Abstract
Following advancements in the field of genotoxicology, it has become widely accepted that 3D models are not only more physiologically relevant but also have the capacity to elucidate more complex biological processes that standard 2D monocultures are unable to. Whilst 3D liver models have been developed to evaluate the short-term genotoxicity of chemicals, the aim of this study was to develop a 3D model that could be used with the regulatory accepted in vitro micronucleus (MN) following low-dose, longer-term (5 days) exposure to engineered nanomaterials (ENMs). A comparison study was carried out between advanced models generated from two commonly used liver cell lines, namely HepaRG and HepG2, in spheroid format. While both spheroid systems displayed good liver functionality and viability over 14 days, the HepaRG spheroids lacked the capacity to actively proliferate and, therefore, were considered unsuitable for use with the MN assay. This study further demonstrated the efficacy of the in vitro 3D HepG2 model to be used for short-term (24 h) exposures to genotoxic chemicals, aflatoxin B1 (AFB1) and methyl-methanesulfonate (MMS). The 3D HepG2 liver spheroids were shown to be more sensitive to DNA damage induced by AFB1 and MMS when compared to the HepG2 2D monoculture. This 3D model was further developed to allow for longer-term (5 day) ENM exposure. Four days after seeding, HepG2 spheroids were exposed to Zinc Oxide ENM (0–2 µg/ml) for 5 days and assessed using both the cytokinesis-block MN (CBMN) version of the MN assay and the mononuclear MN assay. Following a 5-day exposure, differences in MN frequency were observed between the CBMN and mononuclear MN assay, demonstrating that DNA damage induced within the first few cell cycles is distributed across the mononucleated cell population. Together, this study demonstrates the necessity to adapt the MN assay accordingly, to allow for the accurate assessment of genotoxicity following longer-term, low-dose ENM exposure.
Introduction
In recent years, 3D in vitro liver models have become increasingly useful tools for the genotoxic assessment of hazardous materials as they more accurately represent the physiological environment of the liver (1–5). It is widely accepted that the traditional in vitro 2D monocultures poorly represent the intricacies of the liver cells metabolic activity in vivo and, therefore, are limited in their ability to elucidate complex biological processes (5–7). It has been shown that the expression of liver-specific enzymes and drug metabolism in primary human hepatocytes (albumin and cytochrome P450) were at much lower levels in 2D culture (8,9). Recent advances in tissue engineering allow researchers to better simulate the liver environment using 3D cultures, providing a valuable tool for advancing the study of hepatotoxicity. At the International Workshop on Genotoxicity Testing (IWGT) it was recognised that 3D models, including those representing the liver, were deemed more physiologically relevant than the standard 2D monocultures (1).
Primary 2D human hepatocytes have been widely used in hepatotoxicity; however, they are not without limitations; for example, phenotypic variation between donors, rapid dedifferentiation and common loss of liver functionality (as indicated by reduced albumin and urea production) (7,10). The more physiologically relevant primary hepatocyte 3D liver model outperforms the standard 2D in vitro system, demonstrating long-term stability and the ability to maintain similar in vivo liver functionality (10). Primary hepatocyte 3D models have been reported to be efficacious in assessing longer-term toxicity using a variety of endpoints, such as cytotoxicity, cytokine production and DNA damage induction, following repeated exposure to engineered nanomaterials (ENMs) (11). Whilst 3D primary hepatocytes are relevant for longer-term toxicity testing, they are not suitable for use with the regulatory accepted micronucleus (MN) assay. The success of this assay relies on cellular proliferation; however, many primary or differentiated 3D hepatic models are static (12).
Genotoxicity assessment involves the need for a battery of tests that are capable of quantifying the induction of point mutation, structural chromosomal damage and numerical changes in chromosome number. The key in vitro assays utilised to evaluate these endpoints include the bacterial reverse mutation test, the MN assay and mammalian cell gene mutation tests (e.g. using the HPRT and xprt genes). Other DNA damage reporter assays that are used for screening purposes include the comet assay, gamma-H2AX staining and, in more recent years, the use of ‘-omics’ technologies, that is, transcriptomics have been applied to screen for modifications and alternations in gene expression profiles following DNA damage induction. The MN assay is a reliable technique that measures fixed chromosomal damage demonstrated by the frequency of MN in cells that have undergone cell division, which results from aneugenic or clastogenic damage (13) and is, therefore, recommended as the ‘benchmark’ technique for assessing DNA damage and genotoxicity in vitro (14,15). The MN assay has previously been adapted to accommodate advanced 3D tissue culture models for the assessment of hazardous materials. For example, Curren et al. successfully modified the traditional MN assay for use with 3D EpiDerm Skin model, measuring genotoxicity using the reconstructed skin MN (RSMN) assay following chemical exposures for up to 72 h (16). Similarly, Wills et al. have also adapted the in vitro MN assay for genotoxic assessment of the EpiDerm Skin model and TK6 cells following 24 h nanoparticle exposure (17). Shah et al. have recently developed a cost-effective, low-maintenance 3D HepG2 liver model that has been shown to maintain liver functionality and can be used for genotoxicity assessment using the cytokinesis-block MN assay (CBMN). The use and applicability of this 3D liver genotoxicity approach has been demonstrated with the chemicals benzo[a]pyrene (BaP) and 2-amino-1-methyl-6-phenylimidazo(4,5-b) pyridine (PhIP), where interestingly, the 3D liver spheroid system was more sensitive to the induction of damage induced by these pro-genotoxins than the comparative 2D HepG2 monocultures (3). The reason for this difference was the substantially higher expression levels of metabolic enzymes in the 3D liver spheroid models than in the 2D HepG2 cultured cells. Thus, the capability of the 3D models to more efficiently metabolise the BaP and PhIP exposures into their genotoxic metabolites resulted in greater genotoxicity being reported in the more complex culture system than when the standard CBMN assay was applied in 2D cell cultures.
The efficacy of the MN assay in 3D culture systems following exposure to ENMs must, however, also be considered, as it has long been recognised that the Organisation for Economic Co-operation and Development (OECD) test guidelines, which were primarily developed for chemicals, are not wholly appropriate for ENMs (13). For example, the CBMN assay requires the addition of cytochalasin B (Cyto B), a cytostatic agent that will halt cell division after one cell cycle and block cytokinesis, therefore resulting in the formation of clearly identifiable bi-nucleated cells (15). When exposing cells to chemicals, Cyto B can be added during or after the addition of the chemical. However, for ENM exposures, co-treatment of Cyto B with ENM has been shown to prevent cellular uptake of ENM; therefore, it is critical that Cyto B is added post-ENM exposure (13,18). More recently, a review of published data resulted in a series of recommendations on the genotoxic assessment of ENM (19). When evaluating the MN assay, of the 36 studies that satisfied the inclusion criteria, the conclusion was that there was a large degree of variation amongst assay approach and that standardisation of the MN assay to evaluate ENM is required (19). The MN assay is routinely used to evaluate genotoxicity associated with chemical compounds. However, unlike chemical compounds which often can have short half-lives, many ENMs are biopersistent. They, therefore, do not always readily breakdown in biological systems and can result in a gradual intracellular accumulation with prolonged and/or repeated exposures (18).To further develop our understanding and accurately assess the impact of more realistic longer-term, low-dose ENM exposure using the MN assay, it is necessary to have biological test systems and assays that facilitate this. The aim of this study, therefore, was to develop an advanced 3D liver spheroid model that could be used with the MN assay for genotoxicity assessment following longer-term exposure to test agents at low doses that are more representative to an in vivo exposure system. This will be achieved via the modification of the protocol previously described in Shah et al. (3) to sustain extended culture periods of HepG2 and HepaRG liver spheroids in vitro while maintaining liver functionality and also adapt the approach to support the analysis of fixed DNA damage using the MN assay following longer-term ENM exposures.
