Abstract
Introduction: Mesenchymal stem cells (MSCs) are able to differentiate into several cell lineages including skeletal muscle. In addition to their differentiation capacities, they have the ability to transfer their content genomic information horizontally through their exosomes and fusion abilities, as we have shown in our previous clinic study on Duchenne Muscular Dystrophy (DMD) patients, dystrophin expression increased after MSC treatment. Therefore, this study aimed to compare the transcriptomic properties of Wharton’s jelly derived (WJ-) MSC and Adipose tissue (AT-) derived MSC, which are the two most preferred sources in MSC treatments applied in DMD. Methods: Both MSC cell lines obtained from ATCC (PCS-500-010; PCS-500-011) were characterized by flow cytometry then WJ-MSC and AT-MSC cell lines were sequenced via RNA-SEQ. R language was used to obtain the differentially expressed genes (DEGs) and differentially expressed miRNAs, respectively. Additionally, in order to support the results of our study, a gene expression profile data set of DMD patients (GSE1004) were acquired from Gene Expression Omnibus (GEO) database. Results: Here, we demonstrated that activated WNT signaling and downregulated TGF-β pathways under the control of decreased mir-24 which are involved in myogenic differentiation are differentially expressed in WJ-MSC. We have shown that the expression of mir-199a-5p, which is known to increase in exosomes of DMD patients, is less in WJ-MSC. Additionally, we have shown activated PI3K/Akt pathway, which is controlling mitochondria transfer via Tunnelling Nanotube as a new perspective in cellular therapies in myodegenerative diseases, in WJ-MSC more than in AT-MSCs. Conclusion: Summing up, WJ-MSC, which we recommend as an appropriate source candidate due to its immune-regulation properties, stands forward as a preferable source in the cellular treatment of DMD patients due to its transcriptomic aspect.
Keywords: Duchenne muscular dystrophy, gene expression, mesenchymal stem cells, cellular therapy
Introduction
Duchenne Muscular Dystrophy (DMD) is an X recessive disease that affects 1/3500 males [1]. Boys with normal growth refer to the clinic at around 3 or 5 years old with symptoms such as difficulty in climbing stairs and early fatigue. The basic pathogenesis is explained by the formation of fibrous tissue by means of covering the connective tissue because the muscle tissue does not function due to the absence of dystrophin [2]. Dystrophin gene is localized at Xp21 and it is the largest gene defined in the human genome. The dystrophin protein is the sarcolemma protein at the skeletal muscle. It is in tight contact with cell membrane proteins and muscle fibers. It is a tight bridge between the cytoplasm and cell membrane; in the absence of dystrophin muscle cannot contract and fibrous [3,4].
In a large quantity of variations, large deletions, duplications or point mutations, in the gene affect functional dystrophin protein production and cause DMD [5-8]. There is no conventional therapy that targeting all these variations. Treatment options were determined based on the type of variation. Exon-skipping is one of the most promising therapeutic approaches that aim to restore the expression of a shorter but functional dystrophin protein [9,10] rather than a truncated and nonfunctional protein. Allele specific oligonucleotides (ASO) are paired with the exonic splice site to removing mutated exon from pre-mRNA and produce a shorter but functional dystrophin protein in translation [11]. However, ASO therapy offers a treatment option only to the 15% of patients. Converting the nonsense mutation to missense mutation is another treatment option to produce functional protein [12,13]. The therapy is based on the altering 3rd amino acid to get rid of the early stop codon to continue transcription. This treatment option is known as Ataluren treatment and provides a 70% reduction in plasma CK level.
There is a requirement for alternative treatment solutions, owing to mutation-specific treatment options are targeting just for a small group of patients. The goal for this would be to send the wild type mRNA for functional protein, rather than correcting the mutation and forming a truncated protein. Hence, gene transferring could be a treatment option that can reach all patients, nevertheless there is no suitable vector transferring the dystrophin gene. Herein, cellular therapies are a good option for the persistence of treatment in the long term for of all patients via horizontal gene transfer by cellular fusions [14,15] or transferring exosome [16-18]. Mesenchymal Stem Cells (MSCs) promises a treatment hope for untreatable diseases with the conventional treatment protocols, such as autoimmune diseases, lung diseases, cardiovascular diseases and neuromuscular diseases. Their importance in clinic stand out providing the regeneration of damaged cells and suppression of inflammation via secreting many bioactive molecules [19].
MSCs can be derived from many sources such as Wharton’s Jelly (WJ), placenta tissue, amniotic fluid, cord blood, adipose tissue (AT), dermis and tooth pulp [20,21]. For the allogenic transplantations, WJ-MSC and AT-MSCs are most preferable sources for DMD. Although they are the same type of cells, gene expression profiles are differentiated according to the tissue origin from which they are obtained. Different transcriptomic features will cause differentiation in mRNA, miRNA or protein cargoes that will be sent to the host cell by horizontal transfer. As a result, the effectiveness of treatment actually depends on the characteristics of these transfers of cells. In this study, we have compared transcriptomes of WJ- and AT- sources of MSCs to determine the most effective source that can be used in DMD treatment in order to show which of them is more effective as a transcriptomic content in cellular treatments (Figure 1).
