Significance
Penguins have long been of interest to scientists and the general public, but their evolutionary history remains unresolved. Using genomes, we investigated the drivers of penguin diversification. We found that crown-group penguins diverged in the early Miocene in Australia/New Zealand and identified Aptenodytes (emperor and king penguins) as the sister group to all other extant penguins. Penguins first occupied temperate environments and then radiated to cold Antarctic waters. The Antarctic Circumpolar Current’s (ACC) intensification 11.6 Mya promoted penguin diversification and geographic expansion. We detected interspecies introgression among penguins, in some cases following the direction of the ACC, and identified genes acting on thermoregulation, oxygen metabolism, and diving capacity that underwent adaptive evolution as they progressively occupied more challenging thermal niches.
Keywords: penguin, Antarctica, genome, ancestral niche, ancestral distribution
Abstract
Penguins are the only extant family of flightless diving birds. They currently comprise at least 18 species, distributed from polar to tropical environments in the Southern Hemisphere. The history of their diversification and adaptation to these diverse environments remains controversial. We used 22 new genomes from 18 penguin species to reconstruct the order, timing, and location of their diversification, to track changes in their thermal niches through time, and to test for associated adaptation across the genome. Our results indicate that the penguin crown-group originated during the Miocene in New Zealand and Australia, not in Antarctica as previously thought, and that Aptenodytes is the sister group to all other extant penguin species. We show that lineage diversification in penguins was largely driven by changing climatic conditions and by the opening of the Drake Passage and associated intensification of the Antarctic Circumpolar Current (ACC). Penguin species have introgressed throughout much of their evolutionary history, following the direction of the ACC, which might have promoted dispersal and admixture. Changes in thermal niches were accompanied by adaptations in genes that govern thermoregulation and oxygen metabolism. Estimates of ancestral effective population sizes (Ne) confirm that penguins are sensitive to climate shifts, as represented by three different demographic trajectories in deeper time, the most common (in 11 of 18 penguin species) being an increased Ne between 40 and 70 kya, followed by a precipitous decline during the Last Glacial Maximum. The latter effect is most likely a consequence of the overall decline in marine productivity following the last glaciation.
Few organisms have been as successful at colonizing the globe as seabirds, a large ecological assemblage of oceanic and nearshore species that undergo some of the most remarkable foraging and migratory journeys on Earth (1, 2). Despite their ubiquitous presence, surprisingly little is known about the mechanisms that spurred their diversification and allowed their adaptation to diverse and often dynamic oceanic habitats.
As the only living clade of flightless diving birds, penguins (order Sphenisciformes) occupy both terrestrial and marine habitats. They forage across a wide range of ocean temperatures and depths, from Antarctic to tropical waters (3). Our understanding of penguin diversification and adaptation is hampered by disagreements about their phylogenetic relationships (4–11) and the chronology of their radiation. Estimates made using few genetic markers from different parts of the genome (4–9, 11) recover discordant results, as genomic regions vary in their mutation rates and evolutionary histories, including unknown patterns of gene introgression when different species hybridize (12). The divergence times of ordinal crown age also vary, spanning the Miocene and Eocene (9.9 to 47.6 Mya) (4, 5, 8, 9), yet the earliest crown group penguin fossil dates to the late Miocene (13).
Reconstructions of ancestral distributions and climatic niches are critical to our understanding of penguin diversification. Existing hypotheses conflict: positing either an Antarctic origin with later expansion toward warmer areas (5) or a sub-Antarctic origin with subsequent colonization of Southern Ocean islands and Antarctica (7, 11). Testing these alternative hypotheses on a broad scale requires accurate knowledge of the pattern and chronology of penguin phylogeny. Candidate drivers of diversification have been hypothesized and provide an explicit framework for testing. The extent of ice and changes in the currents of the Southern Ocean during repeated glacial cycles likely played a significant role in the structuring and lineage diversification of seabird populations (14), including penguins (15, 16). Population sizes of several penguin species contracted during glaciation and expanded during postglacial periods (17). Sharp changes in these effective population sizes suggest that the diversification of penguins has been sensitive to climate shifts.
Biogeographic boundaries in the Southern Ocean, particularly the Antarctic Polar Front (APF) and the Subtropical Front (STF), serve as barriers to dispersal for some penguin species (15, 16, 18, 19). Differences in abiotic (e.g., temperature and salinity) and biotic (e.g., types of food resources) variables on either side of these two fronts may promote local adaptation and enable niche divergence among penguins (20). Furthermore, the associated currents have varied significantly over time in latitude and strength in response to changing global circulation patterns (21). These changes have been implicated in the colonization, isolation, and extinction of some penguin populations and species (4, 9).
We report here on our reconstruction of the history of penguin diversification and adaptation using 22 newly sequenced genomes representing 18 extant species and one outgroup. The aim of this work is to address current uncertainties about the phylogenetic relationships of penguins and to uncover the relationships among the timing of penguin diversification, past climate changes, and oceanographic characteristics. Toward this end, we reconstructed the biogeographical areas of diversification and environmental niche through space and time for all penguin species and studied the adaptations of penguins across environmental gradients. We used reconstructions of ancestral effective population sizes to evaluate sensitivity to past climate changes and hence the potential that such changes influenced penguin diversification. Finally, we assessed the extent of introgression between species, a factor that might have contributed to previous disagreements over phylogenetic reconstructions.
Results
The penguin and petrel outgroup genomes were sequenced at ∼30× coverage (SI Appendix, Tables S1–S3). All but one genome sequence contained >90% of the conserved single-copy genes identified across avian genomes (complete Benchmarking Universal Single-Copy Orthologs [BUSCOs] >90%) (SI Appendix, Table S4 and Figs. S1–S4). From the assembled genomes, we extracted 23,108 loci: ultra-conserved elements (UCE; 4,057 loci), coding sequences (CDS; 11,011), and introns (8,040). We applied species tree and concatenation methods to construct a phylogenetic hypothesis for extant penguins using all loci combined and for each set of loci independently (Fig. 1 and SI Appendix, Figs. S5–S8 and Tables S5 and S6).
