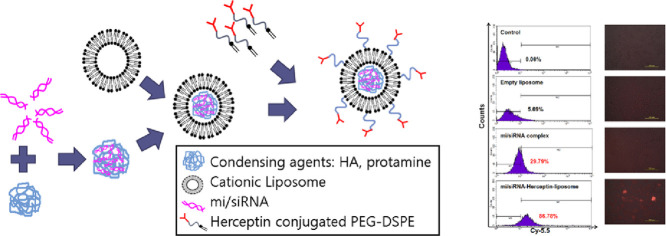
Abstract
Recently, breast cancer stem cells (BCSCs) have rapidly emerged as a novel target for the therapy of breast cancer as they play critical roles in tumor growth, maintenance, metastasis, and recurrence. Let-7 miRNA is known to be downregulated in a variety of cancers, especially BCSCs, whereas CDK4 being overexpressed in human epidermal growth factor receptor 2 (HER-2) overexpressing tumor cells. In this study, let-7 miRNA and CDK4-specific siRNA were chosen as therapeutic agents and co-encapsulated in Herceptin-conjugated cationic liposomes for breast cancer therapy. Particle size, zeta potential, and encapsulation efficacy of mi/siRNA-loaded PEGylated liposome conjugated with Herceptin (Her-PEG-Lipo-mi/siRNA) were 176 nm, 28.1 mV, and 99.7% ± 0.1%, respectively. Enhanced cellular uptake (86%) was observed by fluorescence microscopy when SK-BR-3 cells were treated with Her-PEG-Lipo-mi/siRNA. Also, the increased amount of let-7a mRNA and decreased amount of cellular CDK4 mRNA were observed by qRT-PCR when SK-BR-3 cells were treated with Her-PEG-Lipo-mi/siRNA, which was even more so when SK-BR-3 stem cells were used (197 vs 768 times increase for let-7a, 62% vs 68% decrease for CDK4). Growth inhibition (65%) and migration arrest (0.5%) of the cells were achieved by the treatment of the cells with Her-PEG-Lipo-mi/siRNA, but not with mi/siRNA complex or other formulations. In conclusion, an efficient liposomal delivery system for the combination of miRNA and siRNA to target the BCSCs was developed and could be used as an efficacious therapeutic modality for breast cancer.
Keywords: Let-7 miRNA, CDK4 siRNA, Liposomes, Breast cancer stem cells
Graphical abstract
Herceptin-conjugated cationic liposome can enhance the cellular uptake of let-7 miRNA and CDK4 siRNA toward the HER-2 expressing breast cancer stem cells.
1. Introduction
Breast cancer shows the highest rate of cancer incidence in women in many countries including the USA and the resistance against chemotherapy and radiation therapy makes it very hard to treat the breast cancer even with aggressive surgery [1], [2]. One of the reasons for the occurrence of these resistances is known to be due to the presence of breast cancer stem cells (BCSCs) which exist in small population, usually less than 5%, within the tumor mass [3], [4]. BCSCs exert self-renewal and differentiation properties, like other cancer stem cells, with a characteristic of CD44+CD24−/low cells and are regarded as culprit of tumor generation, metastasis, and recurrence [5].
RNA interference (RNAi) is a naturally occurring phenomenon that silences the expression of specific genes in mammalian cells and the small-interfering RNA (siRNA), the most representative synthetic RNAi material, can silence target genes by degrading a specific mRNA post-transcriptionally [6]. Micro-RNA (miRNA) is a small noncoding RNA molecule that consists of 22 nucleotides in general and plays an central role in the regulation of gene expression either as oncogenes or tumor suppress genes [7].
Let-7 miRNA is known to be downregulated in a variety of cancers including breast cancer and the restoration of let-7 expression to normal levels restricts several mitogenic pathways and the expression of oncogenes, resulting in the reduction of cancer-related proteins such as H-RAS, high mobility group AT-hook 2 (HMGA2), and c-MYC [8], [9], [10]. Let-7 mediates apoptosis and differentiation of cancer stem cells (CSCs) to reduce the stem-like properties of the cells [11], [12], [13]. Let-7a, one of the members of let-7 family, is usually selected as the representative mRNA [14], [15]. Moreover, deficiency of let-7a is required for tumor development and self-renewal of BCSC as the increased let-7a not only suppresses cell proliferation and self-renewal but also converts metastatic BCSCs to unaggressive cells [16]. Cyclin-dependent kinase 4 (CDK4) and Cyclin D1 are known to be overexpressed in breast cancer, specifically in human epidermal growth factor receptor 2 (HER-2) overexpressing tumor cells and to play a critical role in cell growth and tumorigenesis in a form of CDK4/cyclin D1 complex [17].
Application of miRNA and/or siRNA for the treatment of breast cancer is possible only when these genetic materials are effectively delivered to the cells because their large molecule weights (over 10 kDa) and negative charges make them hard to penetrate into the cells [18]. Cationic liposome is a representative non-viral gene delivery system owing to its high transfection efficiency and low toxicity [19], [20]. However, lipoplex, which is a complex of cationic liposome and genetic materials, often exerts unstable properties and low delivery efficiency due to the low charge density and high stiffness of miRNA or siRNA [21]. These problems could be overcome in part by using condensing agents such as hyaluronic acid (HA) and protamine.
In this study, Herceptin-conjugated cationic liposomes consisting of DC-chol and DOPE were used to co-encapsulate the let-7 miRNA and CDK4-specific siRNA with the help of HA and protamine as condensing agents and tested for the anticancer effect both in SK-BR-3 cells and BCSCs.