Figure 1.
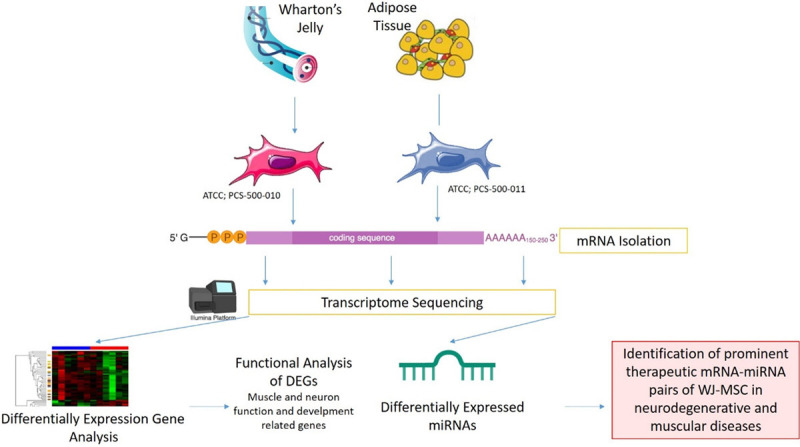
Graphical introducing of the study.
Materials and methods
Samples and study design
Two distinct sources derived MSC cell lines, which are WJ-MSC (PCS-500-010) and AT-MSC (PCS-500-011) were obtained from ATCC (Manassas, VA). Two of these cell lines were grown in culture according to suppliers’ instructions.
In order to increase the significance of the analysis on cell lines, the filtering of DEGs was generated via comparing the open source gene expression data which are the muscle biopsies derived from 12 DMD patients and 12 healthy controls (GSE1004) [22]. These examples were also evaluated with the GEOR2 tool [23].
Cell culture
WJ and AT derived MSC lines were cultured in T175 flasks with MSC NutriStem® XF Basal Medium and MSC NutriStem® XF Supplement Mix (Biological Industries, Cromwell, CT, USA) cell culture media supplied with 10% fetal bovine serum (FBS), 2 mM L-glutamine and streptomycin (100 mg/mL) solution at 37°C in a humidified 5% CO2 incubator and subcultured until the 3rd passage. After reaching 70% to 80% confluency, adherent cells were harvested with trypsinization by 0.05% trypsin-EDTA (Gibco, Germany).
Characterization of WJ-MSC and AT-MSC with cell surface marker screening
For characterization of MSCs with their cell surface markers, the cells were harvested at their third passages (P3), and were labeled with CD90, CD105, CD44 and CD73 (BD). The characterization data were acquired with Flow Cytometry analysis at FACS calibur (BD).
RNA extraction, library preparation and sequencing
Total RNA was extracted from WJ- and AT- derived MSC lines with Purelink RNA kit (Invitrogen) according to the manufacturer’s instructions. Concentration and the Integrity Number of RNA (RIN) samples were determined with the Agilent 2100 Bioanalyzer and suitable RNA samples were selected with the RIN ≥ 7. Libraries were prepared with Illumina TruSeq RNA library kit (Illumina) and were sequenced by HiSeq2000 with 100 bp paired-end reads per sample.
Annotation and differentially expressed genes
The quality control of the raw sequence reads were analysed with FastQC v. 0.11.3 [24]. Low-quality sequences and overrepresented sequences like adapters were trimmed by fastq-mcf module (v 1.04) [25]. Raw reads were mapped to the GRCh38 reference genome by RSEM 25 (v. 1.2.22) with TopHat aligner. For each transcript, the fragments per kilobase per million (FPKM) were detected and the reads were normalized. For the differentially expressed genes (DEGs) analysis, the threshold was determined as log2 FPKM and P≤0.05. DEGs were analysed in R environment EBSeq package.
Functional analysis
The result of DEG lists were submitted to Database for Annotation, Visualization, and Integrated Discovery (DAVID v.6.7) and IPA (Qiagen) software to identify their functions and the pathways that are attended.
Differentially expressed miRNAs and their target prediction
Trimmed raw sequence reads were realigned by using SHiMPS aligner [26]. The counts read associated to mature miRNAs and detection of differentially expressed miRNAs were analyzed by using the R package of DESeq2 [27].
