Significance
Changes in the extent of ice sheets through evolutionary timescales have influenced the connectivity of soil invertebrate populations across the Antarctic landscape. We use genetic divergences to estimate isolation times for soil invertebrates along the Transantarctic Mountains. Four species of Collembola (Arthropoda) each showed genetically distinct populations at locations likely isolated for millions of years. Two further species were less genetically diverse although also range restricted. Our genetic data corroborate climate reconstructions and estimates of past warm periods of reduced ice and absent ice shelf in the Ross Sea region, during which time open seaways would have facilitated dispersal of Collembola, and possibly other taxa.
Keywords: climate change, microarthropods, molecular clock, terrestrial biodiversity, phylogeography
Abstract
During austral summer field seasons between 1999 and 2018, we sampled at 91 locations throughout southern Victoria Land and along the Transantarctic Mountains for six species of endemic microarthropods (Collembola), covering a latitudinal range from 76.0°S to 87.3°S. We assembled individual mitochondrial cytochrome c oxidase subunit 1 (COI) sequences (n = 866) and found high levels of sequence divergence at both small (<10 km) and large (>600 km) spatial scales for four of the six Collembola species. We applied molecular clock estimates and assessed genetic divergences relative to the timing of past glacial cycles, including collapses of the West Antarctic Ice Sheet (WAIS). We found that genetically distinct lineages within three species have likely been isolated for at least 5.54 My to 3.52 My, while the other three species diverged more recently (<2 My). We suggest that Collembola had greater dispersal opportunities under past warmer climates, via flotation along coastal margins. Similarly increased opportunities for dispersal may occur under contemporary climate warming scenarios, which could influence the genetic structure of extant populations. As Collembola are a living record of past landscape evolution within Antarctica, these findings provide biological evidence to support geological and glaciological estimates of historical WAIS dynamics over the last ca. 5 My.
During the Pliocene (5.3 Mya to 2.6 Mya), global mean sea surface temperatures were between 1.9 °C and 3.6 °C higher than the present day (ref. 1 and references therein). These warmer temperatures are thought to have caused a total collapse of the West Antarctic Ice Sheet (WAIS) and retreat of the East Antarctic Ice Sheet (EAIS) marine margins, as well as the Greenland Ice Sheet, resulting in an estimated 14-m rise in global mean sea level as compared to present-day levels (2–4). Other more recent periods of glacial retreat occurred during the Pleistocene (<2 Mya), although transitions between fully glaciated and completely collapsed states of the WAIS are thought to be much more rapid, and the collapsed states more short lived, than during the Pliocene (4, 5).
Much of Antarctica’s terrestrial biodiversity, including microinvertebrates (mites, springtails, nematodes, tardigrades, and rotifers), is concentrated along the Transantarctic Mountains where the WAIS and EAIS converge (Fig. 1). Many taxa in these ice-free areas have presumably survived repeated glacial cycles in local refugia, with some taxa dating back to before isolation of the Antarctic continent and establishment of the Antarctic Circumpolar Current ∼34 Mya to 36 Mya (6, 7). The distribution of many Antarctic soil taxa such as lichens, nematodes, and microarthropods is disjunct (8–10) and does not conform to conventional patterns of diversity which usually show decreasing trends with increasing latitude (i.e., the Latitudinal Diversity Gradient; ref. 11). Instead, areas with equivalent, and sometimes higher, levels of biological diversity can be found at more southern locations (8, 12). Rather than latitude, local environmental factors (including those associated with elevation and distance from the coast), local microhabitats, historical glacial events, and geographic barriers have largely dictated the distribution and diversity of present-day biological communities in Antarctica (12, 13).
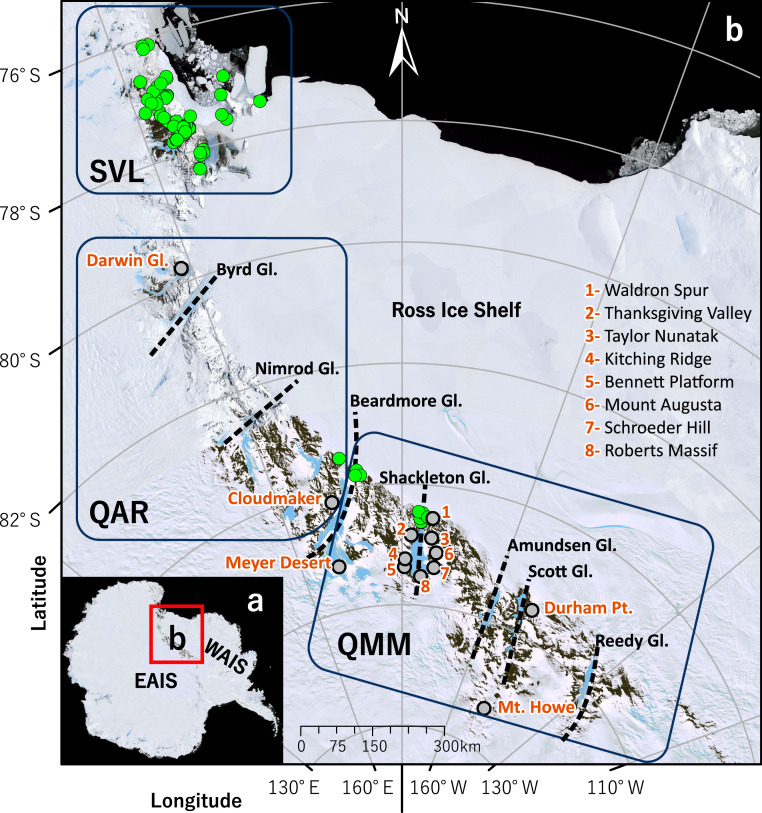
Fig. 1.
(A) Location of study area within the Ross Sea sector of continental Antarctica, showing general area of WAIS and EAIS. (B) Collection sites where six Collembola species were found (green circles; total n = 78 sites) from SVL, the QAR, and the QMM. Locations where no Collembola were found in the QAR−QMM regions (n = 13) are identified with gray circles.
Among the largest terrestrial animals in Antarctica are the microarthropod Collembola (springtails) and Acari (mites), at generally <2 mm in length (10), as well as Diptera (flies/midges) from the Antarctic Peninsula. Their long-term persistence in Antarctica would have necessitated survival in local refugia during glacial maxima followed by recolonization during more favorable interglacial periods (14). As a result of repeated glacial cycles, present-day populations are highly isolated and locally unique (15–17). DNA sequencing of local invertebrate populations, including Collembola, has provided fine-scale patterns of genetic evidence at levels that support speciation and population structuring as a result of landscape changes through time (18, 19).
Linking the structure of natural populations with past geological events or features such as crustal uplift or glacial cycling has long been of interest to biologists (20). The isolation of once contiguous populations into geologically disparate habitats can provide information on the timing of isolation events. For example, ancient and vicariant species of Antarctic Peninsula and Scotia Arc endemic midges have been separated for an estimated 49 My, since before the opening of the Drake Passage between South America and the South Shetland Islands 33 Mya to 30 Mya (21). Furthermore, based on biodiversity patterns and genetic assessments of marine taxa (bryozoans, gastropods, and bivalves), collapse of the WAIS likely connected the Ross Sea and Weddell Sea (near the Antarctic Peninsula) (22).
