Significance
Our work challenges the existing paradigm that marine Arctic ecosystems are depauperate extensions of southerly (temperate) communities established in the wake of recent glaciation, fundamentally changing how these systems should be viewed and interpreted. We forward hypotheses regarding the history of Arctic marine systems, particularly with regards to endemism being an integral feature of Arctic biomes, and present a firm framework for future evolutionary research in this system typically viewed as “ecologically immature.”
Keywords: Pleistocene glaciation, Last Glacial Maximum, species distribution models, endemism, species-pump
Abstract
The Arctic is experiencing a rapid shift toward warmer regimes, calling for a need to understand levels of biodiversity and ecosystem responses to climate cycles. This study presents genetic data for 109 Arctic marine forest species (seaweeds), which revealed contiguous populations extending from the Bering Sea to the northwest Atlantic, with high levels of genetic diversity in the east Canadian Arctic. One-fifth of the species sampled appeared restricted to Arctic waters. Further supported by hindcasted species distributions during the Last Glacial Maximum, we hypothesize that Arctic coastal systems were recolonized from many geographically disparate refugia leading to enriched diversity levels in the east Canadian Arctic, with important contributions stemming from northerly refugia likely centered along southern Greenland. Our results suggest Arctic marine biomes persisted through cycles of glaciation, leading to unique assemblages in polar waters, rather than being entirely derived from southerly (temperate) areas following glaciation. As such, Arctic marine species are potentially born from selective pressures during Cenozoic global cooling and eventual ice conditions beginning in the Pleistocene. Arctic endemic diversity was likely additionally driven by repeated isolations into globally disparate refugia during glaciation. This study highlights the need to take stock of unique Arctic marine biodiversity. Amplification of warming and loss of perennial ice cover are set to dramatically alter available Arctic coastal habitat, with the potential loss of diversity and decline in ecosystem resilience.
The Arctic is characterized by a turbulent climatic history and the prospect of further change. Repeated glaciations over the past 2.6 Ma had a lasting impact on biological communities, forcing populations to repeatedly contract and expand with the formation and retreat of ice sheets (1). Today, warming in the Arctic is significantly exceeding the Northern Hemisphere average (2), and boreal and temperate regimes are expected to shift northwards as a result (3). A prescient need exists to understand the responses of Arctic marine communities to climate change, a need that will inherently depend on understanding levels of biodiversity, the recent history of Arctic ecosystems, and ultimately the potential for adaptation.
Marine forests can provide insight on anticipated responses of Arctic marine communities to environmental changes. Marine forests are structurally complex seascapes created by seaweeds, are ubiquitous worldwide, and provide valuable ecosystem and economic services in the forms of habitat, nursery grounds, primary productivity, and harvesting resources (4 and 5). Arctic marine forests are broadly distributed, with circumpolar species distributions extending from the Pacific through to the Atlantic with large gaps along the Siberian Arctic coastline because of unsuitable soft substrate (6 and 7) (see SI Appendix, Fig. S1 for exemplar species). Marine forests in the Arctic can also grow to incredible depths in some locations, particularly off the coasts of Greenland where macroalgae have been recorded as deep as 60 m (8). Annual ice cover is often a limiting factor in the Arctic, preventing a lush intertidal community from flourishing, but nonetheless allowing annuals and hardier flora to take advantage of the short growing season (6). Some species are particularly resilient to these conditions. The kelp Laminaria solidungula, for example, completes nearly all its growth under ice, using the summer months when photosynthetic rate is high to focus exclusively on carbon capture and storage (9). Unsurprisingly, climate change is expected to alter Arctic marine forests, although the exact nature of these impacts depends on many interplaying factors. While northerly distributions are projected to expand in some taxa (3), increased turbidity and declines in salinity may impact survival (7). Given the foundational nature of marine forests and the anticipated responses to a shifting climate, Arctic assemblages provide a framework to assess the resilience of polar marine ecosystems to ongoing environmental changes.
The presence of species that are finely tuned to marine Arctic conditions raises an interesting paradox, in that Arctic marine forests were historically viewed as entirely derived from southerly European refugia following the Last Glacial Maximum (LGM) (10 and 11). Under this view, Arctic marine forests were regarded as an extension of cold-tolerant temperate species, precluding the notion of Arctic endemism. While the notion of survival in refugia south of ice sheets persists, alternative hypotheses regarding the recent origins of Arctic marine forests have been brought forward, emphasizing contributions from the northern Pacific (12–14). A few genetic studies suggest there may have been multiple recolonization pathways for marine Arctic communities, and have also revealed cryptic diversity in the Arctic, reviving the notion of Arctic endemism (12, 15, and 16). Clarifying the recent history of marine forests marks a critical step toward characterizing levels of biodiversity along Arctic coastlines and the subsequent resilience to anticipated environmental changes (17).
The notion of Arctic marine forests as a depauperate extension of temperate communities is an antiquated view. Genetic signature at odds with the hypothesis of recolonization, predominantly out of European refugia, and the presence of cryptic diversity in the Arctic, challenge researchers to revise our understanding of Arctic marine forests and how they persist through cycles of glaciation. Our objective was to clarify the recent origin of Arctic marine forests, emphasizing a community-level approach by summarizing genetic data results across many species, and using species distribution models to identify likely refugial locations during the LGM. We evaluated several hypotheses, employing two lines of evidence in the forms of genetic surveys (DNA barcoding) and hindcasting species distributions using ecological niche modeling. If Arctic marine populations were recently recolonized from southerly refugia, particularly from Europe, then we should observe a decline in genetic diversity along the recolonization pathway (east to west and south to north). In addition, recolonized areas should be genetically closer to the source basin as compared to conspecifics in alternate basins because of vicariance during glaciation (i.e., Atlantic vs. Pacific). Finally, if refugia were only available in southerly modern-day temperate areas, then hindcasting species distributions should reflect this; that is, areas of likely species occurrences lacking persistent ice cover during the LGM should be restricted to southern coastlines of the North Atlantic and North Pacific. If these hypotheses are rejected, our approach will otherwise yield insight on alternate locations of persistence, subsequent dynamics of recolonization following glaciation, and standing levels of biodiversity in contemporary assemblages.
Results
Analysis of Sequence Data.
