Significance
This study assessed the effects of myelin decompaction on motor behavior and brain-wide structural and functional connectivity, and the effect of nitric oxide synthase inhibition by N-nitro-l-arginine methyl ester (L-NAME) on these imaging measures. We report that inducible oligodendrocyte-specific inactivation of the Nf1 gene, which causes myelin decompaction, results in reduced initial motor coordination. Using diffusion-based magnetic resonance imaging (MRI), we show reduced myelin integrity, and using functional MRI, we show reduced functional connectivity in awake passive mice. L-NAME administration results in rescue of the pathology at the mesoscopic level, as measured using imaging procedures that can be directly applied to humans to study treatment efficacy in clinical trials.
Keywords: connectome, resting state, NF1, oligodendrocytes
Abstract
Neurofibromin gene (NF1) mutation causes neurofibromatosis type 1 (NF1), a disorder in which brain white matter deficits identified by neuroimaging are common, yet of unknown cellular etiology. In mice, Nf1 loss in adult oligodendrocytes causes myelin decompaction and increases oligodendrocyte nitric oxide (NO) levels. Nitric oxide synthase (NOS) inhibitors rescue this pathology. Whether oligodendrocyte pathology is sufficient to affect brain-wide structure and account for NF1 imaging findings is unknown. Here we show that Nf1 gene inactivation in adult oligodendrocytes (Plp-Nf1fl/+ mice) results in a motor coordination deficit. Magnetic resonance imaging in awake mice showed that fractional anisotropy is reduced in Plp-Nf1fl/+ corpus callosum and that interhemispheric functional connectivity in the motor cortex is also reduced, consistent with disrupted myelin integrity. Furthermore, NOS-specific inhibition rescued both measures. These results suggest that oligodendrocyte defects account for aspects of brain dysfunction in NF1 that can be identified by neuroimaging and ameliorated by NOS inhibition.
Neurofibromatosis type 1 (NF1) is one of the most common autosomal dominant diseases in humans, present in 1 in 2,000 to 3,000 births (1, 2). NF1 results from mutation in a single allele of the Neurofibromin 1 (NF1) gene. This can cause manifestations including skin hyperpigmentation and susceptibility to tumors of the central nervous system (CNS) and peripheral nervous system (PNS) that correlate with biallelic NF1 loss of function (1). Individuals with NF1 are also at risk for nonneoplastic brain disease manifestations, including attention and learning deficits, autism-like features (3, 4), and delayed acquisition of motor skills (5). Affected children often display impairments in fine and/or gross motor function, and 40% receive remedial education to improve their cognitive, social, and motor performance (6–8).
White matter abnormalities are frequent in NF1 and may be associated with cognitive impairment, as reviewed previously (4). For example, children with NF1 often show enlargement of the corpus callosum, the brain white matter tract that spans the brain hemispheres and contains oligodendrocytes and myelinated neural axons (9, 10). Brain magnetic resonance imaging (MRI) findings in NF1 include diffuse and focal areas of T2 hyperintensity, speculated to represent myelin defects (9, 11, 12). Diffusion tensor imaging (DTI) has been used to examine white matter microstructure in NF1. Fractional anisotropy (FA) is higher in heavily myelinated fiber tracts than in other brain regions (13), and is reduced in NF1 children compared to typically developing children; 60% to 70% of NF1 children exhibit abnormalities in white matter, including reduced FA (14–19). All these changes lack treatment options.
A histological evaluation of three NF1 brains suggested myelin edema to explain vacuolar and spongiform findings (20). Consistent with the possibility that altered myelin exists in the NF1 brain, inactivation of a single Nf1 allele in adult oligodendrocytes caused progressive myelin decompaction (21, 22). Numbers of myelin lamellae were unchanged, and demyelination was not observed. Inflammation was not detected. The nitric oxide synthase (NOS) inhibitor l-arginine methyl ester (L-NAME) rescued myelin decompaction in the mouse model. In humans, whether behavioral/cognitive deficits result from Nf1 mutation/loss in neurons, glial cells, or a combination has not been defined (23).
Oligodendrocyte pathology is associated with decreased motor performance in humans and in mice (24–26). Changes in myelin itself also may be correlated with altered behavior (26–28). DTI and functional connectivity MRI (i.e., resting-state fMRI [rs-fMRI]) are used to assess structural integrity and functional connectivity in humans and rodents (29–35). In NF1, functional connectivity abnormalities correlate with cognitive, social, and/or behavioral deficits in both humans and Nf1+/− mice (4, 35–37). Using human MRI techniques, with mouse-specific adaptations that allow scanning of animals in an awake passive state and probing of macroscopic brain-wide changes noninvasively over time, bridges the gap between microscopic and macroscopic levels, as well as that between basic research and clinical trials (38, 39).
Here we tested whether oligodendrocyte-specific Nf1 mutation causes brain-wide structural and/or functional changes that impact motor areas of the brain/motor coordination, which might be rescued by NOS inhibition. Using DTI, we found that Plp-Nf1fl/+ mice display reduced FA across the brain. Using rs-fMRI, we identified disrupted functional connectivity in motor areas connected via the corpus callosum that were rescued by brief inhibition of NOS.
Results
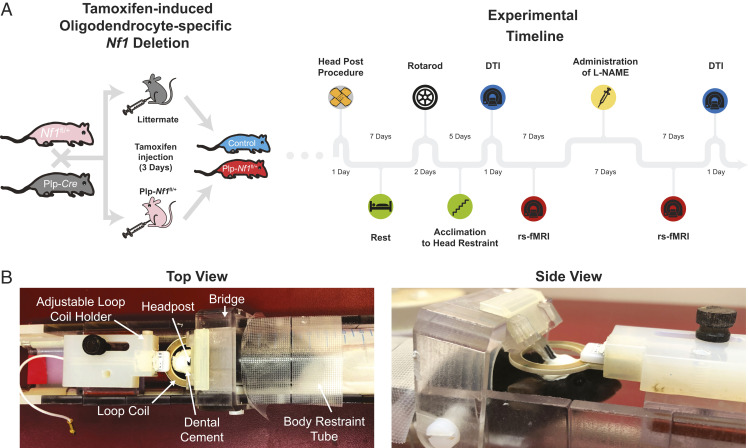
We induced oligodendrocyte-specific Nf1 inactivation in mice and compared structural and functional differences from littermate controls (Plp-Cre or Nf1fl/+). We induced Nf1 inactivation in 8-wk-old adult mice and 8 to 12 mo later evaluated motor coordination using an accelerating rotating rod and brain-wide connectivity using both DTI and rs-fMRI, and then evaluated the impact of inhibition, using L-NAME, on brain-related measures (Fig. 1).