The concept of a universal, standardized, clock-like approach for dating evolutionary divergences was first proposed by Brower (23), who calculated a clock rate of 2.3% My−1. This rate was based on DNA sequence data from neotropical butterflies, combined with other invertebrate groups and calibrated using relatively recent geological events (<3.25 Mya). This rate had been considered universal and appropriate for estimating invertebrate divergence times based on DNA sequence variation, including for Antarctic Collembola (16, 24). However, this rate has been reevaluated (25, 26), and a recalibrated rate of 3.54% My−1 has been proposed (27). This revised rate was based on the separation of tenebrionid beetle populations by the formation of the mid-Aegean Trench (12 Mya to 9 Mya) and has since been applied to Collembola globally (28–30). Here, we use the revised rate as a more conservative estimate of divergence times for Antarctic Collembola of the Ross Sea region.
Collembola are ametabolous (no larval stage), and the larger individuals (with more food reserves) have greater potential for successful long-distance dispersal (31). Possible dispersal methods for Collembola include walking (or jumping), aerial drift, animal-mediated (“hitch-hiking”), or flotation on water. While aerial transport for Cryptopygus antarcticus has been documented to occur at the Antarctic Peninsula (32), taxa in more arid environments, such as ice-free areas of the Ross Sea region, face a greater threat of desiccation while in transit (33). As a result, long-range dispersal events for Collembola are limited and most likely rely on flotation (rafting) on freshwater (i.e., melt streams) or seawater (i.e., open seaways), when and where available (34).
Long-range dispersal of Collembola, particularly in the vicinity of the Ross Ice Shelf (Ross Sea region), would have only been possible with access to liquid water during periods of glacial retreat, as survival under shelf ice is extremely unlikely. With open water during warmer periods promoting dispersal and population connectivity, increases in gene flow would have homogenized populations. Subsequent glaciation, as well as the reformation of the Ross Ice Shelf and WAIS, would have isolated previously contiguous populations. These events would serve to “reset the molecular clock,” where newly isolated populations would begin accumulating local nucleotide substitutions (35). The opportunity to use biological organisms to test hypotheses of landscape evolution is relatively rare, and Antarctic Collembola provide such an opportunity.
We assessed levels of mitochondrial cytochrome c oxidase subunit 1 (COI) sequence divergence for the six known species of Collembola found in southern Victoria Land (SVL), the Queen Alexandra Range (QAR), and the Queen Maud Mountains (QMM), within the Ross Sea region of continental Antarctica (Fig. 1). We then applied a molecular clock rate to estimate divergence times of populations. By comparing these estimated divergence times to estimates of past glaciological events in this region (3, 4), we suggest that warm periods, with reduced ice and open water, within the last 5 My were at least partially responsible for the present-day patterns of Collembola diversity observed in the Ross Sea region of Antarctica. We hypothesize that species with high levels of genetic divergence (>5%) indicate populations that have been isolated for between 5 My and 3 My or longer, and may even represent speciation events, whereas species with lower levels of population divergence (<5%) were isolated more recently (e.g., within the last 2 My). We suggest that these biological data provide evidence to corroborate hypothesized local geological/glaciological events and enhance our understanding of terrestrial and glaciological landscape evolution in Antarctica.
Results
We generated mitochondrial COI sequences (n = 132) and combined these with publicly available sequences (n = 734) for analysis of all six species of Collembola in SVL, the QAR and the QMM. Two species had relatively low intraspecific divergences (<2% mean uncorrected p distance; Fig. 2 and Table 1), each with very limited distributions (Fig. 3 B and C). Biscoia sudpolaris (n = 35 sequences; h = 5 haplotypes; <1.6% divergence) was found on both sides of the Beardmore Glacier (83.5°S; ∼20 km across), while Tullbergia mediantarctica (n = 50; h = 3; <0.4% divergence) was found at sites farther south, on both sides of the Shackleton Glacier (84.5°S; ∼10 km across).
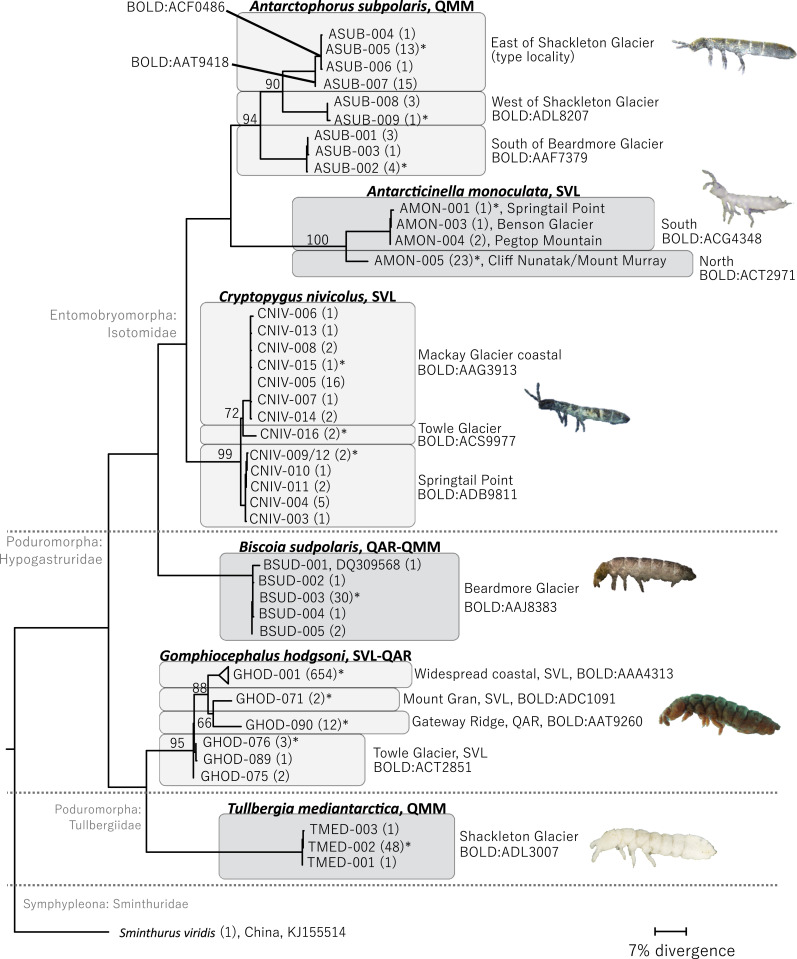
Fig. 2.
Maximum likelihood tree (560 bp; n = 866 sequences) for the six Collembola species from throughout SVL, the QAR, and the QMM, generated in IQ-Tree with TPM2u+F+I+G4 and 5,000 bootstrap replicates. BOLD BINs are provided, and representatives selected for subsequent analyses are denoted with asterisks. Refer to SI Appendix, Tables S3 and S4 for haplotype details.
Table 1.