Results
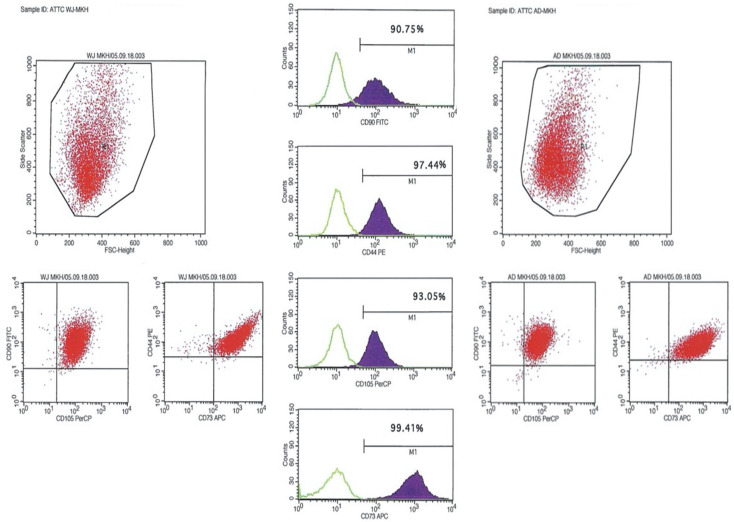
Flow cytometry analysis of cell-surface markers for WJ- and AT-MSC at P3. They all expressed MSC markers including CD90, CD105, CD44 and CD73 (Figure 2).
Figure 2.
Characterization of both WJ-MSC and AT-MSC by flow cytometry. Positive expressions of CD90, CD105, CD44 and CD73 markers are presenting.
RNA-Seq analysis
An average of 43 million 100-base long reads from each source of MSCs were mapped to the human reference genome and FPKM values were generated, which provided a measure of expression levels for each gene mapped to the transcriptome. 92 genes between WJ-MSC and AT-MSC were identified as DEGs which passed the |log2 (fold-change)|>1.5 and p Value < 0.05 filtering.
WJ-MSC has a higher regenerative capacity
We analyzed the functions of 92 DEGs. Consequently, the important features for stem cell therapies that regulation of muscle cells, muscle development, extra cellular matrix (ECM) organization, response to growth factor and the MSC differentiation which are important for neurodegenerative diseases are significantly increased for WJ-MSCs. 35 of 92 DEGs were associated with these features and 23 of these 35 genes were also detected in DMD patients expressed contrarily. Filtering was applied with the comparison of the 35 DEGs and the DEGs of DMD patient muscle biopsy vs healthy muscle biopsy retrieved from GEO1004. Twenty-three of DEGs overlapped (Figure 3). The expression of 12 genes in WNT signalling and 5 genes that are play an important role in myogenic differentiation involved in TGF-β signalling were detected differentially expressed. Logarithmic fold changes and p-values of the differentially expressed genes in these pathways are presented in Table 1 in detail.
Figure 3.
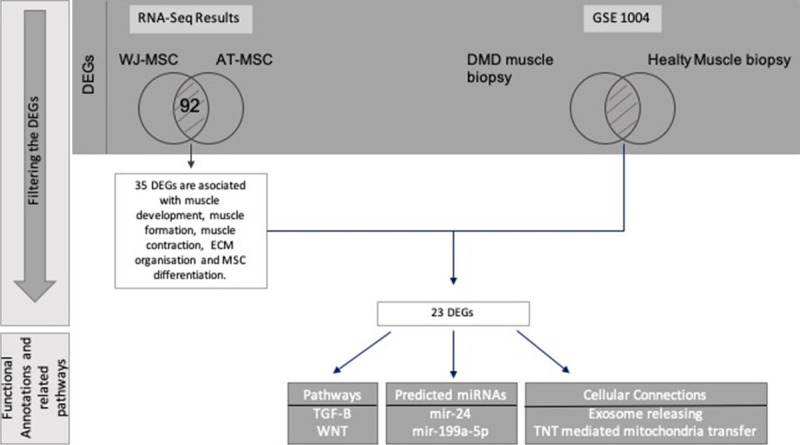
Filtering workflow of whole transcriptomic data. The DEGs that both passed the filtering and placed at the intersection of MSC and DMD patients were annotated to their functional annotations and their attended pathways.
Table 1.
