SUMMARY
Molecular mechanisms underlying plasticity at brain inhibitory synapses remain poorly characterized. Increased postsynaptic clustering of GABAA receptors (GABAARs) rapidly strengthens inhibition during inhibitory long-term potentiation (iLTP). However, it is unclear how synaptic GABAAR clustering is maintained to sustain iLTP. Here, we identify a role for miR376c in regulating the translation of mRNAs encoding the synaptic α1 and γ2 GABAAR subunits, GABRA1 and GABRG2, respectively. Following iLTP induction, transcriptional repression of miR376c is induced through a calcineurin-NFAT-HDAC signaling pathway and promotes increased translation and clustering of synaptic GABAARs. This pathway is essential for the long-term expression of iLTP and is blocked by miR376c overexpression, specifically impairing inhibitory synaptic strength. Finally, we show that local de novo synthesis of synaptic GABAARs occurs exclusively in dendrites and in a miR376c-dependent manner following iLTP. Together, this work describes a local post-transcriptional mechanism that regulates inhibitory synaptic plasticity via miRNA control of dendritic protein synthesis.
Graphical Abstract
In Brief
Clustering of GABAARs at inhibitory synapses is crucial for synaptic inhibition. Rajgor et al. discover that synaptic GABAAR expression is controlled by their local translation, regulated by miR376c. During inhibitory synaptic potentiation, miR376c is downregulated, relieving its translational repression of GABAAR mRNAs and leading to de novo synthesis of dendritic GABAARs.
INTRODUCTION
Neuronal inhibition in the central nervous system is essential for information processing, cognition, and behavior, and its disruption contributes to numerous neurological disorders (Klausberger and Somogyi, 2008; Smith and Kittler, 2010; Tremblay et al., 2016). GABAergic synapses are the major sites of inhibition in the brain, and changes in their strength and plasticity profoundly affect the activity of neuronal networks (Chiu et al., 2019). Inhibitory synapses are dynamic structures, capable of undergoing multiple forms of long-term plasticity (Castillo et al., 2011). Regulating the number of GABAA receptors (GABAARs) at inhibitory postsynaptic sites is a key determinant of synaptic strength (Nusser et al., 1997), and changes in GABAAR trafficking, clustering, and stability have major effects on synaptic efficacy (Barberis, 2020; Luscher et al., 2011). For example, induction of inhibitory long-term potentiation (iLTP) promotes synaptic strengthening via the rapid recruitment and synaptic clustering of GABAARs and the postsynaptic scaffold gephyrin (Marsden et al., 2007; Petrini et al., 2014). However, the mechanisms that maintain iLTP-induced synaptic GABAAR clustering over extended time periods remain largely unknown and are likely to be critical for sustaining iLTP and controlling long-term neuronal excitability.
Local protein synthesis near excitatory synapses is crucial for meeting synaptic demands to support long-term and rapid synapse-specific changes (Biever et al., 2019; Cajigas et al., 2010). However, little is known about translational regulation of GABAergic synapses and how local protein synthesis may affect their structure and function. MicroRNAs (miRNAs) have been identified as key regulators of the local synaptic protein translation that mediates excitatory synaptic plasticity (Hu and Li, 2017; Rangaraju et al., 2017; Schratt, 2009). These small non-coding RNAs inhibit translation by recruiting RNA-inducing silencing complexes (RISCs) to target mRNAs through complementary base pairing in 3′ untranslated regions (UTRs; Pratt and MacRae, 2009). In the brain, miRNAs regulate an array of processes, including neuronal development and learning and memory (McNeill and Van Vactor, 2012; Wang et al., 2012), and their dysfunction is implicated in neurological disease (Rajgor, 2018; Thomas et al., 2018). miRNAs are expressed in neuronal dendrites, and their functions can be controlled by synaptic activity, allowing highly regulated and localized translation near synapses (Sambandan et al., 2017). Moreover, multiple independent miRNA pathways can converge to control the structural plasticity of dendritic spines, the translation and trafficking of AMPARs, and excitatory synaptic transmission following excitatory LTP and long-term depression (LTD) stimulation (Gu et al., 2015; Hu et al., 2014; Olde Loohuis et al., 2015). In contrast, the role of miRNAs in modulating the plasticity of inhibitory synapses remains unexplored.
Here, we find that local miRNA-dependent translational mechanisms are essential for persistent strengthening of inhibitory synapses following iLTP stimulation. We demonstrate that miR376c silences GABRA1 and GABRG2, which encode for the critical synaptic GABAAR subunits α1 and γ2, respectively. Transcriptional repression of miR376c via NFATc3 and histone de-acetylases (HDACs) reduces miR376c-mediated GABRA1 and GABRG2 silencing during iLTP, promoting de novo synthesis of the α1 and γ2 subunits specifically in dendrites, providing a local post-transcriptional mechanism by which synaptic GABAAR expression can be increased to sustain iLTP.
RESULTS
Protein Synthesis Is Required for Maintaining Surface GABAAR Clustering after iLTP Induction
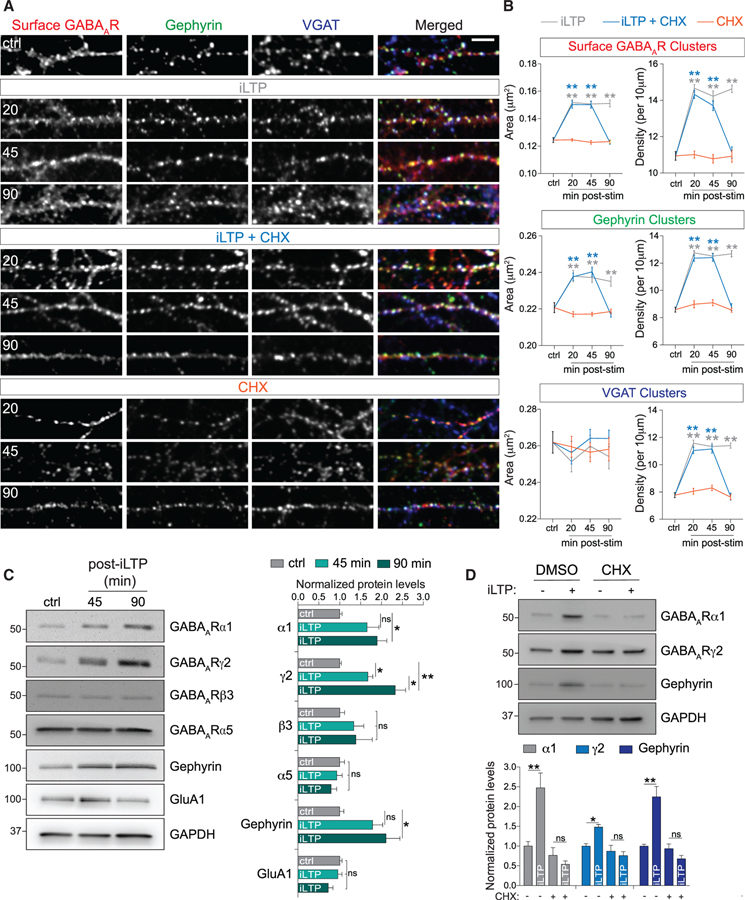
GABAARs are rapidly trafficked to inhibitory synapses within the first 20 min of iLTP induction to increase GABAAR synaptic clustering and synapse strength (Marsden et al., 2007; Petrini et al., 2014). Yet how long this increase in synaptic strength persists remains unknown. To induce iLTP in hippocampal neurons, we used a well-established mild NMDAR activation protocol (iLTP: 20 μM NMDA and 10 μM CNQX for 2 min), which increases GABAAR surface expression and GABAergic currents (Marsden et al., 2007; Petrini et al., 2014). We examined levels of synaptic GABAARs (surface γ2) in dendrites at 20, 45, and 90 min post-stimulation (Figure 1A) and co-labeled with antibodies to gephyrin and the inhibitory presynaptic marker vesicular GABA transporter (VGAT). In all conditions, ~90% of surface γ2-containing GABAARs clusters were positive for gephyrin or VGAT (Figure S1A). Consistent with previous work (Petrini et al., 2014), we observed increased synaptic GABAAR and gephyrin cluster area and density in dendrites at 20 min post-iLTP stimulation, accompanied by increased VGAT puncta density (Figures 1A and 1B). These increases were maintained at 45 and 90 min post-stimulation, indicating that maximal steady state of synaptic surface GABAAR expression was achieved within the first 20 min of iLTP induction and maintained for at least 90 min (Figures 1A and 1B). Somatic inhibitory synapses within the same neurons were unaffected (Figures S1B and S1C), suggesting that only dendritic inhibitory synapses are affected by iLTP stimulation.
Figure 1. Protein Synthesis Is Required to Maintain Dendritic Surface GABAARs after iLTP.
(A) Representative dendritic segments of hippocampal neurons labeled with antibodies to surface GABAAR(γ2) and intracellular gephyrin and VGAT. Neurons were fixed at 20, 45, and 90 min post-treatment: iLTP induction, iLTP induction in the presence of cycloheximide (CHX), or CHX alone. Scale bar, 10 μm.
(B) Quantification of cluster area and density from (A). n = 15 or 16 cells per condition. Colored stars compare statistical significance to the control (ctrl).
(C) Hippocampal lysates analyzed at 45 and 90 min post-iLTP induction. Protein levels were normalized to GAPDH; n = 5.
(D) α1, γ2, and gephyrin upregulation can be blocked by cycloheximide (CHX) at 90 min post-iLTP. Protein levels were normalized to GAPDH; n = 5.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, one-way ANOVA, Bonferroni post hoc test (C) or two-way ANOVA, Bonferroni post hoc test (B and D).
De novo protein synthesis may maintain potentiated inhibitory synapses over extended time periods. To explore this possibility, we induced iLTP in the presence of the translational inhibitor cycloheximide (CHX; 10 μg/mL) and measured levels of dendritic surface GABAARs, gephyrin, and VGAT. CHX had no effect on GABAAR clustering at 20 and 45 min post-iLTP (Figures 1A and 1B), but by 90 min post-iLTP, clustering of all three proteins was reduced to basal levels. In unstimulated neurons, dendritic synaptic GABAAR, gephyrin, and VGAT levels were unaffected by the presence of CHX at all time points, indicating that blocking protein synthesis does not influence basal clustering and synapse numbers (Figures 1A and 1B). CHX also did not affect somatic inhibitory synapses in stimulated or unstimulated neurons (Figures S1B and S1C). To complement these imaging data, we performed surface biotinylation assays, which also showed increased surface GABAAR expression at 20 and 90 min post-iLTP induction, and blockade of this increase at 90 min by CHX (Figure S1D). We also observed a reduction in surface GluA1 AMPARs from the same neuronal population, confirming that LTD of excitatory synapses occurs alongside iLTP (Figure S1D). Together, these data suggest that protein synthesis maintains increased dendritic GABAAR-gephyrin clustering and synapse density over extended time periods of iLTP expression.
iLTP-Induced De Novo Protein Synthesis of Inhibitory Synaptic Proteins
To determine whether key postsynaptic inhibitory proteins are upregulated following iLTP stimulation, we examined hippocampal lysates at 45 and 90 min post-iLTP induction (Figure 1C). We measured increased expression of gephyrin and GABAAR subunits α1 and γ2 at 90 min, but no increase in the β3 subunit, the extra-synaptic α5 subunit, or the AMPAR GluA1 subunit (Figure 1C). Importantly, CHX abolished the iLTP-induced increases in α1, γ2, and gephyrin expression observed at 90 min, demonstrating that core inhibitory synaptic components undergo de novo protein synthesis following iLTP induction (Figure 1D).