Materials and Methods
Chemicals
Aflatoxin B1 (AFB1; Sigma Aldrich, UK), Cyto B and methyl-methanesulfonate (MMS; Sigma Aldrich, UK) were prepared according to manufacturer’s instructions. Stock solutions of AFB1 (3 mM), MMS (1 mM) and Cyto B (1.5 mg/ml) were prepared in DMSO and stored at −20°C. ZnO ENM (JRC Nanomaterials Repository, Belgium) stock solutions (2.56 mg/ml) were prepared and dispersed as per the NanoGenoTox Dispersion Protocol (grant agreement no. 20092101, 2018) (20). Working stocks of both chemicals and ENMs were subsequently made fresh for each experiment.
Cell culture and maintenance
The human Caucasian hepatocellular carcinoma-derived epithelial cell line HepG2 (ECACC 85011430) was cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/l D-glucose and L-glutamine (GIBCO, Paisley, UK) supplemented with 10% foetal bovine serum and 1% penicillin/streptomycin (GIBCO, Paisley, UK). HepG2 cells were sub-cultured with trypsin/EDTA (0.05%) solution (GIBCO, Paisley, UK). HepG2 cells were sub-cultured every 3–5 days when 80% confluency was reached.
HepaRG cells (Biopredic International, HPR116) were thawed in HepaRGTM Thawing/Plating/General Purpose medium (Biopredic International, MIL600C with ADD670C), counted and seeded immediately into spheroids using HepaRGTM Maintenance/Metabolism medium (Biopredic International, MIL600C with ADD620C). Both cell types were examined for morphology using a Zeiss Axiovert 25 light microscope at ×40 objective.
3D spheroid liver models for acute exposures
HepG2 monolayers were used to form spheroids using the previously described hanging drop method (3). In short, HepG2 cells were trypsinised and a cell stock (2.0 × 105 cells/ml) was prepared. HepaRG cells were thawed straight from liquid nitrogen and viability assessed using the trypan blue cell exclusion (TB) assay prior to preparing a cell stock of 2.0 × 105 cells/ml. A total of cell suspension (~4000 cells per 20 μl drop) was pipetted onto an inverted 9.4-cm square petri dish lid (Greiner Bio-One, UK). Approximately 100 drops were placed on each inverted petri dish lid. Subsequently, 20 ml of phosphate buffered saline (PBS) was added to the base petri dish to prevent the drops from drying out during the culture period. The petri dish was placed into the incubator at 37°C and 5% CO2 atmosphere. To maintain cell viability in the hanging drop, 6 μl of media was added to each drop on Day 3. Assessment of the spheroid morphology was examined using a Zeiss Axiovert 25 light microscope at ×40 objective.
3D spheroid liver models for longer-term exposures
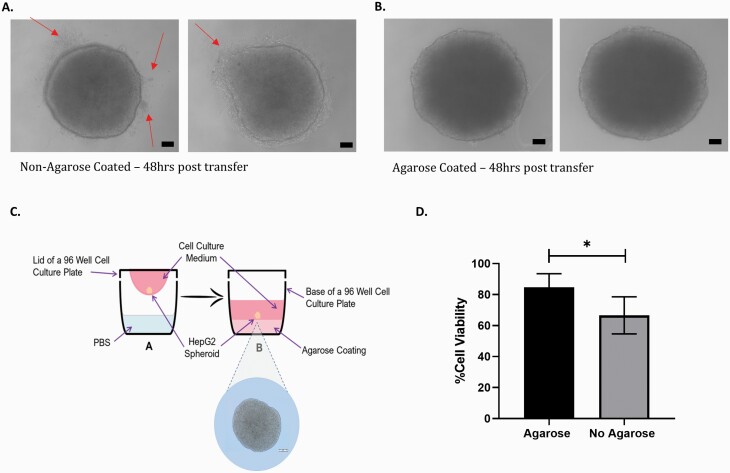
To support the growth of 3D spheroids over longer time periods, we have adapted the protocol previously described by Shah et al. (3). Following trypsinisation of 2D HepG2 cells, a stock solution of cells was prepared. Next, 20 µl of the cell suspension (4000 HepG2 cells per 20 μl hanging drop) was pipetted onto the inverted side of a 96-well tissue culture plate. To prevent the hanging drops from drying out, 100 µl PBS was transferred into the wells of the 96-well plates. The lid of the 96-well plate was gently inverted and placed onto the 96-well plate. The plate was then placed in the incubator at 37°C with 5% CO2. Three days after seeding (into hanging drops), the PBS was removed from each of the wells. The base of each well was coated with 50 µl of 1.5% agarose gel (Figure 1). Once dried, 100 µl fresh media was added to the wells. The spheroids were then transferred into the wells by centrifugation at 200g for 3 min. The HepG2 spheroids, suspended in the cell culture medium, were left to settle for 24 h, after which, they were ready to be exposed. To maintain cell viability over extended culture periods, the cell culture medium was refreshed every 3 days, whereby 50 μl of media was aspirated and replaced with a fresh 50 μl of DMEM. Assessment of the spheroid morphology was examined using a Zeiss Axiovert 25 light microscope at ×40 objective.
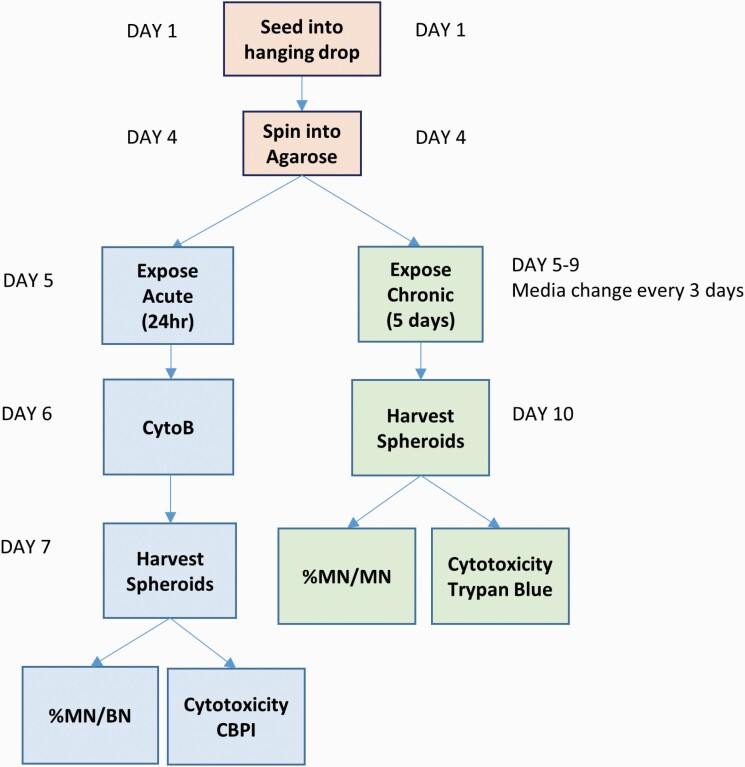
Fig. 1.