2. Materials and methods
2.1. Materials
Human breast cancer cells, SK-BR-3, were purchased from the Korean Cell Line Bank (Seoul, Korea). RPMI 1640, Dulbecco's phosphate-buffered saline (DPBS), penicillin-streptomycin, fetal bovine serum (FBS), and trypsin-ethylenediaminetetraacetic acid (EDTA) were purchased from WelGENE Inc. (Daegu, Korea). Forward and reverse primers of CDK4 and c-Myc, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and let-7a miRNA and siRNAs corresponding to the CDK4 (siRNA 179, 185, and 189) were chemically synthesized and modified using Cy5.5 fluorescence probe by Bioneer Inc. (Daejeon, Korea). RNA to cDNA EcoDry™ Premix (Oligo dT) was purchased from TAKARA BIO Inc. (Shiga, Japan). DSPE-PEG(2000)-Maleimide, 3ß-[N-(N',N'-dimethyl-aminoethane)-carbamoyl] cholesterol hydrochloride (DC-chol), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Dimethyl sulfoxide (DMSO), hyaluronic acid (HA) sodium salt, protamine sulfate (fraction X from salmon sperm), chloroform (CHCl3), methanol (MeOH), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and 2-propanol, diethylpyrocarbonate (DEPC) were purchased from Sigma-Aldrich Chemical Co. (St. Louis, USA). Anti-Ras and Anti-HMGA2 antibodies were purchased from Abcam (Cambridge, UK). Lipofectamine™2000 transfection reagent and TRIzol® reagent were purchased from Invitrogen (Carlsbad, USA). All other reagents and solvents were of analytical grade or higher.
2.2. Preparation of RNAs and reagents
2.2.1. Preparation of miRNA and siRNAs
Let-7a miRNA was chemically synthesized as follows:
Sense primer of let-7a: UGA GGU AGU AGG UUG UAU AGU U
Antisense primer of let-7a: AAC UAU ACA ACC UAC CUCA
Three different siRNAs (179, 185, and 189) corresponding to the CDK4 gene were designed and synthesized as follows:
Sense primer of siRNA 179: CUG ACU UUU AAC CCA CAC A
Antisense primer of siRNA 179: UGU GUG GGU UAA AAG UCA G
Sense primer of siRNA 185: CCA GAA UCU ACA GCU ACC A
Antisense primer of siRNA 185: UGG UAG CUG UAG AUU CUG G
Sense primer of siRNA 189: CCG ACC AGU UGG GCA AAA U
Antisense primer of siRNA 189: AUU UUG CCC AAC UGG UCG G
2.2.2. Preparation of mi/siRNA complex and cationic liposome
To prepare the mi/siRNA, HA, and protamine mixture (mi/siRNA complex), 2.5 µl of protamine (700 µg/ml) and 4 µl of mi/siRNA mixture, and HA (700 µg/ml) were mixed in a 1.5 ml tube (1:1.6, w/w). The mixture was incubated at ambient temperature for 10 min before analyzing the size distribution and zeta potential.
Small unilamellar liposomes composed of DOPE and DC-chol (DOPE:DC-chol molar ratio = 1:0.5, 1:1, and 1:1.5) were prepared using a thin film hydration method. Briefly, DOPE and DC-chol mixtures were dissolved in chloroform. After evaporating the solvent under nitrogen gas at room temperature using a rotary evaporator (Laborota 4000; Heidolph, Italia), the formed lipid film was rehydrated in HEPES buffer (pH 7.5) and sonicated (Laboratory Supplies Co. Inc., NY, USA) for 30 s. Then, liposomes were downsized by extrusion through 0.2 and 0.1 µm polycarbonate membranes five times using a Lipex™ extruder (Avestin Inc., Toronto, Canada).
2.2.3. Preparation of cationic liposome co-encapsulating mi/siRNA complex
To prepare cationic liposome co-encapsulating mi/siRNA complex (Lipo-mi/siRNA), the mi/siRNA complex was mixed with cationic liposome (DOPE/DC-chol, total lipid concentration = 20 mM) by electrostatic interaction. The mixture was incubated at ambient temperature for 20 min.
2.2.4. Preparation of Herceptin-conjugated liposome co-encapsulating mi/siRNA complex
For antibody conjugation, Herceptin (1 mg/ml) was thiolated with 2-iminothiolane (1 mg/ml) for 2 h at ambient temperature. The thiolated Herceptin was purified by dialysis kit (Spin DIALYZERS™; Harvard Apparatus, MA, USA) overnight at ambient temperature. After 24 h purification, the thiolated Herceptin was mixed with DSPE-PEG(2000) Maleimide (DSPE-PEG-Mal) at room temperature for 2 h.
The Herceptin-conjugated liposome co-encapsulating mi/siRNA complex (Her-PEG-Lipo-mi/siRNA) was prepared using a post-insertion method [22]. The Herceptin-conjugated DSPE-PEG(2000) (10 mg/ml) was incubated with Lipo-mi/siRNA at 50 °C for 10 min.
2.3. Determination of size distribution, zeta potential analysis, and encapsulation efficiency
Size distribution and zeta potential of the liposomes were measured using a dynamic laser-light scattering (DLS) system (DLS, NICOMP 380XLS; Inc., Santa Barbara, CA, USA) using a He–Ne laser light source.
Quant-iT™ RiboGreen® RNA assay kit (Molecular Probes®, Warrington, UK) was used to measure the encapsulation efficiency of siRNA and miRNA in the liposomes. When the RiboGreen interacts with RNA, it exhibits fluorescence quantitatively. Briefly, 50 µl of the liposomes was diluted with Tris-EDTA buffer and added to 100 µl of 200-fold diluted RiboGreen reagent in 96-well plates. After 5 min incubation in a dark place, the fluorescence intensity was measured using a spectrofluorometer at excitation wavelength (λex) = 480 nm and emission wavelength (λem) = 520 nm. The encapsulation efficiency was calculated using the following equation:
Where, Eun and E0 are the unencapsulated and loading amounts of the miRNA plus siRNA, respectively.
2.4. SK-BR-3 cell culture
Human breast cancer cells, SK-BR-3, were cultured in RPMI 1640 media with 100 unit/ml penicillin/streptomycin, L-glutamine, and 10% FBS at 37 °C in a 5% CO2 incubator (Sanyo Electric Co. Ltd., Osaka, Japan). When the cells reached approximately 80% confluency, they were rinsed with DPBS, detached, and harvested. After centrifugation at 2000 rpm for 5 min, the cells were resuspended with culture media and seeded in culture flasks. The culture media was changed to fresh one periodically.
2.5. SK-BR-3 sphere cell culture
SK-BR-3 sphere cells were cultured in MammoCult® media (Gibco®, USA) with fresh hydrocortisone and heparin at 37 °C in a 5% CO2 incubator. When confluence reached approximately 70%, cells were harvested. After centrifugation at 2000 rpm for 5 min, cells were dissociated with accutase (Sigma-Aldrich, St. Louis, USA) for 10 min at 37 °C. After centrifugation, cells were resuspended with fresh media and seeded in polyHEMA-coated culture dishes.