GABRA1 and GABRG2 Undergo Post-transcriptional Regulation after iLTP Induction
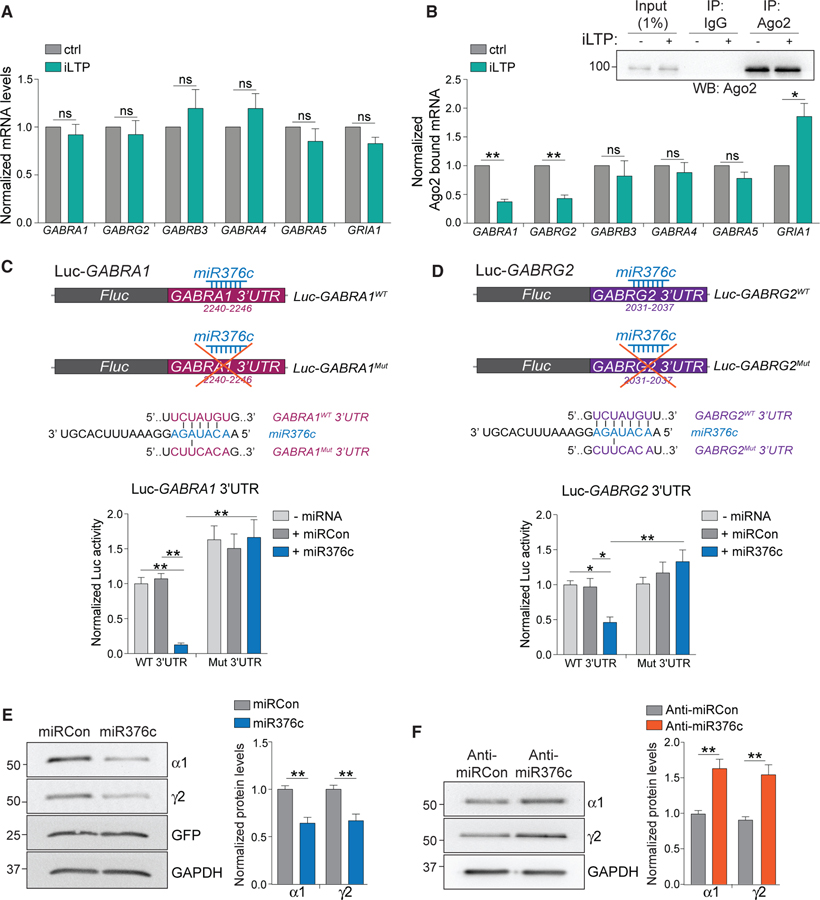
The de novo synthesis of α1 and γ2 GABAAR subunits following iLTP stimulation could be due to transcriptional or post-transcriptional regulation of their mRNAs, GABRA1 and GABRG2, respectively. qRT-PCR showed no change in GABRA1 or GABRG2 levels 90 min post-iLTP compared with controls (Figure 2A), indicating that synthesis of α1 and γ2 following iLTP is not due to changes in their gene transcription. There were also no changes in the mRNA levels of GABRB3, GABRA4, GABRA5, and GRIA1, which encode for the GABAAR β3, α4, α5, and AMPAR GluA1 subunits, respectively (Figure 2A).
Figure 2. miR376c Silences GABRA1 and GABRG2.
(A) qRT-PCR measuring mRNA levels 90 min post-iLTP induction. Readings were normalized to U6 small nuclear RNA (snRNA) levels; n = 5.
(B) Ago2 was immunoprecipitated equally from control neurons and from neurons 90 min post-iLTP induction. qRT-PCR measured mRNA bound to Ago2. Readings were normalized to total levels from (A); n = 5.
(C) Schematic of the Luc-GABRA1 luciferase reporters. miR376c seed site is mutated in Luc-GABRA1Mut. Graph of Luc-GABRA1 activities in HEK293T cells co-expressing miR376c, miRCon, or no miRNA. Firefly activities were normalized to Renilla activity; n = 9.
(D) As in (C) but for GABRG2; n = 10.
(E) Levels of α1 and γ2 in neurons expressing miRCon or miR376c; n = 6. miRNA expression constructs contain a GFP reporter.
(F) Levels of α1 and γ2 in neurons expressing anti-miRCon or anti-miR376c; n = 5.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, t test (A, B, E, and F) and two-way ANOVA, Bonferroni post hoc test (C and D).
As miRNAs represent likely candidates to regulate activity-dependent translation of GABAARs, we performed Argonaute-2 (Ago2) RNA immunoprecipitation assays to determine if GABRA1 and GABRG2 are regulated by miRNAs during iLTP. The amount of these mRNAs incorporated into RISCs is indicative of mRNA silencing in a miRNA-dependent manner. RISCs were isolated by immunoprecipitation of Ago2 (Figure 2B), and bound RNAs were eluted and analyzed using qRT-PCR. Significantly less GABRA1 and GABRG2 were bound to Ago2 at 90 min post-iLTP induction compared with controls (Figure 2B), indicating that miRNA-mediated gene silencing of GABRA1 and GABRG2 is likely reduced following iLTP. In contrast, there was more GRIA1 bound to Ago2 following stimulation, which is in line with reduced synaptic AMPAR expression following excitatory LTD (Hu et al., 2015). Importantly, there were no changes in the levels of GABRA4, GABRA5, and GABRB3 bound to Ago2 (Figure 2B), signifying that these transcripts are not regulated in a miRNA-dependent manner following iLTP.
miR376c Is a Translational Regulator of GABRA1 and GABRG2
To identify miRNAs that might regulate the iLTP-dependent translation of GABRA1 and GABRG2, we screened for miRNAs exhibiting expression changes in response to the mild NMDAR activation that induces iLTP. A next-generation sequencing (NGS) study demonstrated miRNA expression changes in response to this treatment in hippocampal brain slices (Hu et al., 2014) and identified 36 miRNAs to be downregulated at 90 min post-stimulation. Any targets of these miRNAs should undergo enhanced translation following stimulation. Surprisingly, one of these miRNAs, miR376c, was predicted to contain conserved seed sites in the 3′ UTR of both GABRA1 and GABRG2 as determined by TargetScan 7.0 and miRDB prediction algorithms (Lewis et al., 2003; Wong and Wang, 2015), suggesting that miR376c might mediate translational repression of two synaptic GABAAR subunits.
To determine the functionality of the predicted miR376c seed sites in GABRA1 and GABRG2, we fused their 3′ UTRs to a Firefly luciferase reporter construct (Luc-GABRA1WT and Luc-GABRG2WT, respectively) and performed dual luciferase reporter assays in HEK293 cells (Figures 2C and 2D). Co-transfection of Luc-GABRA1WT or Luc-GABRG2WT with miR376c significantly attenuated luciferase activity of both reporter constructs compared with a control miRNA (miRCon) or no miRNA co-expression (Figures 2C and 2D), indicative of increased silencing. However, miR376c co-expression did not silence reporter activity of GABRA1 or GABRG2 constructs with their predicted miR376c seed sites mutated (Luc-GABRA1Mut or Luc-GABRG2Mut; Figures 2C and 2D), strongly suggesting that GABRA1 and GABRG2 each contain a single functional miR376c binding site. This strong silencing of the GABRA1 and GABRG2 wild-type (WT) reporter activity could be due to miR376c promoting endonucleolytic cleavage of GABRA1 and GABRG2. However, miR376c overexpression did not alter the luciferase mRNA levels for Luc-GABRA1WT or Luc-GABRG2WT (Figure S2A), indicating that miR376c does not induce mRNA endonucleolytic cleavage and degradation.
Next, we examined the ability of miR376c to regulate endogenous GABAAR α1 and γ2 expression in hippocampal neurons. We found that miR376c overexpression reduced protein levels of both subunits by ~50% compared with miRCon (Figure 2E) without altering their mRNA levels (Figure S2B), further indicating a mechanism of miR376c-mediated translational repression rather than of mRNA degradation. In contrast, an inhibitory anti-miR designed to block the function of endogenous miR376c (anti-miR376c) significantly increased protein levels of both subunits by ~50% compared with a control inhibitor (anti-miRCon; Figure 2F). Neither miR376c overexpression or inhibition changed the protein levels of β3, α5, gephyrin, or GluA1 (Figures S2C and S2D), confirming that miR376c specifically regulates GABRA1 and GABRG2 translation in hippocampal neurons. To further validate the specificity of anti-miR376c, we examined its regulation of α1 and γ2 expression compared with an anti-miR for miR134, a dendritic miRNA that controls the translation of Lim kinase 1 (LIMK1) (Schratt et al., 2006). Anti-miR376c increased expression of α1 and γ2, but had no effect on LIMK1 levels. Furthermore, anti-miR134 induced upregulation of LIMK1, but not α1 and γ2 GABAAR subunits (Figure S1E), emphasizing the specificity of these miRNAs for their targets.
miR376c Affects Synaptic GABAAR Levels
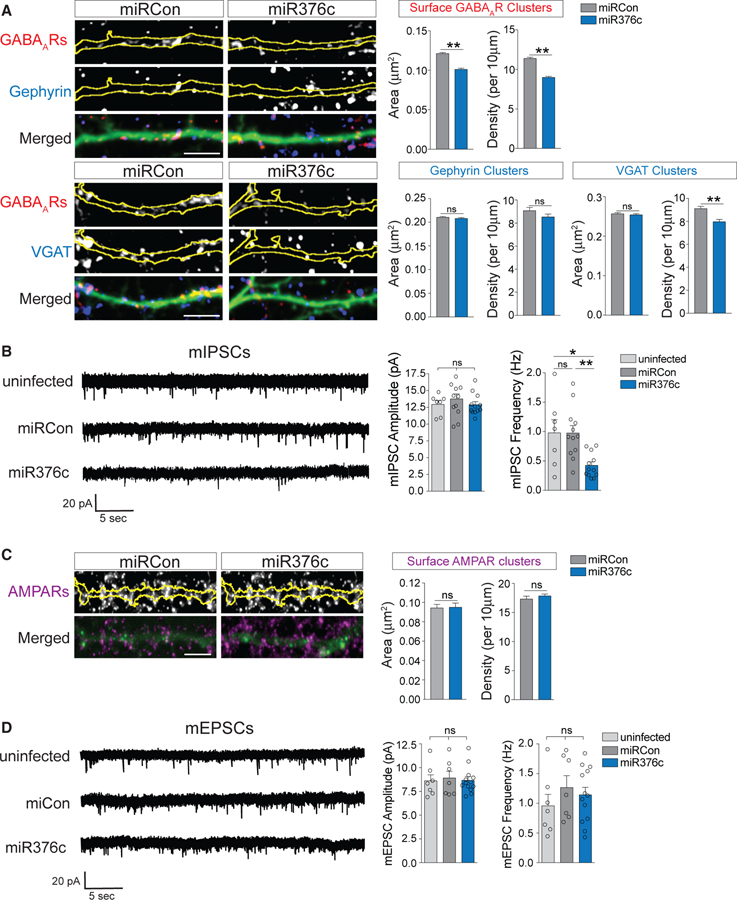
As miR376c manipulation had significant effects on total α1 and γ2 protein expression, we then asked whether GABAAR abundance at synapses was also affected. We first performed immunocytochemistry experiments with neurons expressing miRCon or miR376c to detect surface γ2 and either gephyrin or VGAT. Cluster analysis of dendritic surface γ2 revealed that overexpression of miR376c reduced surface GABAAR cluster area and density and VGAT cluster density, with no effect on gephyrin clusters (Figure 3A). In contrast, overexpression of miR376c had no effect on the area of somatic surface GABAAR clusters, although it slightly increased their density (Figure S3A), indicating potentially region-specific effects. To verify these results using another method, we performed electrophysiological recordings of miniature inhibitory postsynaptic currents (mIPSCs) from miRCon- or miR376c-expressing neurons. miR376c expression specifically reduced mIPSC frequency, with no change in amplitude or kinetic properties compared with uninfected neurons or neurons expressing miRCon (Figures 3B and S3B). Together, this shows that increasing repression of α1 and γ2 translation by miR376c expression leads to a reduction in GABAARs at dendritic inhibitory synapses that is manifested by a reduction in the total number of functional synapses. The observation that mean mIPSC amplitude is not affected by miR376c overexpression likely reflects the lack of impact of this manipulation on somatic GABAARs (Figure S3A), which have outsized contributions to mIPSC amplitude measurements made from the soma, due to distance-dependent filtering of dendritic synaptic responses. miR376c overexpression also had no effect on surface AMPAR levels or miniature excitatory postsynaptic current (mEPSC) amplitude or frequency (Figures 3C and 3D), underlining the specificity of miR376c for its inhibitory targets. Surprisingly, expression of the inhibitory anti-miR376c had no impact on dendritic or somatic inhibitory synapse area or density (Figures S3C and S3D) or mIPSC event parameters (Figure S3E) compared with anti-miRCon. This is likely due to the requirement that other essential proteins would also need to be upregulated to form larger and more numerous synapses following iLTP, so although synthesis of GABAAR α1 and γ2 subunits are upregulated by anti-miR376c, these subunits are not being assembled into functional GABAARs and/or delivered to the synapse.
Figure 3. miR376c Regulates Inhibitory Synaptic Strength.
(A) Representative dendritic segments from neurons expressing miRCon or miR376c and labeled with antibodies to GABAARs (surface γ2) and gephyrin or VGAT. Yellow outlines are derived from GFP fill. Graphs show dendritic cluster area and densiy. Scale bar, 5 μm; n = 7–18 cells per condition.
(B) Representative miniature inhibitory postsynaptic current (mIPSC) traces from uninfected neurons or neurons expressing miRCon or miR376c. Graphs show mIPSC amplitude and frequency; n = 7–12 cells per condition.
(C) Representative dendritic segments from neurons expressing miRCon or miR376c, and labeled with antibodies to surface GluA1. Graphs show quantification of dendritic GluA1 cluster area and density. Scale bar, 5 μm; n = 19 cells per condition.