A schematic of the spheroid–ENM exposure timeline and MN assay specific adaptations.
Exposures
Short term
The spheroids were left in the hanging drop positions for 3 days (Figure 1). On Day 4, the hanging drops were treated with either AFB1 or MMS for 24 h, after which, Cyto B [6 μg/ml (Merck)] was added for an additional 24 h. The spheroids were harvested and trypsinised as described below for the TB cytotoxicity assay. After the final centrifugation step, the pellet was re-suspended in 400 µl PBS. Slides were prepared for both cytokinesis-block proliferation index (CBPI) and semi-automated MN scoring as previously described by Shah et al. and Chapman et al. (3,21).
Longer term
As detailed in the NanoGenoTox Dispersion Protocol (grant agreement no. 20092101, 2018) (20), ZnO ENM was dispersed by sonication (Branson Sonifier 250, Ø 13 mm, 400 W output power, 20 kHz) in 0.05% bovine serum albumin. Stock solution of ZnO was prepared at a concentration of 2.56 mg/ml, which was diluted in cell culture media to the required concentration (0.2–2 µg/ml). The protocol for exposing 3D liver spheroids to ENM was recently published by Llewellyn et al. (5). In brief, on Day 5, 50 µl of media was removed from the 96-well plate, taking caution not to disturb the spheroids, and replaced with 50 µl of ZnO ENM or AFB1. The plates were then incubated at 37°C for 5 days (Figure 1). AFB1 (0.1 µM) was used as a positive chemical control. Three days following exposure, the media was replenished. Being careful not to disrupt the settled ENM and spheroids, 50 µl of media was aspirated from each well and replaced with 50 µl of fresh media. On Day 10, 5 days post-ENM exposure, the spheroids were pooled and analysed for relative viable cell count (RVCC) using the TB assay and for genotoxicity using the mononuclear MN assay (as described below).
Cytotoxicity
Cell viability of the spheroids was assessed using the TB assay (GIBCO, Paisley, UK). Pooled spheroids were centrifuged at 230g for 5 min. To remove any residual media, the cell pellet was re-suspended in 1 ml PBS (GIBCO, Paisley, UK) and centrifuged at 230g for 5 min. To dissociate the spheroids, the cell pellet was re-suspended in trypsin/EDTA for 8–10 min. Fresh media was added in equal volumes to neutralise the trypsin. The cells were centrifuged at 230g for 5 min and re-suspended in fresh media. Cell viability was then analysed using the standard TB assay (22). RVCC was calculated in accordance with the OECD guidelines and that previously described by (5,15).
Liver functionality assays
Spheroid albumin and urea levels were examined to assess HepG2 and HepaRG spheroid liver-like functionality (BCG Albumin Assay Kit, MAK124 and Urea Assay Kit, MAK006, Sigma Aldrich, UK). All assays were performed as per manufacturer’s instructions. However, to ensure that samples fell within the standard curve, for the urea assay, the supernatants were diluted 1:10 with urea assay. On the day of harvest, spheroids were pooled and centrifuged at 230g for 5 min. Cell culture supernatant was collected (1.5 ml) for use with the albumin and urea secretion assays. Samples were stored at -−80°C until the assays were performed.
MN assay
Manual slide preparation for CBPI
Manual slides were prepared to calculate the CBPI. Glass slides were cleaned with 70% ethanol and left to air dry. Slides were prepared using the previously described cytospin method (23). In short, 100 µl of cell suspension in PBS was added to the cytospin cassette. The cassettes were centrifuged at 500g for 5 min. Once dried, the slides were fixed with cold 90% methanol for 10 min. The slides were then left to dry overnight at room temperature and stained using 20% Giemsa stain diluted in phosphate buffer (pH 6.8). The CBPI was calculated using the Olympus BH2 microscope at ×100 objective on the first 500 cells according to the OECD guidelines(15).
Preparation of slides for scoring MN frequency using the automated Metafer System
The remaining cells in PBS are then fixed and placed on slides as previously described (23). The cell density on the slide was examined accordingly using the Olympus BH2 microscope at ×100 objective. Once the slides were dry, they were stored at −20°C until scoring. Prior to automated scoring using the Metafer system, 30 µl of Vectashield Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Peterborough, UK) was added in the dark and a coverslip applied. When scoring, detection of MN in binucleated (BN) or mononucleated cells were performed as previously described by Chapman et al. and Manshian et al. (21,23). A minimum of 1000 BN cells or 2000 mononucleated cells were counted per exposure dose per replicate using the principles previously established by Fenech et al. (24) and in accordance with the OECD guidelines (15).
Statistical analysis
All experiments were performed three times independently (n = 3) with data presented as mean ± standard deviation (SD), unless stated otherwise. Statistical analysis was performed using Prism 8, GraphPad Software, Inc. Shapiro–Wilk test was used to calculate normality for each data set. For normally distributed data, either a one-way analysis of variance (ANOVA; HepG2 and HepaRG cell viability, HepG2 Cyto B; 2D and 3D HepG2 AFB1, MMS) or two-way ANOVA (HepG2 vs. HepaRG; cell viability, albumin, urea; 2D vs. 3D HepG2 AFB1, MMS) with Bonferroni post hoc were used. Fisher’s Exact test was used to determine statistical significance of MN/BN% and MN/MN% when compared to the untreated control (P < 0.05). For non-parametric data, a Kruskal–Wallis test was used to calculate significance when there were more than two variables (MN/MN% vs. MN/BN%, RVCC vs. CBPI, ZnO ENM) or to compare to the untreated control (RVCC and CBPI; P < 0.05).
Results
Characterisation of HepG2 and HepaRG 3D models for genotoxic assessment using the MN assay
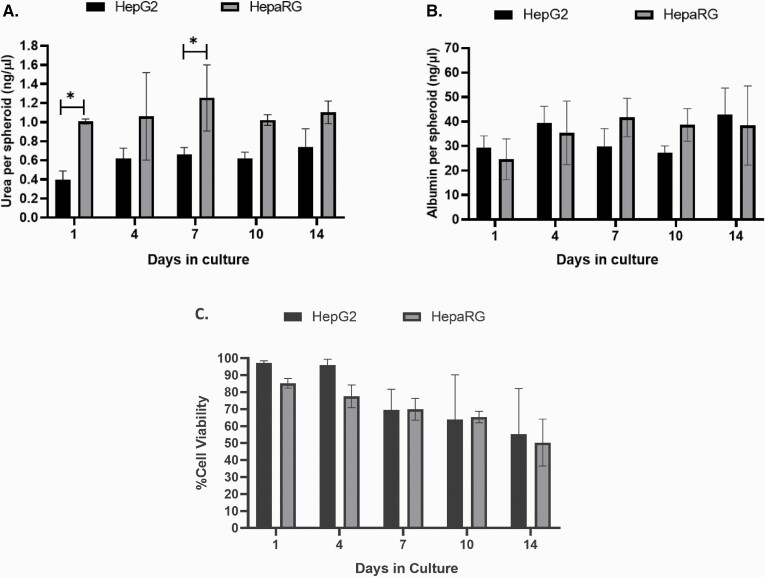
HepG2 and HepaRG cells are commonly used in liver toxicology studies due to their capacity to retain many of the metabolic functions of human liver cells. Urea and albumin production were assessed using the supernatant from both models on Days 4, 7, 10 and 14. As demonstrated in Figure 2A, there was a significant difference between the HepaRG and HepG2 cells (P < 0.001). Bonferroni’s post hoc test further revealed significant differences in the level of urea on Days 1 and 7 between the two cell types (P < 0.033). HepG2 spheroids demonstrated an overall reduced capacity to produce urea compared to HepaRG; however, this difference was less notable after 10 days of culture. Additionally, no significant difference in albumin production was observed between the two cell types over the 14-day period (Figure 2B). Within the HepaRG model, a significant reduction in cell viability was observed on Days 10 and 14 when compared to Day 1 (P < 0.05); no significant differences were observed for HepG2 spheroids over the 14 days when compared to Day 1 (Figure 2C). When then two models were compared, there was a main effect for time (P < 0.001); however, over that time period, post hoc analysis demonstrated no significant difference in viability between the two models (Figure 2C).