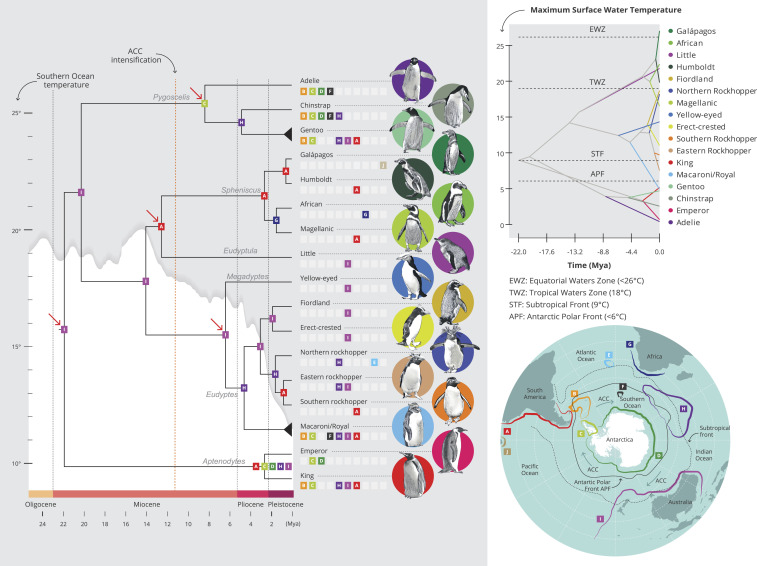
Fig. 1.
Evolutionary history of penguins. Phylogenetic hypothesis of penguin species and divergence time estimates using the UCE dataset. Red arrows represent the four fossil calibration points. The fifth point corresponds to the outgroup node, which is not shown in this figure (see SI Appendix, Fig. S10). Each node is represented by the ancestral distribution before the cladogenesis event using ancestral range reconstruction based on the best-fitting model (DIVALIKE+J) and is associated with one or more of the 10 geographic locations depicted in the map at the bottom right (letters A to J and color codes); areas at branch tips represent the current range of species. Past temperature of the Southern Ocean is represented by the white graph behind the phylogeny (22, 23); onset of the strengthening of the ACC, by the dashed red line (24, 25). (Top Right) Ecological niche disparity through time (DTT) for penguins, with the phylogeny projected onto niche parameter space on the y-axis (maximum surface water temperature) with predicted niche occupancy (PNO) over time (x-axis) reconstructed for internal nodes.
Phylogenetic History and Patterns of Introgression of Crown Group Penguins.
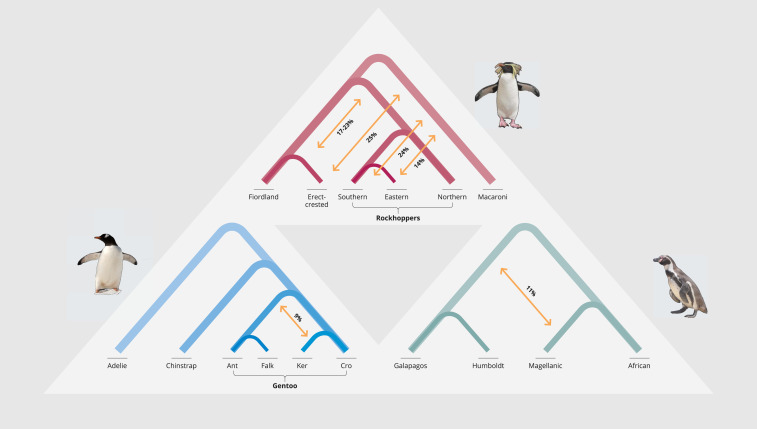
All analyses supported a similar phylogenetic hypothesis, placing Aptenodytes as the sister clade to all other extant penguin species (Fig. 1), distinct from other clades comprising the genera Pygoscelis, Spheniscus+Eudyptula, and Megadyptes+Eudyptes. The phylogeny based on mitogenomes was similar to that retrieved from the genomic datasets. There were a few minor differences among Eudyptes penguins that are likely explained by genome-wide introgression among closely related species (Fig. 2 and SI Appendix, Figs. S7 and S9 and Table S7). Some of the main episodes of genomic introgression were detected among erect-crested and the ancestral rockhopper penguin species (17 to 23%), erect-crested and macaroni/royal penguins (25%), and the Galápagos/Humboldt ancestor and Magellanic penguins (11%) (Fig. 2 and SI Appendix, Fig. S9). Signatures of more recent introgression episodes included genomic contributions from eastern and southern rockhopper penguins to the erect-crested penguin (SI Appendix, Fig. S9). Divergence of the crown group penguins was estimated to have been in the early Miocene, at 21.9 Mya (95% CI, 19.06 to 25.19 Mya) (Fig. 1 and SI Appendix, Fig. S10 and Table S8).
Fig. 2.
Summary of the main introgression episodes among penguin taxa. Arrows represent the percentage of introgression between taxa for all eight combinations of the five taxon statements evaluated. Only episodes involving >2% introgression are shown. More details, including additional signatures of introgression, are provided in SI Appendix, Fig. S9 and Table S7. Species are identified by their common names (and sampled populations in the case of gentoo penguins); see Fig. 1 for correspondence to genus names.
Biogeographic History and Ancestral Niche Reconstruction.