Fig. 1.
Experimental setup for whole-brain imaging of Plp-Nf1fl/+ mice. (A) Oligodendrocyte-specific Nf1 deletion was achieved by crossing of Plp-Cre and Nf1fl/+ mice. Plp-Nf1fl/+ and littermate animals received a 3-d tamoxifen treatment (twice daily) to induce the deletion in Plp-Nf1fl/+ mice and create littermate controls (Plp-Nf1+/+). The experimental timeline is shown for mice that underwent full evaluation including behavior, as well as structural and functional imaging. (B) A custom-built 3D-printed modular cradle allows head fixation for functional scans of awake passive mice. The same cradle allows the addition of a mouse-compatible mask that can be placed around the animal’s nose and mouth to maintain constant anesthesia of the head-fixed mice for long structural scans.
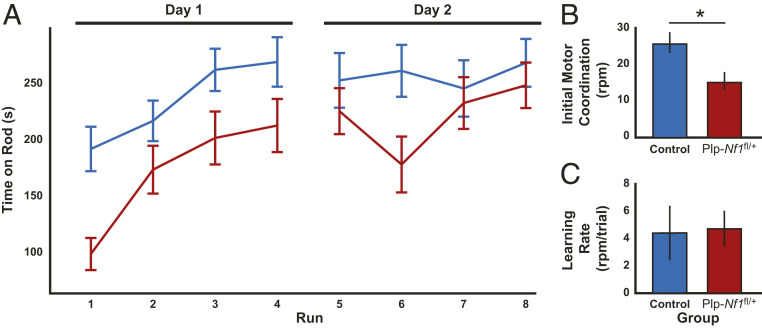
To evaluate motor performance and coordination, we used the accelerating rotating rod experiment over 2 d (four runs per day: control, n = 14; Plp-Nf1fl/+, n = 15). Repeated-measures ANOVA (Fig. 2A) showed main effects of genotype (F1,27 = 6.3, P = 0.018), time (F1,27 = 38.4, P < 0.001), and interaction (F1,27 = 4.65, P = 0.04). Interestingly, Plp-Nf1fl/+ mice demonstrated impaired initial performance (first run; Mann–Whitney U test: U = 31, P = 0.0013) in the test and then caught up to controls (last run: U = 89, P = 0.4956). To quantify this phenomenon, we adapted a previously described analysis (40). In brief, we performed a linear regression from the first run until the run at which the animal first reached maximal performance (top speed of 40 revolutions per minute). From the linear regression, we calculated the intercept and slope that reflect initial motor coordination (Fig. 2B) and learning rate (Fig. 2C), respectively. Plp-Nf1fl/+ mice showed reduced initial motor coordination (U = 42, P = 0.006) but a normal learning rate (U = 73, P = 0.17). Overall, the Plp-Nf1fl/+ animals showed differential performance on the rotating rod, with impaired initial motor coordination but normal learning rate, leading them to learn the task by the end of the eighth run and then perform at the same level as the controls.
Fig. 2.
Plp-Nf1fl/+ animals show differential performance on the rotating rod with impaired initial motor coordination but normal learning rate. (A) Average time on the rod plotted for each group on consecutive testing days. Repeated-measures ANOVA demonstrates an overall significant difference between control (n = 14) and Plp-Nf1fl/+ (n = 15) mice. (B) Initial motor coordination plotted for Plp-Nf1fl/+ demonstrating impaired initial motor coordination. *P < 0.05, Mann–Whitney U test. (C) The learning rate is equivalent for both groups.
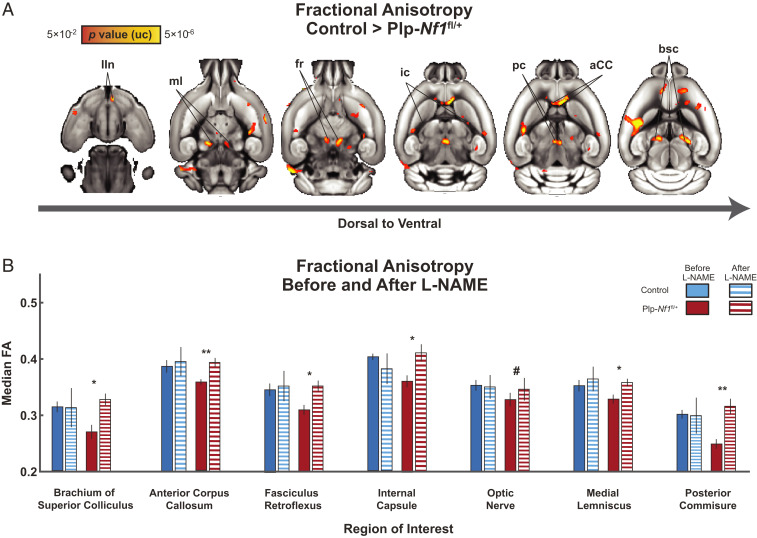
To determine whether brain-wide genotype-specific changes in white matter integrity in this model alter FA, we scanned controls and Plp-Nf1fl/+ mice using DTI. We calculated and coregistered FA maps of each animal onto a common atlas and then compared the genotypes with a whole-brain voxel-based two-sample Student’s t test. We observed a widespread, mostly symmetrical, reduction in FA across the brain (Fig. 3A), similar to that reported in NF1 patients. We focused on selected white matter tracts as regions of interest (ROIs) to quantify effects. To evaluate the effects of treatment, some of the animals (controls, n = 8; Plp-Nf1fl/+, n = 10) underwent intraperitoneal injection of L-NAME for seven consecutive days, followed by a repeated DTI scan to examine the same ROIs (Fig. 3B). We observed a reliable recovery of FA to control levels in six out of seven regions including the brachium of superior colliculus (Z = 2.31, P = 0.01), fasciculus reticulata (Z = 2.03, P = 0.02), internal capsule (Z = 1.75, P = 0.04), medial lemniscus (Z = 1.89, P = 0.03), posterior commissure (Z = 2.45, P = 0.007) and the anterior corpus callosum (aCC) (Z = 2.45, P = 0.007), and a trend toward recovery in the optic nerve (Z = 1.47, P = 0.07). When considered with a conservative correction for multiple comparisons using the Bonferroni method, the aCC and posterior commissure remained statistically significant at α < 0.05. Taken together, the voxel-based analysis of FA shows white matter regions of marked difference between Plp-Nf1fl/+ and controls, with rescue to normal FA levels after inhibition of NOS.
