Significance
Cochlear hair cell loss is a leading cause of deafness in humans and other mammals. In the immature cochlea, lost hair cells are regenerated by neighboring glia-like supporting cells. However, for reasons unknown, such regenerative capacity is rapidly lost as supporting cells undergo maturation. Here we identify a direct link between LIN28B, mammalian target of rapamycin (mTOR) activity, and supporting cell plasticity. Mimicking later developmental stages, we found that loss of the RNA binding protein LIN28B attenuated mTOR signaling and rendered young, immature supporting cells incapable of producing hair cells. Conversely, we found that reexpression of LIN28B reinstated the ability of maturing supporting cells to revert to a progenitor-like state and generate hair cells via activation of mTOR signaling.
Keywords: hair cell regeneration, mTOR pathway, inner ear cochlea, LIN28, let-7 miRNA
Abstract
Mechano-sensory hair cells within the inner ear cochlea are essential for the detection of sound. In mammals, cochlear hair cells are only produced during development and their loss, due to disease or trauma, is a leading cause of deafness. In the immature cochlea, prior to the onset of hearing, hair cell loss stimulates neighboring supporting cells to act as hair cell progenitors and produce new hair cells. However, for reasons unknown, such regenerative capacity (plasticity) is lost once supporting cells undergo maturation. Here, we demonstrate that the RNA binding protein LIN28B plays an important role in the production of hair cells by supporting cells and provide evidence that the developmental drop in supporting cell plasticity in the mammalian cochlea is, at least in part, a product of declining LIN28B-mammalian target of rapamycin (mTOR) activity. Employing murine cochlear organoid and explant cultures to model mitotic and nonmitotic mechanisms of hair cell generation, we show that loss of LIN28B function, due to its conditional deletion, or due to overexpression of the antagonistic miRNA let-7g, suppressed Akt-mTOR complex 1 (mTORC1) activity and renders young, immature supporting cells incapable of generating hair cells. Conversely, we found that LIN28B overexpression increased Akt-mTORC1 activity and allowed supporting cells that were undergoing maturation to de-differentiate into progenitor-like cells and to produce hair cells via mitotic and nonmitotic mechanisms. Finally, using the mTORC1 inhibitor rapamycin, we demonstrate that LIN28B promotes supporting cell plasticity in an mTORC1-dependent manner.
The cochlea, located in the inner ear, contains highly specialized mechano-receptor cells, termed hair cells, which are critical for our ability to detect sound. In mammals, auditory hair cells are only produced during embryonic development and hair cell loss due to aging, disease, or trauma is a leading cause for hearing impairment and deafness in humans. Nonmammalian vertebrates, such as birds, are capable of regenerating hair cells within their auditory and vestibular sensory organs throughout their lifetime (reviewed in ref. 1). The source of the newly generated (regenerated) hair cells are glia-like supporting cells. Sharing a close lineage relationship, hair cells and supporting cells originate from a common pool of progenitor cells, termed prosensory cells (2, 3). The transcription factor SOX2 is essential for the establishment and maintenance of prosensory cells (4) and plays a critical role in the induction of Atoh1 during hair cell formation (5, 6). The transcriptional activator ATOH1 is required for hair cell formation in the developing inner ear (7) and its reactivation in supporting cells is an essential step in the process of hair cell regeneration (8, 9).
In birds, loss of auditory hair cells induces adjacent supporting cells to either directly convert (transdifferentiate) into hair cells (10, 11), or alternatively, to reenter the cell cycle and after rounds of cell division to produce both hair cells and supporting cells (12, 13). In response to hair cell death, cochlear supporting cells in newborn mice have been recently shown to reenter the cell cycle and produce new hair cells (14, 15). The injury-induced regenerative response in the neonatal/early postnatal cochlea can be greatly enhanced by genetic or pharmacologic inhibition of Notch signaling (16–18) and overactivation of the Wnt/β-catenin signaling (19–22). However, mice are born deaf and their cochlear hair cells and supporting cells are not functional (mature) until the onset of hearing at postnatal days 12 to 13 (P12–P13) (23–25). Recent studies uncovered that as early as P5/P6 murine cochlear supporting cells fail to regenerate hair cells in response to injury (14), inhibition of Notch signaling (26), or overactivation of Wnt/β-catenin signaling (27). What causes the rapid decline in supporting cell plasticity during the first postnatal week is currently unknown.
We previously demonstrated that a regulatory circuit consisting of LIN28B protein and let-7 miRNAs modulates the production of new hair cells in stage P2 murine cochlear explants, with LIN28B promoting new hair cell production and let-7 miRNAs suppressing it (28). The closely related RNA binding proteins LIN28A and LIN28B (LIN28A/B) and members of the let-7 family of miRNAs belong to an evolutionarily conserved network of genes, initially identified in Caenorhabditis elegans for their role in developmental timing (heterochrony) (29, 30). Let-7 miRNAs and LIN28A/B are mutual antagonists that repress each other’s expression. LIN28A/B inhibit the biogenesis of let-7 miRNAs through direct binding to primary and precursor let-7 transcripts. In turn, let-7 miRNAs interfere with the translation of Lin28a and Lin28b mRNAs by binding to their 3′UTR (reviewed in ref. 31). While let-7 miRNAs are linked to a postmitotic, terminal differentiated state, LIN28A/B proteins are positive regulators of stemness, organismal growth, and metabolism (reviewed in ref. 32). How LIN28B/let-7 influence the postnatal production of hair cells by supporting cells and whether the LIN28B/let-7 circuit plays a role in the developmental decline of supporting cell plasticity is currently unknown.
Here, we provide evidence that the decline in supporting cell plasticity in the murine cochlea is, at least in part, due to diminished LIN28B-mammalian target of rapamycin complex 1 (mTORC1) activity. Using organoid and explant cultures to model mitotic and nonmitotic hair cell production by supporting cells, we found that diminished LIN28B-mTOR activity, due to let-7g overexpression or targeted deletion of Lin28a/b, accelerated the developmental decline in supporting cell plasticity and rendered, otherwise plastic, immature supporting cells incapable of producing new hair cells. Conversely, we found that reexpression of LIN28B reinstated the ability of maturing supporting cells to generate hair cells and found that rapamycin, a selective inhibitor of the mTORC1 kinase complex, blocked such LIN28B-induced hair cell production.
Results
The Ability of Murine Cochlear Epithelial Cells to Form and Grow Hair Cell Containing Organoids Sharply Declines within the First Postnatal Week.
Cochlear supporting cells, isolated from neonatal mice, reenter the cell cycle and produce hair cells when cultured on a feeder layer of periotic mesenchymal cells in the presence of mitogens. However, mature cochlear supporting cells isolated from stage P14 mice fail to proliferate and produce only few scattered hair cells when cultured under identical conditions (33). A similar decline in the potential to proliferate and self-renew is observed when cochlear epithelial cells, including supporting cells, are propagated using neurosphere culture conditions (34). To address the underlying molecular mechanisms that cause the decline in supporting cell plasticity, we employed a recently developed organoid culture system, which allows Wnt-responsive cochlear epithelial cells (supporting cells) to propagate and differentiate into large hair cell-containing organoids (35).
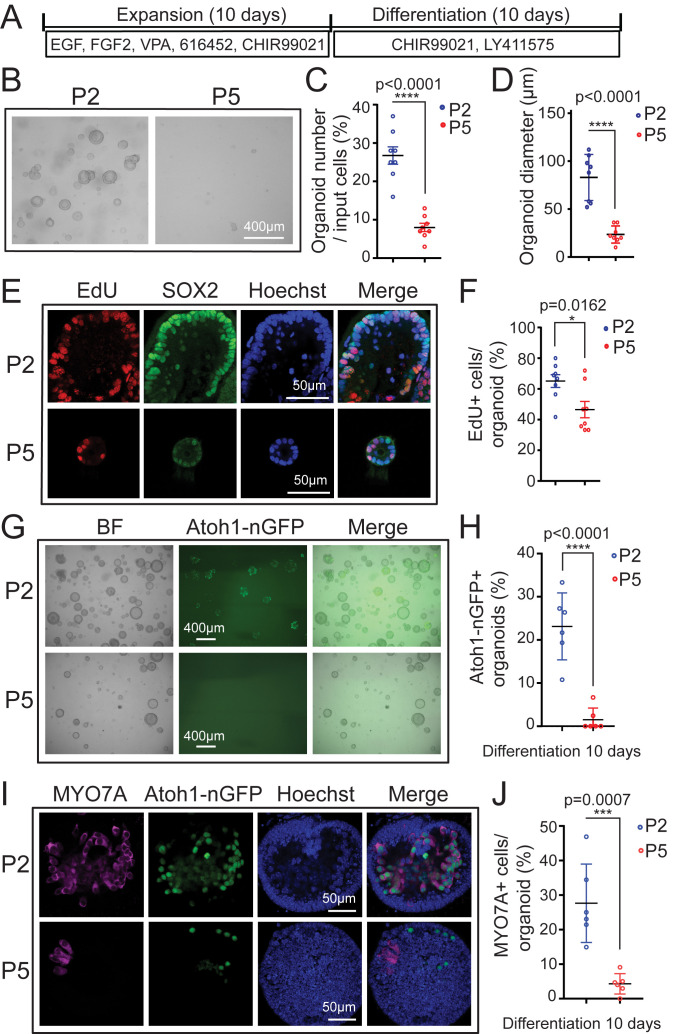
Employing this organoid culture system (Expansion in Fig. 1A), we found that cochlear epithelial cells from stage P2 mice (P2 organoid culture) readily formed large organoids (P2 in Fig. 1B). In contrast, cochlear epithelial cells obtained from stage P5 mice (P5 organoid culture) showed a greatly diminished capacity to form and grow organoids (P5 in Fig. 1B). After 5 d of expansion, P5 organoid cultures contained three times fewer organoids than P2 organoid cultures (Fig. 1 B and C) and were only a quarter the size of organoids in P2 organoid cultures (Fig. 1 B and D). To address potential defects in cell proliferation, P2 and P5 organoid cultures received a 1-h pulse of EdU (5-ethynyl-2′-deoxyuridine), after which organoids were harvested and costained for SOX2. Prior to cochlear differentiation, SOX2 expression identifies prosensory cells and later marks supporting cells and Kölliker’s cells, a transient population of supporting-like cells located at the medial border of the sensory epithelium. In addition, SOX2 is transiently expressed in nascent hair cells (6, 36). We found that SOX2+ cells in P5 organoids proliferated at a 25% lower rate (percent of EdU+ cells) than SOX2+ cells in P2 organoids (Fig. 1 E and F), indicating that the observed defects in P5 organoid formation and growth were the product of reduced cell proliferation.
Fig. 1.