The reconstruction of ancestral distributions identified the coastlines of Australia, New Zealand, and nearby islands as the most likely range of the ancestor of extant penguins (Fig. 1 and SI Appendix, Fig. S11 and Table S9). The first branching event led to the establishment of the genus Aptenodytes in the Antarctic, and reconstructions of the ancestral Pygoscelis species indicate that they colonized the Antarctic Peninsula (area C; Fig. 1 and SI Appendix, Table S9) soon after Aptenodytes, pointing to a long history of Antarctic occupation. In the mid-Miocene, the lineage leading to the Spheniscus/Eudyptula ancestor colonized the South American coast (A), with members of the genera Eudyptes, Eudyptula, Megadyptes, and Spheniscus progressively diversifying and colonizing warmer at-sea environments (Fig. 1).
Consistent with the above findings, our reconstruction of sea surface temperatures (SST) as a proxy for ecological niche disparity through time (DTT) suggests that penguins originated from areas with a maximum SST of 9 °C (Fig. 1 and SI Appendix, Figs. S12 and S13), which broadly corresponds to present-day temperatures of sub-Antarctic waters (26). Southern and eastern rockhopper penguins have retained a thermal preference for a maximum SST of 9 °C, while other closely related species (e.g., northern rockhopper, fiordland, erect-crested) have shifted toward warmer at-sea conditions (SST of 11 to 17 °C) (Fig. 1 and SI Appendix, Fig. S13). This thermal shift was most likely driven by divergence across the STF over the past 4.5 million years. In contrast, macaroni penguins have shifted to occupy colder at-sea conditions near Antarctica to feed in the nutrient-rich sub-Antarctic waters (SI Appendix, Fig. S13). Lower-latitude geographical locations, such as the South African continental coasts (G) and the Galápagos Islands (J), were colonized during the Pleistocene (0.59 Mya). Galápagos penguins are present in Pacific tropical waters near the Equator, where SST reach up to 27 °C, and exhibit a significantly greater thermal tolerance (>6 °C higher) than their sister species, Humboldt penguins (DTT results, Fig. 1).
Genes under Positive Selection.
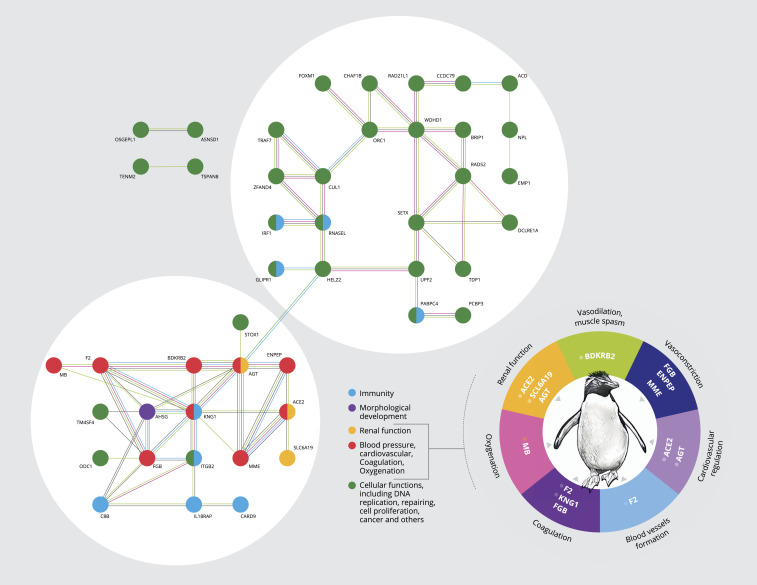
We detected 104 genes under positive selection (empirical Bayes value >0.95; false discovery rate FDR q < 0.1) using a site model across all terminal branches of the Spheniscidae (SI Appendix, Tables S10–S12 and Figs. S14 and S15). Using a gene interaction network analysis, we found that many of these positively selected loci are functionally connected, forming two major clusters (Fig. 3): one primarily related to broad cellular functions, and the other containing genes that affect specific phenotypes including immunity, renal function, and cardiovascular activities (e.g., blood pressure, oxygen metabolism, coagulation). Our gene ontology analysis revealed a concordant pattern of pathway enrichment, including terms related to angiotensin regulation, blood pressure, and oxygen metabolism (Fig. 3 and SI Appendix, Tables S11 and S12). All of these adaptations are important for diving and maintenance of high core body temperatures.
Fig. 3.
Analysis of genes under positive selection in extant penguin lineages. The main panel depicts results from the network analysis of positively selected genes, which retrieved two connected primary clusters, one associated with general cellular functions (green) and the other related to functions associated with osmoregulation (renal function), immunity, thermoregulation (e.g., blood pressure), and diving ability (e.g., oxygen metabolism). All genes under selection that presented two or more connections in the network analysis (46 out of 104 genes) are shown. The functions of the identified loci are further detailed in the pie chart at the bottom right.
Ancestral Effective Population Sizes (Ne).