Fig. 3.
Brain-wide white matter changes in PLP mice show significant differences that are rescued by L-NAME treatment. (A) Voxel-based statistical parametric T map demonstrating reliable differences between Plp-Nf1fl/+ (n = 10) and control (n = 11) mice before L-NAME treatment in multiple white matter structures. bsc, brachium of superior colliculus; fr, fasciculus retroflexus; int, internal capsule; IIn, optic nerve; ml, medial lemniscus; pc, posterior commissure. (B) Response of selected brain structures to pharmacologic inhibition of NOS (by L-NAME treatment) in both genotypes. #P < 0.1; *P < 0.05; **P < 0.01, Wilcoxon signed-rank test.
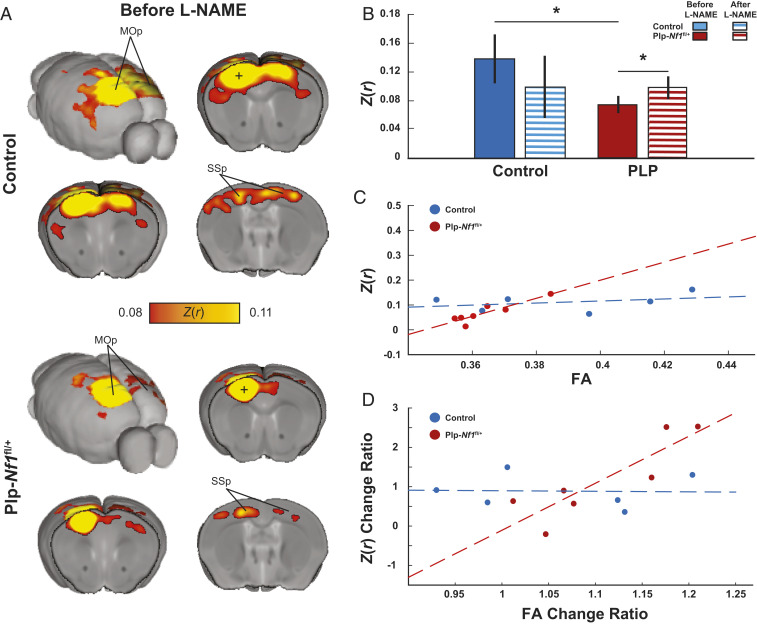
Based on our whole-brain DTI analysis showing structural damage in Plp-Nf1fl/+ mice, together with our previous evidence of disrupted white matter integrity in this model (21, 22), we hypothesized that Plp-Nf1fl/+ mice would also show disrupted functional connectivity. To test this hypothesis, we acclimated 18 animals (controls, n = 7; Plp-Nf1fl/+, n = 11; some DTI sessions were excluded due to image distortions) for awake head-fixed rs-fMRI. We scanned each mouse for five or six sessions (one session per day), administered L-NAME for seven consecutive days, and then scanned for six additional sessions. We focused on the motor cortex, as the aCC connectivity exhibited significantly reduced FA. The aCC contains axons related to the somatomotor system connecting the two hemispheres, and the integrity of this system is reflected in rotarod performance (41). As expected, the control group showed homotopicity (symmetric bilateral correlations), a hallmark of rs-fMRI in humans and rodents (29, 32, 34, 35, 42). In contrast, Plp-Nf1fl/+ mice showed reduced interhemispheric connectivity (Fig. 4 A and B; Mann–Whitney U test: U = 15, P = 0.037). This reduced connectivity was rescued by L-NAME administration (Fig. 4B; Wilcoxon signed-rank test: Z = 2.134, P = 0.033). Collectively, these results indicate that the inhibition of NOS rescues both structural integrity and functional connectivity in Plp-Nf1fl/+ animals.
Fig. 4.
Reduced brain-wide connectivity in the motor network is rescued by L-NAME treatment. (A) Group average correlation maps demonstrate reduced interhemispheric connectivity between control (Top) and Plp-Nf1fl/+ (Bottom) mice. Z(r), Fisher’s Z- transformed r; MOp, primary motor cortex; SSp, primary somatosensory cortex. (B) Correlation of seed-to-seed analysis in the primary motor cortex (denoted as a plus sign in the maps shown in A), plotted for each group before and after L-NAME treatment, demonstrating reduced connectivity between control and Plp-Nf1fl/+ that is rescued by inhibition of NOS. (C) FA in the corpus callosum is plotted as a function of interhemispheric functional connectivity of the motor cortex. The dashed lines represent linear fit for control (blue) and Plp-Nf1fl/+ (red). (D) Structural and functional change due to treatment are plotted. FA change ratio defined as and functional connectivity Z(r) change ratio defined as are plotted. Dashed lines represent linear fit for control and Plp-Nf1fl/+.
To exclude the possibility that increased NO in Plp-Nf1fl/+ animals causes a global change in brain neurovascular coupling that account for these changes in the blood oxygenation level-dependent (BOLD) signal on fMRI, we tested whether the Plp-Nf1fl/+ group shows a nonspecific difference in all interhemispheric connectivity, relative to controls. We divided the cortex into six modules based on Harris et al. (43), further dividing the somatosensory and motor cortices such that the motor cortex is defined as the primary cortex and the somatosensory cortex is all remaining seeds. We calculated the average interhemispheric connectivity between seeds within each module for each animal, then compared each module between genotypes (Mann–Whitney U test). No difference was observed in the anterolateral (U = 38, P = 0.5), somatosensory (U = 26, P = 0.139), visual (U = 24, P = 0.102), medial (U = 30, P = 0.234), or temporal (U = 27, P = 0.16) portions of the cortex. We observed a trend in the prefrontal cortex (U = 20, P = 0.052) and a significant difference in the motor cortex (U = 19, P = 0.032). Within the somatomotor system, rs-fMRI changes were restricted to transcallosal connections (Wilcoxon signed-rank test: Z = 74, P = 0.54). Thus, the observed changes in interhemispheric connectivity are not global and reflect the specific structural changes quantified using DTI.