Fig. 2.
Characterisation of HepG2 and HepaRG liver cells over 14 days in culture. (A) Urea and (B) albumin production and (C) viability of 3D HepG2 and HepaRG spheroids were evaluated over 14 days. Statistical differences between HepG2 and HepaRG spheroids were analysed using two-way ANOVA with Bonferroni post hoc (*P < 0.05). Data shown are expressed as mean ± SD, n = 3.
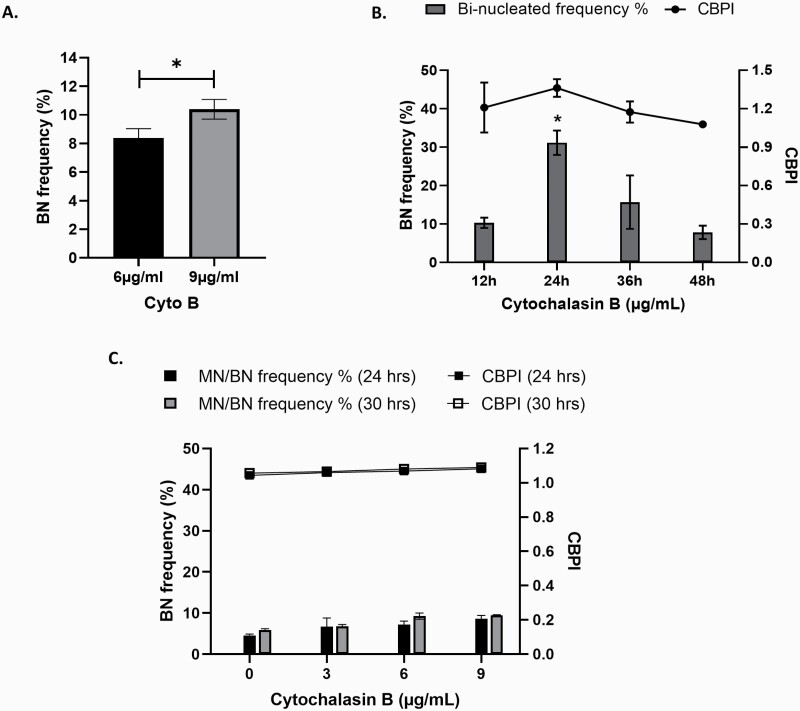
As currently described by the OECD test guideline 487 for the ‘in vitro mammalian cell micronucleus assay’, it is necessary to ensure that, after cells have been exposed to a chemical, they must undergo mitosis prior to being scored. The addition of Cyto B prior to nuclear division blocks the actin ring filaments, preventing the complete formation of two new daughter cells, resulting in the formation of a BN cell. Figure 3A and B demonstrates the optimisation of the incorporation of Cyto B into the CBMN assay in HepG2 3D spheroids. When evaluating the most appropriate Cyto B concentration to use, a small (1.997%) yet statistically significant increase in the frequency of BN cells was recorded when exposed to 9 µg/ml (P = 0.021) when compared to 6 µg/ml of Cyto B (Figure 3A). However, upon visual assessment following exposure to 9 µg/ml, the spheroids displayed irregular morphology demonstrating loss of integrity of the outer layers and evidence of membrane blebbing that was not observed in cells treated with 6 µg/ml (data not shown). Therefore, 6 µg/ml was selected for experiments going forward. In addition to concentration, it is also important to consider the time of Cyto B application. As illustrated in Figure 3B, whilst no significant cytotoxicity was observed at any time point, a significantly higher BN cell frequency was apparent at 24 h post-treatment than that compared with any other time point (P < 0.01).
Fig. 3.
Optimisation of Cyto B application for use with the CBMN assay. HepG2 3D liver spheroids were exposed to (A) 6 and 9 µg/ml Cyto B for 24 h and (B) 6 µg/ml Cyto B for 12, 24, 36 and 48 h and analysed for percentage BN cell frequency. Statistical differences were analysed using (A) unpaired t-test (*P = 0.021) and (B) one-way ANOVA with Bonferroni post hoc (*P < 0.01). (C) HepaRG spheroids were exposed to increasing concentrations of Cyto B (3, 6 and 9 µg/ml) for 24 and 30 h and analysed for percentage BN cell frequency and CBPI. Data shown are expressed as mean ± SD, n = 2.
To determine the proliferative capacity of HepaRG spheroids, cells were exposed to increasing concentrations of Cyto B for 24 and 30 h to capture a full cell cycle. No statistically significant differences were observed between the two time points or at any concentration of Cyto B for either genotoxicity or cytotoxicity (Figure 3C). However, what was apparent was the very low proliferative capacity of differentiated HepaRG 3D spheroids, which regardless of Cyto B concentration or exposure time, generated no more than a 9% BN cell frequency compared to 31% observed in HepG2 spheroids. This low induction of BN cells deems the HepaRG spheroid model unsuitable for genotoxicity assessment using the MN assay. Therefore, HepG2 cell spheroids were selected for further development as a model for longer-term ENM exposures.
HepG2 3D liver model demonstrates increased sensitivity to pro-carcinogens compared to HepG2 monolayers following acute exposures.
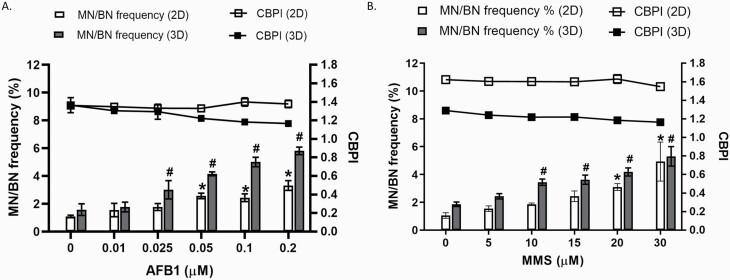
To investigate the sensitivity of 3D spheroids to a known genotoxic insult, both 2D and 3D HepG2 models were exposed to increasing concentrations of the well-known liver carcinogen AFB1 for 24 h and analysed using the CBMN assay. Figure 4A demonstrates a significant dose-dependent increase in MN frequency following exposure to AFB1 for both 2D (P < 0.001; 0.05–0.2 µM) and 3D (P < 0.05; 0.025–0.2 µM) HepG2 models when compared to the untreated control. When both 2D and 3D models were compared, the 3D model demonstrates a greater genotoxic effect. Bonferroni post hoc revealed a significant difference in MN frequency detected between both 2D and 3D HepG2 cells at 0.025–0.2 µM (P < 0.001). At top dose (0.2 µM) in the 2D model, there is 3-fold (3.1) difference in MN when compared to the untreated control; on the other hand, in the 3D model, the MN frequency is nearly 4-fold (3.7) higher than the untreated control. The CBPI demonstrates no cytotoxicity as a result of AFB1 over the concentration range applied. This data demonstrates that 3D HepG2 spheroids are more sensitive to the DNA damage induced by AFB1 than the 2D monoculture. It is suggested that this may be due to the increased metabolic activity of 3D HepG2 models previously demonstrated by Shah et al. (3) and, therefore, they are more efficient at converting AFB1 to the genotoxic metabolite than the same cells grown as 2D monocultures.