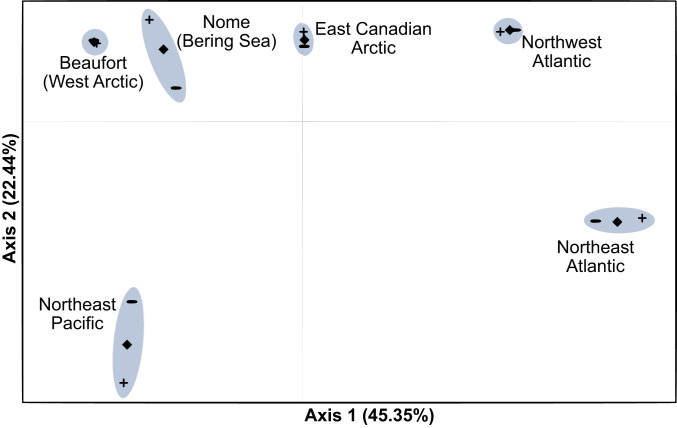
In order to infer refugial locations and recolonization pathways in Arctic marine forest assemblages, we surveyed and sequenced DNA barcode markers in specimens from several areas. For analytical purposes, we pooled specimens according to the following geographic distinctions: the northeast Pacific (British Columbia, Canada, and Washington state in the United States); Nome, Bering Sea, Alaska (proxy for Pacific migrants into the Arctic); Beaufort, Arctic Ocean, Alaska (proxy for the west North American Arctic); east Canadian Arctic (Cambridge Bay, Nunavut, through to Nain, Labrador); northwest Atlantic (Makkovik, Labrador, and southwards, including the Canadian Atlantic provinces and New England States in the United States); and the northeast Atlantic (Europe). In total, 4,631 specimen records were amalgamated, 2,018 of which were collected during this study (2014 to 2018), representing 42 species of red algae, 49 brown algae, and 18 green algae, for a total of 109 marine macroalgal species with Arctic populations. Of these, 21 species (19%) were sampled exclusively within the Arctic basin, predominantly from the east Arctic region (SI Appendix, Fig. S2 and Table S1). The genetic results averaged across 31 species-marker combinations revealed Arctic populations that were typically distinct from conspecifics in the northeast Atlantic and northeast Pacific. A principal coordinates analysis (PCoA) of genetic distances grouped populations extending from the Bering Sea through to the northwest Atlantic, within which the east Arctic fell in the middle (Fig. 1 and Table 1). Northeast Pacific and northwest Atlantic populations featured greater genetic diversity, in contrast to comparable populations in Europe, while genetic diversity was depleted in Nome populations and was lowest in the Beaufort (Table 2). The east Arctic, on the other hand, featured levels of genetic diversity comparable to the northwest Atlantic and northeast Pacific (Table 2). Interestingly, private haplotypes were detected in all of the regions, with the lowest occurrences in the Beaufort. Tajima’s D was negative in southerly populations (northeast Pacific, northwest Atlantic, and northeast Atlantic), and was zero on average for northern populations (Nome, Beaufort, east Arctic). Of the variables analyzed, significant differences were detected across the regions in the number of polymorphic sites, the number of haplotypes, the number of private haplotypes, and Tajima’s D, generally corresponding to large differences in values between the Beaufort and the northwest Atlantic. Post hoc tests, however, were unable to detect differences between specific regions (SI Appendix, Table S2).
Fig. 1.
PCoA results for genetic distances between the sampled regions. Genetic distances are inferred from 31 species-marker combinations. Positive and negative symbols represent the distributions of coordinates with ± SE.
Table 1.
Genetic distances (ΦST) between pairwise populations in 26 species of marine macroalgae with Arctic populations
| Northeast Pacific | Nome, Alaska | Beaufort, Alaska | East Arctic | Northwest Atlantic | Northeast Atlantic | |
| Northeast Pacific | — | 6 | — | 8 | 8 | 4 |
| Nome, Alaska | 0.438 (0.158) | — | 11 | 20 | 17 | 5 |
| Beaufort, Alaska | 0.471 (0.141) | 0.251 (0.089) | — | 15 | 12 | 5 |
| East Arctic | 0.503 (0.124) | 0.300 (0.059) | 0.338 (0.090) | — | 27 | 11 |
| Northwest Atlantic | 0.751 (0.101) | 0.518 (0.085) | 0.675 (0.089) | 0.347 (0.063) | — | 11 |
| Northeast Atlantic | 0.822 (0.141) | 0.741 (0.149) | 0.867 (0.086) | 0.539 (0.109) | 0.437 (0.131) | — |
Genetic distances were evaluated based on four genetic markers (COI-5P, tufA, ITS, ycf35). Note, the pairwise value between Beaufort and the northeast Pacific was “normalized” by taking the average of genetic distances between Nome and the northeast Pacific, and the east Arctic and northeast Pacific. Sample sizes for pairwise distances are in the top right corner of the table. SE for genetic distances are indicated in parentheses.
Table 2.