Reduced FA, an indicator of myelin integrity, is predicted to affect interhemispheric cortical connectivity, which is mediated by axons crossing between sides of the brain in the corpus callosum (44, 45). Therefore, we tested whether the observed FA is correlated with functional connectivity in mice that underwent both DTI and rs-fMRI. Plp-Nf1fl/+ mice, but not controls, showed a significant correlation between FA and rs-fMRI results (control: r = 0.36, P = 0.48; Plp-Nf1fl/+: r = 0.9, P = 0.005), before L-NAME administration (Fig. 4C; control, n = 6; Plp-Nf1fl/+, n = 7). It is important to exclude the possibility that the apparent rescue of reduced interhemispheric connectivity in Plp-Nf1fl/+ mice is caused by a global effect on neurovascular coupling caused by L-NAME. L-NAME blocks all NOS isoforms, reducing NO and impacting neurovascular coupling, and thus potentially the rs-fMRI signal. To exclude this possibility, we tested whether functional connectivity changed in the six cortical modules (43) in controls before and after L-NAME treatment. We observed no difference in functional connectivity (repeated-measures ANOVA; F1,6 = 0.71, P = 0.43), suggesting that L-NAME at this dose and for this time by itself does not introduce a global effect. Thus, Plp-Nf1fl/+ mice treated with L-NAME show specific changes in transcallosal motor system connectivity, which are detectable by rs-fMRI. These results are consistent with the demonstrated effects of L-NAME on FA in Plp-Nf1fl/+ mice noted above.
Changes in FA during normal maturation, aging, and disease are thought to reflect changes in myelin structure at least partially (46, 47). However, there was no significant correlation between FA and rs-fMRI in either group (all: r = 0.43, P = 0.14; control: r = 0.45, P = 0.39; Plp-Nf1fl/+: r = 0.65, P = 0.12). Therefore, we tested whether the changes in FA observed after L-NAME treatment result in a proportional change in functional connectivity. We observed a trend for overall correlation (r = 0.51, P = 0.07) (Fig. 4D). When considered separately, Plp-Nf1fl/+ mice showed a significant correlation (r = 0.87, P = 0.01), but controls did not (r = −0.0332, P = 0.95). We conclude that Plp-Nf1fl/+ animals with a significant difference from baseline demonstrate rescue in interhemispheric functional connectivity, which correlates with the degree of improvement observed in myelin integrity of the corpus callosum based on FA measurement.
Discussion
NF1 is a common disease with multisystem effects, including cognitive/motor impairments and neuroimaging properties, that have been proposed to result from white matter abnormalities but are of unknown etiology. Previous evidence in the PlpCre;Nf1fl/+ mouse model linked NF1 heterozygosity in oligodendrocytes, brain myelin-producing cells, to myelin decompaction. To test whether Nf1 loss in oligodendrocytes is sufficient to disrupt structural and functional myelin integrity in living animals, we investigated the effects of oligodendrocyte-specific Nf1 deletion using two noninvasive longitudinal brain-wide imaging systems. We found that a reduction in FA, a putative marker of myelin structure integrity, is widespread in Plp-Nf1fl/+ animals, and that NOS inhibition by L-NAME rescues this defect in affected white matter tracts. Following evaluation of whole-brain structural disruption using diffusion MRI, we focused a functional connectivity analysis on the somatomotor cortex, which contains neurons whose axons travel in the damaged part of the corpus callosum identified in the DTI analysis. NOS inhibition by L-NAME rescued structural integrity and functional connectivity, and the degree of rescue was correlated in the two independent measures.
Behavioral tests in mice allow measurements of motor performance and dysfunction in a tightly controlled environment and genetic background. Here we used the rotating rod test to examine motor learning. Mice with oligodendrocyte-specific Nf1 deletion showed reduced initial motor coordination but no change in motor-based learning. The peripheral nervous system in the Plp-Nf1fl/+ mice is not detectably abnormal (48), suggesting that this defect is CNS-based. Indicating the specificity of the Nf1 effect in oligodendrocytes, mice overexpressing Plp in oligodendrocytes showed reduced learning and unaltered coordination in a forelimb movement learning task (26). Nf1 heterozygosity in mice is associated with behavioral phenotypes including visuospatial memory and learning deficits, as in humans (49). The human impairments in fine motor coordination, manual dexterity, balance, and hand/eye coordination (6, 50) also have been reported in Nf1+/− mice (51). In contrast to Plp-Nf1fl/+ animals, which exhibited a reduction in initial motor coordination but a normal rate of motor learning, wild-type and Nf1+/− mice showed similar short-term motor performance, but Nf1+/− mice showed reduced long-term motor coordination/learning not observed in the Plp-Nf1fl/+ mice (52). In Nf1+/− mice, all brain cells (neurons, astrocytes, and oligodendrocytes) have reduced Nf1. Our finding of intact motor learning in Plp-Nf1fl/+ mice is consistent with neuron-driven NF1-associated learning impairment and a contribution of oligodendrocytes to motor performance.
Resting-state fMRI measures slow coherent intrinsic fluctuations in oxygenated blood levels that are commonly observed between brain areas connected by monosynaptic or polysynaptic anatomic connections (39). Damage to white matter disrupts functional coupling between regions, suggesting that intact anatomy is necessary for functional coupling (39, 53). Here we observed structural damage in the corpus callosum, the white matter tract that connects the two hemispheres, and disrupted functional connectivity in the motor areas connected via this structure in Plp-Nf1fl/+ mice. These findings are consistent with the motor coordination deficits in these mice being CNS oligodendrocyte-driven.
We found that Plp-Nf1fl/+ mice display reduced FA across the brain, as measured by DTI. While the FA change is global, it was most significant in the aCC. Importantly, inhibition of NOS rescued FA in almost all regions examined. Thus, a readout of myelin integrity recovers in adult animals after only 7 d of treatment, consistent with a similar rapid rescue of myelin integrity following NOS inhibition in this model measured by electron microscopy (21). This rapid response suggests that effects of oligodendrocyte precursor cells on structural change, critical for motor learning, might not be necessary in this setting (54, 55). Interestingly, control mice that received L-NAME showed increased FA variability, indicating a heterogeneous response of the normal CNS to treatment, consistent with the idea that an oxidant response contributes to a normal FA signal. A similar differential response of wild-type mice to L-NAME was noted in motor behavior tests (22).