Enriched pathway analysis and their functional categories
| Functional Categories | Pathway | Regulator Gene Name in Pathway | p-Value | Log Fold change |
|---|---|---|---|---|
| Signaling Transduction | TGF-beta signaling pathway | BAMBI | 8.90E-19 | 5.2684 |
| SMAD7 | 5.91E-06 | 1.6536 | ||
| INHBA | 1.52E-21 | 2.327 | ||
| CDKN2B | 2.19E-14 | 1.74 | ||
| PITX2 | 1.38E-41 | 3.5955 | ||
| WNT signaling pathway | CCND1 | 3.80E-08 | 1.2589 | |
| GPC4 | 1.36E-42 | 4.3748 | ||
| BAMBI | 8.90E-19 | 5.2684 | ||
| NFATC4 | 1.31E-10 | -1.3164 | ||
| SFRP4 | 1.49E-07 | -8.1966 | ||
| DKK1 | 3.20E-09 | 2.8133 | ||
| SFRP2 | 9.50E-27 | -7.8593 | ||
| TCF7L1 | 2.43E-05 | 2.3311 | ||
| DKK2 | 1.88E-08 | 2.1996 | ||
| FZD1 | 5.61E-17 | -2.3299 | ||
| LRP5 | 8.96E-11 | 1.7443 | ||
| TCF7L2 | 0.0030771 | 1.5690 | ||
| Celllular Communication | mTOR Signalling Pathway | VEGFA | 2.66E-10 | 1.0362 |
| PI3K/Akt Signalling Pathway | ITGA3 | 4.61E-14 | 2.9903 | |
| TNC | 1.03E-19 | -1.4597 | ||
| NGFR | 0.00023885 | -7.0205 | ||
| COL5A3 | 6.86E-14 | -1.3267 | ||
| LAMB1 | 1.63E-36 | 1.0533 | ||
| FLT1 | 1.53E-42 | 8.3424 | ||
| COMP | 7.50E-61 | -2.7148 | ||
| VTN | 5.31E-09 | 7.2324 | ||
| CCND1 | 3.80E-08 | 1.2589 | ||
| VEGFA | 2.66E-10 | 1.0362 | ||
| LAMA4 | 1.06E-07 | -1.3721 | ||
| SGK1 | 1.10E-05 | 1.5521 | ||
| LAMA5 | 6.14E-06 | 3.2191 | ||
| COL4A2 | 2.41E-266 | 4.4393 | ||
| EPHA2 | 4.58E-16 | 2.5843 | ||
| OSMR | 1.47E-05 | 1.0598 | ||
| ITGB1 | 1.76E-53 | 1.1654 | ||
| VEGFB | 5.22E-08 | -1.0511 | ||
| THBS2 | 1.95E-63 | 1.092 | ||
| COL4A1 | 8.24E-208 | 5.2997 | ||
| COL4A5 | 2.06E-08 | 3.5296 | ||
| ITGA1 | 1.76E-12 | 3.1042 | ||
| P53 signalling pathway | CCND1 | 3.80E-08 | 1.2589 | |
| CD82 | 3.94E-05 | 1.1701 | ||
| SERPINE1 | 8.52E-299 | 2.1331 | ||
| STEAP3 | 9.73E-08 | -1.7067 | ||
| TP53I3 | 3.91E-07 | -1.2232 | ||
| CCNG2 | 0.00017411 | 1.2924 | ||
| IGFBP3 | 1.62E-168 | -2.411 |
Significant miRNA expressions targeting muscle regulation in WJ-MSC
While mRNA data is mapping, some miRNA might include in the data. While our raw data were aligning, 110 aligned miRNAs were mapped, and we achieved their expression data. 24 of the 110 miRNAs identified differentially expressed. The target match analysis was applied to these 23 miRNAs and 23 DEGs. In this analysis, experimentally observed pairs were selected whose expressions were their opposite. When this target match was investigated for 23 DEGs and 11 miRNAs, 2 miRNAs were targeting the muscle process functions in regeneration of WJ-MSC (Table 2).
Table 2.
Fold change of miRNAs that pass-through filtering that have functions is muscle regeneration
| Predicted miRNAs | Log Fold Change | Functions in the muscle regeneration |
|---|---|---|
| hsa-mir-24 ↑ | 1.987 | Regulation of TGF-B signalling during the skeletal muscle differentiation |
| hsa-mir-199a-5p ↓ | -1.929 | Exosomal miRNA cargo during the muscle fibrosis |
Discussion
MSCs are a group of progenitor cells capable of differentiating into several mesenchymal lineages [28] and an attractive cell source for cellular therapies [29]. MSCs have ability to recognize the biological markers that secreted from the injured cell and have potential to transform into the target cells and forming the healthy tissue [30]. The therapeutic effects of MSCs are promoting by secreting factors and delivering genetic information to tissue-resident cells via either a paracrine effect or a direct cell-to-cell interaction. Each manufactured MSC to be used in clinical are exposed to quality control tests for the key MSC features such as cell surface markers, differentiation capacities, telomerase activity [31]. Although MSCs derived from different sources share basic characteristics, they are transcriptomically different from each other. This transcriptomic difference is also important in terms of the diversity of the secretion and the genetic information that they transfer to the target cell. Therefore, the selection of the appropriate source for MSCs is critical in clinical application. In this study, we identified the transcriptomic patterns of MSCs derived from 2 different sources with RNA-Seq after their characterization by flow cytometry.
The hierarchical clustering that based on whole transcriptome data results demonstrated its regenerative capacity, regulation of extracellular matrix (ECM), immunoregulation and regulation of the musco-skeleton system.
In healthy skeletal muscle is able to regenerate by stem cells and precursor cells that host in its own niche. Unfortunately, DMD patient’s regenerative capacity is depleted and they are facing with the loss muscle mass. MSCs have capable of the regulation of muscle regeneration with pathways and gene expression that came forward with enhancing their regenerative capacities in DMD patients. One of these pathways is transforming growth factor-β (TGF-β) that is also organizing the myogenesis [32]. TGF-β is found upregulated in WJ-MSC cell line related to the AT-MSC. 5 of 80 members are differentially expressed. SMAD7 is upregulated and it controls the BAMBI expression and causes PITX2 upregulation. PITX2, is a transcription factor that belongs to homeobox gene family and controls both the embryonic and adult myogenesis [33]. In the adult myogenesis PITX2 expression was detected in proliferating satellite cells and they promote the differentiation of satellite cell-derived myoblasts [34,35]. As a transcriptional factor, PITX2 can control different pathways simultaneously. One of the negatively regulated mechanisms under the PITX2 is mir-31. The main role is the degradation of the dystrophin mRNA. Since mir-31 expression is repressed, mRNA will be transcripted and resulting in increase of dystrophin protein [36]. Another mechanisms is under the control of PITX2 is the suppression of miR-106b/miR-503/miR-23b/miR-15b pathway. It stimulates the increasing of CCND1, CCND2 and MYF5, that functions in triggering cell proliferation and myogenic commitment [36]. PITX2 revealed as an appropriate candidate regulator in DMD treatments both the compensating the dystrophy expression and increasing the cell proliferation and myogenic commitment. So, these two mechanisms may provide the improvement of muscle regeneration.