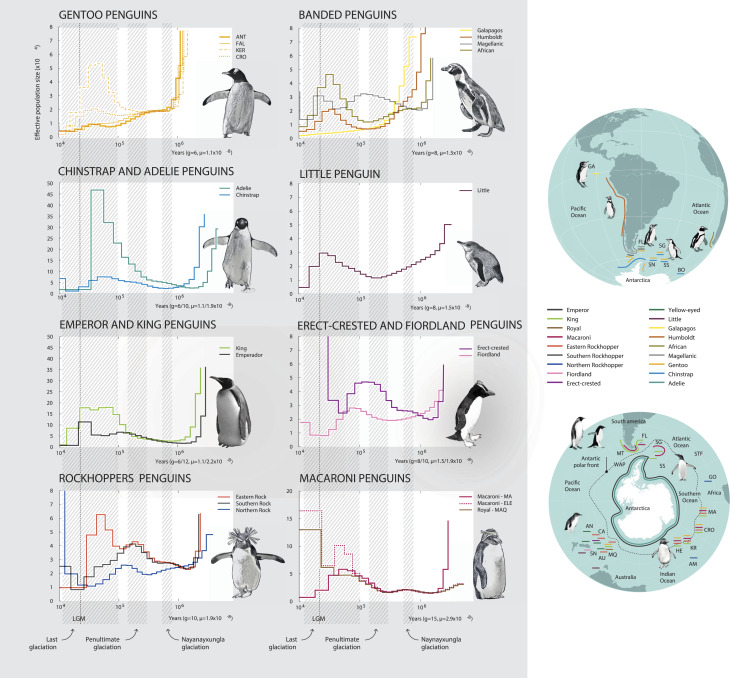
Our reconstruction of changes in ancestral effective population sizes (Ne) recovered three different demographic trajectories in deeper time (Fig. 4). Eleven species of penguin (emperor, king, Adélie, chinstrap, gentoo [Kerguelen Population], Humboldt, Magellanic, African, eastern rockhopper, and little penguin) increased Ne between 40 and 70 kya, with Ne dropping precipitously during the Last Glacial Maximum (LGM; 12–110 kya). In contrast, three species (northern rockhopper, southern rockhopper, and erect-crested) increased Ne during the Penultimate Glaciation Period (PGP) between 130–194 kya but were in decline by 70 kya. Finally, two species (gentoo [Antarctica, Falkland/Malvinas Is. populations], Galápagos) have been in steady decline since at least the Naynayxungla Glaciation (50–72 kya; Fig. 4 and SI Appendix, Fig. S16). Why Galápagos penguins and southern populations of gentoo penguins have been in decline over such a considerable period of time remains uncertain, as there are no large life-history or ecological differences that distinguish these two species from other penguins (SI Appendix, Table S2). One possibility is that both Galápagos penguins and southern populations of gentoo penguins represent recent divergence events (Fig. 1 and SI Appendix, Figs. S7 and S10); hence, the deep time demographic reconstructions may reflect larger ancestral species population sizes before lineage divergence.
Fig. 4.
Demographic history of penguins. Shown are PSMC plots depicting the demographic history of each lineage inferred from genomic data represented by different colored lines (see SI Appendix, Fig. S16 for bootstrapped curves). The periods of the Naynayxungla Glaciation, the Penultimate and Last Glaciations, and the Last Glacial Maximum (LGM) are depicted.
For erect-crested, southern and northern rockhoppers, and Magellanic penguins distributed around the tip of South America and on islands north of the APF, Ne was highest during the PGP with a decline starting in or shortly after the subsequent interglacial. This pattern could reflect changes in ecosystem productivity, but it could also be an artifact of the inability of the pairwise sequential Markovian coalescent (PSMC) method to account for introgression. Our analyses suggest that for these four species, ancestral introgression (SI Appendix, Fig. S9) would have elevated estimates of Ne and could have offset the time interval when a peak in Ne occurred (SI Appendix, Fig. S16). The largest ancestral Ne we observed was for Adélie penguins (>105), a primarily cold-water species (Fig. 1) widely distributed south of the APF; decreases in temperatures and increases in ice cover may have promoted its expansion. Smaller historical peaks for Ne (∼2–3 × 104) were recovered for island endemic species, such as fiordland (southern New Zealand) and northern rockhopper penguins (Tristan da Cunha and Gough Island).
Discussion
Our phylogenetic reconstructions show that the large emperor and king penguins (i.e., Aptenodytes) are sister to all other extant penguins, in agreement with some previous studies (5–7, 11) and a recent study using genomic data (10). The topology refutes the hypothesis that Aptenodytes and Pygoscelis are sister-taxa (4, 8, 9). While the genome-wide data underlying our phylogenetic topology allowed for detection of the deep lineage split between Aptenodytes and all other penguins, the short internal branches recovered by our analysis at this split likely indicate a rapid diversification event (<1 Myr) (27). Such rapid speciation events may explain the discrepancies among the previously proposed phylogenetic hypothesis generated using fewer molecular markers, which may have been insufficient for resolving such short internal branches (28). Where internal branches are longer among penguin taxa, the phylogenetic position of the remaining genera and species are consistent with previous phylogenetic hypotheses (4–10). Reticulated evolution may also have contributed to some inconsistencies in penguin phylogenetic reconstructions. Genome analyses detected deep and shallow introgression events across the phylogeny of extant penguins. For example, species of crested penguins (Eudyptes) appear to have exchanged genes throughout much of their evolutionary history. The directionality of introgression among some linages, especially from the eastern and southern rockhoppers to the erect-crested penguin (SI Appendix, Fig. S9) is consistent with the clockwise direction of the Antarctic Circumpolar Current (ACC) connecting sub-Antarctic islands and promoting dispersal and admixture. We also detected signatures of genomic introgression between an ancestor of the Galápagos/Humboldt penguin and the Magellanic penguin, which is supported by recurrent observations of hybridization and introgression between Magellanic and Humboldt penguins in the wild (29).