Cochlear epithelial cells from stage P5 mice fail to expand and produce hair cells in organoid culture. Organoid cultures were established from cochlear epithelia cells obtained from Atoh1-nGFP transgenic mice, stages P2 and P5. Atoh1-nGFP marks nascent hair cells. (A) Experimental design. (B) Brightfield (BF) images of P2 and P5 cochlear organoid cultures after 5 d of expansion. (C) Organoid forming efficiency in P2 (blue) and P5 (red) cultures in B (n = 8). (D) Organoid diameters in P2 (blue) and P5 (red) cultures in B (n = 8). (E) Cell proliferation in P2 and P5 organoids. A single EdU pulse was given at 5 d of expansion and EdU incorporation (red) was analyzed 1 h later. SOX2 (green) marks supporting cells/prosensory cells and Hoechst (blue) labels cell nuclei. (F) Percentage of EdU+ cells in P2 and P5 organoids in E (n = 8). (G) BF and green fluorescent images (Atoh1-nGFP) of P2 and P5 organoid cultures after 10 d of differentiation. (H) Percentage of Atoh1-nGFP+ organoids in G (n = 6). (I) Confocal images of P2 and P5 organoid cultures after 10 d of differentiation. Newly formed hair cells express Atoh1-nGFP (green) and MYO7A (magenta). (J) Percentage of MYO7A+ hair cells per organoid in I (n = 6). Graphed are individual data points and mean ± SD. Individual data points in D, F, and J represent the average value per animal. n = animals analyzed per group. All data are from two independent experiments. Two-tailed, unpaired Student’s t tests were used to calculate P values.
Next, we analyzed the ability of P2 and P5 organoids to produce hair cells (Differentiation in Fig. 1A). Hair cell formation in P2 and P5 organoid cultures was monitored using Atoh1-nGFP reporter expression (37). Atoh1 (Atoh1-nGFP) expression is high in nascent hair cells, but is absent in hair cells that already underwent maturation, which allows to distinguish between existing and nascent hair cells (37, 38). After 10 d of differentiation, ∼25% of organoids in P2 organoid cultures contained Atoh1-nGFP+ cells (P2 in Fig. 1 G and H), which formed large clusters and coexpressed the hair cell-specific protein myosin VIIa (MYO7A) (P2 in Fig. 1 I and J). In contrast, less than 2% of organoids in P5 organoid cultures contained few, scattered Atoh1-nGFP+ cells and largely lacked myosinVIIa expression (P5 in Fig. 1 G–J). The observed decline in supporting cell plasticity between P2 and P5 correlated with a drop in Lin28b mRNA and LIN28B protein expression within cochlear epithelial cells in vivo (SI Appendix, Fig. S1 A and B).
LIN28B Reactivation Reinstates Cochlear Supporting Cell Plasticity.
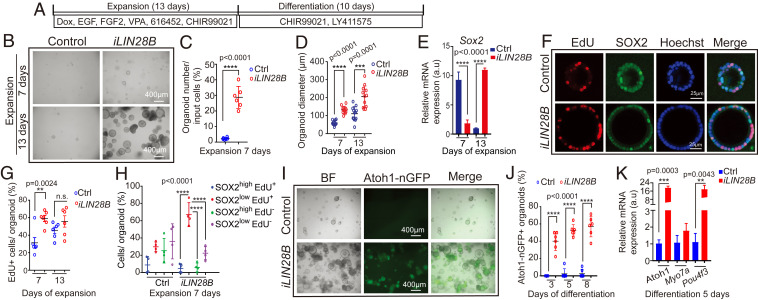
In order to address whether there is a link between LIN28B protein levels and supporting cell plasticity, we reexpressed LIN28B in stage P5 cochlear organoids using iLIN28B transgenic mice. In this double-transgenic mouse model, a flag-tagged human LIN28B transgene is expressed under the control of a TRE promoter (Col1a-TRE-LIN28B) (39), which in the presence of an ubiquitously expressed reverse tetracycline transactivator transgene (R26-rtTA-M2) (40) and doxycycline (dox) allows for robust induction of LIN28B protein (28, 39). First, we expanded cochlear epithelia cells from P5 iLIN28B transgenic mice and control littermates (lack LIN28B transgene) in the presence of dox for 13 d (Fig. 2A). We found that LIN28B overexpression increased the efficiency of organoid formation by more than fivefold, as well as doubled the average organoid size (diameter) compared to control (Fig. 2 B–D). The increase in organoid size in response to LIN28B overexpression was accompanied by a twofold higher rate of cell proliferation during the early phase of expansion (Fig. 2 F and G). Coinciding with the peak of cell proliferation, Sox2 transcript levels in LIN28B overexpressing organoids transiently decreased more than threefold compared to control organoids (Fig. 2E) and the majority of proliferating cells (EdU+) in LIN28B overexpressing organoids expressed SOX2 at a low level (SOX2low, EdU+ in Fig. 2H).
Fig. 2.
LIN28B overexpression promotes cochlear epithelial cell expansion and hair cell production. Cochlear organoid cultures were established from stage P5 Atoh1-nGFP;iLIN28B transgenic mice and control littermates that lacked the LIN28B transgene. Atoh1-nGFP marks nascent hair cells. (A) Experimental strategy. (B) BF images of control and LIN28B overexpressing (iLIN28B) organoid cultures at 7 and 13 d of expansion. (C) Organoid forming efficiency in control (Ctrl, blue) and LIN28B (iLIN28B, red) overexpressing cultures (n = 6, two independent experiments). (D) Organoid diameters in control (Ctrl, blue) and LIN28B (iLIN28B, red) overexpressing cultures after 7 and 13 d of expansion (n = 12, two independent experiments). (E) qRT-PCR analyzing Sox2 mRNA expression in control (Ctrl, blue bar) and LIN28B overexpressing (iLIN28B, red bar) organoids at 7 and 13 d of expansion (n = 3, from one representative experiment, three independent experiments). (F) Cell proliferation in control and LIN28B overexpressing organoids. An EdU pulse was given at 7 d (shown) or 13 d of expansion and EdU incorporation (red) was analyzed 2 h later. SOX2 (green) marks supporting cells/prosensory cells, Hoechst labels cell nuclei (blue). (G) Percentage of EdU+ cells in control (Ctrl, blue) and LIN28B overexpressing (iLIN28B, red) organoids at 7 and 13 d of expansion (n = 6, two independent experiments, n.s. not significant). (H) EdU incorporation in SOX2-high and SOX2-low expressing cells (n = 4, two independent experiments). Note that the majority of EdU+ cells in LIN28B overexpressing organoids expressed SOX2 at a low level (red, SOX2low EdU+). (I) BF and green fluorescent (Atoh1-nGFP) images of control and LIN28B overexpressing organoids after 8 d of differentiation. (J) Percentage of Atoh1-nGFP+ organoids in control (Ctrl) and LIN28B overexpressing (iLIN28B) cultures after 3, 5, and 8 d of differentiation (n = 6, two independent experiments). (K) qRT-PCR analyzing hair cell-specific mRNA expression (Atoh1, Myo7a, Pou4f3) in control (Ctrl, blue bar) and LIN28B overexpressing (iLIN28B, red bar) organoids after 5 d of differentiation (n = 3, from one representative experiment, three independent experiments). Bars in E and K represent mean ± SD, otherwise individual data points and their mean ± SD were plotted. Individual data points in D, G, and H represent the average value per animal. n = animals analyzed per group. Two-tailed, unpaired Student’s t test was used to calculate P values. a.u., arbitrary unit.
Next, we analyzed the capacity of control and LIN28B overexpressing organoids to produce hair cells. We found that after 3 d of differentiation, about 40% of LIN28B overexpressing organoids contained Atoh1-nGFP+ cell clusters, which increased to nearly 60% after 8 d of differentiation (iLIN28B in Fig. 2 I and J). In contrast, less than 5% of control organoids contained Atoh1-nGFP+ cells after 8 d of differentiation (“Ctl” [control] in Fig. 2 I and J). Confirming the presence of hair cells in LIN28B overexpressing organoids, we found that hair cell-specific transcripts (Atoh1, Pou4f3) were more than 10-fold up-regulated in LIN28B overexpressing organoids compared to control organoids (Fig. 2K). Mimicking hair cell development in vivo, nascent Atoh1-nGFP+ hair cells first up-regulated myosin VIIa (MYO7A) (SI Appendix, Fig. S2B), followed by the up-regulation of calretinin, a protein enriched in the inner hair cells (41) (SI Appendix, Fig. S2 A, C, and D). After 7 d of differentiation, Atoh1-nGFP+ hair cells also started to up-regulate prestin, a protein selectively expressed in outer hair cells (SI Appendix, Fig. S2F) and LIN28B overexpressing organoids up-regulated the expression of oncomodulin (Ocm), an outer hair cell-specific gene (42) and Fgf8, an inner hair cell-specific gene (43) (SI Appendix, Fig. S2G). In vivo, cochlear hair cells only transiently express Atoh1 (Atoh1-nGFP) and SOX2 and their expression is lost once cochlear hair cells undergo maturation (6, 44). Similarly, we found that MYO7A+ hair cells initially coexpressed Atoh1-nGFP and SOX2 in LIN28B overexpressing organoids (“Diff. 5 days” in SI Appendix, Fig. S2E). However, at later stages of culture MYO7A+ hair cells often lacked Atoh1-nGFP and SOX2 expression (“Diff. 10 days” in SI Appendix, Fig. S2E). Together, these findings indicate that the newly formed hair cells in LIN28B overexpressing organoids acquired mature characteristics and specialized into inner and outer hair cell-like cells.