Summary statistics of marine macroalgae with Arctic populations
| Location | Northeast Pacific | Nome | Beaufort | East Arctic | Northwest Atlantic | Northeast Atlantic | Overall | P |
| n | 8 | 20 | 15 | 31 | 27 | 11 | 31 | |
| bp | 649 | 639 | 625 | 631 | 632 | 593 | 631 | |
| Npoly | 5.375 | 1.600 | 1.000 | 4.387 | 4.444 | 1.727 | 9.516 | 0.021 |
| 2.187 | 0.461 | 0.390 | 0.931 | 0.894 | 0.740 | 1.433 | ||
| Na | 4.000 | 2.300 | 1.933 | 2.935 | 4.148 | 2.273 | 8.097 | 0.041 |
| 1.102 | 0.242 | 0.330 | 0.321 | 0.574 | 0.702 | 0.995 | ||
| Ne | 1.782 | 1.542 | 1.312 | 1.753 | 1.821 | 1.474 | 2.834 | 0.157 |
| 0.334 | 0.115 | 0.136 | 0.141 | 0.192 | 0.248 | 0.265 | ||
| NPH | 2.500 | 1.000 | 0.625 | 1.323 | 2.655 | 1.538 | — | 0.035 |
| 0.806 | 0.218 | 0.202 | 0.287 | 0.526 | 0.656 | |||
| h | 0.32325 | 0.29515 | 0.14760 | 0.32958 | 0.32674 | 0.20782 | 0.53132 | 0.143 |
| 0.09617 | 0.04919 | 0.05954 | 0.04680 | 0.05140 | 0.07419 | 0.04820 | ||
| θπ | 0.001544 | 0.000744 | 0.000399 | 0.001873 | 0.001383 | 0.000852 | 0.002605 | 0.093 |
| 0.000794 | 0.000268 | 0.000181 | 0.000407 | 0.000569 | 0.000345 | 0.000395 | ||
| D | −1.19122 | −0.02702 | 0.01276 | −0.00114 | −1.06532 | −0.60136 | −0.25835 | 0.012 |
| 0.35562 | 0.29170 | 0.44359 | 0.28512 | 0.21389 | 0.53250 | 0.21314 |
bp = number of base pairs; D = Tajima’s test for neutrality; h = haplotype diversity; n = sample size (number of species); NPH = number of private haplotypes; Npoly = number of polymorphic nucleotide sites; Na = number of haplotypes; Ne = number of effective alleles; θπ = nucleotide diversity; P = Kruskal–Wallis tests for independent distributions of values (Dunn’s post hoc tests with Bonferroni correction did not detect significant pairwise differences). Note sample sizes from northeast Pacific to northeast Atlantic in NPH are 10, 20, 16, 31, 29, 13, and from northeast Pacific to Overall in D, 6, 15, 5, 23, 19, 5, 31. Values for 1 SE are presented below values for each variable.
The presence of private haplotypes in Arctic populations was further highlighted in the haplotype distributions of some species, particularly in the east Arctic. Noteworthy examples included Alaria esculenta (SI Appendix, Figs. S44 and S45), Devaleraea ramentacea (SI Appendix, Figs. S13 and S14), Rhodomela sp. 1virgata (SI Appendix, Fig. S36), and Pylaiella washingtoniensis (SI Appendix, Fig. S74). Some species showed signs of admixture between North Pacific and North Atlantic populations, particularly in Churchill (Hudson Bay) and Northern Labrador (northwest Atlantic); these species included Coccotylus truncatus (SI Appendix, Fig. S12), Eudesme borealis (SI Appendix, Fig. S58), Saccharina latissima (SI Appendix, Fig. S76), Scagelia pylaisaei (SI Appendix, Fig. S39), and possibly Chaetopteris plumosa (SI Appendix, Fig. S49) and Phycodrys fimbriata (SI Appendix, Fig. S26). As well, the genetic make-up of several species populations in the Beaufort was markedly different from conspecifics in Nome (Fig. 1 and Tables 1 and 2), including C. truncatus (SI Appendix, Figs. S11 and S12), Odonthalia dentata (SI Appendix, Fig. S23), P. fimbriata (SI Appendix, Fig. S26), and Rhodomela sibirica (SI Appendix, Figs. S34 and S35).
Hindcasting Species Distributions during the LGM.
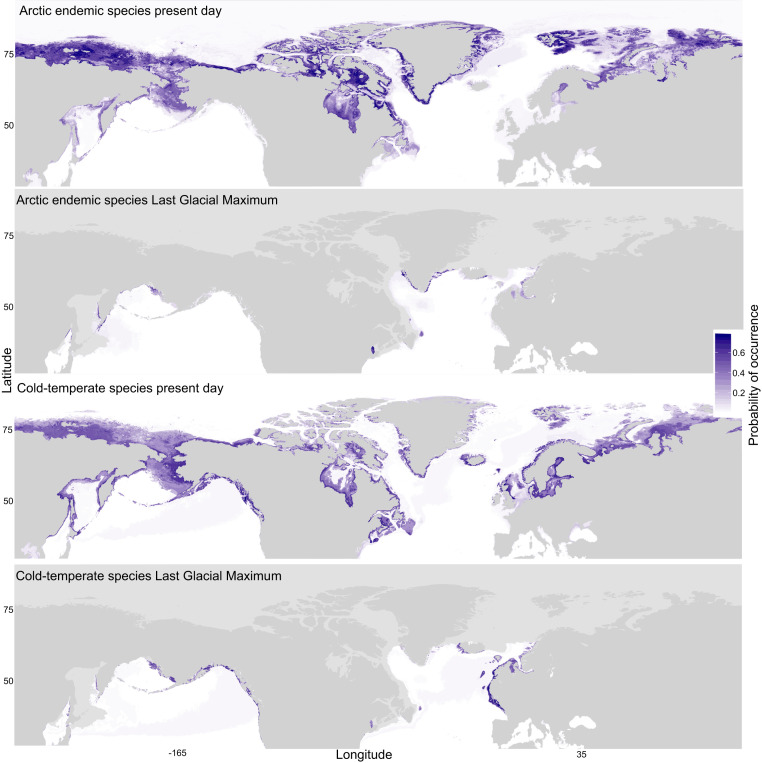
In order to further pinpoint marine refugial locations, we used species distribution models to hindcast the availability of suitable habitat during the LGM (Fig. 2). Hindcasting results differed between Arctic endemic marine forest species (as proxied by the kelp L. solidungula) and cold-temperate species ranging into the Arctic (as proxied by the red alga O. dentata). Cold-temperate species featured a present-day circumpolar distribution with large areas of probable occurrence in the North Pacific and North Atlantic. Projections during the LGM similarly indicated areas of probable occurrence throughout the Pacific and Atlantic basins, although highly reduced in total area compared to the present-day projection and particularly limited in southern Greenland and the northwest Atlantic (Fig. 2). Areas of probable occurrence for Arctic endemics for both time projections were more northerly as compared to cold-temperate species, with clear areas of probable occurrence projected in southern Greenland during the LGM.
Fig. 2.
Present day and LGM distributions of Arctic marine forest species. Arctic endemic species are proxied by the kelp L. solidungula, and cold-temperate species ranging into the Arctic are proxied by the red alga O. dentata. The heatmap represents relative probability of occurrence among global marine locations as proxied by mean annual SSTs and temperatures of the coldest ice-free month. Light gray areas represent persistent ice cover during the LGM.
Discussion
Recolonization of the Arctic Via Multiple Refugial Nuclei.