2.6. Transfection
Transfection of siRNA was performed using the Lipofectamine™2000 reagent (Invitrogen, Carlsbad, USA) in accordance with manufacturer's instructions. Briefly, Lipofectamine™2000 and siRNAs were mixed with Opti-MEM (Gibco®, Warrington, UK). After incubation for 5 min, the solutions were mixed gently and incubated for 20 min at ambient temperature. The cells were washed with serum-free media three times, and placed in the culture dishes with serum-free media, followed by the addition of the mixtures to each dish.
2.7. Cell viability and inhibition of proliferation
Cells were seeded in 96-well plates at 1 × 104 cells/well concentration. After incubation for 24 h at 37 °C, cells were rinsed three times with serum-free media and transfected with siRNAs for 4 h at 37 °C. After then, fresh culture media was treated to the cells and further incubated for 20 h at 37 °C. After transfection, each well was treated with 100 µl of MTT solution (5 mg/ml). After incubation for 4 h at 37 °C, the media was gently removed and 100 µl of DMSO was added to each well. To confirm the dissolution of the formazan crystal, the 96-well plates were incubated for 5 min at 37 °C and agitated using a shaking plate (FINE CR 100; Finemould Precision Ind. Co., Seoul, Korea) for 10 min. The absorbance was determined at 570 nm using an ELISA reader (EL 800; BIO-TEK, Winooski, USA). Cell viability was calculated using the following equation:
Where, OD570(sample) and OD570(control) are the absorbance values of the sample and control, respectively, at 570 nm.
2.8. qRT-PCR
qRT-PCR was performed to evaluate the suppression of CDK4 and c-Myc mRNA by treatment of SK-BR-3 cells with the corresponding siRNAs. Cells were seeded in 6-well plates (1 × 105 cells/well) and incubated for 24 h and treated with three different siRNAs or liposomes and incubated for 6 h. The culture media was replaced with fresh media and incubated for 18 h at 37 °C. After transfection, the RNA was extracted using TRIzol® reagent in accordance with the manufacturer's instructions. The concentration and purity of RNA were detected using a NanoDrop spectrometer (Epoch; BioTeck, Arcugnano, Italy). cDNA was synthesized from the isolated RNA using a PCR thermal cycler Dice (Takara, Shiga, Japan) using RNA to cDNA Ecodry™ (TAKARA Bio Inc., Shiga, Japan). qRT-PCR was performed with 50 ng RNA using Power SYBR® Green (Applied Biosystems, Warrington, UK). To synthesize cDNA from miRNA let-7a, a TaqMan® miRNA assay kit (Applied Biosystems, Warrington, UK) was used. qRT-PCR was performed with 50 ng RNA using Taqman® Universal Master Mix II (Applied Biosystems, Warrington, UK). The designed primer sequences were as follows:
c-Myc (forward): GGA ACG AGC TAA AAC GGA GCT
(reverse): GGC CTT TTC ATT GTT TTC CAA CT
GAPDH (forward): AAC GTG TCA GTG GAC CTG
(reverse): AGT GGG TGT CGC TGT TGA AGT
2.9. Transfection efficiency
The transfection efficiencies of the siRNA and miRNA delivery systems were determined in SK-BR-3 and SK-BR-3 sphere cells. SK-BR-3 cells were seeded in 6-well plates (1 × 105 cells/well). SK-BR-3 sphere cells were seeded in 60 mm dishes (5 × 104 cells/well). After incubation for 24 h, the culture media was aspirated and transfected with Cy5.5-labeled miRNAs in various delivery systems with fresh media. After 6 h of incubation, the cells were rinsed and observed using a fluorescence microscope (IX71IX51; Olympus, Tokyo, Japan). In addition to quantifying the transfection efficiency, Cy5.5 labeled miRNA-transfected cells were examined using cytometric analysis with a FACSCalibur™ system (BD Bioscience, USA).
2.10. Migration assay
SK-BR-3 cells were maintained in a 60 mm dish until they were confluent. Then, scratch wounds were created on the confluent cells using a P200 pipette tip. The wounded cells were carefully washed with DPBS and incubated with DPBS (control), empty liposome (empty-Lipo), naked mi/siRNA, mi/siRNA complex, and Her-PEG-Lipo-mi/siRNA in the media. After incubation for 6 h, the cells were washed two times and the media was replaced with fresh media. Cells were subsequently observed using a microscope after 18 h and the percentage migration was calculated using the following equation:
Where, the initial width is the cell-free area of wounded cells at the time of sample treatment and the final width is the cell-free area of migrated cells after 24 h.
2.11. Western blot
The western blot analysis was performed to determine the inhibition of cancer proteins (RAS and HMGA2) by let-7a. Briefly, 1 × 106 SK-BR-3 cells and 1 × 105 SK-BR-3 sphere cells were seeded in 60 mm dishes. After 2 days of incubation, the cells were transfected with DPBS (control), empty-Lipo, naked mi/siRNA, mi/siRNA complex, Her-PEG-Lipo-mi/siRNA, and PEGylated liposomes encapsulating scrambled siRNA (PEG-Lipo-scrambled siRNA) for 6 h at 37 °C. Then, the media was replaced and the cells were incubated for 42 h at 37 °C and then lysed with radioimmunoprecipitation assay (RIPA) buffer (iNtRON Biotechnology, Seongnam, Korea) and added to 1.5 ml tubes. The lysed cells were vortexed for 1 min and then incubated for 30 min on ice. The western blot analysis was performed using the Power Pac 300 system (Bio-Rad, Hercules, USA).
2.12. Statistical analysis
The results are expressed as the means ± standard deviation (SD). Statistical analysis was carried out using the Student's t-test. Statistical significance was accepted at a P < 0.05 or P < 0.01 (95% and 99% confidence levels, respectively).