Levels of genetic divergence among genetic lineages for each of the six endemic species of Collembola from SVL, the QAR, and the QMM
| No. SVL sequences | No. QAR−QMM sequences | No. genetic lineages | Mean p distance (658 bp), % | |||
| Uncorrected | K2P | TN93+G | ||||
| A. subpolaris | — | 42 (36) | 3 | 11.7 to 13.9 | 12.9 to 16.2 | 14.5 to 19.0 |
| B. sudpolaris | — | 35 (34) | 1 | — | — | — |
| T. mediantarctica | — | 50 (50) | 1 | — | — | — |
| G. hodgsoni | 662 | 12 (12) | 4 | 5.7 to 7.9 | 6.0 to 8.4 | 6.5 to 9.4 |
| A. monoculata | 27 | — | 2 | 10.7 | 11.6 | 12.8 |
| C. nivicolus | 37 | — | 3 | 3.3 to 4.1 | 3.3 to 4.4 | 3.5 to 4.6 |
| Total | 726 | 139 (132) | 14 | |||
Numbers of sequences generated in this study are shown in parentheses. One representative sequence (658 bp) from each divergent lineage was used for calculation of pairwise distances in MEGA 7.0.26. —, not applicable.
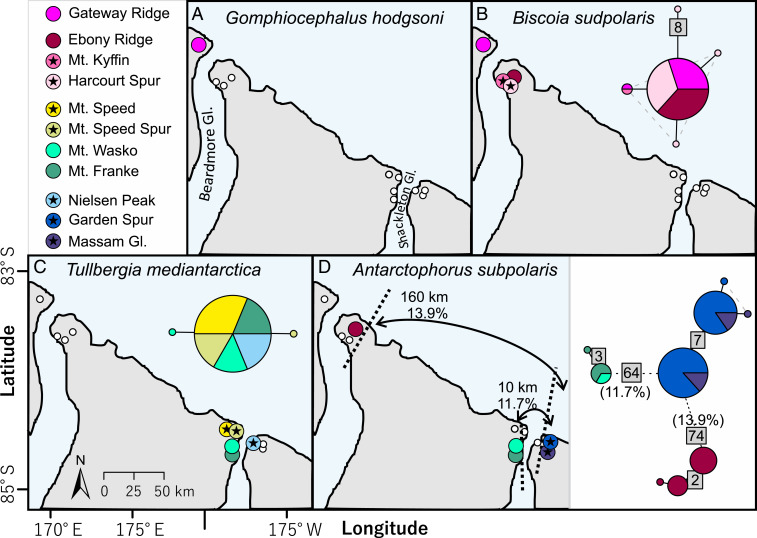
Fig. 3.
Haplotype diversity and distribution of the four Collembola species among sites along the QAR and the QMM, including (A) a new genetic lineage and expanded distributional range for the SVL springtail G. hodgsoni, (B) low divergence (<1.6%) within B. sudpolaris, (C) low divergence (<0.4%) within T. mediantarctica, and (D) spatial isolation of the three genetically diverged lineages (11.7 to 13.9% p distance) of A. subpolaris (type locality: east of Shackleton Glacier). Sites where only one species was found are indicated with stars, while sites where any given species was not found are indicated with small white circles. Numbers in the gray boxes indicate the number of mutational steps between haplotypes.
The remaining four species were more widespread, each comprising between two and four divergent lineages (total n divergent lineages = 14; 3.3 to 13.9% mean uncorrected p distance; Fig. 2 and Table 1). Each genetic lineage had a unique Barcode Index Number (BIN) and was found exclusively at one or a range of geographically isolated locations (Figs. 3 and 4). Tests of substitution saturation were performed separately for each of these four species, and, in all cases, the critical index of substitution saturation (Iss.C) was significantly greater than the index of substitution saturation (Iss) (P < 0.01; SI Appendix, Table S2), confirming that the data were appropriate for molecular clock analyses.
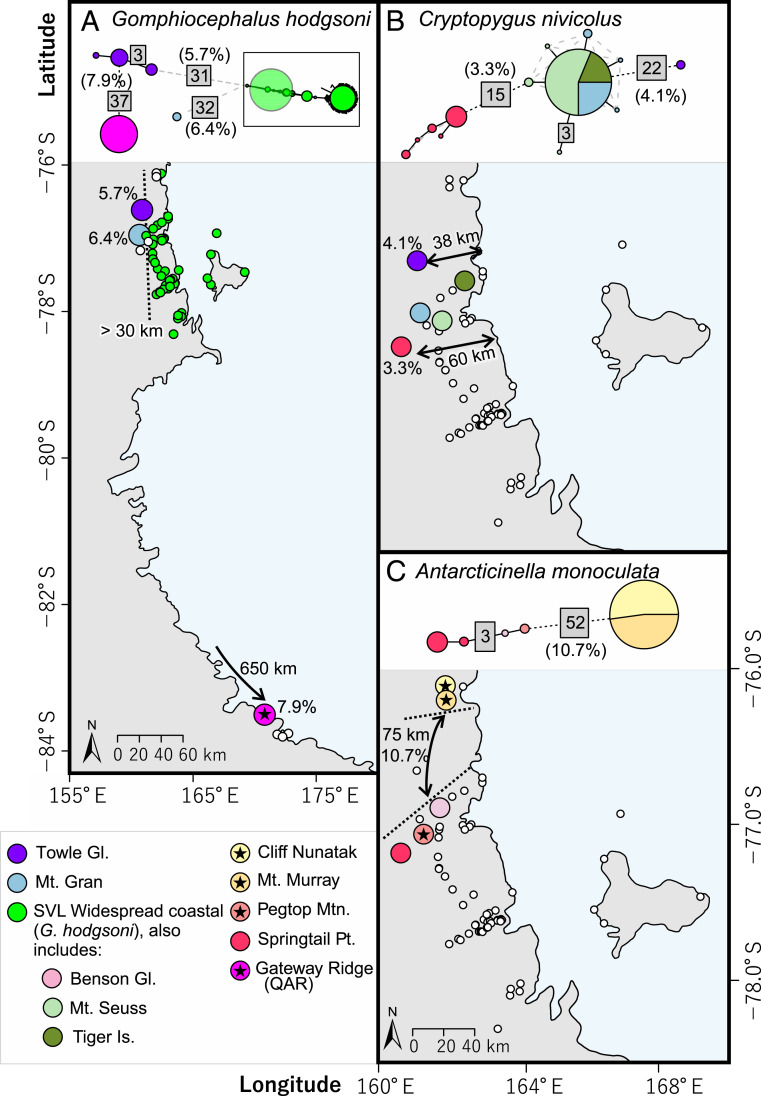
Fig. 4.