Our understanding of biodiversity patterns in Arctic marine forests has lagged compared to marine animals. The reported similarity of Arctic marine floral assemblages to Atlantic communities was an observation historically at odds with the Pacific origins inferred in Arctic marine fauna, particularly in the western Arctic (11). Besides noting the “paradox of marine benthic fauna and flora,” Dunton (11) also predicted the importance of molecular data in resolving such disjunct perspectives. Today, with over 4,500 sequence records for more than 100 species, the increased fidelity to resolve historical events in populations of Arctic marine forests can shed light on not just recent origins, but also the potential for evolution and northern adaptation in Arctic marine communities.
The results presented here further reject the earlier hypothesis that Arctic marine forests are entirely or predominantly derived from European conspecifics (10 and 11). This was first evident in the genetic distances, which indicated a series of populations extending from the northwest Atlantic through to the northern Bering Sea distinct from the northeast Atlantic and northeast Pacific (Fig. 1 and Table 1). Furthermore, a decline in diversity from the Atlantic into the Arctic was not evident, which ought to occur in the wake of recolonization (Table 2). In fact, levels of diversity were typically highest in the northwest Atlantic and maintained in the east Arctic, and Tajima’s D test suggested a general trend toward population expansion in the northwest Atlantic, consistent with the presence of refugia (SI Appendix, Tables S2 and S3). Hindcasting also showcased areas of probable occurrence during the LGM in the Bering Sea, southern Greenland, northwest Atlantic (although highly limited), in addition to Europe (Fig. 2). Finally, haplotype patterns in several species presented here strongly imply the presence of multiple refugia in the northwest Atlantic (e.g., Ceramium virgatum) (SI Appendix, Fig. S6).
Clearly the European recolonization hypothesis, stemming from a legacy of morphological identifications and the persistent perception of inhospitable glacial conditions in the northwest Atlantic (10 and 11), can be put to rest. For one, refugia in the northwest Atlantic have been increasingly recognized, downplaying the importance of postglacial recolonization from European refugia (3, 18, and 19). Trans-Atlantic populations of marine flora also typically exhibit genetic divergence, corresponding to isolation events during the Pleistocene, further supporting the notion that an abundant marine flora was available to recolonize the Arctic out of the northwest Atlantic (20). Assis et al. (3) also used ecological niche modeling to hindcast kelp distributions during the LGM, which suggested several species likely survived glaciation in the northwest Atlantic, a result further supported in the hindcasting results presented here (Fig. 2). Other modeling work indicated Arctic and sub-Arctic Atlantic marine forests have evolutionary origins in the North Pacific because of Pleistocene climate and geography, suggesting recurring recolonization occurred out of the Pacific (14). Crucially, the modeling work of Adey et al. (14) demonstrated subarctic habitat was restricted to the North Pacific (Okhost Sea, Kamchatka, western Bering Sea) during glaciation, while Arctic-equivalent habitat ceased to exist. Our population-level data partially support this first assertion [compelling examples of recent Pacific migrants into the Arctic include Ahnfeltia borealis (SI Appendix, Figs. S3 and S4), R. sibirica (SI Appendix, Figs. S34 and S35), and Ulva fenestrata (SI Appendix, Fig. S91)], however, our results indicate Arctic habitat was widely available during the LGM.
Indeed, the emerging story of Arctic marine recolonization is more nuanced than anticipated, even in more recent genetic studies. Pacific origins to Arctic flora since the LGM have been speculated in the past (6 and 10), interpreted in more recent reviews of floristic surveys (21), and increasingly inferred by molecular data (12 and 13). Molecular work has been slow to recognize the significance of unique Arctic lineages that are difficult to reconcile with the emphasis on recolonization hypotheses stemming from refugia south of continental ice sheets. For example, Neiva et al. (13) report “temperate” and “cold” northwest Atlantic phylogroups of S. latissima, the latter of which was largely restricted to the Canadian Arctic and western Greenland, but invoke back-crossing or genotyping errors as possible explanations for the closely related genetic groups. The hesitation to consider northern refugia as a possible explanation (e.g., southern Greenland) likely stems from a lack of comprehensive geographic coverage in genetic sampling, limited insight from single species, and confusion regarding the extent of seasonal ice cover during the LGM. Here, the position of east Arctic genetic distances between populations sampled in the Bering Sea and northwest Atlantic (Fig. 1) and sustained levels of genetic diversity in east Arctic populations (Table 2) can be attributed to a combination of admixture between Pacific and Atlantic populations and vicariance, the latter of which invoke the presence of far northern refugia. Hindcasting identified the most likely area of occurrence for these refugia along the coastlines of southern Greenland, where ice cover was seasonal during the LGM (Fig. 2) (22), particularly in species specifically outfitted to thrive in Arctic conditions (e.g., L. solidungula). It is worth noting hindcasting results are consistent with the work of Assis et al. (3), although these authors also inferred high Arctic refugia in areas we identified as having persistent ice cover (namely Baffin Island and northern Labrador). In sum, the origin of Arctic marine forests, as revealed by our community-level analysis, forward the west Arctic as likely derived from Bering Sea refugia, while the east Canadian Arctic exhibits a “melting pot” character, with input from the Pacific, the Atlantic (predominantly the northwest), and Arctic refugia.
High Northern Refugia Drive Arctic Endemism.
The conclusion that refugia were abundant and occurred far north offers an explanation for marine Arctic endemism, which is generally assumed to be rare or impossible given the perception of intolerable ice conditions during the LGM. Lee (10) suggested less than 7% of the Arctic flora was confined to the Arctic and perceived low levels of adaptation, characterizing the flora as “ecologically immature.” More recent estimates bump up the level of Arctic endemism to 13.5% (23). Molecular taxonomic work reflects the trend toward recognizing Arctic affinities, for example, in the recently described species A. borealis, Chorda borealis, and E. borealis (https://www.algaebase.org). Here, 21 species (19%) of the sampled flora appeared to be confined to the Arctic, not to mention the cryptic lineages reported by Laughinghouse et al. (15), Küpper et al. (16), and species with distinct Arctic populations reported here (e.g., A. esculenta) (SI Appendix, Figs. S44 and S45). Altogether, these results suggest that a large portion of the Arctic marine flora has persisted far north through cycles of glaciation, raising the possibility that Arctic marine forests do not simply tolerate but have adapted to high-latitude conditions since the development of modern-day polar conditions coincident with global cooling during the Cenozoic (66 Ma onwards). Note that while red and green seaweeds are among the oldest lineages of Eukaryotic life (24 and 25), brown seaweeds (Phaeophyceae) likely emerged early in the Mesozoic and possibly radiated in response to global cooling during the Cenozoic (26 and 27).