We observed correlations between the extent of change in structural and functional MRI measures and their absolute values. Functional connectivity and structural integrity each showed significant rescue before L-NAME treatment, but a structure–function correlation was not preserved after L-NAME treatment. Both functional and structural imaging were acquired after treatment, but these tests were carried out sequentially; functional imaging was acquired closer to the treatment period, and structural imaging was acquired subsequently. These measures might become correlated after NOS inhibition by altering drug schedule, dose, or administration of different drugs. It is also possible that changes in myelination and axonal propagation integrity (which are thought to impact functional connectivity measures) are not rescued at the same rate. Additional studies are needed to characterize the timing of NOS inhibition on motor behavior and structural MRI and fMRI changes in NF1 patients to distinguish these alternatives and estimate the efficacy of interventions.
Previous work identified aberrant functional connectivity in NF1 children and Nf1+/− mice, with a disruption in association network organization, corticostriatal, and corticocortical connectivity without reduced interhemispheric connectivity (4, 35). This discrepancy between Nf1 gene inactivation in all brain cells and oligodendrocyte-specific Nf1 deletion showing reduced interhemispheric connectivity may be explained by Nf1+/− mice having mutant neurons in addition to oligodendrocytes. Nf1+/− mice show aberrant neural activity that is mimicked by Nf1 deletion specifically in neurons (56–59). In contrast, oligodendrocyte-specific Nf1 inactivation impairs myelin-forming cells, disrupting cellular communication and indirectly influencing neurons. Thus, it appears that when Nf1 inactivation is present in both neurons and glia, the net effect is increased cortical interhemispheric connectivity. Use of the Plp-Nf1fl/+ animal model allows us to capture oligodendrocyte-specific features, partially recapitulating the human NF1 condition. Collectively, the impact of myelin decompaction on function is significant; however, this is not the only factor contributing to the cognitive/behavioral phenotypes of this complex disorder.
Both DTI and rs-fMRI are commonly used techniques in humans. DTI may be useful as an outcome measure in future clinical trials, as FA is commonly used to identify white matter abnormalities. However, FA is not a direct measure of myelin integrity generally or myelin decompaction specifically; other factors, such as axon composition and spacing, also may affect this measure (60). With these reservations, given that myelin decompaction is rescued by administration of L-NAME in Plp-Nf1fl/+ animals on the microscopic level (21, 22), we infer that FA, at least partially, reflects the integrity of myelin in this model. With respect to functional connectivity, rs-fMRI networks have been observed repeatedly and reliably under the presence of anesthetic vasodilators (30, 61). Anesthetic vasodilators impact the signal-to-noise ratio, but network organization is preserved. Thus, we propose that both FA and rs-fMRI can be used as outcome measures in clinical trials.
We note that NOS inhibition via L-NAME is not commonly used in humans. Although L-NAME administration to patients in shock did not demonstrate any significant adverse events, it causes systemic and pulmonary hypertension and is teratogenic during pregnancy (62). Possible consequences of chronic L-NAME use in children are unknown. Other safer and/or specific NOS inhibitors or antioxidants (e.g., N-acetyl-cysteine) might be tested in preclinical or clinical trials and be better tolerated (63) than Ras pathway-specific inhibitors, such as MEK inhibitors (64), and any cognitive/motor rescue antioxidant affords may be of value to NF1 patients.
In summary, NOS inhibition recues structural integrity and functional connectivity across the entire brain in the Plp-Nf1fl/+ animal model. Rescued myelin integrity was correlated with rescue in functional connectivity, albeit to varying degrees, and the extent of rescue in FA is a predictor of the degree of recovery observed in interhemispheric connectivity. Given that this measure of structural MRI is widely used in humans, we suggest that it may be used as an outcome measure of future clinical trials aimed at rescuing myelin decompaction.
Materials and Methods
Ethics Statement.
All animal procedures were conducted with the approval of the Institutional Animal Care and Use Committee and in accordance with the NIH Guide for the Care and Use of Laboratory Animals (65).
Animals.
Inducible Plp-Cre (66) and Nf1fl/+ (67) mice were maintained on a pure C57BL/6 background and genotyped by PCR according to publicly available protocols (22). Tamoxifen was administered to the animals at age 8 wk (see below). Twenty-nine animals participated in Rotarod evaluation (control, n = 14; Plp-Nf1fl/+, n = 15), 19 animals in DTI (control, n = 9; Plp-Nf1fl/+, n = 10), and 18 animals in rs-fMRI (control, n = 7; Plp-Nf1fl/+, n = 11). Of the animals that participated in rs-fMRI, 13 animals participated in DTI (control, n = 6; Plp-Nf1fl/+, n = 7), and 14 in the Rotarod test (control, n = 6; Plp-Nf1fl/+, n = 8).
Tamoxifen Injection.
The injection protocol followed previous publications (21, 22). Tamoxifen (100 mg) was dissolved with 1 mL of ethanol and 9 mL of sunflower seed oil (Sigma-Aldrich). At age 8 wk, both controls and Plp-Nf1fl/+ mice were intraperitoneally injected with tamoxifen (100 µL) twice daily for 3 d at the same time of day.
L-NAME Injection.
Fresh solution of L-NAME (L-NG-nitroarginine methyl ester, 100 µM in 1× PBS; Sigma-Aldrich) was administered daily at 0.4 mg/kg (21).
Animal Surgery.
Before any experiment, to use our in-house designed cradle, we implanted headposts on the animals’ skulls as described previously (34). In brief, the mice were anesthetized with isoflurane mixed with oxygen, which was kept at a level of 2% to 3% for the entire procedure. The skin was removed, and 3D-printed plastic headposts were positioned on the skulls and affixed with dental cement (C&B Metabond; Parkell). To prevent image distortions in MRI acquisition resulting from uneven surfaces, the cement was smoothed using a self-cured resin (general purpose acrylic resin; Unifast Trad). The animals were then left to recover and injected once with antibiotics (cefalexin 0.18 mg/10 g; Norbrook) along with systemic and local analgesics (buprenorphine 0.9 µg/10 g and bupavacaine 0.075 mg/10 g, respectively). Each animal was carefully monitored over the next week before entering the experiment.