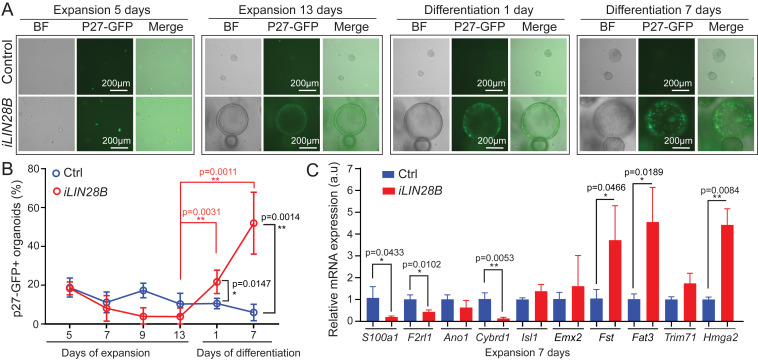
To be able to monitor the behavior of supporting cells within control and LIN28B overexpressing organoids, we generated control and iLIN28B transgenic mice that carried the p27-GFP BAC transgene. GFP expression in p27-GFP transgenic mice is under the control of the p27/Kip1 (cdkn1b) gene locus (45). P27/Kip1, a cyclin-dependent kinase inhibitor, is essential for forcing prosensory cells out of the cell cycle and for maintaining supporting cells in a postmitotic state (46–48). In the cochlear sensory epithelium, p27-GFP is selectively expressed in supporting cells. Other cell types, including hair cells, don’t express p27-GFP or, in the case of Kölliker’s cells, express p27-GFP at a much lower level than supporting cells (33). Importantly, p27-GFP expression is rapidly lost when supporting cells reenter the cell cycle (33), allowing us to pinpoint the peak of cell-cycle reentry of supporting cells in cochlear organoids. There was no significant difference in the number of p27-GFP+ organoids in control and LIN28B overexpressing cultures during early expansion (5 d), suggesting that supporting cells survived equally well in control and LIN28B overexpressing cultures (Fig. 3B). In control cultures the percentage of p27-GFP+ organoids declined during the remainder of the culture (Fig. 3A), and after 7 d of differentiation less than 10% of organoids contained p27-GFP+ supporting cells (Ctrl, blue line in Fig. 3B), suggesting that the majority of supporting cells in control organoids failed to re-enter the cell cycle. In contrast, p27-GFP expression in LIN28B overexpressing organoids, which during the latter half of expansion was near to being absent, dramatically increased upon differentiation (Fig. 3A). After 7 d of differentiation close to 50% of organoids in LIN28B overexpressing cultures contained large clusters of p27-GFP+ cells, indicating that LIN28B overexpression enabled stage P5 supporting cells to propagate in organoid culture (iLIN28B, red line in Fig. 3B).
Fig. 3.
LIN28B overexpression promotes supporting cell de-differentiation. Cochlear organoid cultures were established from stage P5 iLIN28B transgenic mice and control littermates that lacked the LIN28B transgene. Cochlear organoid cultures were maintained as outlined in Fig. 2A. P27-GFP transgene expression was used to monitor postmitotic supporting cells in A and B. (A) Representative BF and green fluorescent images of p27-GFP expression in LIN28B overexpressing and control organoid cultures at 5 and 13 d of expansion and 1 and 7 d of differentiation. (B) Percentage of p27-GFP+ organoids in A (mean ± SD, n = 7, two independent experiments). (C) qRT-PCR analyzing supporting cell-specific (S100a1, F2rl1, Ano1, Cybrd1), prosensory cell-specific (Fst, Fat3, Hmga2, Trim71) mRNA expression in control (Ctrl, blue bar) and LIN28B overexpressing (iLIN28B, red bar) organoids at 7 d of expansion. Emx2 and Isl1 served as controls (mean ± SD, n = 3, from one representative experiment, three independent experiments). n = animals analyzed per group. Two-tailed, unpaired Student’s t test was used to calculate P values.
To address whether LIN28B overexpression facilitates the de-differentiation of supporting cells into prosensory-like cells, we analyzed the expression of supporting cell (S100a1, F2rl1, Cybrd1, Ano1) (33, 49, 50) and prosensory cell-specific genes (Trim71, Hmga2, Fst, Fat3) (28, 51) (Fat3; Allen Developing Mouse Brain Atlas, https://developingmouse.brain-map.org/) in LIN28B overexpressing and control organoids after 7 d of expansion. As controls, we analyzed the expression of Isl1 (52) and Emx2 (53), which are expressed in prosensory cells and continue to be expressed in supporting cells and transiently in hair cells (54). As expected, the expression of Emx2 and Isl1 was not significantly changed in LIN28B overexpressing organoids compared to control. However, consistent with supporting cell de-differentiation, we found that the transcripts of prosensory cell-specific genes Fst, Fat3, and Hmga2 were fourfold higher expressed in LIN28B overexpressing organoids compared to control, whereas supporting cell-specific transcripts for S100a1, F2rl1, and Cybrd1 were two- to sevenfold lower expressed compared to control (Fig. 3C). Once differentiation was induced, LIN28B overexpressing organoids contained both Atoh1-nGFP+ hair cells and Atoh1-nGFP− S100A1+ or Atoh1-nGFP− JAGGED1+ supporting cell-like cells (SI Appendix, Fig. S2 A, H, and I). Together, these results indicate that LIN28B stimulates supporting cells to de-differentiate into prosensory-like cells that are capable of producing both hair cells and supporting cells in organoid culture.
Let-7 Overexpression or Loss of LIN28A/B Accelerates the Age-Dependent Decline in Supporting Cell Plasticity.
LIN28 proteins and let-7 miRNAs are mutual antagonists. To determine whether higher than normal let-7 miRNA levels would diminish the regenerative capacity of stage P2 supporting cells, we made use of iLet-7g transgenic mice (39). In this double transgenic mouse model a LIN28A/B-resistant form of let-7g is expressed under the control of a TRE promoter (Col1a-TRE-let-7S21L), which in the presence of the R26-rtTA-M2 transgene and dox allows for robust let-7g overexpression (28, 39). First, we analyzed whether let-7g overexpression disrupts the capacity of young, immature supporting cells to regenerate hair cells in response to Notch inhibition in cochlear explant culture. To ablate hair cells, control and let-7g overexpressing cochlear explants were treated with gentamicin for 20 h, after which Notch inhibitor (γ-secretase inhibitor) (LY411575) was added to induce supporting cell-to-hair cell conversion (SI Appendix, Fig. S3A). We found that let-7g overexpression reduced the production of hair cell (MYO7A+ SOX2+) in response to Notch inhibition by more than twofold in both intact (PBS, iLet-7) and hair cell-damaged (gentamicin, iLet-7) cochlear explants (SI Appendix, Fig. S3 B and C).
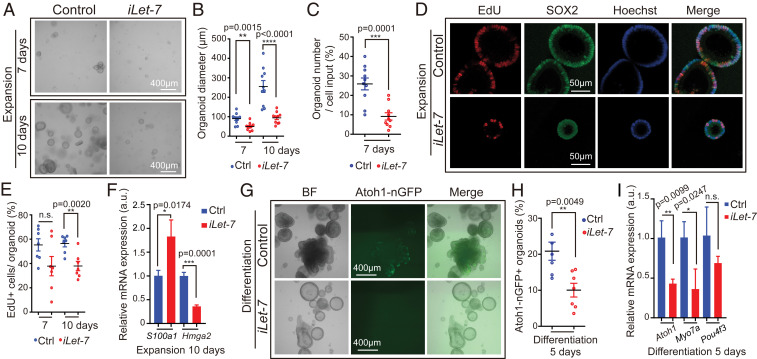
Next, we analyzed whether let-7g overexpression inhibits the capacity of young, immature supporting cells to reenter the cell cycle and propagate in organoid culture. To do so, we expanded cochlear epithelia cells from stage P2 iLet-7g transgenic mice and their nontransgenic control littermates (lack let-7g transgene) in the presence of dox for 10 d (Fig. 1A). We found that overexpression of let-7g decreased the number of organoids (Fig. 4 A and C) and decreased their average size by more than twofold (Fig. 4 A and B). These defects were accompanied by a 30% reduced rate of cell proliferation at 10 d of expansion (Fig. 4 D and E), as well as defects in supporting cell de-differentiation, as indicated by nearly twofold higher S100a1 expression and more than threefold lower Hmga2 expression in let-7g overexpressing organoids compared to control organoids (Fig. 4F). Next, we analyzed the hair cell-producing capacity of control and let-7g overexpressing organoids. To obtain sufficient cells for further analysis, we modified the experimental approach and omitted dox from the early phase of expansion. After 5 d of differentiation, about 20% of control organoids contained Atoh1-nGFP+ cell clusters, whereas only ∼10% of let-7g overexpressing organoids contained Atoh1-nGFP+ cell clusters (Fig. 4 G and H). Moreover, after 5 d of differentiation iLet-7g transgenic organoids expressed hair cell-specific genes (Atoh1, Myo7a) at a twofold lower level than control organoids (Fig. 4I). Together, these findings indicate that let-7 miRNAs inhibit supporting cell proliferation and limit the ability of supporting cells to acquire a hair cell fate. These findings complement our previous finding that let-7g overexpression inhibits progenitor cell proliferation during cochlear development and inhibits transdifferentiation of supporting cells into hair cells in response to Notch inhibition in cochlear explant culture (28).
Fig. 4.
Let-7g overexpression inhibits proliferation and hair cell formation in cochlear organoid culture. Cochlear organoid cultures from stage P2 Atoh1-nGFP;iLet-7 transgenic mice and control littermates that lacked the let-7g transgene were maintained as outlined in Fig. 1A. (A–F) Dox was present throughout expansion. (G–I) Dox was added after 4 d of expansion and was present the remaining 6 d of expansion and 5 d of differentiation. (A–E) Let-7g overexpression inhibits organoid formation and organoid expansion. (A) BF images of control and let-7g overexpressing organoid cultures after 7 and 10 d of expansion. (B) Diameter of control and let-7g overexpressing organoids in A (n = 10, three independent experiments). (C) Organoid forming efficiency of control and let-7g overexpressing organoids (n = 10, three independent experiments). (D) Cell proliferation in control and let-7g overexpressing organoids. A single EdU pulse was given at 7 or 10 d (shown) of expansion and EdU incorporation (red) was analyzed 1 h later. SOX2 (green) marked prosensory cells/supporting cells, Hoechst (blue) staining marked cell nuclei. (E) Percentage of EdU+ cells in D (n = 7, two independent experiments). (F) qRT-PCR analyzing S100a1 and Hmga2 mRNA expression in control and iLet-7 transgenic organoids at 10 d of expansion (n = 3, from one representative experiment, two independent experiments). (G–I) Let-7g overexpression inhibits hair cell formation. (G) BF and green fluorescent images (Atoh1-nGFP) of control and let-7g overexpressing organoid cultures. (H) Percentage of Atoh1-nGFP+ organoids in G (n = 6 for control group, n = 7 for iLet-7 group, two independent experiments). (I) qRT-PCR analyzing hair cell-specific (Atoh1, Myo7a, Pou4f3) mRNA expression in control and let-7g overexpressing organoids after 5 d of differentiation (n = 3, from one representative experiment, two independent experiments). Bars in F and I represent mean ± SD, otherwise individual data points and their mean ± SD were plotted. Note that the individual data points in B and E represent the average values per animal. n = animals analyzed per group. Two-tailed, unpaired Student’s t test was used to calculate P values.