TGF-β has another face as a master regulator of fibrosis [37]. Previous studies indicated that the upregulated TGF-β signaling causes both pathological fibrosis in connective tissue [38-40] and the inflammation in DMD patients [41]. The over expression of both PITX2 and the mir-24 in WJ-MSCs may be serving more anti-inflammatory effects in comparison with the AT-MSC. So, these results can be guide us as strong clues that WJ-MSC might be an appropriate stem cell source for cell treatments [36,42] (Figure 4; Table 1).
Figure 4.
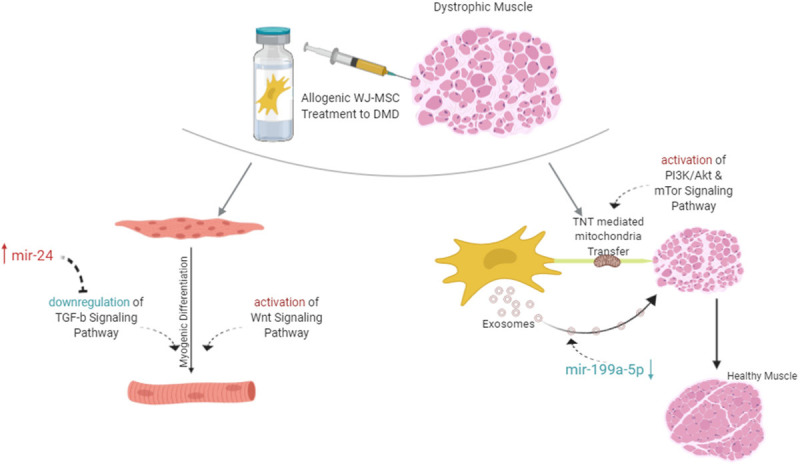
Complete evaluation and functional roles DEGs of whole results of the study.
Another prominent pathway is WNT signaling and it is found upregulated in WJ-MSC cell line related to the AT-MSC. 11 of 140 members are differentially expressed these are on duty in the β-catenin dependent canonical pathway. The effect of canonical pathway on muscle differentiation was determined by evaluating gain of function and loss of function mutations of β-catenin on muscle precursors [43]. In the absence of β-catenin, the muscle differentiation is weakened; in the high amount of β-catenin the premature differentiation was observed. In other words, β-catenin dependent WNT signaling is necessary to muscle tissue repairing and skeletal muscle differentiation. BAMBI, CCND1, DKK1 and TCF7L2 were detected upregulated in WJ-MSCs. These elements both have crucial roles in the canonical WNT pathway and are effective in muscle differentiation. Additionally, MYOD and MYF5 are also key regulators in the satellite cells, so their dysregulated expressions may cause an injury [44]. These two fundamental genes are under the control of the canonical WNT pathway; MYOD is a direct target of BAMBI [45]; MYF5 and CCND1 are co-expressed genes [46]. BAMBI encodes a transmembrane glycoprotein that roles in signaling transduction in myogenesis. Zhang et al. have detected the knockdown of BAMBI via siRNA causes the downregulation of MYOD which forms the myotubes in C2C12 cells [47]. So, the inhibition of BAMBI expression effects the myotube formation and myogenic differentiation by dysregulation the MYOD expression. Another study showed that, upregulation of MYF5 expression in satellite cells also elevating the CCND1 gene and it initiates the myogenesis by providing proliferation [46]. Besides all these, TCF7L2 regulates the cell cycle checkpoints via binding p53 promoter [48]. TCF7L2 has an inhibitory role on p53 expression post-transcriptionally by histone deacetylases [49]. TCF7L2 provides proliferation relatively via evading of apoptosis. Besides apoptosis, TCF7L2 is also important in the organization stabilization of connective tissue. Connective tissue is the well-organized niche for the transition of progenitors to differentiated mature cells. In normal muscle homeostasis, fibroblasts have high TCF7L2 expression and they regulating the muscle fiber development by generating suitable molecular and cellular niche for muscle progenitors [50]. However, when the TCF7L2 expression balance has been disturbed, muscle fibrosis can be observed. So, the high expression of TCF7L2 in WJ-MSC can supply these niche features and it may reverse the muscle injury (Figure 4; Table 1).