Our divergence time estimates are consistent with the fossil record (13), placing the ancestor of crown-group penguins in the early Miocene. Our biogeographic reconstruction for crown-group penguins points to the coastlines of Australia and New Zealand and nearby islands rather than Antarctica (5) as the most likely range of the ancestor of extant penguins, thus supporting an earlier suggestion (7). During the middle Miocene climate transition, 14.2–13.8 Mya, the Southern Ocean cooled 6 °C to 7 °C due to the intensification of atmospheric and oceanic circumpolar circulation, increasing the isolation of Antarctic organisms (30), and coincident with the divergence of Spheniscus+Eudyptula from Megadyptes+Eudyptes (14.08 Mya), and the Spheniscus/Eudyptula split (12.57 Mya). Because of the recency of most diversification events (2–9 Mya), we propose an alternative hypothesis to explain their geographic expansion. Namely, that the opening of the Drake Passage around the tip of South America and the development of a deep, strong ACC by ∼11.6 Mya (24), rather than the initial formation of a weaker ACC ∼34 Mya (ref. 31 and Fig. 1), contributed to the colonization of new areas and the diversification of penguins. Global climate cooling intensified with the strengthening of the ACC and possibly led to the extinction of several species inhabiting Antarctica (21, 32). The ACC also reinforced the isolation of taxa inhabiting Antarctica from those on continental shelves and islands to the north (21, 32), while promoting eastward colonization of available sub-Antarctic islands.
An accelerating decline in SST since the Pliocene (21) may have facilitated the colonization of new areas such as islands in the Indian Ocean. This hypothesis is supported by our analysis of the divergence of the four gentoo penguin lineages across thermal and salinity gradients to the north and south of the APF (SI Appendix, Figs. S5–S8). The Pliocene-Pleistocene cooling, along with the expansion of ice shelves across the Southern Ocean (21), would have reduced connectivity among penguin populations and facilitated speciation across Pygoscelis, Spheniscus, Eudyptes and Aptenodytes between 2 and 5.5 Mya (Fig. 1). More recent Pleistocene speciation of island endemic penguins (<5 Mya) might also have followed the formation of new archipelagos (e.g., Galápagos, Snares, Macquarie, Gough, Antipodes Islands) (9).
During the Quaternary glaciations (1.8 to 0.01 Mya), sea ice is thought to have reached approximately 40°S at the coast of South America (33). This condition would have promoted the northern expansion of Spheniscus to the subtropics and subsequently enabled colonization of the Galápagos Islands, home to rich feeding resources for penguins due to the upwelling of cooler, nutrient-rich waters. Strong north-flowing currents—the Humboldt Current and Benguela Current—would have further facilitated penguin colonization of subtropical habitats in the Pacific (Galápagos) and Atlantic (southern Africa), respectively. Neither of these current systems penetrates beyond the equator; this, together with their cold temperate origin and preference for cooler waters, may explain why crown group penguins never successfully colonized the Northern Hemisphere.
The genes for which we detected signals of positive selection are associated with thermoregulation (e.g., vasoconstriction/vasodilation: ENPEP, MME, BDKRB2), osmoregulation (e.g., balance of fluids and salt: SCL6A19, ACE2, AGT), and diving capacity (e.g., oxygen storage: MB), reflecting adaptations that enabled penguins to colonize areas outside their ancestral thermal maximum of ∼9 °C (Fig. 1 and SI Appendix, Figs. S12 and S13). In Antarctica, emperor penguins are exposed to temperatures as low as −40 °C and forage in waters of −1.8 °C (34). Regulating blood pressure selectively through the constriction of blood vessels can conserve oxygen consumption and facilitate the maintenance of high core body temperatures (35). Galápagos penguins are exposed to SST >27 °C and air temperatures exceeding 40 °C—although these extremes are mostly associated with El Niño events—which may cause heat stress, high mortality, and slow recovery of penguin colonies (36, 37).
Penguins spend most of their lives at sea, often performing prolonged dives while foraging. They store oxygen in their lungs, blood, and muscles (38), and their rates of oxygen consumption can be very low (39). The two largest penguin species, the emperor and king penguins, can achieve depths of >300 m and maximum dive durations of 22 and 8 min, respectively (38, 40, 41). Smaller penguin species, with the exception of chinstrap penguins (40), tend to dive in shallow waters (<50 m) for 1 to 2 min (38, 42). In this sense, nucleotide differences in myoglobin (MB; overall positive selection, Z = 2.645, P = 0.005) across species groups could be associated with differences in diving capacity. For example, we found several nonsynonymous substitutions that were common within Pygoscelis, Eudyptes, and Aptenodytes penguins but differed between genera (SI Appendix, Fig. S15). It is possible that these mutations encode greater oxygen-binding capacity, which would facilitate the deep and prolonged dives performed by Aptenodytes and some species of Pygoscelis penguins compared with Eudyptes (higher dN/dS ratios; SI Appendix, Table S13).
Our results suggest that adaptation across genes involved in multiple interconnected genetic pathways has increased the foraging success and survival of penguin species across diverse temperature and salinity gradients (SI Appendix, Fig. S15). Foraging success is associated with reproductive output (43, 44) and also with survival during long periods of fasting while caring for eggs and chicks (45). Collectively, such adaptations would have promoted the radiation of penguin species across the Southern Hemisphere.
Penguins have a remarkable evolutionary history. Their radiation from the coasts of New Zealand and Australia into other parts of the Southern Hemisphere was facilitated by changes in global circulation patterns over the past 20 million years. Our analysis detected positive selection across several gene networks, suggesting that molecular adaptation promoted the establishment of penguin populations in the Antarctic and tropical regions and enhanced the ability of some species to dive deeply. Demographic reconstructions over the past million years show that most penguin species have declined during the severe ice conditions of the LGM in the Southern Ocean, a result concordant with that recovered for several other bird species (17, 46). Our results suggest that penguins originated from areas with a maximum SST of 9 °C and diversified over millions of years to occupy colder Antarctic and warmer tropical waters. As such, it seems unlikely that locally adapted species will be able to keep pace with the present rapid climate change, a rate far in excess of that observed over geological time, especially because marine species may be more vulnerable to global warming than terrestrial species (47, 48). This vulnerability is especially pertinent to penguins, as illustrated by the recent massive mortality of Adélie penguin chicks and by the relocation of emperor penguins in response to suboptimal sea ice conditions (49, 50). As large-scale genomic studies and sophisticated global climate models become increasingly available, the application of approaches such as those presented here holds significant promise for providing new insight into the evolutionary histories and climatic vulnerability of many of the world’s most enigmatic species.