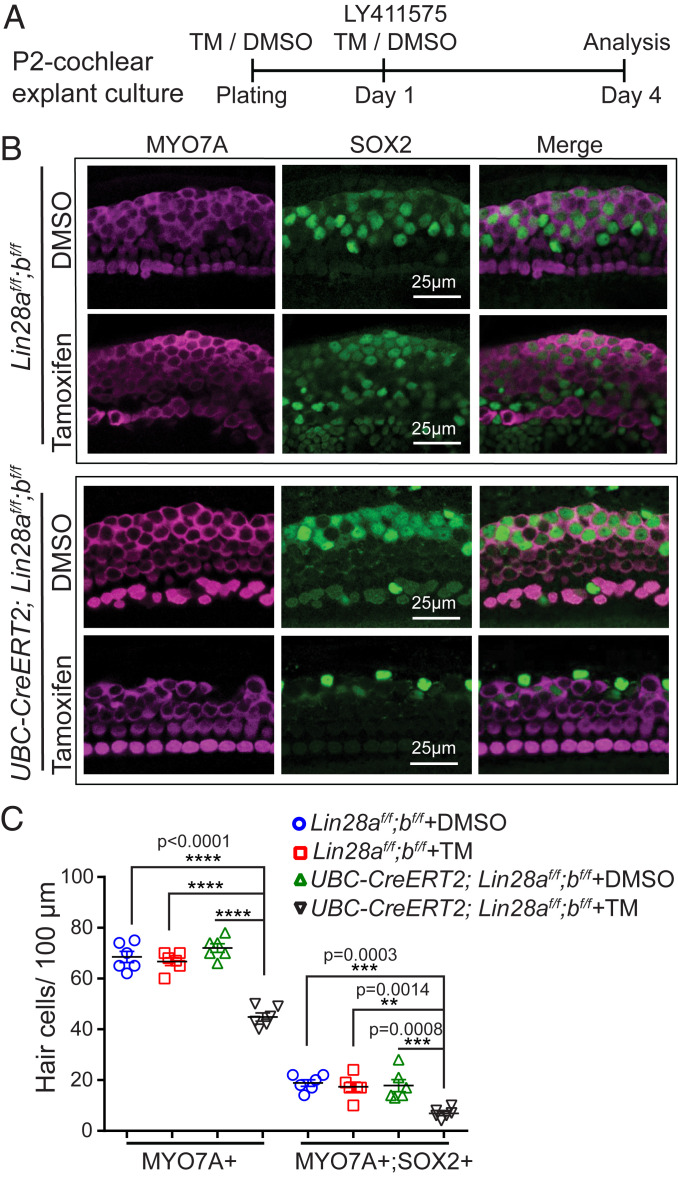
Next, we addressed whether endogenous LIN28B limits the capacity of neonatal supporting cells to form hair cells in vitro. Mice with single deletion of Lin28a or Lin28b are viable and show relatively mild or no overt defects, whereas codeletion of Lin28a and Lin28b result in severe abnormalities and is embryonic-lethal (55). To avoid potential functional redundancy between LIN28B and LIN28A, we conditionally knocked out both Lin28a and Lin28b in vitro using Lin28a and Lin28b floxed (f) mice (55) and UBC-CreERT2 transgenic mice, which ubiquitously express a tamoxifen-inducible form of Cre recombinase (56). First, we examined whether endogenous LIN28A/B are required for the direct conversion (transdifferentiation) of supporting cells into hair cells (Fig. 5A). We generated cochlear explant cultures from stage P2 UBC-CreERT2;Lin28af/f;Lin28bf/f mice and littermates that lacked the UBC-CreERT2 transgene (Lin28af/f;Lin28bf/f). Cultures received 4-hydroxy-tamoxifen (TM) or vehicle control (DMSO) at plating. The next day, cultures received the Notch inhibitor (LY411575) to induce supporting cell-to-hair cell conversion. Three days later, control cochlear explants (DMSO: UBC-CreERT2;Lin28af/f;Lin28bf/f; TM: Lin28af/f; Lin28bf/f; DMSO: Lin28af/f;Lin28bf/f) and Lin28a/b double knockout (dKO) cochlear explants (TM: UBC-CreERT2;Lin28af/f;Lin28bf/f) were analyzed for newly formed hair cells (MYO7A+ SOX2+). Consistent with previous reports, we found that supporting cells in control cochlear explants readily converted into hair cells in response to Notch inhibition, forming close to 20 new hair cells per 100 µm (Fig. 5 B and C, blue, red, green). In contrast, Lin28a/b dKO cochlear explants produced fewer than 7 new hair cells per 100 µm in response to Notch inhibition (Fig. 5 B and C, black).
Fig. 5.
Loss of LIN28A/B inhibits supporting cell-to-hair cell conversion in response to Notch inhibition. (A) Experimental strategy. Cochlear explants from stage P2 UBC-CreERT2;Lin28af/f;Lin28bf/f mice and Lin28af/f;Lin28bf/f littermates were cultured in the presence of 4-hydroxy-tamoxifen (TM) or DMSO (vehicle control). At day 1 of culture, cochlear explants received Notch inhibitor LY411575 and 3 d later cochlear explants were analyzed for new hair cells. (B) Confocal images of midapical turn of control (TM: Lin28af/f;Lin28bf/f; DMSO: UBC-CreERT2;Lin28af/f;Lin28bf/f; DMSO: Lin28af/f;Lin28bf/f) and Lin28a/b dKO cochlear explants (TM: UBC-CreERT2;Lin28af/f;Lin28bf/f) immuno-stained for MYO7A (magenta) and SOX2 (green). (C) Quantification of total number of hair cells (MYO7A+) as well as newly formed hair cells (MYO7A+ SOX2+) for control (blue, red, green) and Lin28a/b dKO (black) in B. Graphed are individual data points, representing average values per animal, and mean ± SD for control (blue, red, green) and Lin28a/b dKO (black), n = 6 animals per group, two independent experiments. Two-way ANOVA with Tukey’s correction was used to calculate P values.
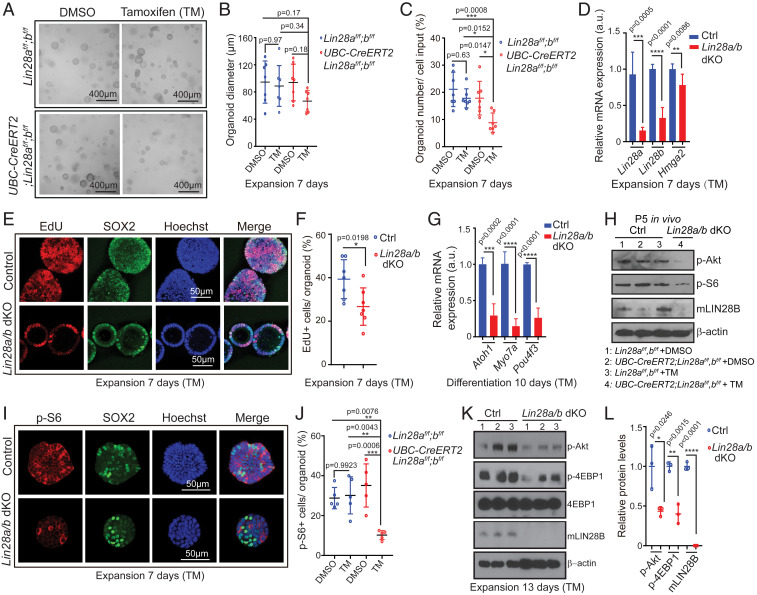
Next, we determined whether endogenous LIN28A/B is required for young, immature supporting cells to propagate and produce hair cells in cochlear organoid culture (Fig. 1A). To do so, we established organoid cultures from stage P2 UBC-CreERT2;Lin28af/f;Lin28bf/f mice and Lin28af/f;Lin28bf/f littermates in the presence of TM or vehicle control (DMSO). The tamoxifen-induced Lin28a/b deletion was confirmed by qRT-PCR analysis, which showed that Lin28a/b dKO organoids expressed Lin28b and Lin28a transcripts at three- to fivefold lower level than control organoids (Fig. 6D). The reduction in Lin28a/b expression was accompanied by the mild, but a significant down-regulation of the let-7 target Hmga2. Moreover, we found that twofold fewer organoids formed in Lin28a/b dKO organoid cultures compared to control cultures (Fig. 6 A and C) and Lin28a/b dKO organoids were on average 30% smaller than control organoids (Fig. 6 A and B). Furthermore, EdU pulse experiments revealed that the fraction of cells that were actively cycling within Lin28a/b dKO organoid was ∼30% lower than in control organoids (Fig. 6 E and F). To address whether LIN28A/B deficiency limits the ability of supporting cells/prosensory cells to generate hair cells, we differentiated control and Lin28a/b mutant organoids for 10 d and analyzed their expression of hair cell-specific genes (Atoh1, Pou4f3, Myo7a) using qRT-PCR. We found that Lin28a/b dKO organoids expressed hair cell-specific transcripts at a more than threefold lower level than control organoids (Fig. 6G). Together, our findings indicate that endogenous LIN28A/B play an essential role in the mitotic and nonmitotic production of cochlear hair cells in vitro.
Fig. 6.
Loss of LIN28A/B attenuates mTOR signaling and limits supporting cell proliferation and hair cell formation in cochlear organoid culture. Cochlear organoid cultures were established from UBC-CreERT2;Lin28af/f;Lin28bf/f mice and Lin28af/f;Lin28bf/f littermates’ stage P2. Cultures received 4-hydroxy-tamoxifen (TM) or vehicle control DMSO at plating. SOX2 (green) marks supporting cells/prosensory cells, Hoechst (blue) staining marks cell nuclei. (A–G) Loss of Lin28a/b inhibits cell proliferation and hair cell production in organoid culture. (A) Representative BF images of control organoids (TM: Lin28af/f;Lin28bf/f; DMSO: UBC-CreERT2;Lin28af/f;Lin28bf/f; DMSO: Lin28af/f;Lin28bf/f) and Lin28a/b dKO organoids (TM: UBC-CreERT2;Lin28af/f;Lin28bf/f) at 7 d of expansion. (B) Diameter of control and Lin28a/b dKO organoids in A (n = 7, two independent experiments). (C) Organoid forming efficiency in control and Lin28a/b dKO cultures (n = 7, two independent experiments). (D) qRT-PCR analyzing Lin28a, Lin28b and Hmga2 mRNA expression in Lin28a/b dKO organoids (red bar) compared to control organoids (blue bar) at 7 d of expansion (n = 5 for control, n = 6 for Lin28a/b dKO, two independent experiments). (E) Cell proliferation in control and Lin28a/b dKO organoids. An EdU pulse was given at 7 d of expansion and EdU incorporation (red) was analyzed 1 h later. (F) EdU incorporation in E (n = 7, two independent experiments). (G) qRT-PCR analysis of Atoh1, Myo7a, and Pou4f3 mRNA expression in Lin28a/b dKO organoids (red bar) compared to control organoids (blue bar) at 10 d of differentiation (n = 4, two independent experiments). (H) Immunoblots for LIN28B, p-Akt, p-S6, and β-actin using protein lysates of acutely isolated control and Lin28a/b dKO cochlear epithelia, stage P5. (I–L) Loss of Lin28a/b attenuates mTOR signaling in cochlear organoids. (I) Immunostaining for p-S6 protein (red) in control and Lin28a/b dKO organoids at 7 d of expansion. (J) Percentage of p-S6+ cells in I (n = 5, two independent experiments). (K) Immunoblots for p-Akt, p-4EBP1, 4EBP1, mLIN28B, and β-actin (loading control) using protein lysates of control and Lin28a/b dKO organoids after 7 d of expansion. (L) Normalized p-Akt, p-4EBP1, and m-LIN28B protein levels in control and Lin28a/b dKO organoids in K (n = 3, from one representative experiment, two independent experiments). Bars in D and G represent mean ± SD, otherwise individual data points and their mean ± SD were plotted. Individual data points in B, F, and J represent the average values per animal. n = animals analyzed per group. Two-way ANOVA with Tukey’s correction was used to calculate P values in B, C, and J. Otherwise, P values were calculated using two-tailed, unpaired Student’s t tests.