Rotating Rod Performance Test.
Gross motor coordination function was evaluated using the Rotarod (Med Associates). Twenty-nine animals (controls, n = 14; Plp-Nf1fl/+, n = 15) underwent one training session and one test session, each consisting of four runs. Every run lasted for 6 min, in which the rod accelerated gradually from 4 to 40 revolutions per minute in the first 5 min and maintained at the final speed for the last minute. Time to fall and peak speed were recorded.
MRI Acquisition.
Imaging was performed with a 9.4-T BioSpec 94/20 USR MRI (Bruker BioSpin MRI). A 20-mm receive-only coil was fixed to the cradle over the animal’s head, and the entire apparatus was inserted into the MRI bore containing an 86-mm quadrature transmit-only coil (Bruker BioSpin MRI).
DTI Acquisition.
To characterize structural connectivity, we scanned 19 animals using DTI (control, n = 9; Plp-Nf1fl/+, n = 10); one control was excluded due to extensive image distortions. Animals were first lightly sedated and fixed to the resting-state cradle with a custom-made mask placed over the nose, which allowed maintenance of anesthesia throughout the DTI scanning session while their respiratory rate was monitored. Data acquisition was performed using a diffusion-weighted spin-echo echo-planar imaging (EPI) pulse sequence (repetition time [TR], 7,000 ms; echo time [TE], 21.68 ms). We used 30 noncolinear diffusion directions, a bfactor of 1,000 s/mm3 with 11 ms duration of the diffusion gradient (δ) and a separation time (Δ) of 2.6 ms. The scan consisted of 24 slices of 400 µm thickness, with an acquisition matrix of 100 × 100 voxels and a field of view of 16 × 16 cm2, resulting in a resolution of 0.1 × 0.1 × 0.4 mm3.
DTI Preprocessing and Analysis.
Diffusion-weighted images were preprocessed using ExplorDTI (68), with tensors calculated using a robust estimation algorithm (69), together with motion and eddy current correction. FA and B0 maps were exported to the NIfTI format. Brain extraction was performed, and a template, B0, for both controls and Plp-Nf1fl/+ animals was created using Advanced Normalization Tools (ANTs) (70). The control template was registered to the Allen Brain Atlas (71), while the Plp-Nf1fl/+ was linearly coregistered to the controls. A final diffeomorphic field from native space to the Allen Brain Atlas space for each animal was calculated and applied in a single step to native scans to prevent data degradation.
Normalized FA maps were submitted to a two-sample Student’s t test between controls and Plp-Nf1fl/+ animals. For ROI analysis, we defined the regions manually based on the t test results comparing genotypes before L-NAME and calculated the mean voxel intensity within each region to assess rescue after treatment (72).
rs-fMRI Acquisition.
Eighteen animals were included in the awake resting-state fMRI experiment (control, n = 7; Plp-Nf1fl/+, n = 11) which followed a previously published protocol (34). To reduce stress, the animals were lightly sedated and fixed to the resting-state cradle. During the 5-d acclimatization period, each animal was placed in the MRI machine and exposed to the different scanning protocols used, while the period of head restraint was increased gradually from 2 to 50 min by the last acclimatization session. Afterward, each animal was scanned before and after the administration of L-NAME for six or seven daily sessions of rs-fMRI at each time period. Each scanning session included structural and functional scanning. The structural scan was acquired using a T1-weighted rapid acquisition process with relaxation enhancement (RARE) sequence in coronal orientation (30 coronal slices; TE, 8.5 ms; RARE factor, 4; flip angle, 180°; in-plane resolution, 150 × 150 μm2; slice thickness, 450 μm). Whole-brain functional scans consisted of 800 repetitions divided over four runs (200 repetitions per run). BOLD imaging was achieved using spin echo-echo planar imaging (SE-EPI) sequence (TR, 2.5 s; TE, 18.398 ms; flip angle, 90°; 30 coronal slices; in-plane resolution, 150 × 150 μm2; slice thickness, 450 μm; the imaged volume was framed with four saturation slices to avoid wraparound artifacts).
rs-fMRI Preprocessing and Analysis.
The preprocessing steps have been reported previously (34, 73). In brief, the first two frames were skipped to eliminate inhomogeneity artifacts and the remaining frames were slice time corrected. To perform motion correction, the whole time series was corrected to the average of the first 200 repetitions using rigid-body correction (74) by calculating a coregistration matrix for each volume. The same averaged volume was then warped automatically, using symmetric diffeomorphic image normalization (SyN) with ANTs (75), to a predefined EPI template that was preregistered with the Allen Brain Atlas. To prevent degradation of image quality, the final displacement field for each volume, including its motion correction displacement matrix, was calculated and applied on each time-corrected volume. Whole-brain and ventricle masks were calculated automatically. Data-scrubbing procedures were as described previously (34, 76), with similar exclusion criteria: frame displacement threshold of 50 µm, temporal derivative mean square root area threshold of 150% the interquartile range above the 75th percentile with exclusion of one frame after the detected motion. Runs with fewer than 50 frames and sessions with fewer than 300 frames were excluded; 29 out of 208 sessions were excluded due to excessive movement or image distortion (control, n = 11; Plp-Nf1fl/+, n = 18). After data scrubbing, the time series underwent demeaning and detrending to remove global mean and nonintrinsic trends, together with regressing out mean ventricles and vascular signals, and six volume-specific motion parameters and their derivatives were regressed out of the signal to remove sources of spurious or regionally nonspecific variance (77). Finally, bandpass filtering (0.009 to 0.08 Hz) and smoothing using convolution with a Gaussian function (FWHM of 0.6 mm) were applied.
To estimate functional connectivity maps, we placed an ROI with a 100-µm radius and extracted the time course from each seed. A seed-to-whole brain correlation map was calculated during each session and averaged to produce a mean Fisher’s Z transform, Z(r), map for that animal. Finally, all the animal-based maps were averaged to produce one map for each genotype, which were displayed on a template registered to the Allen Brain Atlas (78). Seed-to-seed analysis was performed by calculating Pearson’s correlation coefficient between two seeds and transforming it using Fisher’s Z transform to allow for cross-subject comparison.