Haplotype diversity and distribution of the three Collembola species among sites in SVL (and the QAR for G. hodgsoni), showing spatial isolation of the (A) four genetically diverged lineages (5.7 to 7.9% p distance) of G. hodgsoni, (B) three genetically diverged lineages (3.3 to 4.2% p distance) of C. nivicolus, and (C) two genetically diverged lineages (10.7% p distance) of A. monoculata. Sites where only one species was found are indicated with stars, while only two species (of various combinations) were found at each of the remaining named sites (excepting SVL Widespread coastal). Sites where any given species was not found are indicated with small white circles. For G. hodgsoni, the many haplotypes within the “SVL Widespread coastal” cluster were simplified to a scaled-down haplotype network (boxed) for ease of graphical purposes; see ref. 36 for further haplotype and locality detail for this species. Numbers in the gray boxes indicate the number of mutational steps between haplotypes.
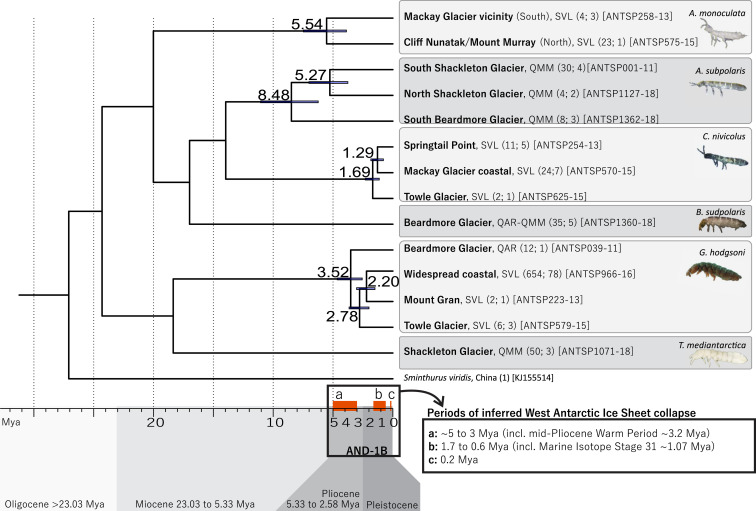
In SVL, three species of Collembola were present (Antarcticinella monoculata, Cryptopygus nivicolus, and Gomphiocephalus hodgsoni), each containing divergent populations, particularly at inland sites (Fig. 4). At three sites in SVL (Cliff Nunatak, Mount Murray, and Pegtop Mountain), only one species was present (A. monoculata), while six other sites had a combination of only two species present. The varying levels of genetic divergence found among these populations reflect these distributional differences; levels of divergence were lowest for C. nivicolus (3.3 to 4.1%), midrange for G. hodgsoni (5.7 to 6.4%), and highest for A. monoculata (10.7%). The range of divergence dates that were estimated for each species also varied accordingly, based on molecular clock analyses (Fig. 5 and Table 2): Divergent populations have been isolated for at least 1.3 My for C. nivicolus, 2.2 My for G. hodgsoni (in SVL), and 5.5 My for A. monoculata (Fig. 5 and Table 2).
Fig. 5.
Dating estimates for the four springtail species with high intraspecific variability (>3.3% divergence) based on the molecular clock rate of 0.0168 (i.e., 3.54% divergence per million years; ref. 37). Estimated dates (in millions of years) of population isolation, and bars representing 95% highest posterior density, are shown at nodes (see also Table 2). Based on the AND-1B core data (3) and simulated modeling (4), collapse of the WAIS likely occurred multiple times during three major periods within the last 5 My (shown as orange boxes on the timeline), facilitating connectivity of Collembola populations. Number of sequences and unique haplotypes, respectively, are shown in parentheses, followed by the BOLD sequence ID of the exact sequences (658 bp) used for molecular clock estimations.
Table 2.
Molecular clock estimations for each geographically isolated lineage of Collembola in SVL, the QAR, and the QMM
| Geographical separation | Posterior age estimates (3.54% My−1) | 5- to 3-My calibration | ||||||
| Divergence time (My) | 95% HPD | ucldMEAN | SD | Divergence rate My−1, % | ||||
| A. subpolaris (3) | ||||||||
| Beardmore Gl. | vs. | Shackleton Gl. | 8.48 | 6.24 to 11.06 | 0.024 | 0.005 | 4.8 | |
| Shackleton Gl. West | vs. | Shackleton Gl. East | 5.27 | 3.76 to 7.01 | 0.016 | 0.004 | 3.2 | |
| G. hodgsoni (4) | 0.0153 | 0.005 | 3.06 | |||||
| SVL | vs. | QAR | 3.52 | 2.56 to 4.64 | ||||
| Towle Gl. | vs. | All other SVL | 2.78 | 2.00 to 3.66 | ||||
| Mt. Gran | vs. | Widespread coastal | 2.20 | 1.49 to 3.03 | ||||
| C. nivicolus (3) | 0.0089 | 0.003 | 1.78 | |||||
| Towle Gl. | vs. | All others | 1.69 | 1.14 to 2.30 | ||||
| Springtail Pt. | vs. | Mackay Gl. coastal | 1.29 | 0.79 to 1.84 | ||||
| A. monoculata (2) | 0.0184 | 0.006 | 3.68 | |||||
| North SVL | vs. | South SVL | 5.54 | 3.88 to 7.48 | ||||
Divergence times ±95% HPD are estimated utilizing the mitochondrial clock rate of 3.54% divergence per My (27). Clock rates were then separately estimated based on calibration to the 5- to 3-My period of WAIS collapse. All estimations were performed on the alignment containing one representative sequence (658 bp) for each lineage (number of lineages is shown in parentheses), implemented in BEAST 2.4.7.
Farther south, within the QMM, we found three genetically distinct populations of Antarctophorus subpolaris each at geographically isolated locations (Figs. 2 and 3D). Either side of the Shackleton Glacier, A. subpolaris was 11.7% divergent, and the other population near the Beardmore Glacier was 13.9% divergent relative to the population on the eastern side of the Shackleton Glacier (type locality). We also found a unique lineage of G. hodgsoni on the northern side of the Beardmore Glacier, within the QAR (7.9% divergent from the SVL widespread coastal lineage; Figs. 3A and 4A), expanding the distributional range for this species by >650 km. This southern population of G. hodgsoni appears to have been isolated from its more northern counterparts for ∼3.52 My (95% highest posterior density [HPD]: 4.64 My to 2.56 My), aligning with the inferred period of WAIS collapse between 5 Mya and 3 Mya (Fig. 5). For A. subpolaris, populations on either side of the Shackleton Glacier have been isolated for 5.27 My (95% HPD: 7.01 My to 3.76 My), aligning with the onset of WAIS collapse during the 5- to 3-Mya timeframe (Fig. 5). However, isolation of the Beardmore Glacier population of A. subpolaris was estimated since 8.48 Mya (95% HPD: 11.06 My to 6.24 My).
To test for the possibility of different rates of molecular evolution within A. subpolaris, we applied the 5- to 3-My WAIS collapse to each of the two node separations, using two separate Bayesian analyses. The resulting calibrated clock rates were estimated at 4.8% divergence per million years for the Beardmore Glacier lineage and 3.2% divergence per My for Shackleton Glacier lineages (Table 2). However, when we performed Tajima’s relative rate analyses, with A. monoculata as an outgroup (as nearest sister taxon in the phylogenetic tree), to specifically test whether it is possible that the northern A. subpolaris population has a different rate of molecular evolution (i.e., clock rate) than the two southern populations, we found no evidence to support a varied clock rate among any of the Antarctophorus sequences (χ2 = 0.01 to 0.13; P > 0.05; SI Appendix, Table S2). It is therefore more likely that the Beardmore Glacier population of A. subpolaris has been isolated for longer than the more southern populations, indicative of lineage independence and possible speciation.