Recent studies uncovered an important link between LIN28A/B and the mTOR signaling pathway (39, 55). Integrating various growth factor-dependent signals and downstream phosphoinositide 3-kinase (PI3K)-Akt signaling, mTOR signaling pathway promotes anabolic processes in aid of cell growth and cell survival (reviewed in ref. 57). Central to the mTOR pathway is the serine-threonine kinase mTOR. The mTOR kinase exists in two distinct protein complexes: mTORC1 and mTORC2, which differ in terms of rapamycin sensitivity, substrate specificity, and functional outputs (reviewed in ref. 58). The rapamycin-sensitive mTORC1 complex plays, among others, a central role in protein synthesis. Commonly used indicators for mTORC1 activity are the phosphorylated forms of the ribosomal protein S6 (Ser240/244) and the eukaryotic initiation factor 4E binding protein 1 (4E-BP1) (Thr37/46) (59–61). Phosphorylation (p) of the kinase Akt on serine 473 (Ser473) by mTORC2 is a readout for maximal Akt activation downstream of PI3K (62).
To determine whether mTOR activity declines during cochlear maturation, we analyzed p-S6 and p-Akt protein levels in cochlear epithelial lysates from stage P2 and P5 wild-type mice. We found that similar to LIN28B, p-S6 and p-Akt protein levels were significantly reduced in P5 cochlear sensory epithelia compared to P2 (SI Appendix, Fig. S1 B and C). Furthermore, LIN28B protein expression as well as mTOR activity (p-4E-BP1) were higher in the apical portion of stage P5 cochlear sensory epithelia than the basal one. However, due to the high variability across samples the apex-base difference in mTOR activity was not statistically significant (SI Appendix, Fig. S1 D–F). To determine whether LIN28A/B positively regulates mTOR activity in cochlear epithelial cells in vivo, we analyzed mTOR activity in acutely isolated cochlear epithelial lysates after postnatal LIN28B overexpression (iLIN28B) or Lin28a/b knockdown (Lin28a/b dKO, iLet-7g). We found that conditional ablation of Lin28a/b genes (Fig. 6H) or let-7g overexpression in neonatal mice (SI Appendix, Fig. S4 A–C) reduced cochlear m-TOR activity (p-Akt and or p-S6 protein), whereas LIN28B overexpression increased it (SI Appendix, Fig. S5 A and B).
Consistent with these in vivo findings, p-Akt, p-S6 and or p-4E-BP1 protein levels were 1.5-fold to twofold lower in lysates of P2 LIN28A/B deficient organoids (Lin28a/b dKO, iLet-7g) than control organoids (Fig. 6 K and L and SI Appendix, Fig. S4 F and G). Furthermore, the percentage of p-S6+ cells in P2 LIN28A/B deficient organoids (Lin28a/b dKO, iLet-7g) was two- to threefold lower than in control organoids (Fig. 6 I and J and SI Appendix, Fig. S4 D and E). Conversely, the levels of p-Akt and p-S6 proteins were more than 1.7-fold higher in lysates of P5 LIN28B overexpressing organoids compared to control organoid lysates (SI Appendix, Fig. S5 C and D). Moreover, we found that P5 LIN28B overexpression increased the percentage of p-S6+ cells within organoids by more than twofold (SI Appendix, Fig. S5 E and F). Costaining with SOX2 revealed that the majority of p-S6+ cells in P5 LIN28B overexpressing organoids expressed SOX2 at a low level (SI Appendix, Fig. S5G, SOX2low, p-S6high, red dots).
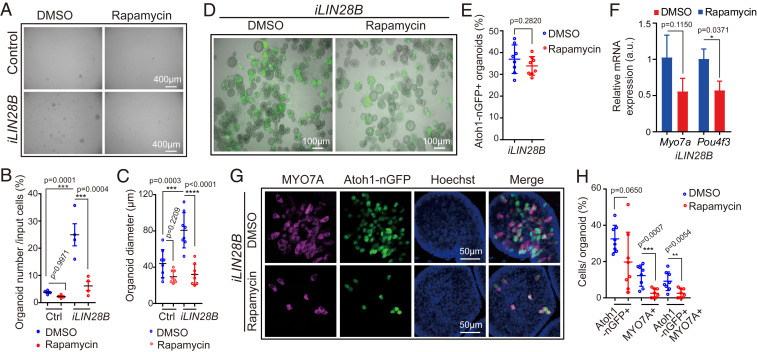
Next, we determined whether the expansion of supporting cells/prosensory cells requires mTOR signaling. To block mTOR signaling we used the mTORC1 complex inhibitor rapamycin (reviewed in ref. 63). Treatment with 50 µM of rapamycin has been reported to induce hair cell death in rat cochlear explants (64). Pilot experiments revealed that rapamycin at 4 ng/mL (4.4 nM) effectively blocked mTOR activity (SI Appendix, Fig. S6 A and B), and attenuated the growth of P2 wild-type cochlear organoids (SI Appendix, Fig. S6 C–E), without increasing cell death within these organoids (SI Appendix, Fig. S6 F and G). In contrast, 50 ng/mL (56 nM) rapamycin did induce cell death in organoid culture (SI Appendix, Fig. S6H). Furthermore, we found that the presence of 4 ng/mL of rapamycin reversed the LIN28B-mediated increase in organoid forming efficiency and organoid growth in P5 organoid culture (Fig. 7 A–C). Next, we investigated whether mTORC1 activity is required for the formation of hair cells in LIN28B overexpressing organoids. To avoid the negative effects of rapamycin on organoid growth, we added rapamycin or vehicle control (DMSO) to LIN28B overexpressing organoids just prior to the first formation of Atoh1-nGFP+ cells at day 10 of expansion. The initial induction of Atoh1-nGFP+ cells within organoids occurred in rapamycin-treated cultures at a similar rate than in DMSO-treated cultures (Fig. 7 D and E). However, LIN28B overexpressing organoids treated with rapamycin contained 3.5-fold fewer Atoh1-nGFP+ MYO7A+ hair cells and expressed hair cell-specific genes (Myo7a, Pou4f3) at a twofold lower level than LIN28B overexpressing organoids treated with DMSO (Fig. 7 F–H). To address whether mTOR signaling plays a role in the initial induction of Atoh1, we modified our experimental approach and pretreated the culture media with rapamycin 1 d prior to the induction of LIN28B overexpression at day 8 of expansion (SI Appendix, Fig. S7A). Using this modified experimental approach, we found that rapamycin treatment significantly reduced the number of Atoh1-nGFP+ organoids produced in response to LIN28B overexpression (SI Appendix, Fig. S7 B and C).
Fig. 7.
Inhibition of mTOR signaling attenuates LIN28B-induced organoid growth and hair cell formation. Cochlear organoid cultures were established from stage P5 Atoh1-nGFP;iLIN28B transgenic mice and Atoh1-nGFP transgenic control littermates and maintained as outlined in Fig. 2A. (A–C) Rapamycin attenuates LIN28B’s positive effect on cell growth in organoid culture. Rapamycin (4 ng/mL) or vehicle control (DMSO) was added at plating and replenished every other day. (A) BF images of control and LIN28B overexpressing (iLIN28B) organoids after expansion in the presence of DMSO or rapamycin for 7 d. (B) Organoid-forming efficiency in A (n = 4, two independent experiments). (C) Organoid diameters in A (n = 8, three independent experiments). (D–H) Rapamycin attenuates LIN28B’s positive effect on hair cell formation in organoid culture. Rapamycin (4 ng/mL) or DMSO was present during the final phase of expansion (10 to 13 d) and during the 5 d of differentiation. (D) Merged BF and green fluorescent images (Atoh1-nGFP) of LIN28B overexpressing organoid cultures treated with DMSO or rapamycin. (E) Percentage of Atoh1-nGFP+ organoids in D (n = 8, two independent experiments). (F) qRT-PCR analyzing Myo7a and Pou4f3 mRNA expression in LIN28B overexpressing organoids treated with rapamycin (red bar) or DMSO (blue bar) (mean ± SD, n = 2, from one representative experiment, two independent experiments). (G) Confocal images of LIN28B overexpressing organoids cultured in the presence of rapamycin or DMSO. Newly formed hair cells are identified by their coexpression of Atoh1-nGFP (green) and MYO7A (magenta). Nuclei were counterstained with Hoechst (blue). (H) Percentage of GFP+ (Atoh1-nGFP), MYO7A+, and GFP+ MYO7A+ hair cells per organoid in G (n = 8, two independent experiments). Graphs in B, C, E, and H show individual data points and the mean ± SD. The individual data points in C, E, and H represent average value per animal. n = animals analyzed per group. Two-way ANOVA with Tukey’s correction was used to calculate P values in B and C. Otherwise, P values were calculated using two-tailed, unpaired Student’s t tests.