(D) Representative miniature excitatory postsynaptic current (mEPSC) traces from uninfected neurons or neurons expressing miRCon or miR376c. Graphs show mEPSC amplitude and frequency; n = 7–13 cells per condition.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, t test (A and C) and one-way ANOVA, Bonferroni post hoc test (B and D).
miR376c-Mediated GABRA1 and GABRG2 Silencing Is Reduced after iLTP Induction
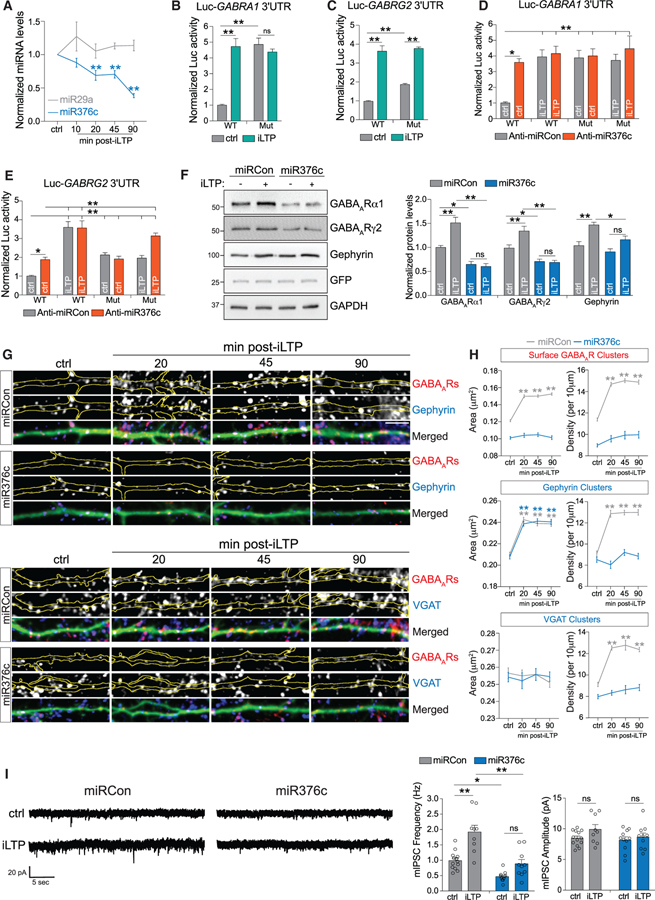
We next assessed whether miR376c levels decrease following iLTP stimulation, as suggested by NGS data (Hu et al., 2014). qRT-PCR showed an ~35% reduction in miR376c levels at 20 and 45 min post-iLTP induction and an ~60% reduction by 90 min (Figure 4A), which parallels increasing α1 and γ2 expression following iLTP (Figure 1C). We observed no change in levels of miR29a, a miRNA previously shown not to respond to NMDA stimulation (Hu et al., 2014). Reporter activities of Luc-GABRA1WT and Luc-GABRG2WT in hippocampal neurons both consequently increased at 90 min post-iLTP (Figures 4B and 4C), indicating that downregulation of miR376c may be sufficient to reduce GABRA1 and GABRG2 silencing. Surprisingly, basal activity of the Luc-GABRA1Mut reporter was increased compared with the WT reporter such that it mimicked and occluded the impact of iLTP (Figure 4B). Although basal Luc-GABRG2Mut reporter activity was also increased, it did not completely occlude the effect of iLTP (Figure 4C), suggesting that additional mechanisms may regulate iLTP-dependent GABRG2 translation. Importantly, anti-miR376c also increased basal activity of both Luc-GABRG1WT and Luc-GABRG2WT reporters, resulting in complete or partial occlusion of their regulation by iLTP, respectively (Figures 4D and 4E). Together, this suggests that although miR376c is likely to be the major post-transcriptional silencing mechanism for GABRA1 in resting neurons, the 3′ UTR of GABRG2 may be regulated by additional post-transcriptional mechanisms. Yet notably, miR376c overexpression completely attenuated the upregulation of endogenous α1 and γ2 protein expression following iLTP (Figure 4F), indicating that downregulation of this miRNA is likely to be the major mechanism promoting de novo synthesis of both α1 and γ2 GABAAR subunits following iLTP. Although miR376c overexpression did not reduce levels of gephyrin under basal conditions, it did attenuate its upregulation following iLTP (Figure 4F), suggesting that there is also likely interplay between regulation of GABAAR and gephyrin protein levels.
Figure 4. miR376c-Mediated Gene Silencing Is Reduced after iLTP.
(A) qRT-PCR showing miRNA expression levels following iLTP induction (normalized to U6 snRNA). Colored stars compare statistical significance to the ctrl; n = 4.
(B) Luc-GABRA1 activity in neurons under control conditions or 90 min post-iLTP induction; n = 5.
(C) As in (B) but for Luc-GABRG2; n = 5
(D) Luc-GABRA1 activity in neurons expressing anti-miRCon or anti-miR376c, under control conditions or 90 min post-iLTP induction; n = 5.
(E) As in (D), but for Luc-GABRG2; n = 5.
(F) α1, γ2, and gephyrin levels 90 min post-iLTP induction in neurons expressing miRCon or miR376c; n = 6.
(G) GFP-filled dendritic segments from miRNA overexpressing hippocampal neurons labeled with antibodies to surface GABAARγ2 and gephyrin or VGAT. Scale bar, 10 μm.
(H) Quantification of cluster area and density from (E); n = 16–18 neurons per condition. Colored stars compare statistical significance with ctrl.
(I) mIPSC traces from neurons infected with miRCon or miR376c. Graphs show mIPSC amplitude and frequency; n = 9–13 cells per condition.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, one-way ANOVA, Bonferroni post hoc test (A) and two-way ANOVA, Bonferroni post hoc test (B–F and I).
miR376c Overexpression Blocks Potentiation of Inhibitory Synapses after iLTP Induction
As miR376c overexpression blocked the de novo synthesis of α1 and γ2 GABAAR subunits following iLTP, we next examined whether it was also able to attenuate the iLTP-induced increases in inhibitory synapse density and surface GABAAR clustering. We observed that miR376c overexpression prevented iLTP upregulation of both inhibitory synapse density (as reflected in no change in cluster density for surface GABAARs, gephyrin, and VGAT) and the level of surface GABAAR clustering (as reflected in no change in surface GABAARs cluster area), without altering gephyrin cluster area (Figures 4G and 4H). Consistent with the finding above that anti-miR376c did not affect basal characteristics of GABAergic synapses (Figures S3C–S3E), expression of anti-miR376c also did not affect upregulation of dendritic surface GABAAR, gephyrin, and VGAT clusters 90 min following iLTP stimulation (Figures S4A and S4B), further supporting the idea that removing translational repression of α1 and γ2 is not sufficient to mimic iLTP. We also measured mIPSCs from separate populations of hippocampal neurons overexpressing miR376c or miRCon both under basal conditions and 20–60 min following iLTP stimulation (Figure 4I). Consistent with our imaging results, we observed a persistent increase in mIPSC frequency during this phase of iLTP in neurons overexpressing miRCon, but not in neurons overexpressing miR376c (Figure 4I). Mean mIPSC amplitude also did not change following iLTP stimulation of miR376c-expressing neurons as expected (Figure 4I). However, perhaps unexpectedly, there was no significant increase in mIPSC amplitude following iLTP in miRCon expressing neurons. This is likely due to the challenges discussed above in detecting changes in the size of smaller dendritic mIPSC responses against a constant background of larger somatic mIPSC responses combined with levels of experimental variation that are inherent when comparing different populations of control and iLTP treated neurons. Nonetheless, together these results show that overexpression of miR376c can prevent the upregulation of surface GABAAR clusters and the number of functional inhibitory synapses following iLTP stimulation.
Increased De Novo Synthesis of Dendritic GABAARs after iLTP Induction
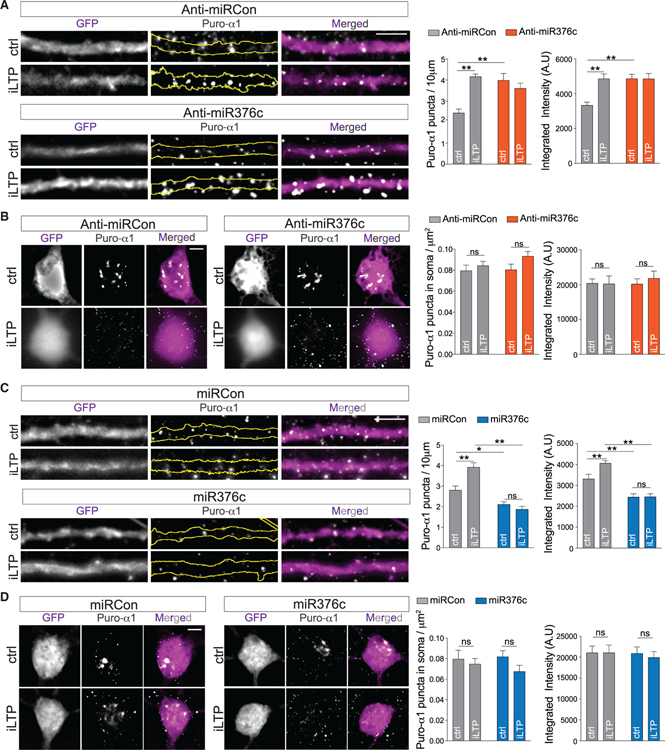
Local transcriptome data indicate that both GABRA1 and GABRG2 are abundantly expressed in the soma and synaptic neuropil (Cajigas et al., 2012), suggesting that these mRNAs may be locally translated in dendrites. To test whether GABAAR mRNAs are translated in dendrites in response to iLTP stimulation, we performed puromycin-proximity ligation assays (puro-PLAs), which enable the direct visualization of de novo protein synthesis in situ (Sambandan et al., 2017). Neurons were pulsed with puromycin for 10 min prior to fixation, allowing both the location and amount of de novo protein synthesis to be determined. We assayed for de novo α1 subunit protein, as a suitable γ2 antibody compatible with puro-PLA is currently unavailable. First, we performed puro-PLAs in neurons expressing a gephyrin intrabody to label endogenous gephyrin (Gross et al., 2013), allowing visualization of α1 synthesis sites relative to inhibitory synapses. The addition of puromycin resulted in detection of nascent puro-α1 in dendritic regions within the vicinity of some inhibitory synaptic sites marked by gephyrin, as well as in the soma (Figures S5A and S5B). In control experiments, puro-α1 PLA signal was not detected when this assay was performed in the presence of CHX or in the absence of puromycin (Figures S5C and S5D).
To determine whether miR376c-dependent GABRA1 silencing occurs in neuronal dendrites and/or the soma, we performed puro-PLAs with neurons expressing the control or anti-miR376c inhibitor (Figures 5A and 5B). In control conditions, miR376c inhibition readily increased puro-α1 labeling in the dendrites, but not in the soma (Figures 5A and 5B). Furthermore, the anti-miR376c occluded any increase in translation following iLTP. Surprisingly, iLTP induction did not affect somatic α1 translation in neurons expressing the control or anti-miR376c inhibitor (Figure 5B), suggesting that miR376c-GABRA1 silencing occurs primarily in the dendrites. In comparison, overexpression of miR376c reduced dendritic puro-α1 labeling in dendrites under basal conditions and blocked the iLTP-induced increase in dendritic puro-α1 (Figure 5C), but had no effect on somatic levels of puro-α1 (Figure 5D), showing that miR376c does not regulate translation of somatic GABRA1 and that increased de novo α1 synthesis following iLTP is restricted to dendrites.
Figure 5. GABAARα1 Is Exclusively Translated in Neuronal Dendrites after iLTP.
(A) Newly synthesized dendritic α1, labeled by puro-PLA in neurons expressing anti-miRCon or anti-miR376c, under control conditions or 90 min post-iLTP induction. Graphs show puro-α1 puncta density or integrated intensity. Scale bar, 5 μm; n = 18 cells per condition.
(B) As in (A), showing representative somata. Scale bar, 5 μm; n = 18 cells per condition.
(C) As in (A), for miRCon or miR376c expression; n = 18 cells per condition.
(D) As in (B), for miRCon or miR376c; n = 18 cells per condition.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, two-way ANOVA, Bonferroni post hoc test.