Discussion
Past climatic and geological changes have constrained terrestrial invertebrate distributions in Antarctica, influencing the genetic structure of extant species (38). Our phylogeographic approach suggests that patterns of population genetic structure were influenced by large-scale glacial events, consistent with past climate reconstructions. Further, our analyses provide independent corroboration of geological, glaciological, and model-based estimates of historical Ross Ice Shelf retreat and potential WAIS collapses such as those of refs. 3 and 4.
Two of the six species we studied, B. sudpolaris and T. mediantarctica, show limited genetic variability among sites and are the most range restricted of the taxa (each found within a point-to-point distance of <40 km). The loss of genetic diversity due to random genetic drift has a greater relative effect on smaller populations (39), and reductions in the gene pool can lead to decreased fitness for the population overall (40). However, these two species were found in relatively high abundance (more than eight sequences from most sites, as well as additional specimens that were collected and not sequenced). We also highlight that B. sudpolaris and T. mediantarctica may be species of special conservation concern, given their currently limited distributions and low genetic diversity.
The four Collembola species with more widespread distributions (A. subpolaris, A. monoculata, C. nivicolus, and G. hodgsoni) each have highly divergent populations that are geographically isolated. If these isolated populations are not highly abundant (e.g., only two individuals were found for the Mount Gran lineage of G. hodgsoni), they would be greatly affected by genetic drift and would therefore also be of special conservation concern. In some cases, genetic divergence occurred between sites that were distantly spaced (A. subpolaris, A. monoculata, and G. hodgsoni). Indeed, the theory of isolation by distance describes how increasing genetic differences between populations are often found as the geographic distance increases (41). However, divergent populations for A. subpolaris, G. hodgsoni, and C. nivicolus also occurred at sites that are within close proximity but separated by prominent geographical features such as the Shackleton and Mackay Glaciers. High levels of genetic divergence have also been found across small spatial scales (<20 km) among mite populations on either side of sub-Antarctic Marion Island (42) as well as among continental Antarctic populations of Friesea propria and Friesea gretae (previously known as Friesea grisea; ref. 43) in northern Victoria Land (36), attributed to historical events and geographical barriers preventing dispersal.
A single genetic lineage for G. hodgsoni is widespread among coastal sites throughout SVL, whereas all three distinct A. subpolaris populations were at coastal sites within the QMM. These findings are consistent with the idea that dispersal is currently possible via open seaways in SVL, similar to what would be expected along the coastline of the more southern QAR and QMM regions during times of WAIS and Ross Ice Shelf collapse, where grounded ice has regularly blocked dispersal of Collembola since the Pliocene. Furthermore, genetically distinct populations of G. hodgsoni and C. nivicolus were located within SVL at sites farther inland (>30 km from the coast) and at higher elevations (>1,000 m above sea level [a.s.l.]), suggesting currently limited dispersal opportunities among these inland sites. No Collembola occurred at sampled sites located farther inland within the QAR and QMM regions. This is likely a direct consequence of the more extreme conditions, including limited availability of liquid water and inhospitable soil habitat (1, 44). However, under warmer climate scenarios, harsh soil environments may become ameliorated, facilitating expansion of Collembola populations and colonization of these inland sites.
Based on our molecular clock estimates, divergent populations of A. subpolaris, A. monoculata, and G. hodgsoni have been isolated since 5.54 Mya to 3.52 Mya. During this time when conditions were warmer, multiple collapses of the WAIS likely occurred (45, 46), lasting for extended periods, and this would have drastically improved dispersal opportunities for Collembola in the Ross Sea region, facilitated by flotation along coastal margins. However, it is unlikely that the Beardmore and Shackleton Glaciers (two of the major outlet glaciers that drain the EAIS; ref. 47) would have fully retreated during these periods of WAIS collapse (48), with the EAIS interior possibly being stable for the last 14 My (2). Therefore, dispersal among locations farther inland in the QAR and QMM vicinities and available habitat would have remained limited during the past 5 My.
More recently (<2 Mya), many glacial cycles have occurred during the Pleistocene, particularly during the period between 1.7 Mya and 0.6 Mya (4, 49) which encompasses the well-documented Marine Isotope Stage 31 between 1.085 Mya and 1.055 Mya (50), as well as other significant warming events since 0.6 Mya. Additional WAIS collapses proposed by refs. 3 and 51 occurred from ∼423 Kya to 362 Kya and from 210 Kya to 205 Kya. Inferred collapses since 2 Mya were likely more rapid (4), in which case opportunities for Collembola dispersal would have been more limited, relative to the longer-duration WAIS collapses between 5 Mya and 3 Mya.
Partial collapses of the WAIS could have provided further dispersal opportunities for the more northern species. For G. hodgsoni, warm periods 3.2 Mya to 2.7 Mya (52) may have provided dispersal opportunities for this species among SVL locations, after which the Mount Gran and Towle Glacier populations probably remained isolated to the present day owing to their remote positioning (>30 km from the coast and >1,000 m a.s.l.). The remaining two SVL species have differing isolation date estimates compared to G. hodgsoni, as a result of differences in their distributional patterns and levels of genetic divergence. For example, A. monoculata was the only species found at Pegtop Mountain, which may have served as a “stepping stone” location for the southern lineage to reach Springtail Point, whereas a unique lineage of C. nivicolus was found at Springtail Point. The finer-scale genetic variation in G. hodgsoni among the widespread coastal populations (<1.6% divergence) could further be attributed to the most recent warm events within the last 2 My, as has been considered for three species of Collembola from the South Shetland Islands (53).
It is entirely possible that some or all of the 14 divergent lineages considered here represent new species. In particular, as noted above, one population of A. subpolaris may have been isolated for up to 8.48 My, beyond the influence of any WAIS collapse that occurred during the 5- to 3-My time period addressed here. While Antarctic species are very slow in developing morphological differences, subtle but valid distinguishing features do arise (43). Our analyses provide clear evidence of lineage independence, that the lineages have unique origins, and have maintained independent evolutionary trajectories for millions of years (54).
Overall, our analyses suggest that the presence of an ice sheet in the Ross Sea is a formidable barrier to gene flow, preventing dispersal along coastal margins, and the extent and duration of ice expansion and contraction have been the primary driver in structuring the lineages of contemporary Antarctic Collembola. Major dispersal events occurred during periods of proposed WAIS collapse but were otherwise restricted by the prevailing extent of ice cover. Rapid ice loss for these regions is predicted in the near future which could have a profound effect on the structure of these populations. Our biological data provide an independent estimate of the frequency and magnitude of historical ice sheet dynamics and corroborate recent models and paleoclimate reconstructions.
Methods
Sample Collection.