Aberrant miRNA expression may cause fibrogenesis [38] by dysregulation the key signaling pathways. Regulation of both TGF-β and WNT signaling is controlling not only by the members of the pathway but also regulating with epigenetic regulators. mir-24 has been upregulated in WJ-MSCs and regulates the signaling transductions as well [51]. Its effect on myogenesis was described via mir-24 antisense treatment on myoblasts. When mir-24 was suppressed in myoblasts, both the myotubule formation and the differentiation was decreased. When the treatment reversed to elevation of mir-24, the expression of myogenic markers returned to normal level and differentiation was compensated (Figure 4).
miRNAs are the one of the most important bio-cargos transferred by exosomes that affecting gene expression [52,53]. Zanotti et al. compared with the healthy myoblasts and DMD fibroblasts derived exosomes and they demonstrated high level expression of miR-199a-5p which has profibrotic effect via dysregulation collagen production [54]. Again in the same study, it has also been suggested that increased mir-199a-5p may be an early prognostic marker for DMD patients due to its fibrotic properties [55]. Parallelly, mir-199a has been reduced in WJ-MSCs. In cellular therapies, the effects of exosomes are known to be through horizontal transfers they send to host cells [56]. In our previous study, we have also demonstrated this transfer with increased dystrophin expression in DMD patients at the post WJ-MSC transplant term [31]. Besides the exosome transfer, Tunneling Nanotube (TNT) organelle transfer between the host and transplanted cells approach is emerging [57]. So, it was understood that horizontal transfer consists of not only exosomes but also included TNT-mediated mitochondria transfer. During fibrosis formation, cells get stressed and p53 pathway is activated. Activation of p53 triggered the activation of PI3K/AKT pathway and then the mTOR pathway like the domino effect and induced the tubular formation [58]. Mitochondria transfer via TNT, is contributing to alterations in the bioenergetic dysfunctions of recipient cells [59] which is important for the DMD patient [60]. In cellular therapies, MSCs are a suitable candidate for mitochondria transfer via TNT [61]. As our transcriptomic finding, WJ-MSC has upregulated p53, PI3K/Akt and mTOR signaling and it is an appropriate candidate source of MSC for cellular therapy for DMD patients (Figure 4).
In our previous study, we also suggested WJ-MSCs as an alternative source for clinical use which is a readily available tissue without an ethical concerns and the weak immunosuppressive properties [62] and we demonstrated its effectiveness as a therapeutic on DMD patients in our another study too [31]. As a conclusion, we supported our previous finding that WJ-MSC is a suitable candidate in the cellular treatment of DMD patients with transcriptomic and pathway analyses. As the study was performed on cell lines, repeating these findings with patients in a large cohort would strengthen our results.
Acknowledgements
We thank Elif Sozen Kucukkara her excellent technical assistance at cell culture and flow cytometry.
Disclosure of conflict of interest
None.
References
- 1.Emery AE. Population frequencies of inherited neuromuscular diseases-a world survey. Neuromuscul Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Klingler W, Jurkat-Rott K, Lehmann-Horn F, Schleip R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012;31:184–195. [PMC free article] [PubMed] [Google Scholar]
- 3.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 4.Gao QQ, McNally EM. The dystrophin complex: structure, function, and implications for therapy. Compr Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, Liu S, Wei T, Yong J, Mao Y, Lu X, Xie J, Ke Q, Jin F, Qi M. Development of a comprehensive real-time PCR assay for dystrophin gene analysis and prenatal diagnosis of Chinese families. Clin Chim Acta. 2013;424:33–38. doi: 10.1016/j.cca.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Guo R, Zhu G, Zhu H, Ma R, Peng Y, Liang D, Wu L. DMD mutation spectrum analysis in 613 Chinese patients with dystrophinopathy. J Hum Genet. 2015;60:435–442. doi: 10.1038/jhg.2015.43. [DOI] [PubMed] [Google Scholar]
- 7.Prior TW, Bridgeman SJ. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J Mol Diagnostics. 2005;7:317–326. doi: 10.1016/S1525-1578(10)60560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Yang Y, Liu J, Chen XC, Liu X, Wang CZ, He XY. Whole dystrophin gene analysis by next-generation sequencing: a comprehensive genetic diagnosis of Duchenne and Becker muscular dystrophy. Mol Genet Genomics. 2014;289:1013–1021. doi: 10.1007/s00438-014-0847-z. [DOI] [PubMed] [Google Scholar]
- 9.Pramono ZA, Takeshima Y, Alimsardjono H, Ishii A, Takeda S, Matsuo M. Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochem Biophys Res Commun. 1996;226:445–449. doi: 10.1006/bbrc.1996.1375. [DOI] [PubMed] [Google Scholar]