3. Results and discussions
3.1. Optimization of lipoplex formulation
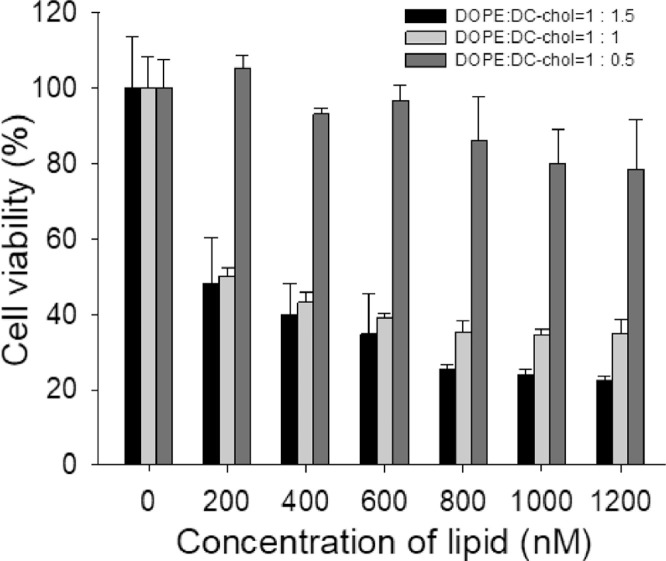
RNAi technology has gained much attention as a new therapeutic modality since its finding in 1990s but physiological barriers including instability and low cellular uptake have remained unsolved [23], [24]. Various approaches to overcome these huddles were introduced by using appropriate gene delivery systems including micelles, lipid-based nanoparticles, cyclodextrins, etc. [25], [26]. Cationic immunoliposome could serve as a good delivery system owing to numerous advantages such as target-specific delivery, biocompatibility and safety [27], [28], [29]. To formulate a lipoplex with the mi/siRNA complex, cationic liposomes were prepared with DOPE:DC-chol at 1:0.5, 1:1 and 1:1.5 molar ratios. Various ratios of neutral lipid DOPE and cationic lipid DC-chol were tested for cytotoxicity and 1: 0.5 molar ratio (DOPE: DC-chol) was used to form the lipoplex as they showed the lowest cytotoxicity among tested (Fig. 1). The mean diameter and zeta potential of the 1:0.5 cationic liposomes were 126 ± 58 nm and 28.0 ± 0.2 mV, respectively (Fig. S2).
Fig. 1.
Cell viability assay of SK-BR-3 cells with treatment of cationic liposomes prepared with DOPE:DC-chol at 1:0.5, 1:1, and 1:1.5 molar ratios.
3.2. Cell viability and qRT-PCR assay of three different siRNAs
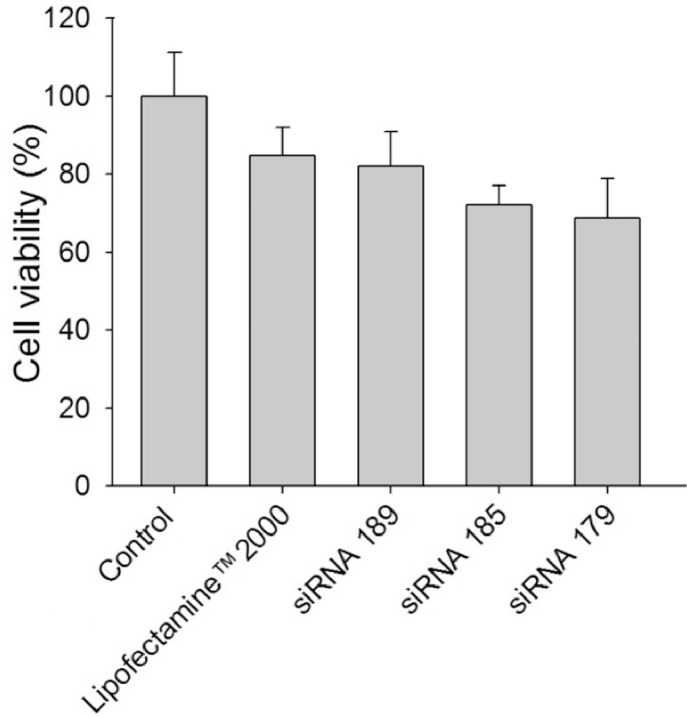
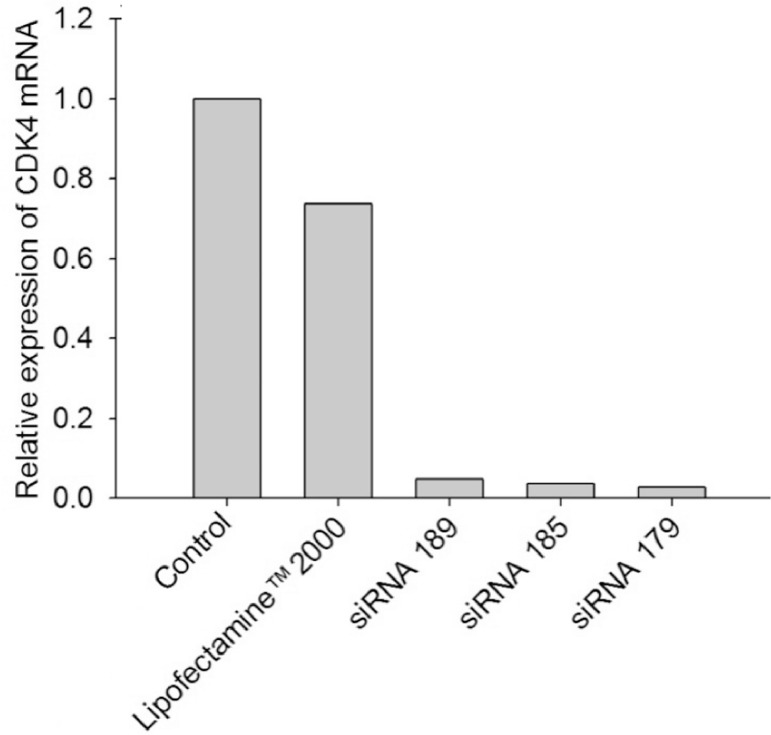
To select the best sequence for the designed siRNAs, a cell viability assay was carried out. To test the downregulation effect of siRNAs, the well-known transfection reagent Lipofectamine™2000 was used. Among the three different sequences of the CDK4-specific siRNA (siRNA 179, 185 and 189), siRNA 179 was found to be the most effective in the inhibition of proliferation of SK-BR-3 cells as revealed by MTT assay (Fig. 2). qRT-PCR analysis was also carried out to test the suppression of CDK4 mRNA in SK-BR-3 cells by siRNA 189, siRNA 185, and siRNA 179. The suppression of CDK4 mRNA by siRNA 179 was most significant (>90% down-regulation) among tested (Fig. 3).