Materials and Methods
More detailed information on the materials and methods used in this study are provided in SI Appendix.
Genome Sequencing and Assembly.
The genomes of 18 extant penguin species (22 individuals) as well as the southern giant petrel (Macronectes giganteus) were sequenced to ∼30× coverage with 150-bp paired-end reads using the Illumina HiSeq X platform at MedGenome (SI Appendix, Table S1). We used the giant petrel as an outgroup for the analyses reported below. In brief, we removed duplicate sequences using Super Deduper (https://github.com/dstreett/Super-Deduper) and then filtered the reads. We aligned the resulting cleaned reads of each individual to the emperor penguin reference genome (gigadb.org/dataset/100005; scaffold-level assembly) using LAST (last.cbrc.jp/). We assessed the extent to which genome assemblies were complete using the BUSCO v2 dataset (51) (SI Appendix, Fig. S2) and KAT spectra-cn plots (52) (SI Appendix, Figs. S3 and S4). We extracted the CDS, intron, and mitogenome sequences for each genome using the reference genome GFF. We extracted 120-bp UCE loci with 750 bp of flanking sequence padded to each side with scripts from the PHYLUCE pipeline (53) and aligned sequences using MAFFT (54).
Estimation of Phylogeny, Divergence Times, and Interspecific Introgression.
To account for potential genome-wide incompatibilities between taxa and loci, we used Astral III (55) to estimate species tree phylogenies for each of the UCE, intron, and CDS datasets and for all three datasets combined (SI Appendix, Fig. S8). We used RAxML-NG (v. 0.5.1b beta) (56) to generate independent gene trees for each locus of the UCE, intron, and CDS alignments. As input to Astral III, we merged all of the “best” trees generated by RAxML-NG into a single file. For the phylogenomic concatenated analysis using all the data from UCEs, CDS, intron, and the mitogenome, we carried out maximum likelihood analyses in IQ-TREE (57), with 1,000 bootstrap replicates (SI Appendix, Figs. S5–S7). We estimated divergence times in BEAST v2.5.2 (58) using the computer resources available through the CIPRES Science Gateway (SI Appendix, Figs. S7 and S10) (59), calibrating the topology with five fossils (SI Appendix, Table S8) using previously published parameters (9). This phylogeny was used for subsequent analyses. Analyses to investigate the extent of interspecific introgression across the phylogeny were performed using a partitioned D-statistics approach implemented in DFOIL (60) for eight combinations of taxa while respecting the priors of a symmetrical tree composed of four taxa and an outgroup, with one ingroup clade younger than the other (Fig. 2 and SI Appendix, Fig. S9 and Table S7). To perform the tests, we split the genome-wide alignments into 100-kb nonoverlapping windows with Bedtools and custom scripts.
Ancestral Distribution and Niche Reconstruction.
For the historical biogeographic analysis, we estimated the ancestral range of the extant penguin species using the R package BioGeoBEARS (61), implementing three models of ancestral area reconstruction with and without long-distance dispersal (the parameter j: “jump dispersal”). We subdivided the extant penguin geographic distribution into 10 different areas and used occurrence records for all penguin species and six marine variables to created raw models with MaxEnt–Javascript (62). Niche overlap was estimated between all penguin species for the set of variables considered. We used the “Phyloclim” package to generate predicted niche occupancy (PNO) profile values and plots. We then combined the PNO profiles with the phylogenetic tree to generate the DTT plots and climatic tolerance chronograms depicted in Fig. 1.
Detection of Signatures of Positive Selection.
We performed a dN/dS ratio test using the CodeML algorithm implemented in ETE3 (63, 64) for all species and subgroups in the phylogeny. Due to the large number of analyzed genes, we performed a multiple comparisons (FDR) test implemented in R. Genes that persisted on the list (i.e., remained significantly different from neutral expectations, supporting a positive selection regime) were then used to perform a Gene Ontology analysis using WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) (65) and a network analysis using StringDB (66) (SI Appendix, Figs. S14 and S15).
Demographic History.
To address questions about how climate may have influenced effective population size (Ne) for each penguin species, we performed a demographic analysis using the PSMC method (67). PSMC was run with parameters -N25 -t15 -r5 -p 4 + 25*2 + 4 + 6 and with an estimated generation time for the different penguin species (Fig. 4 and SI Appendix, Fig. S16 and Table S2) using a substitution rate derived from chicken pedigrees (68).
Supplementary Material
Acknowledgments
We thank Andrea Polanowski, Claudia Godoy, and Klemens Pütz for assistance with sample acquisition; Claudio González-Weber, Luiz Rocha, and Barbara Ustanko for suggestions; and Juan Pablo Bravo for help with illustration design. Financial support for this work was provided by Instituto Antártico Chileno (INACH RT_12–14), Fondecyt Project 1150517, Genomics Antarctic Biodiversity/Programa de Investigación Asociativa/Comisión Nacional de Investigación Científica y Tecnológica (GAB PIA CONICYT ACT172065), NSF DEB-1441652, French Polar Institute Paul-Emile Victor (IPEV; Progs. 137 and 354), a Lakeside grant from the California Academy of Sciences, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Brazil and Instituto Nacional de Ciência e Tecnologia - Ecologia, Evolução e Conservação da Biodiversidade (INCT-EECBio) Brazil.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006659117/-/DCSupplemental.
Data and Materials Availability.