Acknowledgments
We thank the Technion Biological Core Facilities and Edith Suss-Toby for assistance with MRI and the Technion Preclinical Research Authority for assistance with animal care. This work was supported by the Israel Science Foundation (770/17, to I.K.), the Adelis Foundation (to I.K.) and the NIH (1R01 NS091037, to N.R. and I.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data Availability.
All imaging raw data and the relevant codes used in this study are available in brain imaging data structure (BIDS) format on OpenNeuro (https://openneuro.org/datasets/ds003027).
References
- 1.Gutmann D. H. et al., Neurofibromatosis type 1. Nat. Rev. Dis. Primers 3, 17004 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Shofty B., Constantini S., Ben-Shachar S., Advances in molecular diagnosis of neurofibromatosis type 1. Semin. Pediatr. Neurol. 22, 234–239 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Garg S. et al., Autism spectrum disorder profile in neurofibromatosis type I. J. Autism Dev. Disord. 45, 1649–1657 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Baudou E. et al., Can the cognitive phenotype in neurofibromatosis type 1 (NF1) be explained by neuroimaging? A review. Front. Neurol. 10, 1373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine T. M., Materek A., Abel J., O’Donnell M., Cutting L. E., Cognitive profile of neurofibromatosis type 1. Semin. Pediatr. Neurol. 13, 8–20 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Krab L. C. et al., Impact of neurofibromatosis type 1 on school performance. J. Child Neurol. 23, 1002–1010 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Johnson B. A. et al., Motor proficiency in children with neurofibromatosis type 1. Pediatr. Phys. Ther. 22, 344–348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krab L. C. et al., Motor learning in children with neurofibromatosis type I. Cerebellum 10, 14–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting L. E. et al., Megalencephaly in NF1: Predominantly white matter contribution and mitigation by ADHD. Neurology 59, 1388–1394 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Dubovsky E. C. et al., MR imaging of the corpus callosum in pediatric patients with neurofibromatosis type 1. AJNR Am. J. Neuroradiol. 22, 190–195 (2001). [PMC free article] [PubMed] [Google Scholar]
- 11.Pride N. et al., Corpus callosum morphology and its relationship to cognitive function in neurofibromatosis type 1. J. Child Neurol. 25, 834–841 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Williams V. C. et al., Neurofibromatosis type 1 revisited. Pediatrics 123, 124–133 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Feldman H. M., Yeatman J. D., Lee E. S., Barde L. H. F., Gaman-Bean S., Diffusion tensor imaging: A review for pediatric researchers and clinicians. J. Dev. Behav. Pediatr. 31, 346–356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsgodt K. H. et al., Alterations in white matter microstructure in neurofibromatosis-1. PLoS One 7, e47854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aydin S. et al., Relationship between the corpus callosum and neurocognitive disabilities in children with NF-1: Diffusion tensor imaging features. Clin. Imaging 40, 1092–1095 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Brown J. A. et al., Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum. Mol. Genet. 19, 4515–4528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koini M., Rombouts S. A. R. B., Veer I. M., Van Buchem M. A., Huijbregts S. C. J., White matter microstructure of patients with neurofibromatosis type 1 and its relation to inhibitory control. Brain Imaging Behav. 11, 1731–1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemmi F. et al., Discriminating between neurofibromatosis-1 and typically developing children by means of multimodal MRI and multivariate analyses. Hum. Brain Mapp. 40, 3508–3521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Vaart T. et al.; NF1-SIMCODA Study Group , Behavioral and cognitive outcomes for clinical trials in children with neurofibromatosis type 1. Neurology 86, 154–160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiPaolo D. P. et al., Neurofibromatosis type 1: Pathologic substrate of high-signal-intensity foci in the brain. Radiology 195, 721–724 (1995). [DOI] [PubMed] [Google Scholar]
- 21.López-Juárez A. et al., Oligodendrocyte Nf1 controls aberrant notch activation and regulates myelin structure and behavior. Cell Rep. 19, 545–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayes D. A. et al., Nf1 loss and Ras hyperactivation in oligodendrocytes induce NOS-driven defects in myelin and vasculature. Cell Rep. 4, 1197–1212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutmann D. H., Parada L. F., Silva A. J., Ratner N., Neurofibromatosis type 1: Modeling CNS dysfunction. J. Neurosci. 32, 14087–14093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nomura T., Bando Y., Nakazawa H., Kanemoto S., Yoshida S., Pathological changes in mice with long term cuprizone administration. Neurochem. Int. 126, 229–238 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Mandolesi G. et al., Voluntary running wheel attenuates motor deterioration and brain damage in cuprizone-induced demyelination. Neurobiol. Dis. 129, 102–117 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Kato D. et al., Motor learning requires myelination to reduce asynchrony and spontaneity in neural activity. Glia 68, 193–210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franco-Pons N., Torrente M., Colomina M. T., Vilella E., Behavioral deficits in the cuprizone-induced murine model of demyelination/remyelination. Toxicol. Lett. 169, 205–213 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Scott R. et al., Loss of Cntnap2 causes axonal excitability deficits, developmental delay in cortical myelination, and abnormal stereotyped motor behavior. Cereb. Cortex 29, 586–597 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Liska A., Galbusera A., Schwarz A. J., Gozzi A., Functional connectivity hubs of the mouse brain. Neuroimage 115, 281–291 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Zerbi V., Grandjean J., Rudin M., Wenderoth N., Mapping the mouse brain with rs-fMRI: An optimized pipeline for functional network identification. Neuroimage 123, 11–21 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Dodero L. et al., Neuroimaging evidence of major morpho-anatomical and functional abnormalities in the BTBR T+TF/J mouse model of autism. PLoS One 8, e76655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sforazzini F. et al., Altered functional connectivity networks in acallosal and socially impaired BTBR mice. Brain Struct. Funct. 221, 941–954 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Grandjean J. et al., Early alterations in functional connectivity and white matter structure in a transgenic mouse model of cerebral amyloidosis. J. Neurosci. 34, 13780–13789 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergmann E., Zur G., Bershadsky G., Kahn I., The organization of mouse and human cortico-hippocampal networks estimated by intrinsic functional connectivity. Cereb. Cortex 26, 4497–4512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shofty B. et al., Autism-associated Nf1 deficiency disrupts corticocortical and corticostriatal functional connectivity in human and mouse. Neurobiol. Dis. 130, 104479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loitfelder M. et al., Functional connectivity changes and executive and social problems in neurofibromatosis type i. Brain Connect. 