Fig. 2.
Cell viability assay with three different siRNAs with Lipofectamine™2000 (**P < 0.01).
Fig. 3.
CDK4 mRNA expression following treatment with siRNA and Lipofectamine™2000 by qRT-PCR.
3.3. Optimization of mi/siRNA complex
Condensation of the siRNA or miRNA before encapsulation in the liposome was performed by using HA and protamine as condensing agents, which resulted in much smaller size of mi/siRNA complex with increased stability (Fig. S1A). By considering the smallest size (212 nm) and appropriate negative charge (−32.8 mV), the optimal ratio of (mi/siRNA+HA)/protamine was fixed as 1.6 (w/w).
3.4. Optimization of liposome co-encapsulating mi/siRNA complex
Cationic liposomes were formulated with the mi/siRNA complex and analyzed for their particle size and zeta potential (Fig. S1B). The molar ratios of mi/siRNA complex and cationic liposomes were established from 1:500 to 1:6000. The amount of the cationic liposome was correlated with the zeta potential. When the cationic liposomes were mixed with the mi/siRNA complex, the particle size was decreased and the zeta potential was changed to a neutral charge. This observation suggests that the cationic liposomes and mi/siRNA complex formed condensed particles through electrostatic interactions. We chose a molar ratio of 5000:1 as this ratio exhibited a stable particle size of 150 nm.
3.5. Optimization of Herceptin-conjugated liposome co-encapsulating mi/siRNA complex
Conjugation of Herceptin to the liposomes was performed via the post-insertion method, followed by the co-encapsulation of let-7a miRNA and CDK4-specific siRNA in the liposome. The particle size and zeta potential of Her-PEG-Lipo-mi/siRNA particles were 176 ± 54 nm and 28.1 ± 1.8 mV, respectively. The encapsulation efficiency of miRNA and siRNA was measured using the RiboGreen dye-binding assay. The amount of unencapsulated miRNA and siRNA was determined from the calibration curve of standard RNA solution. The encapsulation efficiency of the produced liposomes was 99.7% ± 0.1% (Table 1).
Table 1.
Characterization of various liposomes.
| Formulation | Mean diameter (nm) | Zeta potential (mV) | Encapsulation efficiency (%) |
|---|---|---|---|
| Empty-Lipo | 126 ± 58 | 28.0 ± 0.2 | – |
| PEG-Lipo-mi/siRNA | 135 ± 16 | 26.4 ± 1.23 | 99.6 ± 0.1 |
| Her-PEG-Lipo-mi/siRNA | 176 ± 54 | 28.1 ± 1.8 | 99.7 ± 0.1 |
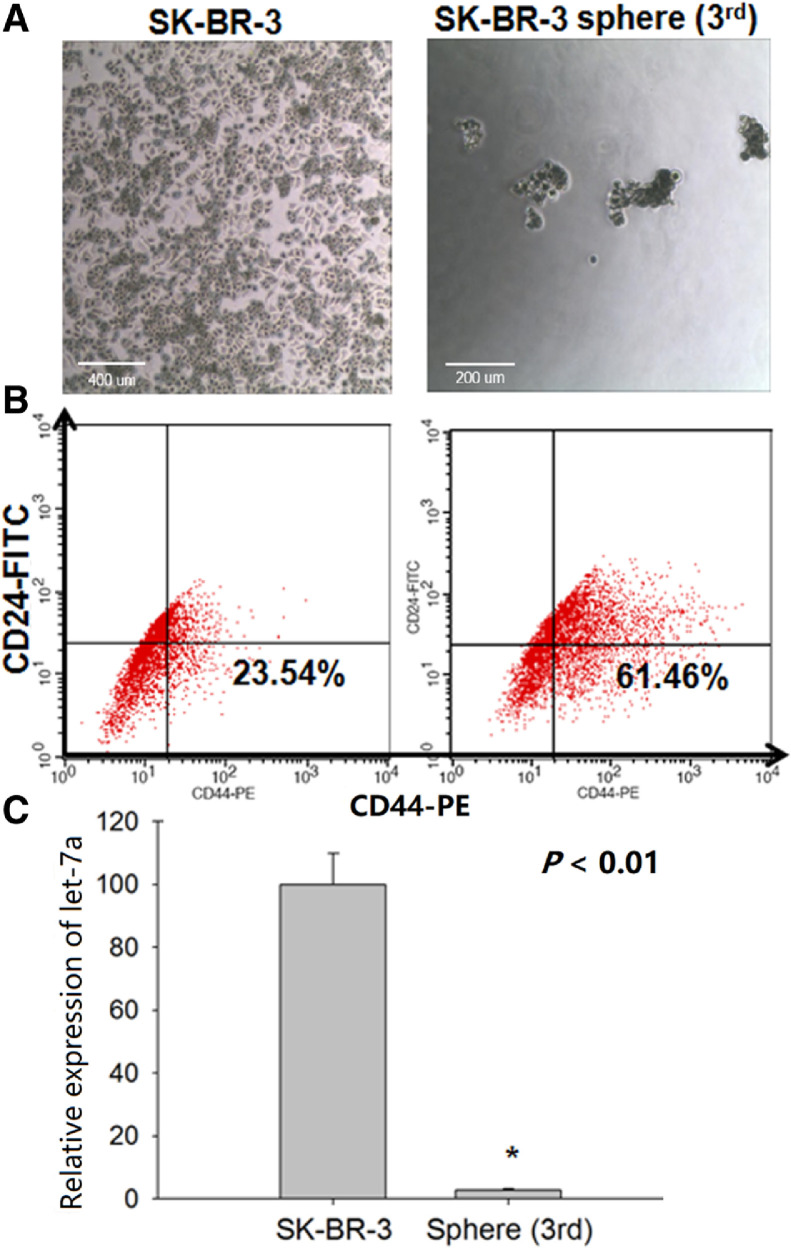
3.6. Characterization of SK-BR-3 and SK-BR-3 sphere cells
SK-BR-3 sphere cells or BCSCs were obtained from the SK-BR-3 breast cancer cells by sphere formation and not only the high CD44/low CD24 expression but low let-7a expression were identified as stem cell-like properties of BCSCs. Fig. 4A shows the shapes of the SK-BR-3 and SK-BR-3 sphere cells. SK-BR-3 cells were small adherent cells, whereas the SK-BR-3 sphere cells were grape-shaped suspended cells. As shown in Fig. 4B, CD44 was expressed in SK-BR-3 sphere cells by approximately 40% more than in SK-BR-3 cells. The CD24 expression showed no difference between SK-BR-3 and SK-BR-3 sphere cells. Fig. 4C shows that a relative let-7a expression in SK-BR-3 sphere cells was only 2% of that in SK-BR-3 cells.