Penguin and giant petrel raw fastq reads, reconstructed genomes (BioProject PRJNA530615; BioSample accession nos. SAMN11566608 to SAMN11566630) and mitogenomes (MK760983 to MK761004, MK761006) have been deposited in the GenBank database. All UCE, CDS, intron and mitogenome alignments, dated phylogenies, and scripts for data analyses are available at Dryad (https://doi.org/10.5061/dryad.pk0p2ngj2) (69). All other data needed to evaluate the conclusions in this paper are provided in either the main text or the SI Appendix.
References
- 1.Egevang C. et al., Tracking of Arctic terns Sterna paradisaea reveals longest animal migration. Proc. Natl. Acad. Sci. U.S.A. 107, 2078–2081 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Péron C., Grémillet D., Tracking through life stages: Adult, immature and juvenile autumn migration in a long-lived seabird. PLoS One 8, e72713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas D. B., Fordyce R. E., Biological plasticity in penguin heat-retention structures. Anat. Rec. 295, 249–256 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Gavryushkina A. et al., Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 66, 57–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker A. J., Pereira S. L., Haddrath O. P., Edge K. A., Multiple gene evidence for expansion of extant penguins out of Antarctica due to global cooling. Proc. Biol. Sci. 273, 11–17 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke J. A. et al., Paleogene equatorial penguins challenge the proposed relationship between biogeography, diversity, and Cenozoic climate change. Proc. Natl. Acad. Sci. U.S.A. 104, 11545–11550 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ksepka D. T. B. S., Giannini N., The phylogeny of the living and fossil Sphenisciformes (penguins). Cladistics 22, 412–441 (2006). [Google Scholar]
- 8.Subramanian S., Beans-Picón G., Swaminathan S. K., Millar C. D., Lambert D. M., Evidence for a recent origin of penguins. Biol. Lett. 9, 20130748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole T. L. et al., Mitogenomes uncover extinct penguin taxa and reveal island formation as a key driver of speciation. Mol. Biol. Evol. 36, 784–797 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Pan H. et al., High-coverage genomes to elucidate the evolution of penguins. Gigascience 8, giz117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertelli S., Giannini N. P., A phylogeny of extant penguins (Aves: Sphenisciformes) combining morphology and mitochondrial sequences. Cladistics 21, 209–239 (2005). [Google Scholar]
- 12.Figueiró H. V. et al., Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci. Adv. 3, e1700299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degrange F. J., Ksepka D. T., Tambussi C. P., Redescription of the oldest crown clade penguin: Cranial osteology, jaw myology, neuroanatomy, and phylogenetic affinities of Madrynornis mirandus. J. Vertebr. Paleontol. 38, e1445636 (2018). [Google Scholar]
- 14.Lombal A. J., O’dwyer J. E., Friesen V., Woehler E. J., Burridge C. P., Identifying mechanisms of genetic differentiation among populations in vagile species: Historical factors dominate genetic differentiation in seabirds. Biol. Rev. 95, 625–651 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Vianna J. A. et al., Marked phylogeographic structure of Gentoo penguin reveals an ongoing diversification process along the Southern Ocean. Mol. Phylogenet. Evol. 107, 486–498 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Frugone M. J. et al., Contrasting phylogeographic pattern among Eudyptes penguins around the Southern Ocean. Sci. Rep. 8, 17481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole T. L. et al., Receding ice drove parallel expansions in Southern Ocean penguins. Proc. Natl. Acad. Sci. U.S.A. 116, 26690–26696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frugone M. J. et al., More than the eye can see: Genomic insights into the drivers of genetic differentiation in Royal/Macaroni penguins across the Southern Ocean. Mol. Phylogenet. Evol. 139, 106563 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Clucas G. V. et al., Comparative population genomics reveals key barriers to dispersal in Southern Ocean penguins. Mol. Ecol. 27, 4680–4697 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Pertierra L. et al., Cryptic speciation in gentoo penguin is driven by geographic isolation and regional marine conditions: Unforeseen vulnerabilities to global change. Divers. Distrib. 26, 958–975 (2020). [Google Scholar]
- 21.Halanych K. M., Mahon A. R., Challenging dogma concerning biogeographic patterns of Antarctica and the Southern Ocean. Annu. Rev. Ecol. Evol. Syst. 49, 355–378 (2018). [Google Scholar]
- 22.Hansen J., Sato M., Russell G., Kharecha P., Climate sensitivity, sea level and atmospheric carbon dioxide. Philos. Trans. A Math. Phys. Eng. Sci. 371, 20120294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zachos J. C., Dickens G. R., Zeebe R. E., An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 451, 279–283 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Dalziel I. W. D. et al., A potential barrier to deep Antarctic circumpolar flow until the late Miocene? Geology 41 (2013). [Google Scholar]
- 25.Pearce J. A. et al., Composition and evolution of the Ancestral South Sandwich Arc: Implications for the flow of deep ocean water and mantle through the Drake Passage Gateway. Global Planet. Change 123, 298–322 (2014). [Google Scholar]
- 26.Dunstan P. K. et al., Global patterns of change and variation in sea surface temperature and chlorophyll a. Sci. Rep. 8, 14624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisrock D. W., Harmon L. J., Larson A., Resolving deep phylogenetic relationships in salamanders: Analyses of mitochondrial and nuclear genomic data. Syst. Biol. 54, 758–777 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Oliveros C. H. et al., Earth history and the passerine superradiation. Proc. Natl. Acad. Sci. U.S.A. 116, 7916–7925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simeone A. et al., Heterospecific pairing and hybridization between wild Humboldt and Magellanic penguins in southern Chile. Condor 111, 544–550 (2009). [Google Scholar]