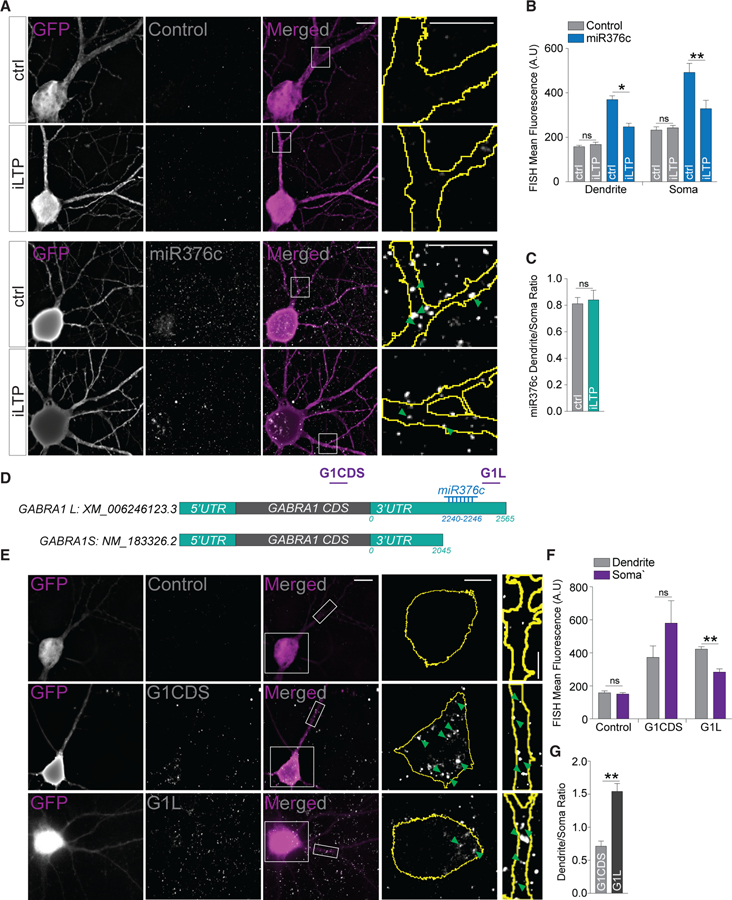
miR376c Targets a Dendritically Localized GABRA1 mRNA Variant
As our data suggest that miR376c silences GABRA1 in dendrites, we then asked whether miR376c was present in the dendritic compartment. We assessed the neuronal localization of miR376c using miRNA fluorescent in situ hybridization (FISH) using double-labeled digoxygenin locked-nucleic acid (LNA) probes. In agreement with the above puro-PLAs, we observed robust labeling of miR376c in the dendrites; however, we also detected miR376c in the soma (Figure 6A). Consistent with our qRT-PCR data, the miR376c FISH signal decreased in both dendritic and somatic compartments at 90 min post-iLTP induction, whereas the control probe signal was not altered (Figures 6A and 6B). We observed no change in the ratio of dendritic to somatic labeling of miR376c following iLTP, indicating this miRNA is not re-distributed following stimulation (Figures 6A and 6C).
Figure 6. miR376c Represses Dendritically Enriched GABRA1 mRNA.
(A) Neurons labeled with double digoxygenin control or miR376c FISH detection probes in control conditions or 90 min post-iLTP induction. Magnifications of dendritic segments are from boxes in merged images. Scale bars, 10 μm for whole images and 5 μm for magnifications.
(B) Mean FISH signals in soma and dendrites from (A); n = 13–21 cells per condition.
(C) Dendrite/soma ratio of miR376c labeling; n = 13–21 cells per condition.
(D) Schematic of GABRA1 mRNAs with NCBI accession numbers. The relative position of the LNA FISH probes G1CDS and G1L are shown in purple.
(E) Neurons labeled with double digoxygenin control, G1CDS or G1L FISH detection probes. Magnification of somatic and dendritic segment from boxed areas in merged images are shown. Scale bars, 10 μm for whole images and 5 μm for magnifications.
(F) Mean FISH signals from the soma and dendrites from (E); n = 12–16 cells per condition.
(G) Dendrite/soma ratio of G1CDS and G1L labeling showing dendritic enrichment of G1L; n = 12–16 cells per condition.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, t test (B, C, F, and G).
Our data suggest that although miR376c is present in both the soma and dendrites, miR376c-GABRA1 silencing is restricted to the dendrites, pointing to potentially unique mechanisms regulating GABRA1 translation in each compartment. For instance, distinct GABRA1 variants may exist that are differentially targeted by miR376c. To explore this, we screened databases for GABRA1 mRNAs and identified a transcript with a truncated 3′ UTR, presumably through the use of an alternative polyadenylation signal, and therefore lacks the miR376c seed site (Figure 6D). To determine whether this shorter GABRA1 (GABRA1S) has differential localization compared with its longer counterpart (GABRA1L), we used LNA FISH to delineate differences in their subcellular localization. We created a FISH probe to the GABRA1 coding sequence (G1CDS) to detect all GABRA1 variants and a FISH probe to a region downstream of the miR376c seed site (G1L) to detect only GABRA1L (Figure 6D). Our FISH experiments revealed that the G1CDS probe strongly labeled the soma and dendrites, whereas the G1L probe predominantly labeled the dendrites (Figures 6E and 6F). Dendritic/somatic ratios demonstrated that G1CDS displayed somatic enrichment, while G1L was significantly enriched in the dendrites (Figure 6G). Together with our local translation assays, these data strongly indicate that de novo α1 synthesis increases specifically in dendrites following iLTP and is likely due to miR376c-dependent de-repression of a dendritically enriched GABRA1 variant.
iLTP Induces Rapid Transcriptional Repression and Turnover of miR376c
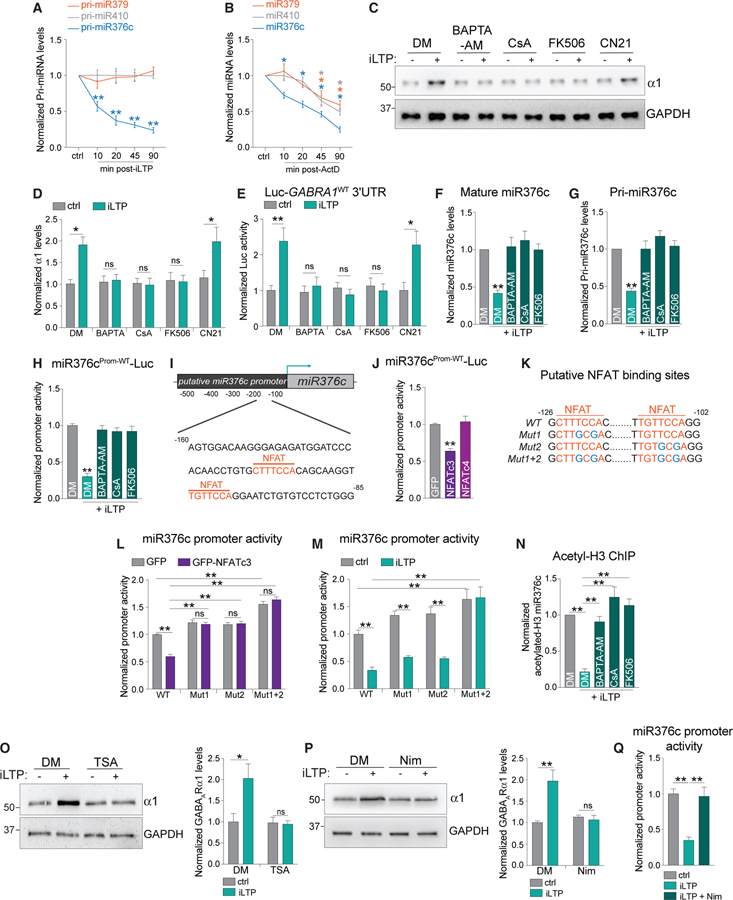
The downregulation of miR376c is key in maintaining increased GABAAR expression and synaptic clustering following iLTP induction. But what mechanisms lead to iLTP-induced miR376c downregulation? To determine whether miR376c downregulation could be due to transcriptional repression of the miR376c gene, we used qRT-PCR to assess levels of its primary transcript (pri-miR376c) following iLTP stimulation. Pri-miR376c levels were significantly decreased by ~40% at 10 min and by ~60% at 90 min post-iLTP induction (Figure 7A), with no change in the levels of mature or primary forms of miR379 and miR410 (two miRNAs within the same gene cluster as miR376c) following iLTP-induction (Figures S6A and 7A). These data indicate that transcription of pri-miR376c is reduced following stimulation. To definitively show that transcriptional repression could account for a rapid reduction in mature miR376c, we treated hippocampal neurons with actinomycin D (ActD) for 10, 20, 45, and 90 min to inhibit transcription and found that mature miR376c levels significantly decreased by 25% following 10 min exposure to ActD and by 60% after 90 min exposure (Figure 7B). In comparison, miR379 and miR410 levels decreased significantly less over this same timescale (Figure 7B). Thus, miR376c undergoes rapid turnover, and its abundance and function are likely to be highly dependent on the continued transcription and processing of pri-miR376c.
Figure 7. miR376c Is Downregulated through a CaN-NFATc3-HDAC Pathway after iLTP.
(A) qRT-PCR showing levels of primary miRNAs following iLTP induction; n = 4.
(B) qRT-PCR showing levels of mature miRNAs following actinomycin-D (ActD) treatment; n = 4.
(C) Levels of GABAARa1 from neurons under control conditions and at 90 min post-iLTP-induction, in the presence of DMSO (DM), BAPTA-AM, CsA, FK506, or TAT-CN21 (CN21).
(D) Quantification of α1 levels from (C); n = 6.
(E) Luc-GABRA1WT activity in neurons under control conditions and at 90 min post-iLTP induction, in the presence of DM, BAPTA-AM, CsA, or FK506; n = 5.
(F) qRT-PCR showing levels of mature miR376c 90 min post-iLTP induction with the inhibitors used in (E); n = 5.
(G) qRT-PCR showing levels of pri-miR376c 90 min post-iLTP induction with the inhibitors used in (E); n = 5.
(H) Putative miR376c promoter reporter (miR376cProm-WT-Luc) activity with the inhibitors used in (E); n = 3.
(I) Predicted NFAT binding sites in the putative miR376c promoter are shown in orange.
(J) GFP-NFATc3 expression significantly reduces miR376cProm-WT-Luc activity; n = 3.
(K) Schematic showing mutations introduced into miR376cProm-WT-Luc (blue).
(L) Putative miR376cProm reporter activity in neurons expressing GFP or GFP-NFATc3; n = 3.
(M) miR376cProm reporter activity in control conditions and 90 min post-iLTP induction; n = 4.
(N) qPCR readout of acetyl-histone H3 chromatin immunoprecipitation (ChIP) from neurons to show the acetylation status of the miR376c promoter with the inhibitors used in (E); n = 4.
(O) Trichostatin A (TSA) inhibits iLTP-induced upregulation of α1; n = 6.
(P) Nimodipine (Nim) inhibits iLTP-induced upregulation of α1; n = 3.
(Q) Nim blocks the iLTP-induced decrease of miR376cProm-WT-Luc activity; n = 3.
All values represent mean ± SEM. *p < 0.05 and **p < 0.01, one-way ANOVA, Bonferroni post hoc test (A, B, F, G, J, O, and P) and two-way ANOVA, Bonferroni post hoc test (D, E, L–N, and Q).
A Calcineurin-Dependent Pathway Reduces miR376c Expression after iLTP Induction
As iLTP induction involves Ca2+ influx through activated NMDARs (Petrini et al., 2014), we hypothesized that Ca2+ signaling through plasticity enzymes is likely to be upstream of increased de novo synthesis of α1 and γ2 following iLTP. To investigate this, we induced iLTP in the presence of the intracellular Ca2+ chelator BAPTA-AM (20 μM) or drugs that either inhibit calcineurin (CaN; CsA 5 μM or FK506 5 μM) or Ca2+/calmodulin-dependent protein kinase II (CaMKII; TAT-CN21 5 μM), two key signaling enzymes that initiate the major NMDAR-dependent excitation-transcription (E-T) coupling pathways (Barbado et al., 2009; Hagenston and Bading, 2011). BAPTA-AM and CaN inhibitors blocked the upregulation of α1 and γ2 protein levels following iLTP stimulation (Figures 7C, 7D, S6B, and S6C), whereas CaMKII inhibition had no effect. Furthermore, Ca2+ chelation and CaN inhibition blocked the iLTP response of the Luc-GABRA1WT and Luc-GABRG2WT reporters (Figures 7E and S6D). Together these data show that CaN-dependent signaling pathways are essential for promoting de novo synthesis of α1 and γ2 subunits via the regulation of their 3′ UTRs. CaN inhibition also prevented downregulation of both the mature and pri-miR376c following iLTP (Figures 7F and 7G), suggesting that CaN is important in transcriptional repression of miR376c gene. To determine if CaN signaling directly affects miR376c transcription, we fused the ‘‘putative’’ promoter region of miR376c (500 bp upstream of pre-miR376c) to luciferase, to generate a miR376cProm-WT-Luc reporter construct that would provide a readout of miR376c transcriptional activity. miR376cProm-WT-Luc had ~5-fold increase in Luc activity compared with an empty luciferase vector with no functional promoter, suggesting that a promoter region is located within these 500 bp (Figure S6E). At 90 min post-iLTP induction, we observed a ~70% reduction in miR376cProm-WT-Luc activity, which was fully blocked by BAPTA-AM, CsA, or FK506 application (Figure 7H), indicating that iLTP-induced CaN signaling reduces miR376c expression through repression of the miR376c promoter.