5, 312–320 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibrahim A. F. A. et al., Spatial working memory in neurofibromatosis 1: Altered neural activity and functional connectivity. Neuroimage Clin. 15, 801–811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori S., Zhang J., Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51, 527–539 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Buckner R. L., Krienen F. M., Yeo B. T. T., Opportunities and limitations of intrinsic functional connectivity MRI. Nat. Neurosci. 16, 832–837 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Rothwell P. E. et al., Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 158, 198–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao V. Y. et al., Motor learning consolidates Arc-expressing neuronal ensembles in secondary motor cortex. Neuron 86, 1385–1392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panzeri S. et al., Homozygous loss of autism-risk gene CNTNAP2 results in reduced local and long-range prefrontal functional connectivity. Cereb. Cortex 28, 1141–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Harris J. A., et al. , Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeter A., Grandjean J., Schlegel F., Saab B. J., Rudin M., Contributions of structural connectivity and cerebrovascular parameters to functional magnetic resonance imaging signals in mice at rest and during sensory paw stimulation. J. Cereb. Blood Flow Metab. 37, 2368–2382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roland J. L. et al., On the role of the corpus callosum in interhemispheric functional connectivity in humans. Proc. Natl. Acad. Sci. U.S.A. 114, 13278–13283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geeraert B. L., Lebel R. M., Lebel C., A multiparametric analysis of white matter maturation during late childhood and adolescence. Hum. Brain Mapp. 40, 4345–4356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teipel S. J. et al., Longitudinal changes in fiber tract integrity in healthy aging and mild cognitive impairment: A DTI follow-up study. J. Alzheimers Dis. 22, 507–522 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Mayes D. A. et al., Perinatal or adult Nf1 inactivation using tamoxifen-inducible PlpCre each cause neurofibroma formation. Cancer Res. 71, 4675–4685 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva A. J. et al., A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat. Genet. 15, 281–284 (1997). [DOI] [PubMed] [Google Scholar]
- 50.North K. et al., Specific learning disability in children with neurofibromatosis type 1: Significance of MRI abnormalities. Neurology 44, 878–883 (1994). [DOI] [PubMed] [Google Scholar]
- 51.Robinson A., Kloog Y., Stein R., Assaf Y., Motor deficits and neurofibromatosis type 1 (NF1)-associated MRI impairments in a mouse model of NF1. NMR Biomed. 23, 1173–1180 (2010). [DOI] [PubMed] [Google Scholar]
- 52.van der Vaart T., van Woerden G. M., Elgersma Y., de Zeeuw C. I., Schonewille M., Motor deficits in neurofibromatosis type 1 mice: The role of the cerebellum. Genes Brain Behav. 10, 404–409 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Magnuson M. E., Thompson G. J., Pan W.-J., Keilholz S. D., Effects of severing the corpus callosum on electrical and BOLD functional connectivity and spontaneous dynamic activity in the rat brain. Brain Connect. 4, 15–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monje M., Myelin plasticity and nervous system function. Annu. Rev. Neurosci. 41, 61–76 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Bells S. et al., White matter plasticity and maturation in human cognition. Glia 67, 2020–2037 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Costa R. M. et al., Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 415, 526–530 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Shilyansky C. et al., Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc. Natl. Acad. Sci. U.S.A. 107, 13141–13146 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomson S. N. et al., Resting state functional MRI reveals abnormal network connectivity in neurofibromatosis 1. Hum. Brain Mapp. 36, 4566–4581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui Y. et al., Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 135, 549–560 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scholz J., Tomassini V., Johansen-Berg H., “Individual differences in white matter microstructure in the healthy brain” in Diffusion MRI, Johansen-Berg H., Behrens T. E. J., Eds. (Academic Press, San Diego, 2009), chap. 11, pp. 237–249. [Google Scholar]
- 61.Grandjean J., Schroeter A., Batata I., Rudin M., Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage 102, 838–847 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Kiehl M. G., Ostermann H., Meyer J., Kienast J., Nitric oxide synthase inhibition by L-NAME in leukocytopenic patients with severe septic shock. Intensive Care Med. 23, 561–566 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Wong V., Chong W., Lerner E., Nitric oxide inhibition strategies. Futur. Sci. 1, FSO35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Broman K. K., Dossett L. A., Sun J., Eroglu Z., Zager J. S., Update on BRAF and MEK inhibition for treatment of melanoma in metastatic, unresectable, and adjuvant settings. Expert Opin. Drug Saf. 18, 381–392 (2019). [DOI] [PubMed] [Google Scholar]
- 65.National Research Council , Guide for the Care and Use of Laboratory Animals, (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 66.Doerflinger N. H, Macklin W. B., Popko B., Inducible site-specific recombination in myelinating cells. Genesis 35, 63–72 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Zhu Y. et al., Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15, 859–876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leemans A., Jeurissen B., Sijbers J., Jones D., “ExploreDTI: A graphical toolbox for processing, analyzing, and visualizing diffusion MR data” in Proceedings of the 17th Scientific Meeting International Society of Magnetic Resonance in Medicine, (ISMRM, Hawaii, 2009), p. 3537. [Google Scholar]
- 69.Plaisier A. et al., Choice of diffusion tensor estimation approach affects fiber tractography of the fornix in preterm brain. AJNR Am. J. Neuroradiol. 35, 1219–1225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avants B. B., Epstein C. L., Grossman M., Gee J. C., Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oh S. W. et al., A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenkinson M., Beckmann C. F., Behrens T. E. J., Woolrich M. W., Smith S. M., FSL. Neuroimage 62, 782–790 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Kahn I. et al., Characterization of the functional MRI response temporal linearity via optical control of neocortical pyramidal neurons. J. Neurosci. 31, 15086–15091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jenkinson M., Bannister P., Brady M., Smith S., Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002). [DOI] [PubMed] [Google Scholar]
- 75.Avants B. B., A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Power J. D. et al., Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grandjean J. et al., Common functional networks in the mouse brain revealed by multi-centre resting-state fMRI analysis. Neuroimage 205, 116278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dorr A. E., Lerch J. P., Spring S., Kabani N., Henkelman R. M., High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage 42, 60–69 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All imaging raw data and the relevant codes used in this study are available in brain imaging data structure (BIDS) format on OpenNeuro (https://openneuro.org/datasets/ds003027).