Fig. 4.
Assessment of micronuclei frequency in 2D monocultures and 3D HepG2 spheroids following a genotoxic insult. 2D HepG2 monocultures and 3D spheroids were exposed to increasing concentrations of (A) Aflatoxin B1 (0–0.2 µM) and (B) MMS (0–30 µM) for 24 h and analysed for MN in BN cells (MN/BN%) and CBPI. Statistical significance was compared to the untreated control and was analysed by one-way ANOVA with Bonferroni post hoc (*P < 0.007). Data shown are expressed as mean ± SD, n = 3.
To investigate whether 3D HepG2 spheroids also demonstrated increased sensitivity to DNA damage induced by direct-acting genotoxins, both cell culture formats were exposed to increasing concentrations of the alkylating agent and direct-acting genotoxin MMS for 24 h (Figure 4B). Similarly, as demonstrated with AFB1, Figure 4B demonstrates a significant dose-dependent increase in MN frequency for both 2D (P < 0.05; 20–30 µM) and 3D (P < 0.001; (10–30 µM) HepG2 models when compared to the untreated control. No cytotoxicity was observed following MMS treatment. In contrast, Bonferroni post hoc testing revealed a significant difference in MN frequency when comparing the induction of DNA damage between the 2D and 3D model formats at 10 and 15µM (P < 0.05). Generally, however, there was little difference between the fold difference in the induction of genotoxicity over the control when comparing the 2D and 3D model formats, thus indicating that they had a very similar sensitivity to the induction of DNA damage by the direct-acting genotoxin MMS.
Modified hanging drop approach supports longer-term viability of HepG2 spheroids
To maintain cell viability in the hanging drop during short-term exposures, 6 μl of media was added to each 20 µl drop on Day 3; however, for longer-term exposures, continued addition of media to the droplet would result in the droplet becoming too large and, thus, would fall from the lid of the petri dish. In an effort to overcome this, spheroids were seeded onto the lid of a 96-well plate. After 3 days, 100 µl of fresh media was added to the base of each well of the plate. The spheroids were then transferred into each of the wells containing fresh media by centrifugation. As seen in Figure 5A, after 72 h, the HepG2 cells within the spheroid begin to elongate and form protrusions in an effort to reform a monolayer on the base of the 96-well plate. To combat this issue and extend the longevity of the spheroid model, a layer of agarose (1.5% agarose in DMEM without phenol red) was added to the base of the 96-well plate (Figure 5C). Following the addition of the agarose layer, the spheroid forms a defined round spheroid, with no protrusions (Figure 5B).
Fig. 5.
Modification of the hanging drop method to support longer-term ENM exposures. Spheroids were grown on the lid of a 96-well plate for 3 days; on Day 4, they were transferred by centrifugation into the base of the plate containing media that was either (A) not coated in agarose or (B) coated in agarose. Images of the spheroids were obtained using light microscopy (×40 objective) after 72 h. Changes to spheroid structure are denoted by red arrows. Scale bar 20 µM. (C) Schematic of the modified hanging drop method with the addition of a 1.5% agarose layer that is used for the remainder of the study. (D) Viability of HepG2 spheroids was calculated using the Trypan Blue assay in the presence and absence of agarose. Statistical significance was determined using an unpaired t-test (*P <0.01). Data shown are expressed as mean ± SD, n = 3.
Table 1 below demonstrates the results of a comparison study of spheroid viability and urea and albumin production in both the presence and absence of an agarose layer added to the base of the 96-well plate. A significant increase in urea production was observed on day 10 in the presence of agarose (P = 0.014). No difference was observed between the two methods for albumin. Similarly, when comparing the viability at each time point, no significance was observed; however, a significant difference is demonstrated in the total mean difference with cell viability in the presence and absence of agarose (P = 0.013; Figure 5D). To maintain the structural integrity of the spheroids and prolong viability of the culture over 10 days, it was concluded that the addition of a 1.5% agarose layer to the base of the 96-well plates prior to the addition of media was necessary to support longer-term exposures.
Table 1.
Characterisation of 3D liver spheroids in the presence and absence of agarose.
| HepG2 cells | No Agarose | Agarose | ||
|---|---|---|---|---|
| Day 7 | Day 10 | Day 7 | Day 10 | |
| % Viability (95% CI) | 73.5% (90.07–56.03) | 60.09% (93.99–26.20) | 89.98% (102.03–77.95) | 79.6% (102.42–56.82) |
| Albumin per spheroid (ng/µl) (95% CI) | 32.56 (39.17–25.94) | 29.83 (33.50–26.15) | 34.225 (63.99–4.46) | 48.505 (94.820–2.19) |
| Urea per spheroid (ng/µl) (95% CI) | 0.64 (0.70–0.58) | 0.63 (0.68–0.58) | 0.66 (0.79–0.52) | 0.82* (1.09–0.54) |
*Significant at P < 0.014. CI, confidence interval.
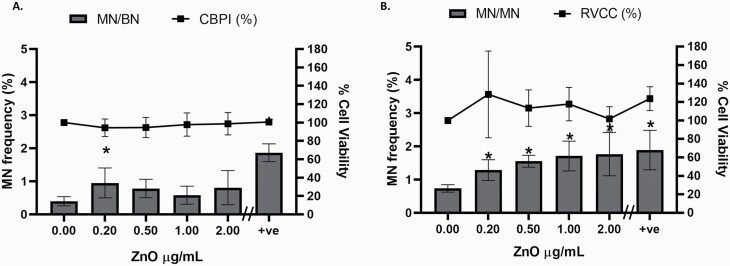
In vitro MN assay for longer-term ENM genotoxicity assessment
As demonstrated previously, the CBMN version of the MN assay with CBPI was performed, where, after a 24-h exposure, Cyto B was added for an additional 24 h, which results in the formation of BN cells. However, it was postulated that the damage encountered following longer-term exposure may occur at an earlier stage; therefore, the addition of Cyto B and counting MN in BN cells after a 5-day exposure could fail to accurately capture the true genotoxic insult. To investigate this, firstly using the CBMN approach, all MN in BN cells (MN/BN%) were scored with CBPI calculated as a measure of cytotoxicity (Figure 6A). A non-dose-dependent induction of genotoxicity was observed, with the only significant difference noted between the untreated control and the lowest concentration 0.2 µg/ml of ZnO (P = 0.037) and, as expected, with the positive control (0.1 µM AFB1, P < 0.001). Second, in the absence of Cyto B, cells were scored for MN in mononucleated cells (MN/MN %) and RVCC calculated as the measure of cytotoxicity (Figure 6B). A significant increase in MN frequency was observed in all concentrations of ZnO ENM compared to the untreated control (P < 0.007, 0.025–30 µg/ml). Additionally, when compared, a significant difference (P = 0.021) was observed between the two methods for assessing MN frequency (MN/BN% and MN/MN%). No significant difference was observed between the two methods for cytotoxicity (CBPI and RVCC).