- 10.Long C, Li H, Tiburcy M, Rodriguez-Caycedo C, Kyrychenko V, Zhou H, Zhang Y, Min YL, Shelton JM, Mammen PPA, Liaw NY, Zimmermann WH, Bassel-Duby R, Schneider JW, Olson EN. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv. 2018;4:eaap9004. doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aartsma-Rus A, Krieg AM. FDA approves eteplirsen for Duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27:1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. NCT01826487. Phase 3 study of ataluren in patients with nonsense mutation duchenne muscular dystrophy. Https://ClinicaltrialsGov/Show/Nct01826487 2013. [DOI] [PMC free article] [PubMed]
- 13.Politano L, Nigro G, Nigro V, Pilus G, Papparella S, Paciello O, Comi LI. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol. 2003;22:15–21. [PubMed] [Google Scholar]
- 14.Nygren JM, Jovinge S, Breitbach M, Säwén P, Röll W, Hescheler J, Taneera J, Fleischmann BK, Jacobsen SE. Bone marrow-derived hematopoietic cells generate cardiomyocytes at a low frequency through cell fusion, but not transdifferentiation. Nat Med. 2004;10:494–501. doi: 10.1038/nm1040. [DOI] [PubMed] [Google Scholar]
- 15.Kouris NA, Schaefer JA, Hatta M, Freeman BT, Kamp TJ, Kawaoka Y, Ogle BM. Directed fusion of mesenchymal stem cells with cardiomyocytes via VSV-G facilitates stem cell programming. Stem Cells Int. 2012;2012:414038. doi: 10.1155/2012/414038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karp JM, Leng GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Porada CD, Almeida-Porada G. Mesenchymal stem cells as therapeutics and vehicles for gene and drug delivery. Adv Drug Deliv Rev. 2010;62:1156–1166. doi: 10.1016/j.addr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Meregalli M, Andrea F, Torrente Y. Mesenchymal stem cells as muscle reservoir. J Stem Cell Res Ther. 2011:01. [Google Scholar]
- 20.Karaoz E, Ayhan S, Okçu A, Aksoy A, Bayazit G, Osman Gürol A, Duruksu G. Bone marrow-derived mesenchymal stem cells co-cultured with pancreatic islets display β cell plasticity. J Tissue Eng Regen Med. 2011;5:491–500. doi: 10.1002/term.342. [DOI] [PubMed] [Google Scholar]
- 21.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 22.Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, Beggs AH, Kunkel LM. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci U S A. 2002;99:15000–15005. doi: 10.1073/pnas.192571199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sean D, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and bioconductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 24.Andrews S. FastQC: a quality control tool for high throughput sequence data. Http://WwwBioinformaticsBabrahamAcUk/Projects/Fastqc/ 2010; http://www.bioinformatics.babraham.ac.uk/projects/
- 25.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma J. 2013;7:1–8. [Google Scholar]
- 26.David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics. 2011;27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 27.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caplan AI. Mesenchymal stem cells. J Orthop Res. 1991;9:641–50. doi: 10.1002/jor.1100090504. [DOI] [PubMed] [Google Scholar]
- 29.Conrad C, Huss R. Adult stem cell lines in regenerative medicine and reconstructive surgery. J Surg Res. 2005;124:201–208. doi: 10.1016/j.jss.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Karbaat L, Wu L, Leijten J, Both SK, Karperien M. Trophic effects of mesenchymal stem cells in tissue regeneration. Tissue Eng Part B Rev. 2017;23:515–528. doi: 10.1089/ten.TEB.2016.0365. [DOI] [PubMed] [Google Scholar]
- 31.Dai A, Baspinar O, Yeşilyurt A, Sun E, Aydemir Çİ, Öztel ON, Capkan DU, Pinarli F, Agar A, Karaöz E. Efficacy of stem cell therapy in ambulatory and nonambulatory children with Duchenne muscular dystrophy - Phase I-II. Degener Neurol Neuromuscul Dis. 2018;8:63–77. doi: 10.2147/DNND.S170087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirskanen A, Kiefer JC, Hauschka SD. IGFs, insulin, Shh, bFGF, and TGF-β1 interact synergistically to promote somite myogenesis in vitro. Dev Biol. 2000;224:189–203. doi: 10.1006/dbio.2000.9784. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Torres F, Rodríguez-Outeiriño L, Franco D, Aranega AE. Pitx2 in embryonic and adult myogenesis. Front Cell Dev Biol. 2017;5:46. doi: 10.3389/fcell.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knopp P, Figeac N, Fortier M, Moyle L, Zammit PS. Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev Biol. 2013;377:293–304. doi: 10.1016/j.ydbio.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Vallejo D, Hernández-Torres F, Lozano-Velasco E, Rodriguez-Outeiriño L, Carvajal A, Creus C, Franco D, Aránega AE. PITX2 enhances the regenerative potential of dystrophic skeletal muscle stem cells. Stem Cell Rep. 2018;10:1398–1411. doi: 10.1016/j.stemcr.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 38.Bowen T, Jenkins RH, Fraser DJ. MicroRNAs, transforming growth factor beta-1, and tissue fibrosis. J Pathol. 2013;229:274–285. doi: 10.1002/path.4119. [DOI] [PubMed] [Google Scholar]