Sampling for soil invertebrates was undertaken during austral summer field seasons between January 1999 and February 2018, at 91 locations throughout SVL, the QAR, and the QMM, including from the southernmost terrestrial habitats in Antarctica (Fig. 1). Several samples were taken from each location, with over 200 samples from the QAR−QMM area alone. Collembola were either collected directly using modified aspirators (55) or isolated from soil samples by flotation (56) while in Antarctica. All samples were preserved in either liquid nitrogen or 100% ethanol and, once back at the laboratory, stored at −80 °C or +4 °C, respectively.
Sequence Analyses.
In the laboratory, photographs were taken of each specimen prior to being individually processed at either the University of Waikato, New Zealand, or the Canadian Centre for DNA barcoding (CCDB), Canada. The DNA barcoding region (658 base pairs [bp]) of the mitochondrial cytochrome c oxidase subunit I (COI) gene was amplified using the universal primers HCO2198 and LCO1490 (57) at the University of Waikato, and the primers LepF1 and LepR1 (58) at CCDB. See SI Appendix for further sequencing details. In total, 132 COI sequences from along the QAR and the QMM were generated (SI Appendix, Table S1), and combined with the existing database of 1,030 COI sequences for Collembola from throughout the study area (Table 1).
Including the 132 COI sequences that were generated here (available via Barcode of Life Datasystems (BOLD) dataset DS-TAMS: http://dx.doi.org/10.5883/DS-TAMS; GenBank accession numbers MN619477 to MN619610), a total of 1,162 mitochondrial COI sequences were available for the study. The final alignment (n = 866 sequences) was generated using Multiple Sequence Comparison by Log- Expectation (MUSCLE) 3.8.425 (59) as implemented in Geneious 11.1.5 (https://www.geneious.com) and was trimmed to 560 bp to maximize available individuals for these analyses. We also included a representative COI sequence for Sminthurus viridis (GenBank accession KJ155514; ref. 60) to root the tree, based on ref. 61.
Using the final alignment, a phylogenetic tree was generated of only unique sequences (n = 113; 560 bp), including the outgroup S. viridis, using the maximum likelihood analysis as implemented in IQ-Tree (62) and the in-built ModelFinder Plus (63) for identification and automatic utilization of the most appropriate model of sequence evolution (TPM2u+F+I+G4). Ultrafast bootstrap approximation was carried out for 5,000 replicates using UFBoot (64). In FigTree 1.4.2 (available at http://tree.bio.ed.ac.uk/software/figtree/), the resulting tree was visualized and processed to produce Figs. 2 and 5. The best-fit model of sequence evolution was separately estimated for each species, following the same method as outlined above.
Sequences were grouped into genetic lineages based on considerations of biogeographical distributions, levels of genetic divergence (3.3 to 14.3% uncorrected mean p distance), and congruence with BINs (65). This was straightforward, as all 14 lineages identified were found at distinct geographic locations. To represent these lineages, Figs. 3 and 4 include haplotype networks that were generated in R v3.5.1 utilizing the package “pegas” (37) in RStudio, from the final 560-bp alignment. Distributional maps were also generated in R v3.5.1 utilizing the packages “mapdata” (66) and “mapplots” (67).
Four of the six Collembola species contained high levels of diversity. As higher levels of genetic diversity can lead to an increasing likelihood of substitution saturation, particularly at the third and more variable codon position, we assessed nucleotide substitution saturation individually for each of the four genetically variable species at codon positions one and two combined, and for the third codon position separately, using the entropy-based Iss test (68) as implemented in Data Analysis for Molecular Biology and Evolution (DAMBE) 7.0.48 (69). For the analysis of codon positions one and two, the proportion of invariant sites was estimated for each species based on the in-built neighbor-joining tree construction method, and resulting P(inv) values (SI Appendix, Table S2) were incorporated into the tests of substitution saturation.
Application of the Molecular Clock Rate.
For molecular clock analyses, one representative sequence encompassing the full 658-bp DNA barcoding COI gene region was randomly selected from each of the 14 distinct genetic lineages and aligned with the outgroup S. viridis using MUSCLE in Geneious (representative sequences are noted in Fig. 2 and SI Appendix, Tables S3 and S4). Pairwise uncorrected (p distance) and corrected (K2P and TN93+G) distances were generated in Molecular Evolutionary Genetics Analysis (MEGA) 7.0.26 (70) based on these 658-bp sequences.
The best-fit model of sequence evolution was identified as HKY+G+I by bModelTest (71) in Bayesian Evolutionary Analysis Sampling Trees (BEAST 2.4.7). Therefore, Bayesian analyses were conducted with the following parameters: substitution rate estimated; four gamma categories, proportion invariant set at 0.1, estimated; HKY substitution model; strict clock rate set to 0.0168 (3.54% divergence per My; ref. 27); Yule tree prior; monophyletic constraint prior on all ingroup taxa; Markov chain Monte Carlo (MCMC) for 100 million generations; and sampling trees and statistics every 1,000 generations.
Molecular Clocks Calibrated to Warming Events and Past Ice Sheet Collapses.
Based on sediment cores obtained from beneath the McMurdo Ice Shelf as part of the Antarctic Drilling Project (ANDRILL) program (e.g., AND-1B), and ice sheet modeling of these data, confident estimations of past extents of the WAIS and the marine margin of the EAIS throughout the past 5 My have been made (3, 4). Here, we use the large-scale ice retreat event that occurred during the Pliocene to calibrate divergence dates for Antarctic Collembola. During this warmer time (5 Mya to 3 Mya) the WAIS had completely collapsed, and coastal thinning and retreat of the EAIS had occurred (2–4). The increase in water movements during these melting events, and exposure of new ice-free habitat, may have provided many more opportunities for Collembola dispersal as compared to the present-day climate. The subsequent reformation of ice sheets and resulting population isolation was used to calibrate molecular clock rates and divergence time estimates for Antarctic Collembola.
Bayesian analyses were conducted on the 658-bp alignment of representative sequences (n = 15) with the following parameters: automatic set clock rate unchecked; substitution rate estimated; HKY substitution model; relaxed clock log normal; Calibrated Yule tree prior; monophyletic constraint prior on all ingroup taxa; MCMC for 10 million generations; and sampling trees and statistics every 1,000 generations. Separate runs were performed, where each of the four species was separately constrained as a monophyletic group and the species node was calibrated with the 5- to 3-My isolation event. Where relevant, we also performed Tajima’s relative rate tests (72) in MEGA to assess likelihood of equal rates of molecular evolution at each branch.