Fig. 6.
Optimisation of the in vitro micronucleus assay for longer-term ENM genotoxicity assessment. 3D HepG2 spheroids were exposed to increasing concentrations of ZnO ENM (0–2.0 µg/ml) and 0.1 µM AFB1 (positive control, +ve) for 5 days and analysed for MN in (A) BN cells (MN/BN %, n = 2) with CBPI (n = 2) and (B) mononucleated cells (MN/MN %, n = 3) with RVCC. Statistical significance compared to the untreated control was analysed by Fisher’s exact (*P < 0.05). Data shown are expressed as mean ± SD.
Discussion
The purpose of this study was to develop a 3D liver spheroid model that would support ENM genotoxicity assessment following exposure over a prolonged period (i.e. up to 5 days). Over the last decade, there have been a number of protocols published demonstrating the development of 3D liver models. However, a model that supports DNA damage testing using the MN assay following longer-term ENM exposure and associated genotoxic assessment using the MN assay is yet to be established. Herein describes a method that supports the growth and maintenance of 3D liver spheroids over a total of 10, allowing for an extended exposure period of 5 days.
In an effort to replace, reduce and refine the reliance on animal experimentation, substantial efforts have been placed on developing 3D in vitro model systems that demonstrate greater physiological relevance and that mimic the liver-like functionality and metabolism seen in vivo. It is thought that the enhanced features of 3D models are due to the compact density of the spheroid, complex cell to cell interactions and signalling, making them an ideal candidate to improve the state-of-the-art for genotoxicological testing (25). Using liver-specific functionality assays, our data demonstrates that 3D HepG2 spheroids maintain functionality for up to 14 days in culture as fully formed spheroids. HepaRG liver cells are widely used in drug development and toxicology, as they retain liver-like functionality. Using the same protocol, we generated a differentiated HepaRG 3D model and the two models showed comparable liver-like functionality. However, there was some variance between the two models in urea production, with the HepaRG model demonstrating the ability to maintain consistently high levels of urea over the 14 days, albeit only significant at Days 1 and 7. There was no significant difference in albumin levels over the 14 days between the two models; however, HepG2 levels appear to be lower. No differences in cell viability were observed over the 14 days. Based on this metabolic data, it would appear that the HepaRG model exhibits a slight advantage over HepG2, which agrees with observations by others, although they are largely comparable (12).
An important feature of the MN assay is the requirement for cells to demonstrate a high proliferative capacity. This study demonstrates a basal-level BN cell frequency following treatment with 6 µg/ml Cyto B in 3D HepG2 cells, indicating that the cells within the spheroid (particularly at the periphery) are actively proliferating. Whilst this is lower than typically observed in 2D culture, this level of proliferation is sufficient to support the MN assay. We found that 24 h was the optimal time point to capture cells that have been arrested after one cell cycle and presented as BN cells. In contrast to this, our data showed that HepaRG cells exhibit a very low proliferation rate. Despite adjusting the Cyto B concentration and exposure duration, the BN frequency for 24 h was 7.2%, whilst, for HepG2 spheroids, it reached 31.13%. The very low proliferative capacity of differentiated HepaRG 3D spheroids deems them unsuitable for genotoxicity assessment using the MN assay. Interestingly, as demonstrated by others, this does not seem to be the case when HepaRG cells are cultured in 2D (26,27). However, for 3D spheroids, this data agrees with Mandon et al., who demonstrated that HepaRG 3D spheroids had low levels of DNA topoisomerase II, a specific marker for cell division (2), confirming the quiescent nature of differentiated HepaRG spheroids. Therefore, if the endpoint for analysis is the MN assay, HepaRG in 3D spheroid format are not a practical cell model to be used. Jossé et al. previously described the adaptation of the HepaRG cell line to the MN assay for genotoxicity testing, which was facilitated by treating the HepaRG cells in 2D with epidermal growth factor to stimulate cell proliferation (27). To the best of our knowledge, this has not be explored in 3D HepaRG models but offers a potential strategy to overcome the quiescent nature of 3D HepaRG spheroids.
Shah et al. have previously determined that HepG2 spheroids demonstrate increased efficacy over the standard 2D monoculture and can readily metabolise the pro-carcinogens B[a]-P and PhIP into their genotoxic metabolites, thus initiating an enhanced genotoxic response compared to 2D cultures (3). Similarly, our data confirms that 3D HepG2 spheroids detect greater levels of genotoxicity at low concentrations of the liver pro-carcinogen AFB1 compared to 2D monocultures. This may be due to the increase in metabolic function observed in 3D models (3,28,29). We also establish that the direct-acting genotoxin, MMS, induced similar levels of genotoxicity in both 2D and 3D models. The data demonstrates that the elevated number of MN observed in the 3D models with pro-carcinogens is not an artefact of the 3D system (e.g. DNA damage induced through enhanced metabolism). Although no significant cytotoxicity was observed between the two models, the CBPI was lower in the 3D spheroid model than the 2D monoculture following both AFB1 and MMS exposure. This coincides with that observed by Shah et al. who suggested that, due to the compact nature of the spheroid, it is primarily the outer layer of cells that are rapidly dividing, whereas the cells within the spheroid core have a lower proliferation index (3).
In an effort to increase the longevity of the 3D culture, spheroids were seeded onto the lid of a 96-well plate as opposed to the lid of a square petri dish as previously described by Shah et al. (3). Following a 3-day incubation period to allow for spheroid formation, the spheroids were dropped by centrifugation into each well of the 96-well plate, which contained 100 µl media. However, during visual examination of the HepG2 spheroids, it was apparent that the naturally adherent nature of the HepG2 cells began affecting the structural integrity of the spheroid. The spheroids began to form protrusions in an effort to reform a monolayer on the base of each well. HepG2 cells are, by nature, an adherent cell line and, when given the opportunity to attach to a surface, they will try to form a monolayer as demonstrated in the present study. To overcome this obstacle, 1.5% agarose was added to the base of each well prior to the addition of media. A small change was demonstrated in urea production on day 10 that was in favour of the presence of agarose. This agrees with numerous studies that demonstrate the use of agarose in 3D spheroid formation (30–32). Interestingly, our data also showed an increase in cell viability when agarose was added, similarly to that observed by Friedrich et al. who noted that spheroid formation and growth were superior when wells were coated in agarose (31). It was also clear that the integrity and structure of the spheroids were not compromised. Therefore, it was determined for this reason that we would use the modified hanging drop setup going forward. Agarose not only inhibits cell adhesion and is non-toxic but also offers a cost-effective alternative to ultra-low adhesion plates.