- 39.Ishitobi M, Haginoya K, Zhao Y, Ohnuma A, Minato J, Yanagisawa T, Tanabu M, Kikuchi M, Iinuma K. Elevated plasma levels of transforming growth factor β1 in patients with muscular dystrophy. Neuroreport. 2000;11:4033–4035. doi: 10.1097/00001756-200012180-00026. [DOI] [PubMed] [Google Scholar]
- 40.Song Y, Yao S, Liu Y, Long L, Yang H, Li Q, Liang J, Li X, Lu Y, Zhu H, Zhang N. Expression levels of TGF-β1 and CTGF are associated with the severity of duchenne muscular dystrophy. Exp Ther Med. 2017;13:1209–1214. doi: 10.3892/etm.2017.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YW, Nagaraju K, Bakay M, McIntyre O, Rawat R, Shi R, Hoffman EP. Early onset of inflammation and later involvement of TGFβ in Duchenne muscular dystrophy. Neurology. 2005;65:826–834. doi: 10.1212/01.wnl.0000173836.09176.c4. [DOI] [PubMed] [Google Scholar]
- 42.Ismaeel A, Kim JS, Kirk JS, Smith RS, Bohannon WT, Koutakis P. Role of transforming growth factor-β in skeletal muscle fibrosis: a review. Int J Mol Sci. 2019;20:2446. doi: 10.3390/ijms20102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudolf A, Schirwis E, Giordani L, Parisi A, Lepper C, Taketo MM, Le Grand F. β-catenin activation in muscle progenitor cells regulates tissue repair. Cell Rep. 2016;15:1277–1290. doi: 10.1016/j.celrep.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 44.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 45.Fujimaki S, Kuwabara T, Takemasa T. Wnt regulates satellite cell conversion after voluntary running. Adv Exerc Sport Physiol. 2014;20:89. [Google Scholar]
- 46.Panda AC, Abdelmohsen K, Martindale JL, Di Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E, Dudekula DB, De S, Becker KG, White EJ, Wilson GM, De Cabo R, Gorospe M. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016;44:2393–2408. doi: 10.1093/nar/gkw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Shi XE, Song C, Sun S, Yang G, Li X. BAMBI Promotes C2C12 myogenic differentiation by enhancing wnt/β-catenin signaling. Int J Mol Sci. 2015;16:17734–17745. doi: 10.3390/ijms160817734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Zhang E, Berggreen C, Jing X, Osmark P, Lang S, Cilio CM, Göransson O, Groop L, Renström E, Hansson O. Survival of pancreatic beta cells is partly controlled by a TCF7L2-p53-p53INP1-dependent pathway. Hum Mol Genet. 2012;21:196–207. doi: 10.1093/hmg/ddr454. [DOI] [PubMed] [Google Scholar]
- 49.Roose J, Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 50.Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Q, Zhang Y, Yang G, Chen X, Zhang Y, Cao G, Wang J, Sun Y, Zhang P, Fan M, Shao N, Yang X. Transforming growth factor-β-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008;36:2690–2699. doi: 10.1093/nar/gkn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 53.Zomer A, Vendrig T, Hopmans E, van Eijndhoven M, Middeldorp J, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010;3:447–450. doi: 10.4161/cib.3.5.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanotti S, Gibertini S, Blasevich F, Bragato C, Ruggieri A, Saredi S, Fabbri M, Bernasconi P, Maggi L, Mantegazza R, Mora M. Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol. 2018;74:77–100. doi: 10.1016/j.matbio.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Hrach HC, Mangone M. miRNA profiling for early detection and treatment of duchenne muscular dystrophy. Int J Mol Sci. 2019;20:4638. doi: 10.3390/ijms20184638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 57.Rustom A, Saffrich R, Markovic I, Walther P, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Cui J, Sun X, Zhang Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011;18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torralba D, Baixauli F, Sánchez-Madrid F. Mitochondria know no boundaries: mechanisms and functions of intercellular mitochondrial transfer. Front Cell Dev Biol. 2016;4:107. doi: 10.3389/fcell.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hughes MC, Ramos SV, Turnbull PC, Rebalka IA, Cao A, Monaco CMF, Varah NE, Edgett BA, Huber JS, Tadi P, Delfinis LJ, Schlattner U, Simpson JA, Hawke TJ, Perry CGR. Early myopathy in Duchenne muscular dystrophy is associated with elevated mitochondrial H2O2 emission during impaired oxidative phosphorylation. J Cachexia Sarcopenia Muscle. 2019;10:643–661. doi: 10.1002/jcsm.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Li H, Yao Y, Zhao T, Chen YY, Shen YL, Wang LL, Zhu Y. Stem cell-derived mitochondria transplantation: a novel strategy and the challenges for the treatment of tissue injury. Stem Cell Res Ther. 2018;9:106. doi: 10.1186/s13287-018-0832-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karaoz E, Cetinalp Demircan P, Erman G, Gungorurler E, Eker Sariboyaci A. Comparative analyses of immunosuppressive characteristics of bone-marrow, Wharton’s Jelly, and adipose tissue-derived human mesenchymal stem cells. Turk J Haematol. 2017;34:213–225. doi: 10.4274/tjh.2016.0171. [DOI] [PMC free article] [PubMed] [Google Scholar]