Fig. 4.
Characterization of SK-BR-3 and SK-BR-3 sphere cells. (A) Shape of SK-BR-3 and SK-BR-3 sphere cells. (B) Expression of CD44/CD24 analyzed by flow cytometry. (C) Relative expression of let-7a in SK-BR-3 cells and SK-BR-3 sphere cells analyzed by qRT-PCR.
3.7. Transfection efficiency
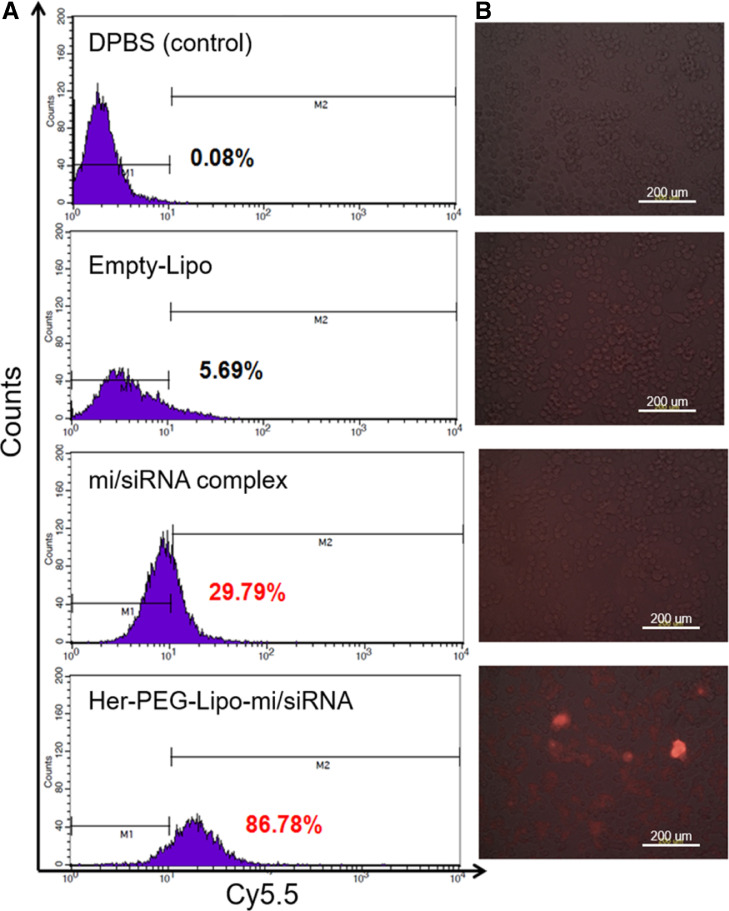
Enhanced cellular uptake of mi/siRNA was obtained by using Herceptin-conjugated liposomal formulation compared with non-formulated one as revealed by fluorescence microscopy. As shown in Fig. 5A, the quantitative flow cytometric analysis revealed that cell uptake efficiency of Her-PEG-Lipo-mi/siRNA was 86.78%. By contrast, naked mi/siRNA and mi/siRNA complex showed < 20% uptake. Then, the samples were observed by fluorescence microscopy. As shown in Fig. 5B, the mi/siRNA complex was hardly taken up by the cells, whereas a higher amount of Cy5.5-labeled miRNA was internalized into the SK-BR-3 cells when Her-PEG-Lipo-mi/siRNA was used as a delivery system.
Fig. 5.
Transfection efficiency of Her-PEG-Lipo-mi/siRNA. Cellular uptake of Cy5.5-labeled let-7 miRNA in SK-BR-3 cells was (A) quantified by flow cytometry and (B) observed under a fluorescence microscope (×10 magnification). The assay was performed three times.
3.8. Let-7a expression following treatment with liposomal formulation
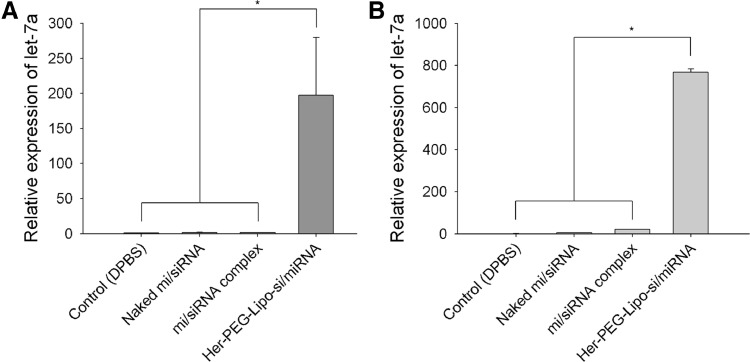
Expression of let-7a miRNA by treatment with Her-PEG-Lipo-mi/siRNA was elevated than the DPBS control in SK-BR-3 cells and this increase was intensified when the BCSCs were used. As shown in Fig. 6A, the amount of let-7a in the Her-PEG-Lipo-mi/siRNA-treated group was increased 197 times more than that in the control group. In SK-BR-3 sphere cells, the amount of let-7a in the Her-PEG-Lipo-mi/siRNA-treated group was increased 768 times more than that of the control group (Fig. 6B). The naked mi/siRNA- or mi/siRNA complex-treated groups did not show a significant increase.
Fig. 6.
Relative expression of let-7a in (A) SK-BR-3 and (B) SK-BR-3 sphere cells after treatment with various formulations of mi/siRNA (*P < 0.05).