It is evident from our data that using the CBMN form of the MN assay to measure genotoxicity following longer-term exposures (over 24 h) underestimates the true level of genotoxicity. A significant difference between the two methods was observed and there is a clear trend of a higher MN frequency in mononucleated cells. When compared to the untreated control, there was a significant increase in MN frequency in mononucleated cells at all ZnO ENM concentrations. In contrast, a significant increase in MN frequency in BN cells was only observed at the lower concentration (0.2 µg/ml). It was hypothesised that the damage encountered is occurring early on and, therefore, the addition of Cyto B and counting of MN in BN cells on the final day is missing the initial genotoxic insult. Following longer-term exposures, cells will have undergone numerous cell divisions and, thus, the addition of Cyto B will only capture the cells that have divided in the last cell cycle. According to the OECD test guideline 487, measuring cytotoxicity by CBPI should only be used following the addition of Cyto B (15). Therefore, we suggest that, as an alternative, when performing longer-term exposures, cytotoxicity is measured using an alternative method, such as RVCC using the TB assay instead. It is noted that the OECD test guideline 487 suggests relative population doubling (RPD) or relative increase in cell count (RICC) as alternative methods when Cyto B is not used to ensure that the cells have undergone cell division after extended treatment periods. However, based on the nature of the experimental setup of this 3D spheroid assay, it would be impractical and costly to set up a satellite plate of spheroids for every concentration to perform the RPD. The RICC was also considered, however, as the cells are replicating and dividing in the hanging drop during the spheroid formation phase, that is, the first 4 days post-seeding and prior to exposure, this would also underestimate cytotoxicity. Consequently, due to the complex structure of the 3D liver models, the RVCC was calculated using the TB assay. This method was previously described by Roy et al. as a measure of cytotoxicity in the 3D RSMN assay (33). The authors measure cytotoxicity using both the CBPI and RVCC, identifying that the RVCC was the more sensitive measure of cell viability for treatment with N-ethyl-N-nitrosourea and 2-ethyl-1,3-hexanediol in their 3D skin model. Our data demonstrates no significant difference between the RVCC and CBPI and, therefore, is put forward as a feasible method for measuring cytotoxicity when assessing longer-term genotoxicity using the MN assay in 3D liver spheroid models.
In recent decades, there has been a rapid development and implementation of a variety of ENMs used in fields such as food and agricultural industry, medical devices and drug delivery. The investigation of the potential genotoxicity of these materials over acute and longer-term exposure scenarios is a vital component of their safety assessment. The in vitro MN assay is deemed the gold standard when assessing chromosomal damage induction (specifically clastogenicity and/or aneugenicity); however, this method was originally established for use with chemicals and 2D culture systems. There are many limitations of 2D systems for genotoxicity assessment, which have previously been extensively reviewed (34). In addition, the regulated genotoxicity in vitro testing methods (i.e. for pharmaceuticals) demonstrate ‘low specificity’ and ‘misleading false positives’ which, at present, results in the requirement for additional animal tests (1,35). It is vital that there is continued development of novel advanced in vitro test systems and robust protocols, such as 3D in vitro models, that can be used as reliable alternatives to animal testing. More importantly, the European directive (Directive 2010/63/EU) driving the ‘3Rs’ to reduce, replace and refine in vivo animal-based experiments aims to advance in vitro-based systems as alternative methods to provide predictive and reliable results for hazard assessment (36). With advancements in 3D in vitro systems and continuous development of novel ENMs, it is essential that we also adapt our current methodologies to evolve with the field. Investigative studies into the use of the MN assay for short-term ENM exposures using the CBMN assay have identified that Cyto B can halt cellular uptake of the novel materials (13,18). Therefore, it was established that Cyto B should only be added to the cells post-ENM exposure and not as a co-exposure with the ENM, a method that is commonly performed with chemical compounds. Recently, Wills et al. have successfully adapted the RSMN version of the MN assay for the genotoxic assessment of silica nanoparticles in both 2D and 3D in vitro models following 24-h exposures (17). Having modified the 3D liver spheroid system and accompanying MN assay to support a longer-term ENM exposure regime, this study demonstrates a dose-dependent increase in the frequency of MN following 5 days of exposure to ZnO ENM. This would not have been observed had methodology alterations not been made. Careful consideration must be taken when using the ‘cytokinesis-block’ version of this assay for longer-term exposure regimes, as any DNA damage induced within the first few cell cycles is distributed across the mononucleated cell population as opposed to being retained and scored within the BN cells. This study demonstrates the potential to score false negatives, as the DNA damage accumulated over the period of chronic exposure can be masked when using the CBMN version of the MN assay.
In conclusion, this study has developed a physiologically relevant, advanced 3D HepG2 hepatocyte model that demonstrates increased efficacy for genotoxicity over 2D monoculture. It also maintains stability and liver functionality over extended culture periods. Herein, the present study has demonstrated the suitability of this model for assessing genotoxicity of direct and indirect acting mutagens using the regulatory approved in vitro MN assay. Additionally, taking into consideration the necessary adaptations, our study demonstrates that this model can also be used for the genotoxic assessment of ENMs using the MN assay.
Acknowledgements
The authors would like to acknowledge that this research has received funding from the Horizon 2020 Research and Innovation Programme for the PATROLS project, under grant agreement no. 760813.
Conflict of interest statement: The authors declare no competing interests.
Availability of data and material
The authors will freely release all data underlying the published paper upon direct request to the corresponding author.
Author contributions
S.H.D and G.J. J conceived the project and designed the experiments. U.K.S, S.L., T.C., S.J.E., A.S.A.A. and G.E.C. performed the experiments and collected and analysed the data. G.E.C., U.K.S., S.L., S.H.D and M.J.D.C co-wrote the paper. All authors discussed the results and reviewed the manuscript.