The implications of these findings extend beyond marine forests. Patterns in accompanying hard-bottom marine fauna, some of which thrive in the habitat provided by macroalgae, are readily resolved by invoking northern refugia. A phylogeographic review of Arctic marine fauna highlighted substantial geographical subdivision in the COI-5P complex of the polychaete Harmothoe imbricata, with lineages that appeared to be restricted to the east Canadian Arctic (28). The presence of cryptic Arctic lineages was further revealed in polychaetes (29), molluscs (30), and amphipods (31). In concert with the results for Arctic forests, these studies demonstrate the resilience of Arctic marine communities to cycles of glaciation and the potential for adaptation to extreme-cold environments. Given the relative scarcity of genetic surveys in the Arctic, these results also suggest levels of Arctic marine endemism remain underestimated, a disparity that is likely further exacerbated at the population level.
The species pump hypothesis, initially forwarded by Haffer (32), proposes that isolation of populations into highly localized refugia during glaciation drives speciation. Theoretically, this concept should find strong evidence in the marine environment. Eustatic sea-level fluctuations, which precipitated a >100-m drop in sea level and exposed continental shelves during the LGM, led to drastic shifts in the distribution of coastal marine ecosystems, pushing benthic habitats, such as coral reefs and marine forests to narrow strips of the upper continental slopes (33) (Fig. 2). Our results can be interpreted within the context of a species pump mechanism, driving diversity in high-latitude marine benthic environments. Several of the species surveyed displayed compelling examples of unique lineages seemingly restricted to the Arctic (e.g., D. ramentacea (SI Appendix, Figs. S13 and S14) and A. esculenta (SI Appendix, Fig. S44)), and the total number of genotypes sampled had a clear “Arctic-only” component (SI Appendix, Fig. S2). The timeframe for cycles of glaciation (41- to 100-ka cycles over the past ∼2.6 Ma) (1), however, may not be sufficient to produce novel species in marine benthic taxa. In fact, the species pump has seen little support in other marine systems, such as coral-reef fishes (34) and tropical rocky shore assemblages (35), and in marine forests the species pump appears to be limited to driving population differentiation and incipient speciation (13 and 36). Nonetheless, taking stock of diversity at high latitudes ought to consider cryptic lineages born out of cycles of glaciation, regardless of whether these constitute novel species or population genotypes, as both serve to bolster the resilience of ecosystems in the face of environmental change (17).
On a final note, it is possible northern refugia are not confined to the southern coast of Greenland. Alhough the western Arctic was locked in multiyear sea ice, portions of the Siberian coastline as far east as the Laptev Sea appear to have remained seasonally ice-free at least as early as 16 ka (37). This was due to warm Atlantic water entering the Arctic through the Fram Strait, which may have provided Arctic refuge for marine forests. The role of katabatic winds and the formation of polynyas, recurrent areas of ice-free water, also may have played a role maintaining a seasonally ice-free shoreline (37). The modern-day Arctic features numerous polynyas, particularly along the margins of the Arctic basin, enhancing early spring productivity and creating biodiversity hotspots (38). Lee (10) even describes seaweed communities in a polynya near Brock island (78°N), including L. solidungula, Desmarestia viridis, and Turnerella pennyi. Hypotheses regarding the role of polynyas maintaining Arctic refugial locations during the LGM may need to be invoked to explain the population structure of the Beaufort, which is oddly more genetically distant from Atlantic conspecifics than Nome populations (which are geographically farther away) (Fig. 1). We do caution in interpreting this pattern, however, given the limited sampling of the west Arctic/Bering Sea, lack of sampling from Russia, and “founder-takes-all” effects, which can result in sharp demarcations in macroalgal population structure, even at small spatial scales (39). Even so, if history is an indication, we need to temper our assumptions regarding the inability for marine forests to flourish in areas with “harsh” ice conditions.
Conclusions
Over the decades, our understanding of Arctic marine forests has gradually evolved from a one-vector ecosystem stemming from European refugia to many melding pathways, and from a biologically depauperate expanse into a complex genetic landscape. Ultimately, our goal was to provide a checkpoint in our understanding of Arctic marine diversity, particularly by sampling Arctic locations where DNA barcode surveys are rare or absent (i.e., Bering Sea, east Arctic, northern Europe) and by summarizing genetic patterns across macroalgal species with sequence data from the Arctic. We emphasize the significance of summarizing insight across species, with over 100 species examined through this work spanning 3 phyla and 2 kingdoms of life, and extensively so in 26 species (SI Appendix, Fig. S1 and Table S1). Single-species studies have struggled to reconcile postglacial dynamics, particularly where findings do not align with prevailing views (e.g., refs. 13 and 15). As well, a population genetic approach to resolving these patterns in Arctic marine communities oftentimes falls short of conclusive because of the inherent difficulty in sampling any individual location and subsequent limited geographical coverage for analysis. Ecological niche modeling can close these gaps in our knowledge by highlighting likely locations of persistence during glaciation and aiding in the interpretation of genetic patterns.
We forward the view that the marine Arctic environment was recolonized from numerous and globally distributed source populations, including unrecognized far northern refugia that additionally contribute to endemism in polar waters. This work further supports the view that complex evolutionary processes are born out of the Arctic environment, particularly the interplay between incipient speciation and secondary contact (species-pump), and suggests Arctic marine benthic ecosystems are adapted to polar environments (evolution of Arctic assemblages throughout the Cenozoic and into the Pleistocene) rather than exclusively being cold-tolerant extensions of temperate flora/fauna. Most prescient, however, our results indicate Arctic coastal systems are unique in their composition, insight that comes at a time when the Arctic is most at risk from climate change impacts. Amplification of warming trends (2) and the rapid decline of perennial ice cover has left an uncertain path forward for the Arctic biome. For the marine realm, the answer is not simple; while increased temperatures and the loss of multiyear sea ice will likely open new Arctic habitat, unstable coastlines due to permafrost melt and changes to water chemistry are likely to hamper potential gains in marine forests, with these impacts unevenly distributed throughout the Arctic ocean (7). Net gains and losses of diversity in the Arctic therefore remains an area of active research (3), and will continue to improve as DNA sequence data enhance our knowledge of cryptic taxa and high-latitude endemic species. As such, taking stock of biodiversity remains among the first steps toward monitoring the resilience of Arctic marine ecosystems to ongoing environmental changes.