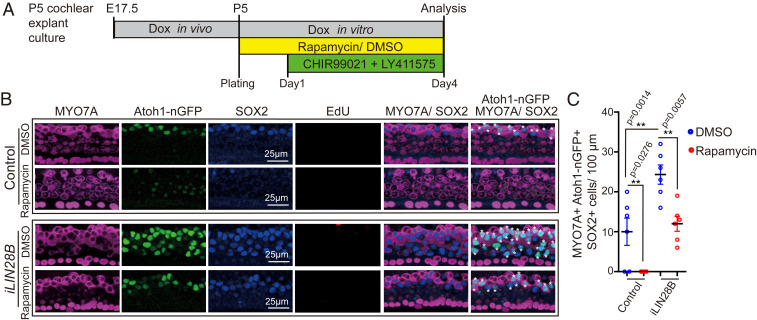
Next, we analyzed whether elevated LIN28B-mTOR levels would promote the production of new hair cells in stage P5 cochlear explants. To ensure robust LIN28B transgene expression, we administered dox in vivo starting at embryonic day 17.5 (E17.5) and provided dox throughout the duration of the experiment. At plating, control and LIN28B overexpressing cochlear explants received rapamycin or DMSO (vehicle control). The next day, the Notch inhibitor LY411575 and Wnt-activator (GSK3-β inhibitor) CHIR99021 was added to the culture media and 3 d later explants were harvested and analyzed for newly generated hair cells (Atoh1-nGFP+ SOX2+ MYO7A+) (Fig. 8A). Inhibition of Notch signaling in combination with Wnt-activation is highly effective in stimulating hair cell production in the neonatal cochlea in vitro and in vivo (22, 65). However, the effectiveness of such combined treatment has not yet been tested at later stages. We found that combined Notch inhibition and Wnt activation yielded on average 10 new hair cells per 100 µm in stage P5 control cochlear explants, a modest but significant improvement over the previously reported failure to produce hair cells with Notch inhibitor treatment alone (26) (control and DMSO in Fig. 8 B and C). As anticipated, LIN28B overexpression significantly increased the production of new hair cells in P5 cochlear explants, yielding on average 25 new hair cells per 100 µm, a more than twofold improvement compared to control cochlear explants (iLIN28B and DMSO in Fig. 8 B and C). The increase in hair cell formation in response to LIN28B overexpression was not due to an increase in cell proliferation as new hair cells were produced by nonmitotic mechanisms (EdU in Fig. 8B). The ability to produce new hair cells was completely abolished when control cochlear explants were cultured in the presence of rapamycin (control and rapamycin in Fig. 8 B and C). Furthermore, we found that rapamycin treatment reversed the gain in hair cell production observed in response to LIN28B overexpression (iLIN28B and rapamycin in Fig. 8 B and C).
Fig. 8.
LIN28B promotes nonmitotic hair cell production in an mTOR-dependent manner. (A) Experimental strategy. Cochlear explant cultures were established from stage P5 Atoh1-nGFP iLIN28B transgenic and Atoh1-nGFP transgenic control littermates. Pregnant dams were fed dox containing food starting at E17.5 and dox was present during culture. EdU (3 µM) and rapamycin (4 ng/mL) or vehicle control DMSO was added at plating. CHIR99021 (3 μM) and LY411575 (5 μM) were added the next day. (B) Rapamycin treatment inhibits hair cell generation in control and LIN28B overexpressing cochlear explants. Shown are representative confocal images of the midapical turn of control and LIN28B overexpressing cochlear explants stained for MYO7A (magenta), Atoh1-nGFP (green), SOX2 (blue), and EdU (red). White asterisks mark newly formed hair cells. (C) Quantification of newly formed hair cells (MYO7A+ Atoh1-nGFP+ SOX2+) in B. Graphed are average values for each animal and the mean ± SD, n = 6 animals per group, two independent experiments, two-way ANOVA with Tukey’s correction was used to calculate P values.
Discussion
The developing embryo and neonate show a remarkable capacity for regeneration in response to injury; however, only few tissues retain such regenerative plasticity into adulthood (reviewed in ref. 66). Likewise, the capacity of murine cochlear supporting cells to produce hair cells sharply declines during the first postnatal week (27, 49) and little to no hair cell production/regeneration is observed within the cochlea of adult mice (14, 17, 67). The reason for the decline in supporting cell plasticity is unknown.
Here, we provide evidence that the decline in supporting cell plasticity during cochlear maturation is, at least in part, the result of diminishing LIN28B-mTORC1 activity. We show that in vivo, coinciding with the abrupt decline in supporting cells plasticity, cochlear LIN28B protein levels and Akt-mTORC1 kinase activity sharply decline. We establish a regulatory link between cochlear Akt-mTORC1 activity and LIN28B/let-7 expression levels in vitro and in vivo, with Akt-mTORC1 kinase activity being augmented by LIN28B overexpression but suppressed by loss of LIN28A/B or overexpression of let-7g. Using cochlear organoid and explant culture systems, we found that diminished LIN28B-mTORC1 activity due to let-7g overexpression, or targeted deletion of Lin28a/b, accelerated the developmental decline in supporting cell plasticity. Conversely, we found that LIN28B overexpression halted the decline and enabled supporting cells that were undergoing maturation to activate a progenitor-like state and to produce new hair cells in response to regenerative cues. Finally, using the mTORC1 inhibitor rapamycin, we uncovered that LIN28B-induced supporting cell reprogramming required mTORC1-dependent signaling.
Rapamycin-sensitive mTOR signaling supports a wide range of regenerative processes, including the regrowth of axons, injury-induced cell proliferation, and stem cell-based muscle regeneration (68–71). Activation of mTORC1 kinase activity in response to injury or other environmental stimuli increases the cell’s synthesis of proteins and other macromolecules and ramps up mitochondrial ATP production, which is critical for the cell’s ability to divide and differentiate (72). Thus, rapamycin-sensitive mTOR signaling may enhance the regenerative capacity of supporting cells by ensuring that supporting cells have the energy and building blocks needed for mitotic or nonmitotic hair cell generation. Indeed, LIN28A/B induced and mTORC1-dependent metabolic reprogramming has been reported in the context of organismal growth and glucose metabolism (39, 55). Moreover, mTOR signaling is known to cross-talk and cross-regulate growth-related signaling pathways, including Wnt/β-catenin signaling (73; reviewed in ref. 74). Future studies will examine both LIN28B-dependent and -independent roles of mTOR signaling in supporting cell plasticity and address how mTOR signaling may alter the strength and transcriptional output of Wnt/β-catenin signaling in the context of hair cell regeneration.
How does LIN28B promote mTORC1 activity in cochlear epithelial cells? Several members of the mTOR pathway and the upstream IGF2/IGF1R and amino acid-sensing pathways are direct let-7 targets (39, 75, 76), suggesting that LIN28B enhances mTOR activity, at least in part, by relieving let-7–mediated repression. In addition, LIN28A/B have been shown to promote IGF2/IGF1R–mTOR signaling through increasing the mRNA translation of key member genes (39, 77, 78). It needs to be noted that treatment with the mTORC1 inhibitor rapamycin attenuated, but did not completely block supporting cell proliferation and hair cell production, suggesting that LIN28B promotes these processes through additional, mTORC1-independent mechanisms. LIN28A/B and let-7 miRNAs regulate self-renewal and regeneration via translational activation/repression of progrowth genes, such as Igf2bp1 (79), Trim71 (80), and Hmga2 (81), as well via the direct targeting of genes critical for oxidative phosphorylation (82) and lipid synthesis (83). Indeed, we found LIN28B activated, whereas let-7g limited, Hmga2 expression in cochlear organoids. In addition, we found that LIN28B overexpression led to a transient down-regulation in Sox2 mRNA expression at the peak of organoid expansion. Recent studies found that SOX2 positively regulates p27/Kip1 transcription and reduction in Sox2 expression primes cochlear supporting cells for proliferation and hair cell regeneration (48, 67). There is precedent for LIN28A/B limiting Sox2 expression. A recent report found that LIN28A overexpression during somitogenesis inhibits Sox2 expression, causing a promesodermal phenotype (84). Furthermore, the transcription factor ARID3A, the expression of which is positively regulated by LIN28A/B and negatively regulated by let-7 miRNAs, has been shown to inhibit Sox2 expression in murine embryonic stem cells (85, 86). Future studies are warranted to address LIN28B’s role in Sox2 expression during hair cell regeneration.
It is important to note that reactivation of LIN28B is, by itself, not sufficient to stimulate supporting cell proliferation and is not sufficient to induce the conversion of supporting cells into hair cell. Additional signals and factors, such as Wnt and Notch signaling, need to be manipulated to stimulate cochlear supporting cell proliferation and their conversion into hair cells. Furthermore, it needs to be emphasized that our study was conducted in vitro and was limited to immature, early postnatal stages. While recent studies showed promise in regenerating vestibular hair cells in adult mice (9, 87), the regeneration of cochlear hair cells in adult mice has proven thus far an impossible task (67; reviewed in ref. 88).
Similar to mammals, auditory supporting cells in birds are highly specialized postmitotic cells. The lack of progenitor-like features has led to the yet to be proven hypothesis that hair cell loss in birds triggers a partial de-differentiation of supporting cells, in which supporting cells revert to a transitional progenitor-like state (reviewed in ref. 89). Here, we provide evidence that supporting cells do undergo a process of de-differentiation in organoid culture and show that supporting cell de-differentiation is regulated by the LIN28B/let-7 axis. We found that overexpression of let-7g in cochlear organoids repressed the down-regulation of supporting cell-specific genes (e.g., S100a1) and limited the up-regulation of prosensory-specific genes (e.g., Hmga2), whereas LIN28B reactivation enhanced it. Such positive role of LIN28B in supporting cell de-differentiation is reminiscent of the role of LIN28A in Müller glia de-differentiation. In zebrafish, Lin28a reactivation is essential for the de-differentiation of Müller glia into multipotent retinal progenitors in response to injury (90, 91). The importance of LIN28-mediated de-differentiation in the context of retinal regeneration is further highlighted by the recent finding that LIN28A reactivation induces Müller glia de-differentiation and proliferation in adult mice (92, 93).
We anticipate that the findings presented here will lead to a similar evaluation of the role of LIN28A/B in injury-induced hair cell regeneration in birds and fish. Furthermore, an evaluation of the role of LIN28A/B in vestibular hair cell regeneration may address why vestibular supporting cells have a higher regenerative capacity than their cochlear counterparts (94). Most importantly, our observation that reactivation of LIN28B facilitates the reprogramming of supporting cells into “progenitor-like” cells may inform future therapeutic strategies that combine LIN28B-mediated reprograming with Wnt-activation and/or Notch inhibition to regenerate lost cochlear hair cells in adult mammals.
Methods
Mouse Breeding and Genotyping.
All experiments and procedures were approved by the Johns Hopkins University Institutional Animal Care and Use Committees protocol, and all experiments and procedures adhered to National Institutes of Health-approved standards. The Atoh1-nGFP transgenic (tg) (MGI:3703598) mice were obtained from Jane Johnson, University of Texas Southwestern Medical Center, Dallas, TX (37). The p27-GFP tg mice (MGI:3774555) were obtained from Neil Segil, University of Southern California, Los Angeles, CA (45). The Col1a-TRE-LIN28B (MGI:5294612), Col1a-TRE-let-7S21L (MGI:5294613), and Lin28afl/fl (MGI:5294611), and Lin28bfl/fl (MG:5519037) mice were obtained from George Q. Daley, Children’s Hospital, Boston, MA (39, 55). UBC-CreERT2 tg (MGI:3707333; stock no. 007001) (56) and R26-M2-rtTA (MGI:3798943; stock no. 006965) (40) mice were purchased from the Jackson Laboratories. Mice were genotyped by PCR and genotyping primers are listed in SI Appendix, Table S1. Mice of both sexes were used in this study. Embryonic development was considered as E0.5 on the day a mating plug was observed. To induce LIN28B or let-7g transgene expression, dox was delivered to time-mated females via ad libitum access to feed containing 2 g of dox per kilogram feed (Bioserv, no. F3893).