3.9. CDK4 and c-Myc mRNA expression following treatment with liposomal formulation
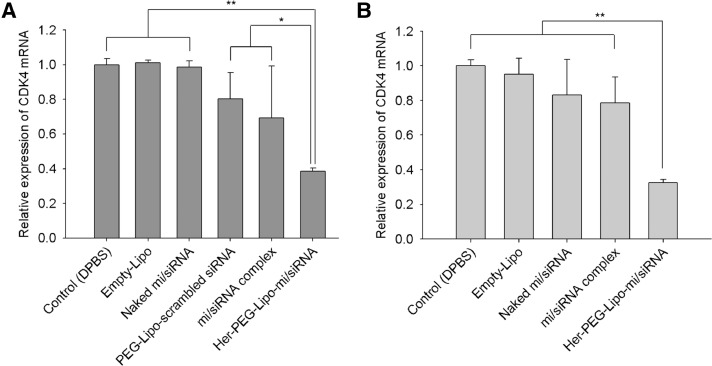
Expression of CDK4 mRNA was inhibited both in SK-BR-3 cells and BCSCs by the treatment of CDK4-specific siRNA in Herceptin-conjugated liposomal formulation. As shown in Fig. 7A, the amount of CDK4 mRNA in the SK-BR-3 cells was decreased by 62% by treatment with Her-PEG-Lipo-mi/siRNA compared with the control group. The group treated with the mi/siRNA complex showed only a slight decline in CDK4 mRNA (∼30%). In the SK-BR-3 sphere cells (Fig. 7B), the amount of CDK4 mRNA was decreased by 68% in the Her-PEG-Lipo-mi/siRNA-treated group compared to the control (DPBS) group. No significant amount of decrease in CDK4 mRNA was observed by other treatment.
Fig. 7.
Relative expression of CDK4 mRNA in (A) SK-BR-3 and (B) SK-BR-3 sphere cells after treatment with various formulations of mi/siRNA (**P < 0.01).
Moreover, expression of c-Myc mRNA was also suppressed by the treatment of CDK4-specific siRNA in Herceptin-conjugated liposomal formulation as the CD4 is known as a target of c-Myc [30]. The amount of c-Myc mRNA was decreased by 50% in the Her-PEG-Lipo-mi/siRNA-treated group compared to the control group (Fig. S3). The empty-Lipo group showed a slight decline of c-Myc mRNA, which is presumably due to the cationic charge of the liposome.
3.10. Inhibition of proliferation and migration of SK-BR-3 cells
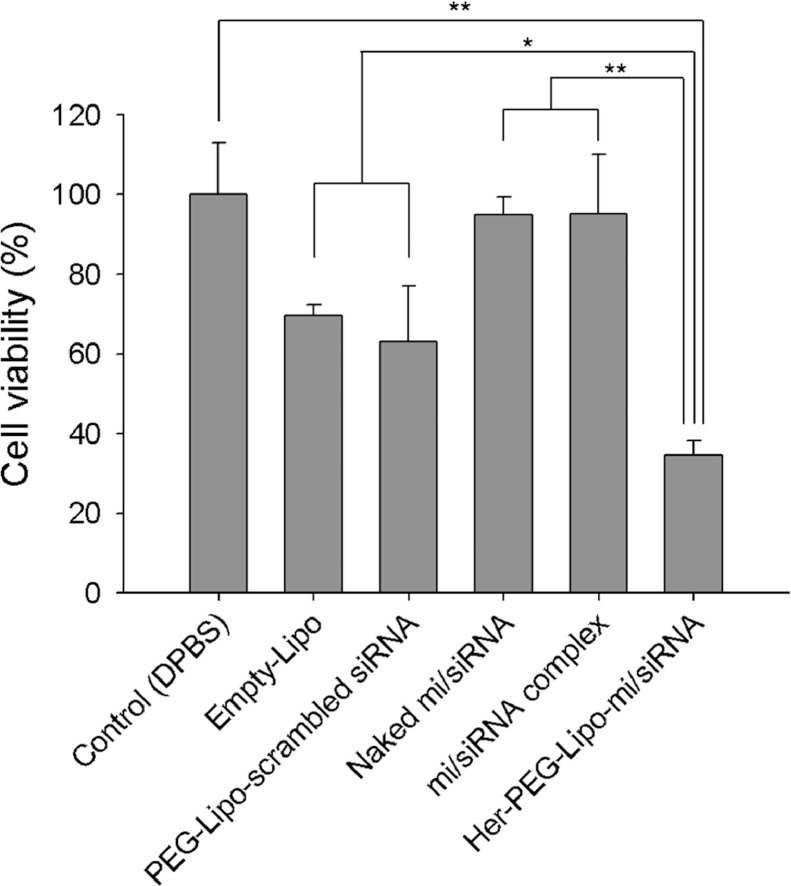
Suppression of CDK4 and c-Myc as well as elevation of let-7a miRNA leads to the cell death and migration arrest of both SK-BR-3 cells and BCSCs. As shown in Fig. 8, the proliferation of naked mi/siRNA and mi/siRNA complex was not dramatically decreased compared with that of the control. The empty-Lipo showed a cationic charge and, therefore, the cell viability of the empty-Lipo was slightly low. In the Her-PEG-Lipo-mi/siRNA-treated group, over 50% inhibition of proliferation was observed.
Fig. 8.
Inhibition of proliferation in SK-BR-3 cells by treatment of liposomal mi/siRNA formulations (**P < 0.01).