Materials and Methods
Genetic Data and Analyses.
Marine macroalgae were sampled in several key locations across the Arctic over the course of 5 y, including Japan; Kamchatka (Russia); Haida Gwaii, British Columbia, Canada; Nome, Alaska (northern Bering Sea); the Beaufort Sea, northern Alaska; Cambridge Bay (Nunavut, Canada); Hudson Bay, Manitoba, Canada; Baffin Island, through northern Labrador to Makkovik, Canada; and Bergen, Norway. Marine macroalgae were generally collected from the intertidal and via scuba, but occasionally via dredge. A portion of each specimen (∼1 cm2) was preserved in silica gel for DNA extraction, while several representatives of putative species were preserved as pressed vouchers. Specimens were brought back to the University of New Brunswick (where specimens are stored) for DNA extraction. Several genes were amplified, including the 5′ end of the cytochrome c oxidase subunit I gene (COI-5P) in red and brown algae, tufA in green algae, and partial reads of the ribulose-1, 5-biphosphate carboxylase large subunit (rbcL-3P) (SI Appendix, Table S4) in red and brown algae. Secondary markers were acquired in select species, including the full-length nuclear internal transcribed spacer region (ITS) and plastid ycf35, in order to further clarify or support COI-5P patterns (SI Appendix, Table S4). Successful PCR products were sent to Genome Quebec for forward and reverse sequencing. All genetic data were edited in Geneious v8.0 (40), and any relevant previously published DNA barcodes were added to the dataset.
Populations were pooled for analysis according to broad geographic regions: northeast Pacific (British Columbia, Canada, and Washington state, United States); Nome, Alaska (proxy for Pacific migrants into the Arctic); Beaufort, Alaska (proxy for the west North American Arctic); east Canadian Arctic with the southern distribution delimited using the 10 °C air temperature isotherm for July (Cambridge Bay, Nunavut, through to Nain, Labrador); northwest Atlantic (Makkovik, Labrador, and southwards, including the Canadian Atlantic provinces and New England States, United States); northeast Atlantic (Europe) (Fig. 2). The northwest Pacific was excluded given the paucity of data. Species generally corresponded to Barcode Index Numbers, a binning system provided through the Barcode of Life Datasystem (BOLD), which utilizes a fluid threshold to proxy species units based on levels of intra- and interspecific genetic variation (41). Species were included in genetic analyses provided they featured in at least one Arctic region for which ≥10 individuals were sampled. Of 109 species sampled with Arctic populations, we analyzed 26 (SI Appendix, Fig. S1), 5 of which had sequence data from multiple markers, for a total of 31 species-marker combinations. All 109 species were considered in additional analyses evaluating total diversity levels in the Arctic, which were used to quantify the number of Arctic endemic species. These analyses are presented as supplementary material, and include accumulation curves of all genotypes sampled (42) (SI Appendix, Fig. S2), a table interpreting haplotype patterns in all 109 species sampled (SI Appendix, Table S1), and haplotype maps and networks (SI Appendix, Figs. S3–S95).
Various populations statistics were calculated for each region, including measures of genetic diversity, genetic differentiation, and Tajima’s D test for neutrality. Sequences were truncated to the shortest length sequence within each species prior to all genetic analyses. GenAlEx 6.51 (43) was used to run an analysis of molecular variance (AMOVA) and derive values of PhiST (ΦST), an analog to Fst that incorporates nucleotide diversity in distance calculations (i.e., haplotypes are not assumed to be equidistant from each other). Calculations for ΦST followed that of Meirmans (44). Pairwise tests for significant ΦST values were conducted using 9,999 permutations of the dataset. Null values for ΦST occurred wherein two populations were monotypic for the same haplotype and were changed to 0 (no genetic differentiation). Species-specific analyses are presented in SI Appendix, Table S5. ΦST values were averaged for each region across all species, and a PCoA was conducted on the pairwise distance matrix using the covariance-standardized method in GenAlEx. Given that notably few measurements were available between the northeast Pacific and the Beaufort (only two species, with two to seven records from the Beaufort, and no genetic differentiation), we “normalized” this pairwise distance by taking the average of genetic distances between the northeast Pacific and Nome, and the northeast Pacific and the east Arctic. The unaltered PCoA figure is presented in SI Appendix, Fig. S96. We also conducted the same analysis with low sample-size populations removed (<10 individuals) (SI Appendix, Fig. S97). Alternate PCoAs revealed the same pattern, except the Beaufort grouped closer to the northeast Pacific when not “normalized.”
Frequency-based parameters were also calculated for each region within each species, including the number of haplotypes (Na), the number of effective alleles (Ne), the number of private haplotypes within populations (NPH), and haplotype diversity (h). Ambiguous sites were removed for these calculations. DnaSP v6 (45) was also used to calculate the number of polymorphic sites, nucleotide diversity (θπ), and Tajima’s D statistic with accompanying P values. These results were again averaged across all species-marker combinations, and differences in measures between populations were assessed using Kruskal–Wallis H tests (46). Dunn’s post hoc tests with Bonferroni corrections were performed for variables yielding significant Kruskal–Wallis H tests. The analyses for individual species are presented in SI Appendix, Table S3, while the post hoc tests are presented in SI Appendix, Table S2.
Hindcasting Species Distributions.