Organoid Culture.
Cochleae from early P2 and P5 mice were harvested and microdissected in HBSS (Corning, no. 21-023-CV). Enzymatic digest with dispase (1 mg/mL; ThermoFisher, no. 17105041) and collagenase (1 mg/mL; Worthington, no. LS004214) was used to isolate cochlear epithelia, as previously described (95). To obtain a single cell suspension, cochlear epithelia were incubated in TrypLE solution (ThermoFisher, no. 2604013), triturated, and filtered through a 35-µm filter (BD, no. 352235). An aliquot of cells was counted to establish the total number of cells isolated per animal. Equal numbers of cells were resuspended in a 1:1 mix of Matrigel (Corning, no. 356231) and expansion medium and plated into prewarmed four-well plates (CELLTREAT, no. 229103). For stage P2, eight organoid cultures and for stage P5, five organoid cultures, placed in individual wells, were established per animal. The expansion medium was prepared with DMEM/F12 (Corning, no. 10-092-CV), N2 supplement (1×; ThermoFisher, no.17502048), B-27 supplement (1×; ThermoFisher, no.12587010), EGF (50 ng/mL; Sigma-Aldrich, no. SRP3196), FGF2 (50 ng/mL; ThermoFisher, no. PHG0264), CHIR99021 (3 μM; Sigma-Aldrich, no. SML1046), VPA (1 mM; Sigma-Aldrich, no. P4543), 616452 (2 μM; Sigma-Aldrich, no. 446859-33-2), and penicillin (100 U/mL; Sigma-Aldrich, no. P3032). Dox hyclate (10 μg/mL; Sigma-Aldrich, no. D9891), EdU (3 μM, ThermoFisher, no. A10044) or TM (20 ng/mL; Sigma-Aldrich, no. H7904) was added to expansion media as indicated. To induce differentiation, expansion media was replaced by a differentiation media composed of DMEM/F12, N2 (1×), B27 (1×), CHIR99021 (3 μM), and LY411575 (5 μM; Sigma-Aldrich, no. SML0506). To block mTORC1 activity, organoids received rapamycin (4 ng/mL; Sigma-Aldrich, no. R0395) or vehicle control DMSO. Culture media was changed every other day.
Quantification of Organoid Forming Efficiency, Organoid Diameter, and GFP Reporter Expression.
Brightfield and fluorescent images of organoid cultures were captured with an Axiovert 200 microscope using 5× and 10× objectives (Carl Zeiss Microscopy). To calculate organoid forming efficiency, the total number of organoids per culture was counted manually and values were normalized to the total number of cells plated. To calculate organoid size, the diameter of organoids in two randomly chosen fields was measured per culture using ImageJ (https://imagej.nih.gov/ij/) and the average value per animal was reported. To establish the percentage of GFP+ organoids per culture, the total number of organoids and the number of GFP+ organoids was counted manually. For each genotype and treatment, three independent organoid cultures were established and analyzed. At a minimum, two independent experiments were conducted and analyzed.
Explant Culture.
Cochlear tissue was collected and microdissected in HBBS. Cochlear sensory epithelia with attached spiral ganglion were placed onto SPI-Pore membrane filters (Structure Probe, no. E1013-MB) and cultured in DMEM-F12, EGF (5 ng/mL), penicillin (100 U/mL), and N2 Supplement (1×). Cultures were maintained in a 5% CO2/20% O2 humidified incubator. To induce let-7g or LIN28B transgene expression, timed pregnant dams received dox-containing feed starting at E17.5/E18.5 until pups were harvested. To maintain let-7g or LIN28B transgene expression in vitro, the culture media contained dox hyclate. To induce hair cell loss, control and let-7g overexpressing cochlear explants received gentamicin sulfate (100 µg/mL; Sigma-Aldrich, no. G1272) at plating. After 20 h the gentamicin-containing media was replaced with culture media containing LY411575 (5 μM). To block mTORC1 activity, control and LIN28B overexpressing cochlear explants received rapamycin (4 ng/mL) or vehicle control DMSO at plating. CHIR99021 (3 μM), LY411575 (5 μM) and EdU (3 μM) were added the following day (day 1). For Lin28a/b dKO experiments, TM or vehicle control DMSO was added to the culture medium at plating. LY411575 (5 μM) or vehicle control DMSO were added the next day. In all experiments, the culture media were exchanged every other day, and cultures were maintained for a total of 4 d.
Immunohistochemistry.
Organoids/explants were fixed with 4% (vol/vol) paraformaldehyde in PBS (Electron Microscopy Sciences, no. 15713) for 30 min at room temperature. To permeabilize cells and block unspecific antibody binding, organoids/explants were incubated with 0.5% (vol/vol) TritonX-100/10% (vol/vol) FBS in 1× PBS for 30 min. Antibody labeling was performed according to manufacturer’s recommendations. Used antibodies are listed in SI Appendix, Table 2. Cell nuclei were stained using Hoechst 33258 solution (Sigma-Aldrich, no. 94403).
Cell Proliferation.
EdU was added to culture media at a final concentration of 3 μM. EdU incorporation was detected using Click-iT Edu Alexa Fluor 555 imaging Kit (ThermoFisher, no. C10338) following the manufacturer’s specifications.
Cell Death.
TUNEL reactions were used to detect dying cells. PFA-fixed and permeabilized organoids were processed using the In Situ Cell Death Detection Kit, Fluorescein (Roche, no.11684795910) according to the manufacturer’s specifications. As positive control, organoids were preincubate 10 min at room temperature in DNase I (3000 U/mL, in 50 mM Tris⋅HCl, pH 7.4, 1 mg/mL BSA). As negative control, organoids were processed without a reaction mixture that lacked terminal deoxynucleotidyl transferase.
Cell Counts.
High-power confocal single-plane and z-stack images of fluorescently immunolabeled organoids and explants were taken with 40× objective using LSM 700 confocal microscope (Zeiss Microscopy). To establish the percentage of cells within an organoid that expressed an epitope of interest, two to three images containing a minimum of three organoids were analyzed and the average value per animal was reported. To establish the number of hair cells within cochlear explants, confocal z-stacks spanning the supporting cell and hair cell layer were exported into Photoshop CS6 (Adobe). Existing (MYO7A+) and newly formed hair cells (MYO7A+ SOX2+) were manually counted. The length of the analyzed cochlear segment was measured using ImageJ. At a minimum, two independent experiments were conducted in which at a minimum three cochlear explants/organoid cultures per genotype and treatment were established and analyzed.
RNA Extraction and qRT-PCR.
Total RNA was extracted using the miRNeasy Micro Kit (Qiagen, no. 217084). The mRNA was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad, no. 1708889). SYBR Green-based qPCR was performed by using Fast SYBR Green Master Mix reagent (ThermoFisher, no. 4385612) and gene-specific primers. qPCRs were carried out in triplicate using a CFX Real-Time PCR System (Bio-Rad). Rpl19 was used as reference gene. Relative gene expression was calculated using the ΔΔCT method (96). qPCR primers are listed in SI Appendix, Table S3.
Immunoblotting.
Cochlear epithelia/organoids were lysed with RIPA buffer (Sigma-Aldrich, no. R0278) supplemented with protease inhibitor (Sigma-Aldrich, no.11697498001), phosphatase Inhibitor mixture 2 (Sigma-Aldrich, no. P5726) and phosphatase inhibitor mixture 3 (Sigma-Aldrich, no. P0044). Equal amounts of protein lysate were resolved on NuPAGE 4 to 12% Bis-Tris gels (ThermoFisher, no. NP0321PK2) and transferred to a PVDF membrane (Bio-Rad) by electrophoresis. Membranes were blocked with 5% (w/vol) nonfat dry milk in TBST and immunoblotted with primary antibodies and HRP-conjugated secondary antibodies (Jackson Immuno Research) according to the manufacturer’s instructions. Primary antibodies are listed in SI Appendix, Table S4. This was followed by washes in TBST and incubation in SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher, no.34580). The resulting chemiluminescence was captured using X-ray films. To quantify protein levels, X-ray films were scanned, digital images converted to gray scale and the relative density (intensity) of bands was analyzed using the Gel Analysis plug-in ImageJ.
Statistical Analysis.
The sample size (n) represents the number of animals analyzed per group. Animals (biological replicates) were allocated into control or experimental groups based on genotype and/or type of treatment. To avoid bias, masking was used during data analysis. Data were analyzed using Graphpad Prism 8.0 (Graphpad Software). Relevant information for each experiment including sample size, statistical tests and reported P values are reported in each figure.