NFATc3 and HDACs Repress miR376c Transcription after iLTP Induction
A major downstream target of CaN is the nuclear factor of activated T cells (NFAT) family of transcription factors, which translocate to the nucleus and induce activity-dependent transcriptional changes (Kipanyula et al., 2016). As the transcriptional repression of miR376c was dependent on CaN activity, we reasoned that NFATs might mediate miR376c transcriptional repression following iLTP. We identified two putative NFAT binding sites in the miR376c promoter region, located at −125 and −109 upstream of the pre-miR376c coding region (Figure 7I). Co-expression of miR376cProm-WT-Luc with GFP-NFATc3 or GFP-NFATc4 (the most highly expressed hippocampal isoforms; Ulrich et al., 2012; Wild et al., 2019) revealed that only GFP-NFATc3 reduced transcriptional reporter activity by ~40%, compared with a GFP control suggesting that NFATc3 may negatively regulate miR376c transcription. In agreement, live imaging revealed increased nuclear localization of GFP-NFATc3 within 10 min of iLTP induction (Figures S6F and S6G). If NFATc3 is a transcriptional repressor of miR376c, disrupting its binding sites within the miR376c promoter should increase the activity of miR376cProm-WT-Luc. Therefore, we mutated the NFAT binding sites individually (miR376cProm-Mut1-Luc and miR376cProm-Mut2-Luc), in addition to creating a construct with both sites mutated (miR376cProm-Mut1+2-Luc; Figure 7K). GFP-NFATc3 overexpression only repressed activity of the WT reporter, indicating both NFAT binding sites are functional in negatively regulating miR376c transcription (Figure 7L). To test the involvement of both binding sites in miR376c repression following iLTP, we compared activity of all three mutant reporters to the activity of the WT in neurons 90 min post-iLTP induction. The activities of both the single mutants were reduced to similar levels as the WT reporter following iLTP (Figure 7M), whereas miR376cProm-Mut1+2-Luc activity was completely insensitive to transcriptional changes following stimulation (Figure 7M). These data demonstrate that NFATc3 can transcriptionally repress miR376c by associating with a single binding site in the miR376c promoter following iLTP induction.
Next we asked how NFATc3 represses miR376c transcription. NFAT proteins interact with HDACs and recruit them to reduce transcription of specific genes in non-neuronal systems (Baksh et al., 2002; Choo et al., 2009). Furthermore, miR376c expression is regulated by the acetylation status of its promoter (Iwaki et al., 2013; Zehavi et al., 2012). Therefore, to determine whether the miR376c promoter is de-acetylated following iLTP (a mark for gene silencing), we performed chromatin immunoprecipitation (ChIP) assays to assess the interaction between acetylated histone H3 and the miR376c promoter. qPCR of acetylated miR376c promoter DNA revealed reduced levels of acetylated H3 histones associated with the miR376c promoter following iLTP, and this effect was blocked by BAPTA-AM or CaN inhibitors (Figure 7N), suggesting the miR376c promoter is de-acetylated by HDACs in a CaN-dependent manner, following iLTP. To assess the involvement of HDACs in GABAAR translation during iLTP, we pre-treated neurons with Trichostatin A (TSA; 1 μm) to block class I and II HDACs. TSA attenuated the synthesis of α1 and γ2 subunits following iLTP stimulation, but not in control conditions (Figures 7O and S6H); demonstrating that HDACs promote the GABAAR protein synthesis that is required for sustained iLTP.
How might signaling in dendrites be rapidly transmitted to somatic NFATc3 and HDACs to induce the repression of miR376c in the nucleus? NFATc3 translocation to the nucleus is dependent on the activation of L-type Ca2+ channels (LTCCs) in response to NMDAR activation in distal dendrites (Murphy et al., 2014; Wild et al., 2019). We asked whether LTCCs might also rapidly transmit signals in the dendrites to repress iLTP-induced miR376c transcription and ultimately increase de novo synthesis of α1 and γ2. Following iLTP, de novo synthesis of α1 and γ2 was attenuated by the LTCC antagonist nimodipine (10 μM), as was the reduced reporter activity of miR376cProm-WT-Luc (Figures 7P, 7Q, and S6I). These data demonstrate that LTCCs are crucial for mediating the iLTP-dependent transcriptional repression of miR376c and increased GABAAR synthesis.
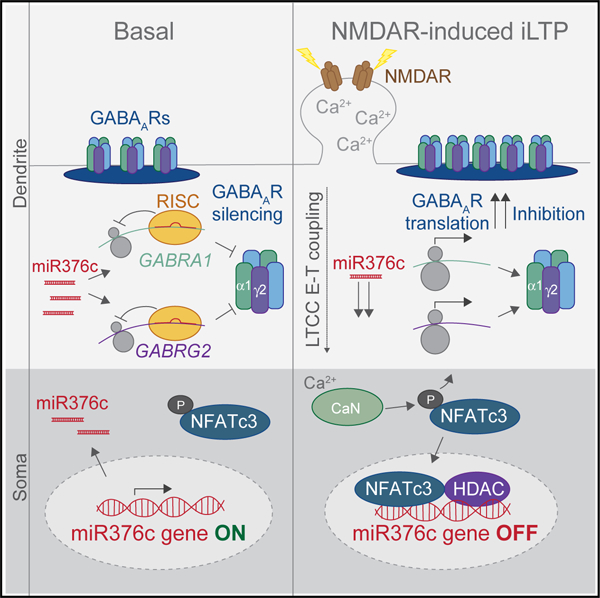
Together, these experiments demonstrate that an LTCC-CaN-NFATc3-HDACs axis, activated by NMDARs, results in the de-acetylation of the miR376c promoter following iLTP-induction. De-acetylation inhibits transcription of miR376c, leading to the rapid clearance of mature miR376c over time and thereby enhancing GABRA1 and GABRG2 protein translation in neuronal dendrites (Figure S7).
DISCUSSION
miRNA Regulation of Inhibitory Synapses
Continuous protein synthesis and turnover influences the composition of the synaptic proteome. miRNAs can dictate the rate of mRNA translation, and their activity can modify synaptic protein content, which controls synaptic transmission and plasticity (Schratt, 2009). We have identified miR376c as a translational repressor of GABRA1 and GABRG2, which encode for synaptic α1 and γ2 subunits and assemble to form one of the most commonly found hippocampal synaptic GABAAR subtypes (Pirker et al., 2000). Overexpression of miR376c reduced GABAAR surface clustering, synapse numbers, and mIPSCs. On induction of iLTP, miR376c levels were reduced over a 90 min time period, relieving miR376c-dependent translational repression of GABRA1 and GABRG2, leading to increased de novo α1 and γ2 protein synthesis and maintaining potentiated surface clustering of GABAARs. Through analysis of pri-mi376c levels and use of ActD, we discovered that the iLTP-induced downregulation of miR376c is due primarily to reduced transcription of the miR376c gene, likely accompanied by rapid cytoplasmic clearance of mature miR376c. Our ActD experiment revealed that miR376c exhibits a short half-life (~45 min) compared with other neuronal miRNAs. This is consistent with miRNAs studied in the retina and human brain that have very short half-lives, enabling their abundance to be quickly altered in response to changes in neuronal activity (Krol et al., 2010; Rüegger and Großhans, 2012; Sethi and Lukiw, 2009).
The upregulation of synaptic GABAARs by reduced miR376c-mediated translational repression is likely one of numerous mechanisms that are involved in the persistent potentiation of inhibitory synapses. We show that although iLTP-dependent control GABRA1 translation via its 3′ UTR is dependent primarily on the activity of miR376c, control of GABRG2 translation is not exclusively mediated by miR376c and may be regulated by additional post-transcriptional mechanisms, such as other miRNAs or RNA binding proteins. Furthermore, gephyrin expression is also increased at 45 and 90 min following iLTP stimulation in a miR376c-independent manner, again suggesting other post-transcriptional events must control gephyrin expression. Notably, miR376c inhibition with anti-miR376c increased total protein expression levels of α1 and γ2, but not surface GABAAR clustering or mIPSCs. Correct GABAAR clustering at synapses also requires upregulation of other subunits and proteins to produce correctly assembled and synaptically expressed receptors; therefore, sequestration of miR376c to induce increased α1 and γ2 expression is not sufficient to mimic iLTP. Moreover, increased expression of synaptic gephyrin is also required to potentiate inhibitory synapses during iLTP (Petrini et al., 2014). As gephyrin expression is insensitive to miR376c inhibition, we conclude that without increased gephyrin expression, anti-miR376c would have little effect on surface GABAAR clustering. These anti-miR376c experiments strongly indicate that other mechanisms must exist (including translational repression by other miRNAs), to upregulate key inhibitory synaptic proteins following iLTP stimulation. Further work is needed to determine these molecular mechanisms and how they may cooperate to potentiate inhibitory synapses during the long-term expression of iLTP.
Local Protein Synthesis of GABAARs during the Expression of iLTP
Inhibitory synapses have great capacity for plasticity, but little is known about how local protein synthesis mechanisms might contribute to their plasticity. Multiple studies have demonstrated that iLTP stimulation leads to increased GABAAR clustering, inhibitory synapse density, and GABAergic currents within a 20 min time period (Chiu et al., 2018; Marsden et al., 2007; Petrini et al., 2014). Within this time range, increased synaptic surface GABAAR clusters arise from increased GABAAR exocytosis and their immobilization at synaptic sites (Marsden et al., 2007; Petrini et al., 2014). We extend these findings by showing that iLTP-induced enhancement of inhibitory synaptic transmission is persistent and maintained for 90 min following stimulation (Figures 1A and 1B). Furthermore, CHX abolished the iLTP-dependent increase in surface GABAAR clustering at 90 min, but not 20 or 45 min following stimulation. These findings indicate that previously described trafficking events likely govern the increase in surface and synaptic GABAAR expression at early time points of iLTP expression, but protein translation is required to maintain potentiated GABAAR clustering over the long term (Figure S7). At 20 min post-stimulation, we detect an ~40% reduction in mature miR376c expression, which likely releases translational repression of GABRA1 and GABRG2 mRNA, giving rise to their subsequent increase in protein expression. GABAAR subunits can be translated, assembled into receptors, and inserted into the membrane within ~15 min ( Joshi et al., 2013; Saliba et al., 2012), therefore this timescale implies that as long as translational de-repression occurs within the first 40 min of stimulation, there is time for newly synthesized receptors to reach and potentiate synapses.
Recent sequencing data show that key GABAAR subunit mRNAs are found in hippocampal neuropil (Cajigas et al., 2012) and therefore are likely to be locally translated in dendrites. Although GABRA1 and miR376c are present in the soma and dendrites, our FISH experiments indicate that GABRA1 variants harboring the miR376c seed site are dendritically enriched (Figure 6). Therefore, only GABRA1 variants enriched in neuronal dendrites are specifically de-repressed and locally translated following iLTP, leading to upregulation of surface GABAARs in the dendrites, but not in the soma. This model matches well with our detection of de novo synthesis of α1 subunits in dendrites using puro-PLA assays and imaging data showing that dendritic GABAAR clusters are exclusively upregulated following iLTP stimulation in a miR376c-dependent manner, with no effect on somatic clusters. We suspect that γ2 is also locally translated in dendrites during iLTP expression and that other GABAAR subunits might be locally translated during other modes of plasticity. Interestingly, the longer 3′ UTR variant of GABRA1 may also contain motifs that are absent from the shorter variant, possibly enabling association with specific RBPs involved in dendritic targeting (Schieweck and Kiebler, 2019). Altogether, dendrite-specific miR376c-dependent enhancement in surface GABAAR clustering is in agreement with recent work demonstrating that iLTP stimulation specifically potentiates somatostatin-positive, dendritic GABAergic synapses and not those located in the somatic region (Chiu et al., 2019) and supports the idea that local translation mechanisms may participate in regulating specific subtypes of inhibitory synapses.
E-T Coupling of miRNAs
We define a signaling pathway that links NMDAR stimulation in dendrites to transcriptional repression of a miRNA in the nucleus. By measuring the activity of the miR376c ‘‘putative’’ promoter, we found that Ca2+, CaN activity, and LTCC activity were required to mediate the iLTP-induced transcriptional repression of miR376c and for mediating the overall upregulation of α1 and γ2 protein levels. We also demonstrate that NFATc3 is a transcriptional repressor of the miR376c gene, as mutating both the predicted NFAT binding sites within miR376cProm-Luc increased transcriptional reporter activity following iLTP, while overexpression of NFATc3 reduced basal reporter activity. Moreover, GFP-NFATc3 translocates to the nucleus within the time frame required to induce transcriptional repression of miR376c following iLTP stimulation. Many E-T signaling pathways depend on LTCCs to communicate synaptic signaling in dendrites to transcriptional regulation in the nucleus (Deisseroth et al., 2003). This includes dendritic NMDAR activity to somatic CaN-NFAT, promoting NFAT translocation to the nucleus (Wild et al., 2019). Our experiments now place these critical regulators of E-T coupling within the context of iLTP stimulation and miR376c regulation, and introduce HDACs as an additional component required for NFAT-mediated transcriptional repression in neurons.
miR376c is located within the miR379–410 gene cluster, and miRNAs from this cluster are thought to be transcribed from a single promoter (Fiore et al., 2009). However, we find that miR376c is independently downregulated following iLTP stimulation (compared with other miRNAs within the miR379–410 gene cluster), indicating that in this context, miR376c is likely transcribed from an alternative and independent promoter. As the precise transcriptional start site of miR376c is unknown, we reasoned that the 500 bp immediately upstream of the pre-miR376c genomic sequence would likely contain a promoter region and dubbed it a ‘‘putative’’ promoter. Indeed, this region possesses promoter-like activity (Figure S6E) and contains functional NFAT binding sites. Further work is needed to fully characterize the miR376c promoter and define additional regulatory elements that may govern miR376c transcription. Here, we also reveal a mechanism whereby the acetylation state of the miR376c gene, determined by HDAC activity, leads to subsequent changes in α1 and γ2 expression. This suggests that iLTP stimulation can lead to de-acetylation of the miR376c gene, thus promoting a more condensed DNA structure that prevents miR376c transcription. As CaN inhibitors block miR376c gene de-acetylation, and NFAT-HDAC de-acetylation mechanisms of gene repression have been reported in non-neuronal cells (Baksh et al., 2002; Choo et al., 2009), we anticipate that NFATs and HDACs may also play a significant role in regulating activity-dependent gene repression in neurons. Future work will investigate these processes, as they may represent a more general gene repression mechanism with importance for many types of activity-dependent neuronal processes.