Supplementary Material
Acknowledgments
We are grateful for the considerable logistical support provided by Antarctica New Zealand and the United States Antarctic Program over the duration of this study and to the New Zealand Antarctic Research Institute, the University of Waikato, and Polar Knowledge Canada for funding to I.D.H. Additional funding and support was provided by NSF Grants OPP-0840979, 1341736 and 1637708, and this study contributes to the McMurdo Dry Valleys Long Term Ecological Research program. Genome Canada and the Ontario Genomics Institute via the Canadian Centre for DNA Barcoding and the International Barcode of Life Project subsidized the DNA sequencing. P.C. is supported by Natural Environment Research Council core funding to the British Antarctic Survey “Biodiversity, Evolution and Adaptation” Team. We thank B. Storey, J. Watson, U. Nielsen, C. Beet, M. Knox, and M. Stevens for their input and/or assistance in the field, A, Fjellberg for confirming the identification of Gomphiocephalus hodgsoni from Beardmore Glacier, and A. McGaughran for advice on statistical analyses. The satellite imagery used to generate Fig. 1 was retrieved from GloVis, courtesy of the NASA Earth Observing System Data and Information System Land Processes Distributed Active Archive Center, US Geological Survey/Earth Resources Observation and Science Center, Sioux Falls, SD (https://lpdaac.usgs.gov/tools/usgs-earthexplorer/). Small portions of the text were adapted from the PhD thesis of G.E.C. This paper contributes to the “State of the Antarctic Ecosystem” and the “Antarctic Thresholds, Ecosystems Resilience and Adaptation” programs of Scientific Committee on Antarctic Research.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007925117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix. BOLD dataset DS-TAMS is available at http://dx.doi.org/10.5883/DS-TAMS; GenBank accession numbers MN619477 to MN619610.
References
- 1.Masson-Delmotte V., et al. , “Information from paleoclimate archives” in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, Stocker T. F., Ed. (Cambridge University Press, Cambridge, United Kingdom, 2013), pp. 383–464. [Google Scholar]
- 2.Barrett P. J., Resolving views on Antarctic Neogene glacial history–The Sirius debate. Earth Environ. Sci. Trans. R. Soc. Edinb. 104, 31–53 (2013). [Google Scholar]
- 3.Naish T. et al., Obliquity-paced Pliocene West Antarctic ice sheet oscillations. Nature 458, 322–328 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Pollard D., DeConto R. M., Modelling West Antarctic ice sheet growth and collapse through the past five million years. Nature 458, 329–332 (2009). [DOI] [PubMed] [Google Scholar]
- 5.DeConto R. M., Pollard D., Contribution of Antarctica to past and future sea-level rise. Nature 531, 591–597 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Barker P. F., Thomas E., Origin, signature and palaeoclimatic influence of the Antarctic Circumpolar Current. Earth Sci. Rev. 66, 143–162 (2004). [Google Scholar]
- 7.Sarkar S. et al., Late Eocene onset of the proto-Antarctic circumpolar current. Sci. Rep. 9, 10125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chown S. L., Convey P., Spatial and temporal variability across life’s hierarchies in the terrestrial Antarctic. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 2307–2331 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van den Hoogen J. et al., Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Wise K. A. J., Collembola (springtails). Antarct. Res. Ser. 10, 123–148 (1967). [Google Scholar]
- 11.Hillebrand H., On the generality of the latitudinal diversity gradient. Am. Nat. 163, 192–211 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Green T. G. A., Sancho L. G., Türk R., Seppelt R. D., Hogg I. D., High diversity of lichens at 84°S, Queen Maud Mountains, suggests preglacial survival of species in the Ross Sea region, Antarctica. Polar Biol. 34, 1211–1220 (2011). [Google Scholar]
- 13.Powers L. E., Ho M., Freckman D. W., Virginia R. A., Distribution, community structure, and microhabitats of soil invertebrates along an elevational gradient in Taylor Valley, Antarctica. Arct. Alp. Res. 30, 133–141 (1998). [Google Scholar]
- 14.Convey P. et al., “Ice bound Antarctica: Biotic consequences of the shift from a temperate to a polar climate” in Mountains, Climate, and Biodiversity, Hoorn C., Perrigo A., Antonelli A., Eds. (Wiley, 2018), pp. 355–373. [Google Scholar]
- 15.Convey P. et al., Antarctic terrestrial life–challenging the history of the frozen continent? Biol. Rev. Camb. Philos. Soc. 83, 103–117 (2008). [DOI] [PubMed] [Google Scholar]
- 16.McGaughran A., Stevens M. I., Hogg I. D., Carapelli A., Extreme glacial legacies: A synthesis of the Antarctic springtail phylogeographic record. Insects 2, 62–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terauds A., Lee J. R., Antarctic biogeography revisited: Updating the Antarctic conservation biogeographic regions. Divers. Distrib. 22, 836–840 (2016). [Google Scholar]
- 18.von Saltzwedel H., Scheu S., Schaefer I., Founder events and pre-glacial divergences shape the genetic structure of European Collembola species. BMC Evol. Biol. 16, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Vuuren B. J., Lee J. E., Convey P., Chown S. L., Conservation implications of spatial genetic structure in two species of oribatid mites from the Antarctic Peninsula and the Scotia Arc. Antarct. Sci. 30, 105–114 (2018). [Google Scholar]
- 20.Wallace A. R., On the phenomena of variation and geographical distribution as illustrated by the Papilionidae of the Malayan region. Trans. Linn. Soc. Lond. 25, 1–71 (1865). [Google Scholar]
- 21.Allegrucci G., Carchini G., Todisco V., Convey P., Sbordoni V., A molecular phylogeny of Antarctic Chironomidae and its implications for biogeographical history. Polar Biol. 29, 320–326 (2006). [Google Scholar]
- 22.Strugnell J. M., Pedro J. B., Wilson N. G., Dating Antarctic ice sheet collapse: Proposing a molecular genetic approach. Quat. Sci. Rev. 179, 153–157 (2018). [Google Scholar]
- 23.Brower A. V. Z., Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. U.S.A. 91, 6491–6495 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolan L., Hogg I. D., Stevens M. I., Haase M., Fine scale distribution of mtDNA haplotypes for the springtail Gomphiocephalus hodgsoni (Collembola) corresponds to an ancient shoreline in Taylor Valley, continental Antarctica. Polar Biol. 29, 813–819 (2006). [Google Scholar]
- 25.Ding Y. H., Yu D. Y., Guo W. B., Li J. N., Zhang F., Molecular phylogeny of Entomobrya (Collembola: Entomobryidae) from China: Color pattern groups and multiple origins. Insect Sci. 26, 587–597 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Quek S.-P., Davies S. J., Itino T., Pierce N. E., Codiversification in an ant-plant mutualism: Stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 58, 554–570 (2004). [PubMed] [Google Scholar]
- 27.Papadopoulou A., Anastasiou I., Vogler A. P., Revisiting the insect mitochondrial molecular clock: The mid-Aegean Trench calibration. Mol. Biol. Evol. 27, 1659–1672 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Katz A. D., Taylor S. J., Davis M. A., At the confluence of vicariance and dispersal: Phylogeography of cavernicolous springtails (Collembola: Arrhopalitidae, Tomoceridae) codistributed across a geologically complex karst landscape in Illinois and Missouri. Ecol. Evol. 8, 10306–10325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukić M., Delić T., Pavlek M., Deharveng L., Zagmajster M., Distribution pattern and radiation of the European subterranean genus Verhoeffiella (Collembola, Entomobryidae). Zool. Scr. 49, 86–100 (2020). [Google Scholar]