References
- 1. Pfuhler, S., van Benthem, J., Curren, R., et al. (2020) Use of in vitro 3D tissue models in genotoxicity testing: strategic fit, validation status and way forward. Report of the working group from the 7th International Workshop on Genotoxicity Testing (IWGT). Mutat. Res., 850–851, 503135. [DOI] [PubMed] [Google Scholar]
- 2. Mandon, M., Huet, S., Dubreil, E., Fessard, V. and Le Hégarat, L. (2019) Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci. Rep., 9, 10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shah, U.K., Mallia, J.de O., Singh, N., Chapman, K.E., Doak, S.H. and Jenkins, G.J.S. (2018) A three-dimensional in vitro HepG2 cells liver spheroid model for genotoxicity studies. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 834, 35–41. doi: 10.1016/j.mrgentox.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 4. Štampar, M., Tomc, J., Filipič, M. and Žegura, B. (2019) Development of in vitro 3D cell model from hepatocellular carcinoma (HepG2) cell line and its application for genotoxicity testing. Arch. Toxicol., 93, 3321–3333. [DOI] [PubMed] [Google Scholar]
- 5. Llewellyn, S. V., Conway, G.E., Shah, U.K., Evans, S.J., Jenkins, G.J.S., Clift, M.J.D. and Doak, S.H. (2020) Advanced 3D liver models for in vitro genotoxicity testing following long-term nanomaterial exposure. J. Vis. Exp. 2020, 1–10. [DOI] [PubMed] [Google Scholar]
- 6. Yamada, K. M. and Cukierman, E. (2007) Modeling tissue morphogenesis and cancer in 3D. Cell, 130, 601–610. [DOI] [PubMed] [Google Scholar]
- 7. Lauschke, V.M., Hendriks, D.F.G., Bell, C.C., Andersson, T.B. and Ingelman-Sundberg, M. (2016)Novel 3D culture systems for studies of human liver function and assessments of the hepatotoxicity of drugs and drug candidates. Chem. Res. Toxicol., 29, 1936–1955. [DOI] [PubMed] [Google Scholar]
- 8. Schwartz, R. E., Fleming, H. E., Khetani, S. R. and Bhatia, S. N. (2014) Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv., 32, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kondo, Y., Iwao, T., Nakamura, K., et al. (2014) An efficient method for differentiation of human induced pluripotent stem cells into hepatocyte-like cells retaining drug metabolizing activity. Drug Metab. Pharmacokinet., 29, 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell, C. C., Hendriks, D. F., Moro, S. M., et al. (2016) Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep., 6, 25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kermanizadeh, A., Løhr, M., Roursgaard, M., Messner, S., Gunness, P., Kelm, J. M., Møller, P., Stone, V. and Loft, S. (2014) Hepatic toxicology following single and multiple exposure of engineered nanomaterials utilising a novel primary human 3D liver microtissue model. Part. Fibre Toxicol., 11, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kammerer, S. and Küpper, J.-H. (2018) Human hepatocyte systems for in vitro toxicology analysis. J. Cell. Biotechnol., 3, 85–93. doi: 10.3233/jcb-179012. [DOI] [Google Scholar]
- 13. Doak, S.H., Manshian, B., Jenkins, G.J.S. and Singh, N. (2012) In vitro genotoxicity testing strategy for nanomaterials and the adaptation of current OECD guidelines. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 745, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doherty, A., Bryce, S.M. and Bemis, J.C. (2016) The In Vitro Micronucleus Assay. Elsevier Inc., Amsterdam, pp. 161–205. [Google Scholar]
- 15. OECD (2016) Test No. 487: In Vitro Mammalian Cell Micronucleus Test. OECD Publishing, Paris, p. 29. [Google Scholar]
- 16. Curren, R. D., Mun, G. C., Gibson, D. P. and Aardema, M. J. (2006) Development of a method for assessing micronucleus induction in a 3D human skin model (EpiDermTM). Mutat. Res., 607, 192–204. [DOI] [PubMed] [Google Scholar]
- 17. Wills, J.W., Hondow, N., Thomas, A.D., et al. (2016) Genetic toxicity assessment of engineered nanoparticles using a 3D in vitro skin model (EpiDermTM). Part. Fibre Toxicol., 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doak, S. H., Griffiths, S. M., Manshian, B., Singh, N., Williams, P. M., Brown, A. P. and Jenkins, G. J. (2009) Confounding experimental considerations in nanogenotoxicology. Mutagenesis, 24, 285–293. [DOI] [PubMed] [Google Scholar]
- 19. Elespuru, R., Pfuhler, S., Aardema, M. J., et al. (2018) Genotoxicity assessment of nanomaterials: recommendations on best practices, assays, and methods. Toxicol. Sci., 164, 391–416. [DOI] [PubMed] [Google Scholar]
- 20. Jensen, K.A. and Thieret, N. (2014) The NANOGENOTOX Dispersion Protocol for NANoREG, 13. http://safenano.re.kr/download.do?SEQ=175 (accessed April 15, 2020). [Google Scholar]
- 21. Chapman, K. E., Thomas, A. D., Wills, J. W., Pfuhler, S., Doak, S. H. and Jenkins, G. J. (2014) Automation and validation of micronucleus detection in the 3D EpiDerm™ human reconstructed skin assay and correlation with 2D dose responses. Mutagenesis, 29, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Louis, K. S. and Siegel, A. C. (2011) Cell viability analysis using trypan blue: manual and automated methods. Methods Mol. Biol., 740, 7–12. [DOI] [PubMed] [Google Scholar]
- 23. Manshian, B. B., Singh, N. and Doak, S. H. (2013) The in vitro micronucleus assay and kinetochore staining: methodology and criteria for the accurate assessment of genotoxicity and cytotoxicity. Methods Mol. Biol., 1044, 269–289. [DOI] [PubMed] [Google Scholar]
- 24. Fenech, M., Chang, W. P., Kirsch-Volders, M., Holland, N., Bonassi, S. and Zeiger, E.; HUman MicronNucleus project . (2003) HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res., 534, 65–75. [DOI] [PubMed] [Google Scholar]
- 25. Otieno, M.A., Gan, J. and Proctor, W. (2018) Status and future of 3D cell culture in toxicity testing. In Chen M., Will Y. (eds.), Drug-Induced Liver Toxicity. Methods in Pharmacology and Toxicology. Humana Press Inc., New York, pp. 249–261. Accessed 29 February 2020. [Google Scholar]
- 26. Le Hégarat, L., Mourot, A., Huet, S., Vasseur, L., Camus, S., Chesné, C. and Fessard, V. (2014) Performance of comet and micronucleus assays in metabolic competent HepaRG cells to predict in vivo genotoxicity. Toxicol. Sci., 138, 300–309. [DOI] [PubMed] [Google Scholar]
- 27. Jossé, R., Rogue, A., Lorge, E. and Guillouzo, A. (2012) An adaptation of the human HepaRG cells to the in vitro micronucleus assay. Mutagenesis, 27, 295–304. [DOI] [PubMed] [Google Scholar]
- 28. Foster, A. J., Chouhan, B., Regan, S. L., et al. (2019) Integrated in vitro models for hepatic safety and metabolism: evaluation of a human Liver-Chip and liver spheroid. Arch. Toxicol., 93, 1021–1037. [DOI] [PubMed] [Google Scholar]
- 29. Bell, C. C., Dankers, A. C. A., Lauschke, V. M., et al. (2018) Comparison of hepatic 2D sandwich cultures and 3D spheroids for long-term toxicity applications: a multicenter study. Toxicol. Sci., 162, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mirab, F., Kang, Y. J. and Majd, S. (2019) Preparation and characterization of size-controlled glioma spheroids using agarose hydrogel microwells. PLoS One, 14, e0211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Friedrich, J., Seidel, C., Ebner, R. and Kunz-Schughart, L. A. (2009) Spheroid-based drug screen: considerations and practical approach. Nat. Protoc., 4, 309–324. [DOI] [PubMed] [Google Scholar]
- 32. Wenzel, C., Otto, S., Prechtl, S., Parczyk, K. and Steigemann, P. (2015) A novel 3D high-content assay identifies compounds that prevent fibroblast invasion into tissue surrogates. Exp. Cell Res., 339, 35–43. [DOI] [PubMed] [Google Scholar]
- 33. Roy, S., Kulkarni, R., Hewitt, N.J. and Aardema, M.J. (2016) The EpiDermTM 3D human reconstructed skin micronucleus (RSMN) assay: historical control data and proof of principle studies for mechanistic assay adaptations. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 805, 25–37. doi: 10.1016/j.mrgentox.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 34. Evans, S. J., Clift, M. J., Singh, N., de Oliveira Mallia, J., Burgum, M., Wills, J. W., Wilkinson, T. S., Jenkins, G. J. and Doak, S. H. (2017) Critical review of the current and future challenges associated with advanced in vitro systems towards the study of nanoparticle (secondary) genotoxicity. Mutagenesis, 32, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Corvi, R. and Madia, F. (2017) In vitro genotoxicity testing—can the performance be enhanced? Food Chem. Toxicol., 106, 600–608. [DOI] [PubMed] [Google Scholar]
- 36. Council Directive 2010/63/EU (2010) Council Directive 2010/63/EU on the protection of animals used for scientific purposes. Off. J. Eur. Union, 53, 84. doi: 10.3000/17252555.L_2010.276.eng. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors will freely release all data underlying the published paper upon direct request to the corresponding author.