We experimentally demonstrate a role for NFAT in miRNA transcriptional regulation, opening up potential avenues by which NFAT signaling could broadly direct the expression levels of multiple groups of proteins in response to different types of neuronal activity. For instance, the same NMDAR stimulation simultaneously leads to iLTP, excitatory LTD, and the up- and downregulation of numerous miRNAs (Hu et al., 2014). Thus, we predict that NFAT signaling may function in both activating and repressing an array of miRNAs that could regulate and co-ordinate protein expression to maintain multiple modes of long-term plasticity.
In summary, we have defined a local, miRNA-dependent translational mechanism that increases de novo synthesis of GABAARs in dendrites, to maintain long-term surface GABAAR clustering during the expression of iLTP. Further characterization of miRNA pathways that regulate different types of inhibitory synaptic plasticity are now needed to paint a detailed picture of how translational control of GABAergic synaptic proteins can shape inhibition. Modulating miRNA levels may allow fine-tuning of E/I balance in neuronal circuits and potentially aid in restoring neuronal function in disorders associated with defects in neuronal inhibition.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Katharine R. Smith (katharine.r.smith@cuanschutz.edu).
Materials Availability
Plasmids generated in this study will be made available on request but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application
Data and Code Availability
This study did not generate any unique datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal procedures were conducted in accordance with National Institutes of Health (NIH)–United States Public Health Service guidelines and with the approval of the University of Colorado, Denver, Institutional Animal Care and Use Committee.
Dissociated Hippocampal Cultures
Rat hippocampal neurons were dissected from postnatal day 1–2 rats and cultures were prepared as previously described (Crosby et al., 2019). Briefly, the hippocampus was dissociated in papain. Neurons were seeded at a density of 150,000 on 18mm glass coverslips in 12 well plates or at 3, 000, 000 on 6cm dishes coated with poly-D-Lysine. Cells were maintained at 37°C, 5% CO2 for 14–18 days before being used for experiments. Neurons were cultured in Neurobasal media (GIBCO) supplemented with B27 (GIBCO) and 2 mM Glutamax. Half of the neuronal media was replaced with fresh media and anti-mitotics at DIV 5, followed by a subsequent media change at DIV12.
METHOD DETAILS
DNA constructs
pEGFP-FUGW was used as a GFP cell fill to distinguish between soma and dendrites in imaging experiments using anti-miRs, Puro-PLA and FISH. pAAV-miR-GFP control (containing a scrambled miRNA sequence) and pAAV-miR376c-GFP vectors were obtained from ABM (See key resource table for further details). GFP-NFATc3 and GFP-NFATc4 were generated as described previously (Wild et al., 2019). Gephyrin-FingR-GFP was purchased from Addgene. The 3′UTRs of GABRA1 and GABRG2 were created as gene fragments by Twist Bioscience and cloned into the SacI and MluI restriction sites of pMIR-REPORT (See key resource table for full list of primers used for cloning). miR376c seed sites were mutated via site directed mutagenesis. The miR376c promoter was also created as a gene fragment by Twist Bioscience and cloned into the SacI and HindIII sites of the promoter-less pGL4.10 firefly luciferase construct (Promega). NFAT binding sites were introduced into miR376cProm-WT-Luc via site directed mutagenesis.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Argonaute-2 (IP) | Millipore | RRID: AB_10695648 |
| Argonaute-2 (WB) | Cell Signaling | RRID:AB_2096291 |
| Digoxygenin | Jackson ImmunoResearch | RRID: AB_2339005 |
| GABAARα1 (PLA) | Synaptic systems | RRID: AB_2232180 |
| GABAARα1 (WB) | NeuroMab | RRID: AB_10697873 |
| GABAARα5 | NeuroMab | RRID: AB_2491075 |
| GABAARβ3 | NeuroMab | RRID: AB_10673389 |
| GABAARγ2 (Imaging) | Synaptic Systems | RRID: AB_10594245 |
| GABAARγ2 (WB) | NeuroMab | RRID: AB_2566822 |
| GAPDH | GeneTex | RRID: AB_11174761 |
| Gephyrin (Imaging) | Synaptic Systems | RRID: AB_887717 |
| Gephyrin (WB) | Synaptic Systems | RRID: AB_887719 |
| GFP | NeuroMab | RRID: AB_10671444 |
| GluA1 | Millipore | RRID: AB_2721164 |
| IgG control | Cell Signaling | RRID: AB_10829607 |
| LIMK1 | Cell Signaling 3842 | RRID: AB_2281332 |
| Puromycin | Kerafast | RRID: AB_2620162 |
| VGAT | Synaptic systems | RRID: AB_887869 |
| HRP conjugated goat anti-mouse | Biorad | RRID: AB_11125547 |
| HRP conjugated goat anti-rabbit | Biorad | RRID: AB_11125142 |
| anti-guinea pig Alexa Fluor-568 | Life Technologies | RRID: AB_141954 |
| anti-mouse AlexaFluor-488 | Life Technologies | RRID: AB_141607 |
| anti-rabbit AlexaFluor-647 | Life Technologies | RRID: AB_2536183 |
| Bacterial and Virus Strains | ||
| AAV-DJ-GFP-miRControl | This paper | N/A |
| AAV-DJ-GFP-miR376c | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Actinomycin D | Tocris | Cat #: 1229 |
| BAPTA-AM | Tocris | Cat #: 2787 |
| Cycloheximide | Sigma | Cat #: C7698 |
| Cyclosporin A | Tocris | Cat #: 1101 |
| CNQX | Tocris | Cat #: 1045 |
| FK506 | Tocris | Cat #: 3631 |
| NBQX | Tocris | Cat #: 0373 |
| Nimodipine | Tocris | Cat #: 0600 |
| NMDA | Tocris | Cat #: 0114 |
| Picrotoxin | Tocris | Cat #: 1128 |
| Puromycin | Tocris | Cat #: 4089 |
| TAT-CN21 | Gift from Dr. Ulli Bayer | N/A |
| Trichostatin | Tocris | Cat #: 1406 |
| TTX | Tocris | Cat #: 1078 |
| Critical Commercial Assays | ||
| Dual-luciferase reporter assay system | Promega | Cat #: E1910 |
| EpiQuik Acetyl-Histone H3 ChIP kit | EpiGentek | Cat #: P-2010 |
| miScript II RT kit | QIAGEN | Cat #: 218160 |
| RNeasy mini kit | QIAGEN | Cat #: 74104 |
| PIERCE Sulfho-NHS-LC-Biotin | ThermoFisher | Cat #: PG82075 |
| Pierce NeutrAvidin | ThermoFisher | Cat #: 29201 |
| PLA mouse/rabbit kit | Sigma | Cat #: DUO92101 |
| miRCURY LNA miRNA ISH kit | QIAGEN | Cat #: 339459 |
| Experimental Models: Cell Lines | ||
| HEK293T cells | ATCC | RRID:CVCL_0045 |
| Experimental Models: Organisms/Strains | ||
| Rat, Sprague Dawley Charles River | Charles Rivers | RRID: RGD_734476 |
| Oligonucleotides | ||
| Luc-GABRA1 3′UTR F: TATATACGAGCTCG TTCTTTTAGTCGTATTCTGTTG | This paper | N/A |
| Luc-GABRA1 3′UTR R: ATATATAAC GCGTGTACTCGAGTCTAATTGTCT | This paper | N/A |
| Luc-GABRG2 3′UTR F: TATATACG AGCTCGGAGGTTTGGGTTTTATCGA | This paper | N/A |
| Luc-GABRG2 3′UTR R: ATATATAAC GCGTCAGAGAGCAGCTGATGATTA | This paper | N/A |
| GABRA1 376c mut F: TATATACGAGCTCGT TCTTTTAGTCGTATTCTGTTG | This paper | N/A |
| GABRA1 376c mut R: ATATATAACGCG TGTACTCGAGTCTAATTGTCT | This paper | N/A |
| GABRG2 376c mut F: F:5′TATATACGAGCTC GGAGGTTTGGGTTTTATCGA | This paper | N/A |
| GABRG2 376c mut R: R:5′ATATATAACGCG TCAGAGAGCAGCTGATGATTA | This paper | N/A |
| Luc-miR376cPromF: TATATAGAGCTC AGTGCCCATGATGGGCTTT | This paper | N/A |
| Luc-miR376cPromR: TATATAAAGCTTA AGATCTAGTCCTGAAGA | This paper | N/A |
| Luc-miR376cPromMut1 F: ACAACCT GTGCTTGCGACAGCAAGGTTGT | This paper | N/A |
| Luc-miR376cProm Mut1 R: ACAACC TTGCTGTCGCAAGCACAGGTTGT | This paper | N/A |
| Luc-miR376cProm Mut2F: ACAGCAAG GTTGTGCGAGGAATCTGTGTC | This paper | N/A |
| Luc-miR376cProm Mut2R: GACACAGA TTCCTCGCACAACCTTGCTGT | This paper | N/A |
| qGABRA1 F: CGGCCATGGACTGGTTTAT | This paper | N/A |
| qGABRA1 R: GCATACCCTCTCTTGGTGAAA | This paper | N/A |
| qGABRA4 F: CAGACGGAAGATGGGCTATTT | This paper | N/A |
| qGABRA4 R: GGCTGGAACAGACTCCTTATTG | This paper | N/A |
| qGABRA5 F: GAATAGGGAGCCCGTGATAAA | This paper | N/A |
| qGABRA5 R: GACCTCTCCTCAGGTTCATTTC | This paper | N/A |
| qGABRA6 F: CTCTACCCAAAGTGTCCTATGC | This paper | N/A |
| qGABRA6 R: GGACTGGAGATTGGTGAAGTAG | This paper | N/A |
| qGABRB3 F: GGGTGTCCTTCTGGATCAATTA | This paper | N/A |
| qGABRB3 R: TCTCGAAGGTGAGTGTTGATG | This paper | N/A |
| qGABRG2 F: ACTTTACCATCCAGACCTACATTC | This paper | N/A |
| qGABRG2 R: GCAGGGACAGCATCCTTATT | This paper | N/A |
| qGRIA1 F: GCCAGATCGTGAAGCTAGAAA | This paper | N/A |
| qGRIA1 R: CTCCGCTCTCCTTGAACTTATT | This paper | N/A |
| qpmirrepLucF: TGAAGAAGTGTTCGTCTTCG | This paper | N/A |
| qpmirrepLucR: TATGTCCGGTTATGTAAACA | This paper | N/A |
| qmiR376cF: AACATAGAGGAAATTTCACGT | This paper | N/A |
| qmiR379F: TGGTAGACTAtGGAACGTAGG | This paper | N/A |
| qmiR410F: AATATAACACAGAtGGCCTGT | This paper | N/A |
| qmiR29aF: ACTGATTTCTTTTGGTGTTCAG | This paper | N/A |
| qPri-376cF: TCTCTTCAGGACTAGATCTT | This paper | N/A |
| qPri-376cR: TTGACACTGAAAACGTG | This paper | N/A |
| qPri-379F: TTGTGCCTTCATGGGCAGT | This paper | N/A |
| qPri-379R: ATACTGAGAGTTAGTGGACC | This paper | N/A |
| qPri-410F: TGCTCCGGTCAACACTGGGT | This paper | N/A |
| qPri-410R: AAAACAGGCCATCTGTGTTA | This paper | N/A |
| qmiR376cPromF: TGGACAAGGGAGAGATGGA | This paper | N/A |
| qmiR376cPromF: ATGAAGAAGACGGCAGACAG | This paper | N/A |
| Recombinant DNA | ||
| pAAV-miR-GFP control | ABM | Cat #: AM00100 |
| pAAV-miR376c-GFP | ABM | Cat #: Amr107900 |
| GFP-NFATc3 | Wild et.al 2019 | N/A |
| GFP-NFATc4 | Wild et. al 2019 | N/A |
| Gephyrin-FingR-GFP | Gross et al., 2013 | Addgene #: 46296 |
| pMIR-REPORT-Luc-GABRA1 | This paper | N/A |
| pMIR-REPORT-Luc-GABRG2 | This paper | N/A |
| pGL4.10-FL-miR376cProm-WT-Luc | This study | N/A |
| Software and Algorithms | ||
| Prism 7 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagej.nih.gov/ij |
| other | ||
| Immobilon Classico Western HRP substrate | Millipore | Cat #: WBLU0100 |
| Immobilon Crescendo Western HRP substrate | Millipore | Cat #: WBLUR0500 |
| Lipofectamine 2000 | Thermofischer | Cat #: 11668027 |
| Protein G Sepharose | Sigma | Cat #: P3296 |
| Biotin solution | PIERCE | N/A |
| miRDIAN Control inhibitor | Horizon | Cat #: IN-001005–01-05 |
| miRDIAN miR376c inhibitor | Horizon | Cat #: IH-320471–04-0002 |
| miRDIAN miR134 inhibitor | Horizon | Cat #: IH-320365–06-0002 |
| LNA miR376c Double DIG probe: AACATAGAGGAAATTTCACGT | QIAGEN | Cat #: 339111 YD00610516-BCG |
| LNA G1CDS Double DIG probe: TAGCTGGTTGCTGTAGGAGCA | QIAGEN | Cat #: 339500 |
| LNA G1L Double DIG probe: TCCTCTATGAGAGTGTATCTT | QIAGEN | Cat #: 339500 |
Drug treatments
Chemical iLTP was induced in DIV14–18 hippocampal neurons via bath application of 20 μM NMDA and 10 μM CNQX in HBS solution (145 mM NaCl, 2 mM KCl, 10 mM HEPES, 2 mM CaCl2, 2 mM MgCl2 10 mM glucose, pH 7.4) for 2 min at 37°C as previously described (Petrini et al., 2014). Neurons were harvested or fixed at specific time points post-stimulation as stated in the figures and/or figure legends. Cycloheximide was supplemented into the bath media and conditioned media at 10 μg/ml and left in the culture media until the neurons were ready to be harvested or fixed. Cyclosporin A (Tocris), FK506 (Tocris) and TAT-CN21 (kind gift from Dr. Ulli Bayer) were used at 5 μM and added to cell culture media 15 minutes prior to iLTP induction and left in the culture media following stimulation until the neurons were ready to be harvested. Likewise, BAPTA-AM (Tocris) was used at 20 μM and Nimodipine at 10 μM. Neurons were treated with Trichostatin A (TSA) for 16 hours at 1 μM prior to iLTP induction.