- 30.Shevenell A. E., Kennett J. P., Lea D. W., Middle Miocene Southern Ocean cooling and Antarctic cryosphere expansion. Science 305, 1766–1770 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Goldner A., Herold N., Huber M., Antarctic glaciation caused ocean circulation changes at the Eocene-Oligocene transition. Nature 511, 574–577 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Crame J. A., Key stages in the evolution of the Antarctic marine fauna. J. Biogeogr. 45, 986–994 (2018). [Google Scholar]
- 33.Glasser N. F., Jansson K. N., Harrison S., Kleman J., The glacial geomorphology and Pleistocene history of South America between 38°S and 56°S. Quat. Sci. Rev. 27, 365–390 (2008). [Google Scholar]
- 34.Williams C. L., Hagelin J. C., Kooyman G. L., Hidden keys to survival: The type, density, pattern and functional role of emperor penguin body feathers. Proc. Roy. Soc. B Biol. Sci. 282, 20152033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler P. J., Jones D. R., Physiology of diving of birds and mammals. Physiol. Rev. 77, 837–899 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Boersma P. D., Population trends of the Galápagos penguin: Impacts of El Niño and La Niña. Condor 100, 245–253 (1998). [Google Scholar]
- 37.Boersma D., “Adaptations of Galápagos penguins for life in two different environments” in The Biology of Penguins, Stonehouse B., Ed. (Macmillan, London, 1975), pp. 101–114. [Google Scholar]
- 38.Ponganis P. J., Kooyman G. L., Diving physiology of birds: A history of studies on polar species. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 126, 143–151 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Williams C. L., Meir J. U., Ponganis P. J., What triggers the aerobic dive limit? Patterns of muscle oxygen depletion during dives of emperor penguins. J. Exp. Biol. 214, 1802–1812 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wienecke B., Robertson G., Kirkwood R., Lawton K., Extreme dives by free-ranging emperor penguins. Polar Biol. 30, 133–142 (2007). [Google Scholar]
- 41.Putz K., Cherel Y., The diving behaviour of brooding king penguins (Aptenodytes patagonicus) from the Falkland Islands: Variation in dive profiles and synchronous underwater swimming provide new insights into their foraging strategies. Mar. Biol. 147, 281–290 (2005). [Google Scholar]
- 42.Kokubun N., Takahashi A., Mori Y., Watanabe S., Shin H. C., Comparison of diving behavior and foraging habitat use between chinstrap and gentoo penguins breeding in the South Shetland Islands, Antarctica. Mar. Biol. 157, 811–825 (2010). [Google Scholar]
- 43.Kowalczyk N. D., Reina R. D., Preston T. J., Chiaradia A., Environmental variability drives shifts in the foraging behaviour and reproductive success of an inshore seabird. Oecologia 178, 967–979 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Berlincourt M., Arnould J. P. Y., Influence of environmental conditions on foraging behaviour and its consequences on reproductive performance in little penguins. Mar. Biol. 162, 1485–1501 (2015). [Google Scholar]
- 45.Thiebot J. B. et al., Adjustment of pre-moult foraging strategies in macaroni penguins Eudyptes chrysolophus according to locality, sex and breeding status. Ibis 156, 511–522 (2014). [Google Scholar]
- 46.Nadachowska-Brzyska K., Li C., Smeds L., Zhang G., Ellegren H., Temporal dynamics of avian populations during Pleistocene revealed by whole-genome sequences. Curr. Biol. 25, 1375–1380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinsky M. L., Eikeset A. M., McCauley D. J., Payne J. L., Sunday J. M., Greater vulnerability to warming of marine versus terrestrial ectotherms. Nature 569, 108–111 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Wiens J. J., Climate-related local extinctions are already widespread among plant and animal species. PLoS Biol. 14, e2001104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cimino M. A., Lynch H. J., Saba V. S., Oliver M. J., Projected asymmetric response of Adélie penguins to Antarctic climate change. Sci. Rep. 6, 28785 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fretwell P. T., Trathan P. N., Emperors on thin ice: Three years of breeding failure at Halley Bay. Antarct. Sci. 31, 133–138 (2019). [Google Scholar]
- 51.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Mapleson D., Garcia Accinelli G., Kettleborough G., Wright J., Clavijo B. J., KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33, 574–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faircloth B. C., PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19 (suppl. 6), 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A., RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bouckaert R. et al., BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller M. A., Pfeiffer W., Schwartz T., “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE) (IEEE, 2010), pp. 1–8. [Google Scholar]
- 60.Pease J. B., Hahn M. W., Detection and polarization of introgression in a five-taxon phylogeny. Syst. Biol. 64, 651–662 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Matzke N., BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts, (University of California, Berkeley, CA, 2013). [Google Scholar]
- 62.Phillips S. J., Anderson R. P., Schapire R. E., Maximum entropy modeling of species geographic distributions. Ecol. Modell. 190, 231–259 (2006). [Google Scholar]
- 63.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z., PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Wang J., Vasaikar S., Shi Z., Greer M., Zhang B., WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szklarczyk D. et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nam K. et al., Molecular evolution of genes in avian genomes. Genome Biol. 11, R68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vianna J., Penguins phylogenome. Dryad. 10.5061/dryad.pk0p2ngj2. Deposited 10 January 2020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Penguin and giant petrel raw fastq reads, reconstructed genomes (BioProject PRJNA530615; BioSample accession nos. SAMN11566608 to SAMN11566630) and mitogenomes (MK760983 to MK761004, MK761006) have been deposited in the GenBank database. All UCE, CDS, intron and mitogenome alignments, dated phylogenies, and scripts for data analyses are available at Dryad (https://doi.org/10.5061/dryad.pk0p2ngj2) (69). All other data needed to evaluate the conclusions in this paper are provided in either the main text or the SI Appendix.