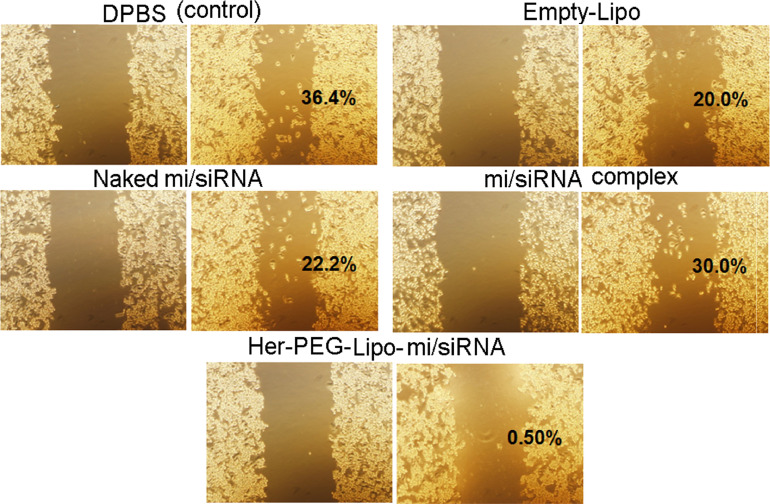
To evaluate the migration of SK-BR-3 cells, a wound-healing assay was carried out. As shown in Fig. 9, the control group showed 36.4% cell migration, whereas the mi/siRNA complex, naked mi/siRNA, and empty-Lipo groups showed 30.0%, 22.2%, and 20.0% migration, respectively. In contrast, the group treated with the Her-PEG-Lipo-mi/siRNA showed only 0.5%, which could be due to the death of the cells.
Fig. 9.
Microscopic observation of inhibition of migration of SK-BR-3 cells (×10 magnification).
3.11. Determination of RAS and HMGA2 by western blot
Increase of let-7a levels reduced the RAS and HMGA2 which are known to be related with self-renewal and differentiation of CSCs, respectively, and overexpressed in breast cancer cells and BCSCs. In western blotting assay, there was no significant decrease in RAS and HMGA2 in the naked mi/siRNA- and mi/siRNA complex-treated group (Fig. S4). The treatment of Her-PEG-Lipo-mi/siRNA reduced the of RAS protein in SK-BR-3 cells. HMGA2 protein was reduced in SK-BR-3 and SK-BR-3 sphere cells by treatment with Her-PEG-Lipo-mi/siRNA. So, treatment with Her-PEG-Lipo-mi/siRNA could regulate the BCSC's stem-like properties through the suppression of additional targets [16].
4. Conclusion
In conclusion, let-7a miRNA and CDK4-specific siRNA were found to be used as therapeutic agents for the regulation of BCSCs as well as breast cancer cells when they are formulated in Herceptin-conjugated cationic immunoliposome with HA and protamine. Further studies are highly expected for the development of new therapeutic modality for breast cancer in the future.
Acknowledgments
Conflicts of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2017R1D1A1B03030849) the National Research Foundation of Korea grant funded by the Korea Government (MEST, No. 2011-0030074).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ajps.2019.03.001.
Appendix. Supplementary materials
References
- 1.DeSantis C., Ma J., Bryan L., Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Ganz P.A., Goodwin P.J. Breast cancer survivorship: where are we today? In: Ganz PA, editor. Improving outcomes for breast cancer survivors: perspectives on research challenges and opportunities. Springer; Cham: 2015. pp. 1–8. [Google Scholar]
- 3.Dalerba P., Cho R.W., Clarke M.F. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58(1):267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 4.Wicha M.S. Cancer stem cells and metastasis: lethal seeds. Clin Cancer Res. 2006;12(19):5606–5607. doi: 10.1158/1078-0432.CCR-06-1537. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Mayr C., Hemann M.T., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W., Liu X., Liu S., Qin Y., Tian X., Niu F. Androgen receptor/let-7a signaling regulates breast tumor-initiating cells. Oncotarget. 2018;9(3):3690–3703. doi: 10.18632/oncotarget.23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang W.P., Kwok T.T. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13(10):1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- 13.Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2(3):219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sureban S.M., May R., Ramalingam S., Subramaniam D., Natarajan G., Anant S. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a MicroRNA-dependent mechanism. Gastroenterology. 2009;137(2):649–659. doi: 10.1053/j.gastro.2009.05.004. 59 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller D.W., Bosserhoff A.K. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27(52):6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 16.Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 17.Dong Y., Sui L., Sugimoto K., Tai Y., Tokuda M. Cyclin D1-CDK4 complex, a possible critical factor for cell proliferation and prognosis in laryngeal squamous cell carcinomas. Int J Cancer. 2001;95(4):209–215. doi: 10.1002/1097-0215(20010720)95:4<209::aid-ijc1036>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 18.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedroso de Lima M.C., Neves S., Filipe A., Duzgunes N., Simoes S. Cationic liposomes for gene delivery: from biophysics to biological applications. Curr Med Chem. 2003;10(14):1221–1231. doi: 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- 21.Jeong J.H., Mok H., Oh Y.K., Park T.G. siRNA conjugate delivery systems. Bioconjug Chem. 2009;20(1):5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 22.Uster P.S., Allen T.M., Daniel B.E., Mendez C.J., Newman M.S., Zhu G.Z. Insertion of poly(ethylene glycol) derivatized phospholipid into pre-formed liposomes results in prolonged in vivo circulation time. FEBS Lett. 1996;386(2–3):243–246. doi: 10.1016/0014-5793(96)00452-8. [DOI] [PubMed] [Google Scholar]
- 23.Saurabh S., Vidyarthi A.S., Prasad D. RNA interference: concept to reality in crop improvement. Planta. 2014;239(3):543–564. doi: 10.1007/s00425-013-2019-5. [DOI] [PubMed] [Google Scholar]
- 24.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9(1):60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundy B.B., Convertine A., Miteva M., Stayton P.S. Neutral Polymeric Micelles for RNA Delivery. Bioconjug Chem. 2013;24(3):398–407. doi: 10.1021/bc300486k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7(4):77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo J., Kim M.J., Jeon S.O., Oh D.H., Yoon K.H., Choi Y.W. Enhanced topical delivery of fish scale collagen employing negatively surface-modified nanoliposome. J Pharm Investig. 2018;48(3):243–250. [Google Scholar]
- 28.Sim T., Lim C., Hoang N.H., Oh K.T. Recent advance of pH-sensitive nanocarriers targeting solid tumors. J Pharm Investig. 2017;47(5):383–394. [Google Scholar]
- 29.Pokharkar V., Patil-Gadhe A., Kaur G. Physicochemical and pharmacokinetic evaluation of rosuvastatin loaded nanostructured lipid carriers: influence of long- and medium-chain fatty acid mixture. J Pharm Investig. 2018;48(4):465–476. [Google Scholar]
- 30.Hermeking H., Rago C., Schuhmacher M., Li Q., Barrett J.F., Obaya A.J. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97(5):2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.