AAV production and infections
AAVs were produced as previously described (Aoto et al., 2013). Briefly, HEK293T cells (obtained from ATCC and maintained under standard conditions: 10% FBS in DMEM, 37°C, 5% CO2) were co-transfected with the pAAV vector and helper plasmids (pDJ and pHelper) using calcium phosphate transfection. 72 h post-transfection, HEK293T cells were harvested and subject to iodixanol gradient (15%, 25%, 40% and 60%) column ultracentrifugation (2 h at 63,500 rpm in a Beckman Type80Ti rotor). Viral particles in the 40% layer were purified through Amicon Ultra-15 columns (Millipore) into a volume of < 300μl. Viral particles were aliquoted and stored at −80°C. Neurons were infected 4 days prior to experiments.
Transfections
DIV 12–14 hippocampal neurons were transfected with plasmid DNA and/or miRDIAN miRNA inhibitors (Horizon; See key resource table for further information) using Lipofectamine 2000 (Invitrogen) and used for experiments at DIV 14–16.
Immunofluorescence microscopy
For surface GABAARγ2 and GluA1 staining, hippocampal neurons grown on coverslips were fixed in 4% paraformaldehyde supplemented with 4% sucrose in PBS at RT for 5 minutes. Next, the coverslips were incubated in blocking solution (3% BSA, 2% normal goat serum in PBS) for 1 hour at RT and surface stained for GABAARγ2 (1:500 Synaptic Systems 224004) or GluA1 (1:200 Millipore ABN241) in blocking solution for 1 hour at RT. Following 3 times 5 minute washes in PBS, the coverslips were incubated for 45 minutes with the appropriate secondary antibodies (1:2000 ThermoFisher Alexa- Fluor 488, 568, 647) before being mounted onto microscope slides using ProLong gold mounting media (ThermoFisher). When necessary, neurons were permeabilized in 0.5% NP-40 for 2 minutes and blocked for 40 minutes prior to Gephyrin (1:500 Synaptic Systems 147011) or VGAT staining (1:1000 Synaptic systems 131003) in blocking solution for 1 hour at RT. Widefield images were acquired using an Axiovert200M microscope (Carl Zeiss) with a 63X plan-apo/1.4 numerical aperture oil objective, 300W Xenon illumination (Sutter Instruments), Coolsnap-HQ2 charge-couples device (CCD) camera, and Slidebook 6.0. Confocal images were acquired using a Zeiss Observer.Z1 inverted microscope equipped with a Yokogawa CSU-X1 spinning disk unit; an EC Plan- Neofluar 40x (1.30 NA) oil immersion objective lens; a Photometrics PVCAM (Dynamic) EMCCD camera with 16-bit dynamic range; and SlideBook 6.0 software. Images were acquired using 2 μm Z stacks of xy planes at 0.2 μm intervals. For widefield images, planes were deblurred using nearest neighbor deconvolution and maximum intensity projected into 2D images. The cluster area and density of surface receptors, gephyrin and VGAT were analyzed in ImageJ on randomly selected dendrites. Cluster analysis was analyzed based on a minimum cluster area threshold of 0.8 μm2. At least three independent experiments were performed, and statistical significance was determined using either t tests or ANOVA, as appropriate.
Fluorescent in situ hybridization (FISH)
RNA FISH was performed using the miRCURY LNA miRNA ISH kit (Exiqon/Qaigen) and double labeled 5′ and 3′digoxygenin probes designed by Exiqon/Qaigen (See key resource table for LNA FISH sequences; a control sequence probe was included with the ISH kit). PFA fixed neurons on coverslips were treated with Proteinase K for 2 minutes at 37°C followed by 3 stringent washes in PBS. 10 μM stock probes were heat denatured at 94°C for 4 minutes and 1 μL of denatured stock was diluted in 999 μL of 1x hybridization buffer and incubated with the neurons at 62°C for 1 hour. The coverslips were then stringently washed at 62°C in 5x saline-sodium citrate (SSC) for 5 minutes, 2 times 1x SSC for 5 minutes each and 2 times 0.2x SSC for 5 minutes each. The coverslips were finally washed in 0.2x SSC for 5 minutes at RT and were then subject to the IF protocol described above. FISH probes were detected with anti-digoxygenin antibody (1:200 Jackson ImmunoResearch 200–002-156). Mean fluorescence signals in the soma and dendrites were determined by ImageJ. At least three independent experiments were performed, and statistical significance was determined using either t tests or ANOVA, as appropriate.
Puromycin-Proximity Ligation Assays (Puro-PLA)
Neurons were treated with 1 μM puromycin for 10 minutes prior to fixation in 4% PFA for 5 minutes. Puro-PLA was performed using the Duolink in situ red PLA mouse/rabbit kit (Sigma) as previously described (Rajgor et al., 2018). Anti-puromycin (Kerafast 3RH11) and anti-GABAARα1 (Synaptic systems 224203) were used at 1:500. The number of PLA positive puncta in dendrites per 10 μm and number of puncta per μM2 area of somatic space was quantified in ImageJ by puncta counting or integrated density as shown in Figure 5. At least three independent experiments were performed, and statistical significance was determined using ANOVA.
Live Imaging
Hippocampal neurons expressing GFP-NFATc3 were imaged pre-stimulation and 10 minutes post-iLTP induction. Live imaging was performed on a Zeiss Observer.Z1 inverted microscope equipped with a Yokogawa CSU-X1 spinning disk unit; an EC Plan- Neofluar 40x (1.30 NA) oil immersion objective lens; a Photometrics PVCAM (Dynamic) EMCCD camera with 16-bit dynamic range; and SlideBook 6.0 software. Images were acquired using 2 μm Z stacks of xy planes at 0.2 mm intervals. Nuclear GFP-NFATc3 levels were normalized to cytoplasmic levels in ImageJ. Five independent experiments were performed, and statistical significance was determined using paired t tests.
Whole cell electrophysiology
DIV 14–16 pyramidal hippocampal neurons were held at −70 mV and recorded from using an intracellular solution containing:130mM cesium methanesulfonate, 3mM Na2ATP, 0.5mM Na3GTP, 0.5mM EGTA, 10mM phosphocreatine, 5mM MgCl2, 2.5mM NaCl, 10mM HEPES (290–300 mOsm) and extracellular solution containing: 130mM NaCl, 5mM KCl, 2mM CaCl2, 1mM MgCl2, 10mM HEPES, 20mM Glucose (pH 7.4). mIPSCs were isolated using 10 μM NBQX and 0.5 μM TTX (Tocris) and mEPSCs were isolated using 50 μM picrotoxin (Tocris) and 0.5 μM TTX extracellularly. For the iLTP experiments, mIPSCs were recorded for ~4 minutes under basal conditions from multiple different GFP-expressing neurons and the average amplitude and frequency were used as ‘control values’. For the iLTP condition, neurons were stimulated and 20 minutes later, mIPSCs were recorded for a 60 min time-period from different infected pyramidal neurons in the same experimental culture. mIPSC parameters were steady state throughout this phase of iLTP and the average amplitude and frequencies across the recording window were used as ‘iLTP’ values. Data were collected using a Digidata 1440A with Axopatch 200B amplifier (Molecular Devices). All data were acquired with pCLAMP software and analyzed in Clampfit 10. A template for the detection of clean and stable mIPSC traces was created in Clampfit10. All traces were manually reviewed prior to analysis. The majority of events were > 5 pA. The decay and rise times were calculated at 10%–90% of the event. The charge transfer was calculated as the readout of area from Clampfit10 which was in pA per ms. At least three independent experiments were performed, and statistical significance was determined using either t tests or ANOVA, as appropriate. All error bars on graphs represent standard error of the mean.
RNA isolation and qRT-PCR
RNA was isolated from neurons using the RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. mRNA and miRNA were reverse transcribed into cDNA using miScript II RT kit (QIAGEN) according to the manufacturer instructions. cDNA was diluted 1/10 in RNAase free water and 1 μL was used per qPCR reaction when measuring mRNA and mature miRNA levels. 1 μL of undiluted cDNA was used when measuring levels of pri-miRNAs. qPCR was performed using gene specific primers or miRNA specific primers with the universal primer provided with the miScript II RT kit in a Biorad CFX384 real time qPCR system. See key resource table for list of primers used for qPCR. All qPCR readings were normalized to the U6 snRNA which was used as a house keeping gene. qPCR primers for U6 snRNA were included with the miScript II RT kit (QIAGEN). Ago2 bound mRNAs were normalized to their respective inputs. qPCR cycling parameters were: 94°C 15min, 55°C 30sec, 70°C 30 s for 40 cycles. At least three independent experiments were performed, and statistical significance was determined using either t tests or ANOVA, as appropriate.
Ago2 RNA-IP
Hippocampal neurons were lysed in 1ml lysis buffer (10mM HEPES pH7.4, 200mM NaCl, 30mM EDTA, 0.5% Triton X-100, 0.5 U/μl SUPERase inhibitor, 1x Complete Protease Inhibitor) and briefly sonicated for 10 s. Cells were left on ice for 10 minutes and the cell debris was pelleted by centrifugation at 13 000 rpm for 10 minutes at 4°C. 50μl was taken as protein input. The remaining lysate was pre-cleared with 100μl or Protein G Sepharose beads (Sigma) for 30 minutes at 4°C. The beads were pelleted and the remaining lysate was equally divided and incubated with 2 μg of Pan-Ago (Millipore; 2A8) or IgG (Cell Signaling 5415) for 1 hour at 4°C. Antibody complexes were captured by incubating with beads for 1 hour at 4°C followed by 5 washes in 1 mL of lysis buffer. 1 quarter of the purified complex was boiled in protein loading buffer and Ago2 enrichment was analyzed by western blotting as described below. The remaining IP was subject to RNA isolation as described above. At least three independent experiments were performed, and statistical significance was determined using t tests.
Western blotting
Whole-cell lysates were prepared by scraping cells in 2X protein loading buffer (4% SDS, 20% glycerol, 120mM Tris pH6.8, 0.02% bromophenol blue, 5% 2-mercaptoethanol) and boiling extracts at 95°C for 5 minutes. Proteins were resolved by SDS–PAGE and transferred to PVDF membrane using a wet transfer apparatus and blocked in 5% milk solution made up in PBS–Tween. The membranes were blotted with the appropriate primary antibody overnight at 4°C: Ago2 (1:1000 Millipore 2A8), GABAARα1 (1:1000 NeuroMab 75136), GABAARα5 (1:1000 NeuroMab 455510), GABAARβ3 (1:1000 NeuroMab 75149), GABAARγ2 (1:1000 NeuroMab 75442), GAPDH (1:10 000 GeneTex 627408), Gephyrin (1:5000 Synaptic Systems 147111), GFP (1:2000 NeuroMab 75131), GluA1 (1:1000 Millipore ABN241), LIMK1 (1:1000 Cell Signaling 3842). Membranes were washed in PBS-Tween and incubated with the appropriate HRP conjugated secondary antibodies for 1 hour at RT (1:10 000 Millipore). Protein bands were visualized using ECL western blotting substrates (Millipore). Where appropriate, the membranes were stripped with stripping buffer and re-probed. For densitometry measurements, western blot images were analyzed in ImageJ. The integrated densities of bands of interest were normalized to GAPDH from the same gel. At least three independent experiments were performed, and statistical significance was determined using either t tests or ANOVA, as appropriate. All error bars on graphs represent standard error of the mean.