The extent of marine forests during the LGM was inferred using maximum entropy modeling of species distributions. This approach utilizes presence-only data to estimate the relationships between species occurrences and accompanying environmental data, which are then used to estimate the most uniform distribution under those constraints (maximizing dispersedness/entropy) (47). Modern and paleo marine environmental data layers were downloaded from MARSPEC (48 and 49), in particular, bathymetry, mean annual sea surface temperatures (SST), and SST of the coldest ice-free month. Seaweed distributions are highly tuned to marine isotherms, and ecological niche modeling consistently indicate SST is the most important variable in modeling macroalgal species distributions (3). Paleo environmental data layers represent average values as derived from six coupled ocean atmosphere general circulation climatic models, including salinity-adjusted CCSM3. Occurrence records were gathered for O. dentata and L. solidungula. These species were selected to proxy distributions in boreal to temperate species with Arctic populations (i.e., the historical view of Arctic marine forests; O. dentata), and putative Arctic endemic species (L. solidungula). These species were also selected given they are reliably distinguished morphologically, and, as such, historical records would not be conflated by cryptic species. Occurrence records were derived from Lüning (6). Distribution maps were scanned and georeferenced, and GPS locations for occurrences were subsequently derived. For continuous distributions, GPS coordinates were haphazardly recorded at ∼250-km intervals. Historical occurrence records were then pooled with locations from DNA barcode records (BOLD), and the Macroalgal Herbarium Portal (https://macroalgae.org/portal/index.php). To correct for sampling bias during training of the ecological niche models, occurrence records in close proximity to each other were randomly removed using the R package spThin (50), with a thinning parameter of 100 km. The resulting datasets kept 111 of 260 occurrence records for O. dentata, and 87 of 164 for L. solidungula (51). The thinned occurrence records and environmental layers were then trained using Maxent (52) and projected onto conditions during the LGM. Models were built using threshold features in order to better reflect lethal temperature limits in macroalgae, and model performance was assessed using cross validation. A regularization multiplier of 1 was used, as was a default prevalence of 0.5 (the probability of presence at average presence locations). Clamping was used to restrict variables outside the training range. Multivariate environmental similarity surfaces were also used to evaluate the distribution of environmental values outside the training data range projected during the LGM, which functioned to indirectly map persistent ice cover and restrict inferences of refugial locations to seasonally ice-free waters. Output asc files were converted to figures in R using ggmap and ggplot2 packages (50).
Supplementary Material
Acknowledgments
We thank the many people who facilitated the collection of specimens: Dr. Meghann Bruce, Dr. Kyatt Dixon, Kirby Morrill, Dr. Amanda Savoie; Dr. Kenneth Dunton at the University of Texas for facilitating our sampling in the Beaufort (northern Alaska); Dr. Yotsukura Norishige at Hokkaido University for providing collections from Hokkaido; Dr. Kjersti Sjøtun at the University of Bergen for facilitating our sampling in Norway; Don Stiles and James Horner for facilitating the collection of samples in Nome, Alaska; Laura Borden for providing collections from Cambridge Bay (Nunavut); and Dr. Selivanova and Dr. Zhigadlova for providing specimens from Kamchatka (Russia). We thank those who helped generate COI-5P data, particularly Tanya Moore, as well as Alex Geoffroy and Line Le Gall for providing trace files; Drs. Jorge Assis and Ester Serrão for providing critical feedback regarding analyses; Dr. Jason Addison for providing critical input leading to the inception of this manuscript; and the health professionals and other essential service providers around the world that have been working on the “front lines” during the SARS-CoV-2 pandemic, which began while this manuscript was under review. We also recognize the Traditional Inhabitants of both ceded and unceded territory on which research is conducted, in particular for this current research the Inuit (Canadian Arctic), the Nunatsiavut (Labrador), and Iñupiat (Northern Alaska). We also acknowledge that gains in contemporary knowledge invariably build on a history of race and gender discrimination. This project was funded by the Northern Scientific Training Program, the Natural Sciences & Engineering Research Council of Canada through a Natural Sciences and Engineering Research Council Post-Graduate Scholarship (to T.T.B.), Discovery Grant 170151-2013 (to G.W.S.), the New Brunswick Innovation Foundation, and the University of Melbourne McKenzie Postdoctoral Fellowship program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002753117/-/DCSupplemental.
Data Availability.
Specimen records, including pictures, collection information, sequence data, and GenBank accessions can be accessed via Figshare (51) and BOLD (53).
References
- 1.Miller G. H. et al., Temperature and precipitation history of the Arctic. Quat. Sci. Rev. 29, 1679–1715 (2010). [Google Scholar]
- 2.Miller G. H. et al., Arctic amplification: Can the past constrain the future? Quat. Sci. Rev. 29, 1779–1709 (2010). [Google Scholar]
- 3.Assis J., Araújo M. B., Serrão E. A., Projected climate changes threaten ancient refugia of kelp forests in the North Atlantic. Glob. Change Biol. 24, e55–e66 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Small E., Kelps: The key to sustainable harvest of marine biodiversity. Biodiversity (Nepean) 19, 1–13 (2018). [Google Scholar]
- 5.Wernberg T., Filbee-Dexter K., Missing the forest for the trees. Mar. Ecol. Prog. Ser. 612, 209–215 (2019). [Google Scholar]
- 6.Lüning K., Seaweeds: Their Environment, Biogeography, and Ecophysiology, (Wiley, New York, 1990). [Google Scholar]
- 7.Filbee-Dexter K., Wernberg T., Fredriksen S., Norderhaug K. M., Pedersen M. F., Arctic kelp forests: Diversity, resilience and future. Global Planet. Change 172, 1–14 (2019). [Google Scholar]
- 8.Krause-Jensen D. et al., Deep penetration of kelps offshore along the west coast of Greenland. Front. Mar. Sci. 6, 1–7 (2019). [Google Scholar]
- 9.Dunton K. H., Schell D. M., Seasonal carbon budget and growth of Laminaria solidungula in the Alaskan high Arctic. Mar. Ecol. Prog. Ser. 31, 57–66 (1986). [Google Scholar]
- 10.Lee R. K. S., General ecology of the Canadian Arctic benthic marine algae. Arctic 26, 32–43 (1973). [Google Scholar]