- 30.Zhang F., Yu D., Stevens M. I., Ding Y., Colouration, chaetotaxy and molecular data provide species-level resolution in a species complex of Dicranocentrus (Collembola : Entomobryidae). Invertebr. Syst. 32, 1298 (2018). [Google Scholar]
- 31.Johnson D. L., Wellington W. G., Dispersal of the collembolan, Folsomia candida Willem, as a function of age. Can. J. Zool. 61, 2534–2538 (1983). [Google Scholar]
- 32.Hawes T. C., Worland M. R., Convey P., Bale J. S., Aerial dispersal of springtails on the Antarctic Peninsula: Implications for local distribution and demography. Antarct. Sci. 19, 3–10 (2007). [Google Scholar]
- 33.Worland M. R., Block W., Desiccation stress at sub-zero temperatures in polar terrestrial arthropods. J. Insect Physiol. 49, 193–203 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Hawes T. C., Rafting in the Antarctic springtail, Gomphiocephalus hodgsoni. Antarct. Sci. 23, 456–460 (2011). [Google Scholar]
- 35.Knowlton N., Weigt L. A., New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. Lond. B Biol. Sci. 265, 2257–2263 (1998). [Google Scholar]
- 36.Collins G. E., Hogg I. D., Convey P., Barnes A. D., McDonald I. R., Spatial and temporal scales matter when assessing the species and genetic diversity of springtails (Collembola) in Antarctica. Front. Ecol. Evol. 7, 76 (2019). [Google Scholar]
- 37.Paradis E., pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Convey P. et al., Exploring biological constraints on the glacial history of Antarctica. Quat. Sci. Rev. 28, 3035–3048 (2009). [Google Scholar]
- 39.Roderick G. K., Geographic structure of insect populations: Gene flow, phylogeography, and their uses. Annu. Rev. Entomol. 41, 325–352 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Vrijenhoek R. C., “Genetic diversity and fitness in small populations” in Conservation Genetics. EXS, Loeschcke V., Jain S. K., Tomiuk J., Eds. (Birkhäuser Basel, Basel, Switzerland, 1994), Vol. vol. 68, pp. 37–53. [Google Scholar]
- 41.Wright S., Isolation by distance. Genetics 28, 114–138 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortimer E., van Vuuren B. J., Phylogeography of Eupodes minutus (Acari: Prostigmata) on sub-Antarctic Marion Island reflects the impact of historical events. Polar Biol. 30, 471–476 (2007). [Google Scholar]
- 43.Carapelli A. et al., Evidence for cryptic diversity in the “pan-Antarctic” springtail Friesea antarctica and the description of two new species. Insects 11, 141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyons W. B. et al., The soil geochemistry in the Beardmore Glacier region, Antarctica: Implications for terrestrial ecosystem history. Sci. Rep. 6, 26189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fielding C. R., Harwood D. M., Winter D. M., Francis J. E., Neogene stratigraphy of Taylor Valley, Transantarctic Mountains, Antarctica: Evidence for climate dynamism and a vegetated Early Pliocene coastline of McMurdo Sound. Global Planet. Change 96–97, 97–104 (2012). [Google Scholar]
- 46.Gasson E., DeConto R. M., Pollard D., Modeling the oxygen isotope composition of the Antarctic ice sheet and its significance to Pliocene sea level. Geology 44, 827–830 (2016). [Google Scholar]
- 47.Hambrey M. J., Webb P. N., Harwood D. M., Krissek L. A., Neogene glacial record from the Sirius Group of the Shackleton Glacier region, central Transantarctic Mountains, Antarctica. Bull. Geol. Soc. Am. 115, 994–1015 (2003). [Google Scholar]
- 48.Marsh O. J., Rack W., Floricioiu D., Golledge N. R., Lawson W., Tidally-induced velocity variations of the Beardmore Glacier, Antarctica, and their representation in satellite measurements of ice velocity. The Cryosphere Discussions 7, 1761–1785 (2013). [Google Scholar]
- 49.Lisiecki L. E., Raymo M. E., A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20, PA1003 (2005). [Google Scholar]
- 50.Beltran C. et al., Southern Ocean temperature records and ice-sheet models demonstrate rapid Antarctic ice sheet retreat under low atmospheric CO2 during Marine Isotope Stage 31. Quat. Sci. Rev. 228, 106069 (2020). [Google Scholar]
- 51.Roberts D. L., Karkanas P., Jacobs Z., Marean C. W., Roberts R. G., Melting ice sheets 400,000 yr ago raised sea level by 13 m: Past analogue for future trends. Earth Planet. Sci. Lett. 357–358, 226–237 (2012). [Google Scholar]
- 52.Miller K. G. et al., High tide of the warm Pliocene: Implications of global sea level for Antarctic deglaciation. Geology 40, 407–410 (2012). [Google Scholar]
- 53.Carapelli A., Convey P., Frati F., Spinsanti G., Fanciulli P., Population genetics of three sympatric springtail species (Hexapoda, Collembola) from the South Shetland Islands: Evidence for a common biogeographic pattern. Biol. J. Linn. Soc. Lond. 120, 788–803 (2017). [Google Scholar]
- 54.Adams B. J., Species concepts and the evolutionary paradigm in modem nematology. J. Nematol. 30, 1–21 (1998). [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens M. I., Hogg I. D., Expanded distributional records of Collembola and Acari in southern Victoria Land, Antarctica. Pedobiologia (Jena) 46, 485–495 (2002). [Google Scholar]
- 56.Freckman D. W., Virginia R. A., Extraction of nematodes from Dry Valley Antarctic soils. Polar Biol. 13, 483–487 (1993). [Google Scholar]
- 57.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R., DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994). [PubMed] [Google Scholar]
- 58.Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H., Hallwachs W., Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A. 101, 14812–14817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crotty F. V. et al., First documented pest outbreak of the herbivorous springtail Sminthurus viridis (Collembola) in Europe. Grass Forage Sci. 71, 699–704 (2016). [Google Scholar]
- 61.Yu D. et al., New insight into the systematics of Tomoceridae (Hexapoda, Collembola) by integrating molecular and morphological evidence. Zool. Scr. 45, 286–299 (2016). [Google Scholar]
- 62.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ratnasingham S., Hebert P. D. N., A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS One 8, e66213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker R., Wilks A., Brownrigg R., mapdata: Extra Map Databases. R Package Version 2.2-6 (2016). https://cran.r-project.org/web/packages/mapdata/. Accessed 12 August 2019.
- 67.Gerritsen H., mapplots: Data Visualisation on Maps. R Package Version 1.5 (2014). https://rdrr.io/cran/mapplots/. Accessed 12 August 2019.
- 68.Xia X., Xie Z., Salemi M., Chen L., Wang Y., An index of substitution saturation and its application. Mol. Phylogenet. Evol. 26, 1–7 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Xia X., DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S., Stecher G., Tamura K., MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouckaert R. R., Drummond A. J., bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 17, 42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tajima F., Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135, 599–607 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix. BOLD dataset DS-TAMS is available at http://dx.doi.org/10.5883/DS-TAMS; GenBank accession numbers MN619477 to MN619610.