Surface biotinylations
Surface biotinylations were performed as previously described (Smith et al., 2014). Briefly, DIV 14–16 hippocampal neurons were incubated on ice with biotin solution (Pierce Sulfho-NHS-LC-Biotin) at 0.5 mg/ml in PBS containing Ca2+ and Mg2+) and quenched with quench buffer (1 mg/ml BSA in PBS containing Ca2+/Mg2+. Neurons were solubilized for 1 hour in RIPA buffer (50 mM Tris pH 7.5, 1 mM EDTA, 2 mM EGTA, 150 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50mM NaF, and 1mM Na3VO4) and neuronal debris was pelleted from lysates by centrifugation at 12000 rpm for 15 minutes at 4°C. 15% of the supernatant was taken to use as a total protein input and the remainder was incubated for 2h with 25 μL Ultralink immobilized Pierce NeutrAvidin 50% slurry at 4°C to precipitate biotin labeled membrane proteins. Beads were washed three times in RIPA buffer and analyzed by western blotting. Surface levels were normalized to their respective inputs. Statistical significance was determined using ANOVA.
Luciferase assays
DIV12–14 hippocampal neurons or HEK293T cells plated in 12 well dishes were co-transfected with 500 μg of the appropriate Firefly luciferase reporter construct and 500 μg of Renilla. 500 μg of GFP, GFP-NFATc3, GFP-NFATc4, miRCon or miR376c, or 1 μL of stock miRNA inhibitor were co-transfected when appropriate. The dual-luciferase reporter assay system (Promega) was used to perform the assays according to the manufacturer’s instructions. Firefly values were normalized to Renilla values for each of at least 3 independent experiments, and statistical significance was determined by ANOVA.
Acetyl-Histone H3 chromatin immunoprecipitation (ChIP)
Acetyl- Histone H3 ChIP was performed using the EpiQuik Acetyl-Histone H3 ChIP kit (EpiGentek) according to the manufacturer’s instructions. The eluted DNA from the IP was normalized to the input DNA using qPCR with a forward primer (5′ TGGACAAGGGAGAGATGGA3′) and reverse primer (5′ ATGAAGAAGACGGCAGACAG3′) targeting the putative miR376c promoter. At least three independent experiments were performed, and statistical significance was determined using ANOVA.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests were performed in Prism7 (GraphPad) using t test or ANOVA, Bonferroni post hoc test. All experiments were performed at least 3 times to ensure rigour and reproducibility. The exact number of repeats and statistical details of experiments can be found in the figure legends. P values were considered significant if < 0.05. All graphs show SEM.
Supplementary Material
Highlights.
miR376c regulates GABAergic synapses by controlling GABAAR translation
Reduced miR367c-mediated GABAAR translational repression is required for iLTP
De novo GABAARs are synthesized specifically in dendrites during iLTP
A CaN-NFAT-HDAC axis represses miR376c transcription leading to GABAAR translation
ACKNOWLEDGMENTS
We are grateful to Dr. Brooke Sinnen for help with making adeno-associated viruses (AAVs), Dr. Joshua Black for sharing equipment, Dr. Ulli Bayer for providing CN21, and Sara Gookin for technical support. We thank Drs. Kevin Crosby and Matthew Kennedy for critical reading of the manuscript. This work was supported by a Brain and Behavior Research Foundation NARSAD Young Investigator Award, a Brain Research Foundation Seed Grant, an AHA Career Development Award, NIH/National Institute of Mental Health (NIMH) grant R01MH119154 to K.R.S., and NIH/National Institute of Neurological Disorders and Stroke (NINDS) grant R01NS040701 and NIH/NIMH grant R01MH102338 to M.L.D. J.D.G. was supported by grant T32GM763540 and an AHA Predoctoral Fellowship. T.M.W. was supported by T32 NS099042. A.M.P was supported by NIH/NINDS grant F31NS096815.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107785.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, and Südhof TC (2013). Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Widlund HR, Frazer-Abel AA, Du J, Fosmire S, Fisher DE, DeCaprio JA, Modiano JF, and Burakoff SJ (2002). NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10, 1071–1081. [DOI] [PubMed] [Google Scholar]
- Barbado M, Fablet K, Ronjat M, and De Waard M (2009). Gene regulation by voltage-dependent calcium channels. Biochim. Biophys. Acta 1793, 1096–1104. [DOI] [PubMed] [Google Scholar]
- Barberis A (2020). Postsynaptic plasticity of GABAergic synapses. Neuropharmacology 169, 107643. [DOI] [PubMed] [Google Scholar]
- Biever A, Donlin-Asp PG, and Schuman EM (2019). Local translation in neuronal processes. Curr. Opin. Neurobiol 57, 141–148. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Will T, and Schuman EM (2010). Protein homeostasis and synaptic plasticity. EMBO J 29, 2746–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, and Schuman EM (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Chiu CQ, and Carroll RC (2011). Long-term plasticity at inhibitory synapses. Curr. Opin. Neurobiol 21, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Martenson JS, Yamazaki M, Natsume R, Sakimura K, Tomita S, Tavalin SJ, and Higley MJ (2018). Input-specific NMDAR-dependent potentiation of dendritic GABAergic inhibition. Neuron 97, 368–377.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Barberis A, and Higley MJ (2019). Preserving the balance: diverse forms of long-term GABAergic synaptic plasticity. Nat. Rev. Neurosci 20, 272–281. [DOI] [PubMed] [Google Scholar]
- Choo MK, Yeo H, and Zayzafoon M (2009). NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone 45, 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby KC, Gookin SE, Garcia JD, Hahm KM, Dell’Acqua ML, and Smith KR (2019). Nanoscale subsynaptic domains underlie the organization of the inhibitory synapse. Cell Rep 26, 3284–3297.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Mermelstein PG, Xia H, and Tsien RW (2003). Signaling from synapse to nucleus: the logic behind the mechanisms. Curr. Opin. Neurobiol 13, 354–365. [DOI] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, and Schratt G (2009). Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J 28, 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, et al. (2013). Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu QH, Yu D, Hu Z, Liu X, Yang Y, Luo Y, Zhu J, and Li Z (2015). miR-26a and miR-384–5p are required for LTP maintenance and spine enlargement. Nat. Commun 6, 6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenston AM, and Bading H (2011). Calcium signaling in synapse-to-nucleus communication. Cold Spring Harb. Perspect. Biol 3, a004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, and Li Z (2017). miRNAs in synapse development and synaptic plasticity. Curr. Opin. Neurobiol 45, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yu D, Gu QH, Yang Y, Tu K, Zhu J, and Li Z (2014). miR-191 and miR-135 are required for long-lasting spine remodelling associated with synaptic long-term depression. Nat. Commun 5, 3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Zhao J, Hu T, Luo Y, Zhu J, and Li Z (2015). miR-501–3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J. Cell Biol 208, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki J, Kikuchi K, Mizuguchi Y, Kawahigashi Y, Yoshida H, Uchida E, and Takizawa T (2013). MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholan-giocarcinoma cell line. PLoS ONE 8, e69496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Keith KJ, Ilyas A, and Kapur J (2013). GABAA receptor membrane insertion rates are specified by their subunit composition. Mol. Cell. Neurosci 56, 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipanyula MJ, Kimaro WH, and Seke Etet PF (2016). The emerging roles of the calcineurin-nuclear factor of activated T-lymphocytes pathway in nervous system functions and diseases. J. Aging Res 2016, 5081021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, and Somogyi P (2008). Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. (2010). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, and Burge CB (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, and Kilpatrick CL (2011). GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron 70, 385–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden KC, Beattie JB, Friedenthal J, and Carroll RC (2007). NMDA receptor activation potentiates inhibitory transmission through GABA receptor-associated protein-dependent exocytosis of GABA(A) receptors. J. Neurosci 27, 14326–14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E, and Van Vactor D (2012). MicroRNAs shape the neuronal landscape. Neuron 75, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JG, Sanderson JL, Gorski JA, Scott JD, Catterall WA, Sather WA, and Dell’Acqua ML (2014). AKAP-anchored PKA maintains neuronal L-type calcium channel activity and NFAT transcriptional signaling. Cell Rep 7, 1577–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser Z, Cull-Candy S, and Farrant M (1997). Differences in synaptic GABA(A) receptor number underlie variation in GABA mini amplitude. Neuron 19, 697–709. [DOI] [PubMed] [Google Scholar]
- Olde Loohuis NF, Ba W, Stoerchel PH, Kos A, Jager A, Schratt G, Martens GJ, van Bokhoven H, Nadif Kasri N, and Aschrafi A (2015). MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Rep 11, 1876–1884. [DOI] [PubMed] [Google Scholar]
- Petrini EM, Ravasenga T, Hausrat TJ, Iurilli G, Olcese U, Racine V, Sibarita JB, Jacob TC, Moss SJ, Benfenati F, et al. (2014). Synaptic recruitment of gephyrin regulates surface GABAA receptor dynamics for the expression of inhibitory LTP. Nat. Commun 5, 3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, and Sperk G (2000). GABA(A) receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience 101, 815–850. [DOI] [PubMed] [Google Scholar]
- Pratt AJ, and MacRae IJ (2009). The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem 284, 17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D (2018). Macro roles for microRNAs in neurodegenerative diseases. Noncoding RNA Res 3, 154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Sanderson TM, Amici M, Collingridge GL, and Hanley JG (2018). NMDAR-dependent Argonaute 2 phosphorylation regulates miRNA activity and dendritic spine plasticity. EMBO J 37, e97943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Tom Dieck S, and Schuman EM (2017). Local translation in neuronal compartments: how local is local? EMBO Rep 18, 693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegger S, and Großhans H (2012). MicroRNA turnover: when, how, and why. Trends Biochem. Sci 37, 436–446. [DOI] [PubMed] [Google Scholar]
- Saliba RS, Kretschmannova K, and Moss SJ (2012). Activity-dependent phosphorylation of GABAA receptors regulates receptor insertion and tonic current. EMBO J 31, 2937–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan S, Akbalik G, Kochen L, Rinne J, Kahlstatt J, Glock C, Tushev G, Alvarez-Castelao B, Heckel A, and Schuman EM (2017). Activity-dependent spatially localized miRNA maturation in neuronal dendrites. Science 355, 634–637. [DOI] [PubMed] [Google Scholar]
- Schieweck R, and Kiebler MA (2019). Posttranscriptional gene regulation of the GABA receptor to control neuronal inhibition. Front. Mol. Neurosci 12, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G (2009). MicroRNAs at the synapse. Nat. Rev. Neurosci 10, 842–849. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, and Greenberg ME (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439, 283–289. [DOI] [PubMed] [Google Scholar]
- Sethi P, and Lukiw WJ (2009). Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett 459, 100–104. [DOI] [PubMed] [Google Scholar]
- Smith KR, and Kittler JT (2010). The cell biology of synaptic inhibition in health and disease. Curr. Opin. Neurobiol 20, 550–556. [DOI] [PubMed] [Google Scholar]
- Smith KR, Davenport EC, Wei J, Li X, Pathania M, Vaccaro V, Yan Z, and Kittler JT (2014). GIT1 and bPIX are essential for GABA(A) receptor synaptic stability and inhibitory neurotransmission. Cell Rep 9, 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KT, Gross C, and Bassell GJ (2018). MicroRNAs sculpt neuronal communication in a tight balance that is lost in neurological disease. Front. Mol. Neurosci 11, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay R, Lee S, and Rudy B (2016). GABAergic interneurons in the neocortex: from cellular properties to circuits. Neuron 91, 260–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich JD, Kim MS, Houlihan PR, Shutov LP, Mohapatra DP, Strack S, and Usachev YM (2012). Distinct activation properties of the nuclear factor of activated T-cells (NFAT) isoforms NFATc3 and NFATc4 in neurons. J. Biol. Chem 287, 37594–37609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kwon EJ, and Tsai LH (2012). MicroRNAs in learning, memory, and neurological diseases. Learn. Mem 19, 359–368. [DOI] [PubMed] [Google Scholar]
- Wild AR, Sinnen BL, Dittmer PJ, Kennedy MJ, Sather WA, and Dell’Acqua ML (2019). Synapse-to-nucleus communication through NFAT is mediated by L-type Ca(2+) channel Ca(2+) spike propagation to the soma. Cell Rep 26, 3537–3550.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, and Wang X (2015). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43, D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, Avni D, and Leibowitz-Amit R (2012). Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol. Cancer 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.