- 11.Dunton K., Arctic biogeography: The paradox of the marine benthic fauna and flora. Trends Ecol. Evol. (Amst.) 7, 183–189 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Saunders G. W., McDevit D. C., DNA barcoding unmasks overlooked diversity improving knowledge on the composition and origins of the Churchill algal flora. BMC Ecol. 13, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neiva J. et al., Glacial vicariance drives phylogeographic diversification in the amphi-boreal kelp Saccharina latissima. Sci. Rep. 8, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adey W. H., Lindstrom S. C., Hommersand M. H., Müller K. M., The biogeographic origin of Arctic endemic seaweeds: A thermogeographic view. J. Phycol. 44, 1384–1394 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Laughinghouse H. D. 4th et al., Evolution of the Northern Rockweed, Fucus distichus, in a regime of glacial cycling: Implications for benthic algal phylogenetics. PLoS One 10, e0143795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Küpper F. C. et al., Arctic marine phytobenthos of northern Baffin Island. J. Phycol. 52, 532–549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver T. H. et al., Biodiversity and resilience of ecosystem functions. Trends Ecol. Evol. (Amst.) 30, 673–684 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Wares J. P., Cunningham C. W., Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55, 2455–2469 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Maggs C. A. et al., Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89 (suppl. 11), S108–S122 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Bringloe T. T., Saunders G. W., Mitochondrial DNA sequence data reveal the origins of postglacial marine macroalgal flora in the northwest Atlantic. Mar. Ecol. Prog. Ser. 589, 45–58 (2018). [Google Scholar]
- 21.Wulff A. et al., Biodiversity, biogeography and zonation of marine benthic micro- and macroalgae in the Arctic and Antarctic. Bot. Mar. 52, 491–507 (2009). [Google Scholar]
- 22.Pflaumann U. et al., Glacial North Atlantic: Sea-surface conditions reconstructed by GLAMAP 2000. Paleoceanography 18, 1–21 (2003). [Google Scholar]
- 23.Wilce R. T., The “Arctic Stamp”, its imprint on endangered marine flora. Pers. Phycol. 3, 155–180 (2016). [Google Scholar]
- 24.Leliaert F. et al., Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 31, 1–46 (2012). [Google Scholar]
- 25.Yang E. C. et al., Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci. Rep. 6, 21361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silberfeld T. et al., A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): Investigating the evolutionary nature of the “brown algal crown radiation”. Mol. Phylogenet. Evol. 56, 659–674 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Starko S. et al., A comprehensive kelp phylogeny sheds light on the evolution of an ecosystem. Mol. Phylogenet. Evol. 136, 138–150 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Hardy S. M. et al., Biodiversity and phylogeography of Arctic marine fauna: Insights from molecular tools. Mar. Biodivers. 41, 195–210 (2011). [Google Scholar]
- 29.Carr C. M., Hardy S. M., Brown T. M., Macdonald T. A., Hebert P. D. N., A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS One 6, e22232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Layton K. K. S., Martel A. L., Hebert P. D. N., Patterns of DNA barcode variation in Canadian marine molluscs. PLoS One 9, e95003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tempestini A., Rysgaard S., Dufresne F., Species identification and connectivity of marine amphipods in Canada’s three oceans. PLoS One 13, e0197174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haffer J., Speciation in Amazonian forest birds. Science 165, 131–137 (1969). [DOI] [PubMed] [Google Scholar]
- 33.Norris R. D., Hull P. M., The temporal dimension of marine speciation. Evol. Ecol. 26, 393–415 (2012). [Google Scholar]
- 34.Rocha L. A., Bowen B. W., Speciation in coral-reef fishes. J. Fish Biol. 72, 1101–1121 (2008). [Google Scholar]
- 35.Williams S. T., Reid D. G., Speciation and diversity on tropical rocky shores: A global phylogeny of snails of the genus Echinolittorina. Evolution 58, 2227–2251 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Bringloe T. T., Saunders G. W., Trans-Arctic speciation of Florideophyceae (Rhodophyta) since the opening of the Bering Strait, with consideration of the “species pump” hypothesis. J. Biogeogr. 46, 694–705 (2019). [Google Scholar]
- 37.Bradley R. S., England J. H., The Younger Dryas and the sea of ancient ice. Quat. Res. 70, 1–10 (2008). [Google Scholar]
- 38.Christine M., “Marine ecosystems” in Arctic Biodiversity Assessment: Status and Trends in Arctic Biodiversity, Barry T., Ed. (Narayana Press, Denmark, 2013), pp. 378–419. [Google Scholar]
- 39.Neiva J., Pearson G. A., Valero M., Serrão E. A., Fine-scale genetic breaks driven by historical range dynamics and ongoing density-barrier effects in the estuarine seaweed Fucus ceranoides L. BMC Evol. Biol. 12, 78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kearse M. et al., Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratnasingham S., Hebert P. D. N., A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS One 8, e66213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colwell R. K., Statistical estimation of species richness and shared species from samples. http://viceroy.colorado.edu/estimates/. Accessed 12 August 2019.
- 43.Peakall R., Smouse P. E., GenAlEx 6.5: Genetic analysis in excel. Population genetic software for teaching and research—An update. Bioinformatics 28, 2537–2539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meirmans P. G., Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60, 2399–2402 (2006). [PubMed] [Google Scholar]
- 45.Rozas J. et al., DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 34, 3299–3302 (2017). [DOI] [PubMed] [Google Scholar]
- 46.IBM Corporation , IBM SPSS Statistics for Windows v25.0, (IBM Corp., Armonk, NY, 2017). [Google Scholar]
- 47.Elith J. et al., A statistical explanation for MaxEnt for ecologists. Divers. Distrib. 17, 43–57 (2010). [Google Scholar]
- 48.Sbrocco E. J., Barber P. H., MARSPEC: Ocean climate layers for marine spatial ecology. Ecology 94, 979 (2013). [Google Scholar]
- 49.Sbrocco E. J., Paleo-MARSPEC: Gridded ocean climate layers for the mid-Holocene and Last Glacial Maximum. Ecology 95, 1710 (2014). [Google Scholar]
- 50.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). https://www.R-project.org/.
- 51.Bringloe T. T., Marine macroalgae DNA barcode specimen data. Figshare. 10.6084/m9.figshare.11301929.v3. Deposited 22 January 2020. [DOI]
- 52.Phillips S. J., Dudík M., Modelling of species distributions with Maxent: New extensions and a comprehensive evaluation. Ecography 31, 161–175 (2008). [Google Scholar]
- 53.Bringloe T. T., Recolonization of Arctic marine forests since the Last Glacial Maximum: Genetic surveys and hindcasting reveal geographically widespread source populations, including high northern refugia. Barcode of Life Data system. http://www.boldsystems.org/index.php/Public_SearchTerms?query=DS-TAMMA. Deposited 19 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Specimen records, including pictures, collection information, sequence data, and GenBank accessions can be accessed via Figshare (51) and BOLD (53).