Supplementary Material
Acknowledgments
We thank the members of the A.D. laboratory for the help and advice provided throughout the course of this study; and Dr. Georg Dailey for essential mouse reagents. The work was supported by National Institute on Deafness and Other Communication Disorders Grants DC011571 (to A.D.) and DC005211 (Sensory Mechanisms Research Core Center); and the David M. Rubenstein Fund for Hearing Research (A.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000417117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Warchol M. E., Sensory regeneration in the vertebrate inner ear: Differences at the levels of cells and species. Hear. Res. 273, 72–79 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Fekete D. M., Muthukumar S., Karagogeos D., Hair cells and supporting cells share a common progenitor in the avian inner ear. J. Neurosci. 18, 7811–7821 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driver E. C., Sillers L., Coate T. M., Rose M. F., Kelley M. W., The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev. Biol. 376, 86–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiernan A. E., et al. , Sox2 is required for sensory organ development in the mammalian inner ear. Nature 434, 1031–1035 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Ahmed M., et al. , Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev. Cell 22, 377–390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempfle J. S., Turban J. L., Edge A. S., Sox2 in the differentiation of cochlear progenitor cells. Sci. Rep. 6, 23293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermingham N. A., et al. , Math1: An essential gene for the generation of inner ear hair cells. Science 284, 1837–1841 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Lewis R. M., Hume C. R., Stone J. S., Atoh1 expression and function during auditory hair cell regeneration in post-hatch chickens. Hear. Res. 289, 74–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks K. L., Wisner S. R., Cox B. C., Stone J. S., Atoh1 is required in supporting cells for regeneration of vestibular hair cells in adult mice. Hear. Res. 385, 107838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberson D. W., Alosi J. A., Cotanche D. A., Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J. Neurosci. Res. 78, 461–471 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Shang J., Cafaro J., Nehmer R., Stone J., Supporting cell division is not required for regeneration of auditory hair cells after ototoxic injury in vitro. J. Assoc. Res. Otolaryngol. 11, 203–222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corwin J. T., Cotanche D. A., Regeneration of sensory hair cells after acoustic trauma. Science 240, 1772–1774 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Ryals B. M., Rubel E. W., Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science 240, 1774–1776 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Cox B. C., et al. , Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816–829 (2014). Correction in: Development141, 1599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu L., et al. , Diphtheria toxin-induced cell death triggers Wnt-dependent hair cell regeneration in neonatal mice. J. Neurosci. 36, 9479–9489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korrapati S., Roux I., Glowatzki E., Doetzlhofer A., Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One 8, e73276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizutari K., et al. , Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W., et al. , Notch inhibition induces mitotically generated hair cells in mammalian cochleae via activating the Wnt pathway. Proc. Natl. Acad. Sci. U.S.A. 112, 166–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai R., et al. , Wnt signaling induces proliferation of sensory precursors in the postnatal mouse cochlea. Proc. Natl. Acad. Sci. U.S.A. 109, 8167–8172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi F., Hu L., Edge A. S., Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl. Acad. Sci. U.S.A. 110, 13851–13856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo B. R., Baldwin E. M., Layman W. S., Taketo M. M., Zuo J., In vivo cochlear hair cell generation and survival by coactivation of β-catenin and Atoh1. J. Neurosci. 35, 10786–10798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ni W., et al. , Wnt activation followed by Notch inhibition promotes mitotic hair cell regeneration in the postnatal mouse cochlea. Oncotarget 7, 66754–66768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szarama K. B., Gavara N., Petralia R. S., Kelley M. W., Chadwick R. S., Cytoskeletal changes in actin and microtubules underlie the developing surface mechanical properties of sensory and supporting cells in the mouse cochlea. Development 139, 2187–2197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legendre K., Safieddine S., Küssel-Andermann P., Petit C., El-Amraoui A., alphaII-betaV spectrin bridges the plasma membrane and cortical lattice in the lateral wall of the auditory outer hair cells. J. Cell Sci. 121, 3347–3356 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Lelli A., Asai Y., Forge A., Holt J. R., Géléoc G. S., Tonotopic gradient in the developmental acquisition of sensory transduction in outer hair cells of the mouse cochlea. J. Neurophysiol. 101, 2961–2973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maass J. C., et al. , Changes in the regulation of the Notch signaling pathway are temporally correlated with regenerative failure in the mouse cochlea. Front. Cell. Neurosci. 9, 110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samarajeewa A., et al. , Transcriptional response to Wnt activation regulates the regenerative capacity of the mammalian cochlea. Development 145, dev166579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden E. J., Benito-Gonzalez A., Doetzlhofer A., The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea. Proc. Natl. Acad. Sci. U.S.A. 112, E3864–E3873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambros V., Horvitz H. R., Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Reinhart B. J., et al. , The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Rehfeld F., Rohde A. M., Nguyen D. T., Wulczyn F. G., Lin28 and let-7: Ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 359, 145–160 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Shyh-Chang N., Daley G. Q., Lin28: Primal regulator of growth and metabolism in stem cells. Cell Stem Cell 12, 395–406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White P. M., Doetzlhofer A., Lee Y. S., Groves A. K., Segil N., Mammalian cochlear supporting cells can divide and trans-differentiate into hair cells. Nature 441, 984–987 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Oshima K., et al. , Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J. Assoc. Res. Otolaryngol. 8, 18–31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLean W. J., et al. , Clonal expansion of Lgr5-positive cells from mammalian cochlea and high-purity generation of sensory hair cells. Cell Rep. 18, 1917–1929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldhaus J., Durruthy-Durruthy R., Heller S., Quantitative high-resolution cellular map of the organ of Corti. Cell Rep. 11, 1385–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumpkin E. A., et al. , Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3, 389–395 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Stojanova Z. P., Kwan T., Segil N., Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development 143, 1632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H. et al.; DIAGRAM Consortium; MAGIC Investigators , The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochedlinger K., Yamada Y., Beard C., Jaenisch R., Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Coppens A. G., Résibois A., Poncelet L., Immunolocalization of calbindin D28k and calretinin in the dog cochlea during postnatal development. Hear. Res. 145, 101–110 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi N., Henzl M. T., Thalmann I., Thalmann R., Schulte B. A., Oncomodulin is expressed exclusively by outer hair cells in the organ of Corti. J. Histochem. Cytochem. 46, 29–40 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Shim K., Minowada G., Coling D. E., Martin G. R., Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev. Cell 8, 553–564 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Lanford P. J., Shailam R., Norton C. R., Gridley T., Kelley M. W., Expression of Math1 and HES5 in the cochleae of wildtype and Jag2 mutant mice. J. Assoc. Res. Otolaryngol. 1, 161–171 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doetzlhofer A., White P., Lee Y. S., Groves A., Segil N., Prospective identification and purification of hair cell and supporting cell progenitors from the embryonic cochlea. Brain Res. 1091, 282–288 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Chen P., Segil N., p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development 126, 1581–1590 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Löwenheim H., et al. , Gene disruption of p27(Kip1) allows cell proliferation in the postnatal and adult organ of Corti. Proc. Natl. Acad. Sci. U.S.A. 96, 4084–4088 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Z., et al. , Regulation of p27Kip1 by Sox2 maintains quiescence of inner pillar cells in the murine auditory sensory epithelium. J. Neurosci. 32, 10530–10540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maass J. C., et al. , Transcriptomic analysis of mouse cochlear supporting cell maturation reveals large-scale changes in Notch responsiveness prior to the onset of hearing. PLoS One 11, e0167286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H. C., et al. , Spontaneous activity of cochlear hair cells triggered by fluid secretion mechanism in adjacent support cells. Cell 163, 1348–1359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son E. J., et al. , Conserved role of Sonic Hedgehog in tonotopic organization of the avian basilar papilla and mammalian cochlea. Proc. Natl. Acad. Sci. U.S.A. 112, 3746–3751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radde-Gallwitz K., et al. , Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J. Comp. Neurol. 477, 412–421 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holley M., et al. , Emx2 and early hair cell development in the mouse inner ear. Dev. Biol. 340, 547–556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kolla L., et al. , Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat. Commun. 11, 2389 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shinoda G., et al. , Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 31, 1563–1573 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruzankina Y., et al. , Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saxton R. A., Sabatini D. M., mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Albert V., Hall M. N., mTOR signaling in cellular and organismal energetics. Curr. Opin. Cell Biol. 33, 55–66 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Hutchinson J. A., Shanware N. P., Chang H., Tibbetts R. S., Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J. Biol. Chem. 286, 8688–8696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Manteuffel S. R., et al. , The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol. Cell. Biol. 17, 5426–5436 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunn G. J., et al. , Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277, 99–101 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Facchinetti V., et al. , The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guertin D. A., Sabatini D. M., The pharmacology of mTOR inhibition. Sci. Signal. 2, pe24 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Leitmeyer K., et al. , Inhibition of mTOR by rapamycin results in auditory hair cell damage and decreased spiral ganglion neuron outgrowth and neurite formation in vitro. BioMed Res. Int. 2015, 925890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ni W., et al. , Extensive supporting cell proliferation and mitotic hair cell generation by in vivo genetic reprogramming in the neonatal mouse cochlea. J. Neurosci. 36, 8734–8745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Godwin J., The promise of perfect adult tissue repair and regeneration in mammals: Learning from regenerative amphibians and fish. BioEssays 36, 861–871 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Atkinson P. J., et al. , Sox2 haploinsufficiency primes regeneration and Wnt responsiveness in the mouse cochlea. J. Clin. Invest. 128, 1641–1656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duan X., et al. , Subtype-specific regeneration of retinal ganglion cells following axotomy: Effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park K. K., et al. , Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willet S. G., et al. , Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 37, e98311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodgers J. T., et al. , mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laplante M., Sabatini D. M., mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao C., et al. , Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12, 781–790 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Vadlakonda L., Pasupuleti M., Pallu R., Role of PI3K-AKT-mTOR and Wnt signaling pathways in transition of G1-S phase of cell cycle in cancer cells. Front. Oncol. 3, 85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang M., et al. , Lin28 promotes the proliferative capacity of neural progenitor cells in brain development. Development 142, 1616–1627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dubinsky A. N., et al. , Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab. 20, 626–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polesskaya A., et al. , Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125–1138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peng S., et al. , Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells 29, 496–504 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Nishino J., Kim S., Zhu Y., Zhu H., Morrison S. J., A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. eLife 2, e00924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin Y. C., et al. , Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol. Biol. Evol. 24, 2525–2534 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Lee Y. S., Dutta A., The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21, 1025–1030 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shyh-Chang N., et al. , Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y., et al. , Lin28 enhances de novo fatty acid synthesis to promote cancer progression via SREBP-1. EMBO Rep. 20, e48115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinton D. A., et al. , The Lin28/let-7 pathway regulates the mammalian caudal body axis elongation program. Dev. Cell 48, 396–405.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Popowski M., et al. , Bright/Arid3A acts as a barrier to somatic cell reprogramming through direct regulation of Oct4, Sox2, and Nanog. Stem Cell Rep. 2, 26–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou Y., et al. , Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J. Exp. Med. 212, 569–580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sayyid Z. N., Wang T., Chen L., Jones S. M., Cheng A. G., Atoh1 directs regeneration and functional recovery of the mature mouse vestibular system. Cell Rep. 28, 312–324.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roccio M., Senn P., Heller S., Novel insights into inner ear development and regeneration for targeted hearing loss therapies. Hear. Res., 10.1016/j.heares.2019.107859 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Wan G., Corfas G., Stone J. S., Inner ear supporting cells: Rethinking the silent majority. Semin. Cell Dev. Biol. 24, 448–459 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramachandran R., Fausett B. V., Goldman D., Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 12, 1101–1107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaur S., et al. , let-7 MicroRNA-mediated regulation of Shh signaling and the gene regulatory network is essential for retina regeneration. Cell Rep. 23, 1409–1423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elsaeidi F., et al. , Notch suppression collaborates with Ascl1 and Lin28 to unleash a regenerative response in fish retina, but not in mice. J. Neurosci. 38, 2246–2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yao K., et al. , Wnt regulates proliferation and neurogenic potential of müller glial cells via a Lin28/let-7 miRNA-dependent pathway in adult mammalian retinas. Cell Rep. 17, 165–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bucks S. A., et al. , Supporting cells remove and replace sensory receptor hair cells in a balance organ of adult mice. eLife 6, e18128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doetzlhofer A., et al. , Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev. Cell 16, 58–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.