Significance
The biogenesis, processing, function, and degradation of cellular RNAs depend critically on their protein interaction partners. Systematic analysis of the protein interactome of specific RNAs of interest inside living cells can therefore enable a better understanding of many biological processes. We developed two complementary methods for tagging endogenous proteins in the vicinity of specific cellular RNAs, for subsequent identification by mass spectrometry. When applied to the human telomerase RNA, our methods recovered known interaction partners as well as unexpected hits, including an enzyme that catalyzes RNA posttranscriptional modification to influence telomerase activity. The technology introduced by our study should facilitate future investigations into RNA–protein interactions in living cells.
Keywords: RNA binding protein, interactome, APEX, proximity labeling, telomerase RNA
Abstract
RNA–protein interactions underlie a wide range of cellular processes. Improved methods are needed to systematically map RNA–protein interactions in living cells in an unbiased manner. We used two approaches to target the engineered peroxidase APEX2 to specific cellular RNAs for RNA-centered proximity biotinylation of protein interaction partners. Both an MS2-MCP system and an engineered CRISPR-Cas13 system were used to deliver APEX2 to the human telomerase RNA hTR with high specificity. One-minute proximity biotinylation captured candidate binding partners for hTR, including more than a dozen proteins not previously linked to hTR. We validated the interaction between hTR and the N6-methyladenosine (m6A) demethylase ALKBH5 and showed that ALKBH5 is able to erase the m6A modification on endogenous hTR. ALKBH5 also modulates telomerase complex assembly and activity. MS2- and Cas13-targeted APEX2 may facilitate the discovery of novel RNA–protein interactions in living cells.
Mapping networks of RNA–protein interactions in living cells is necessary to enable a mechanistic understanding of RNA processing, trafficking, folding, function, and degradation (1, 2). While many protein-centered approaches, such as cross-linking immunoprecipitation sequencing (CLIP-seq) and RNA immunoprecipitation sequencing (RIP-seq), are available for the identification of RNAs bound to specific proteins of interest, few robust methods exist for the reverse problem: RNA-centered identification of protein binding partners of specific cellular RNAs of interest. Previous approaches that rely on aptamer (3–5) or antisense probe (6–8) affinity purification of RNA–protein complexes yield nonspecific hits that bind to denatured RNAs postlysis, and/or miss weak or transient interactions (9–11). In addition, identification of RNAs by widely available sequencing methods is generally easier than unbiased protein detection with mass spectrometry-based proteomics. To address the challenge of RNA-centered interactome mapping, recent studies (12–14) have begun to employ proximity-dependent labeling (15), targeting the promiscuous biotin ligase BioID (16) to specific MS2- or BoxB stem-loop-tagged RNA targets. These methods represent an important technological advance and have yielded biological insights. However, the long labeling time window of BioID is not optimal for the study of dynamic processes (for example, rapid changes in RNA interactomes in response to cellular stress), while the stem-loop tags could affect the function of target RNAs.
Here, we present alternative methodologies for mapping RNA–protein interactions inside living cells, using both MS2 and CRISPR-Cas13 (17, 18) to target the proximity labeling enzyme APEX2 (19) to specific RNAs. The rapid 1-min promiscuous biotinylation catalyzed by APEX enables proteomic identification (20–24) of endogenous interaction partners with much faster temporal resolution than BioID and its relatives (BioID2, BASU) (25). We applied these methods to the human telomerase RNA (hTR), which plays a critical role in regulating cellular senescence and oncogenesis by serving as the template for reverse transcription of telomeres (26, 27). While hTR’s interaction with the telomerase complex has been extensively characterized (28), hTR is present in stoichiometric excess over telomerase in cancer cells (29) and is broadly expressed in tissues lacking telomerase protein (30). These observations suggest that hTR could also function outside of the telomerase complex (31), and uncharacterized telomerase-independent hTR interactors may exist to regulate hTR function (32, 33). We performed APEX labeling on recombinant hTR in HEK293T cells followed by validation of one of the hits, ALKBH5, on endogenous hTR. Our study highlights the utility of RNA-targeted APEX for unbiased discovery.
Results
Using MS2-MCP to Target APEX to Tagged hTR.
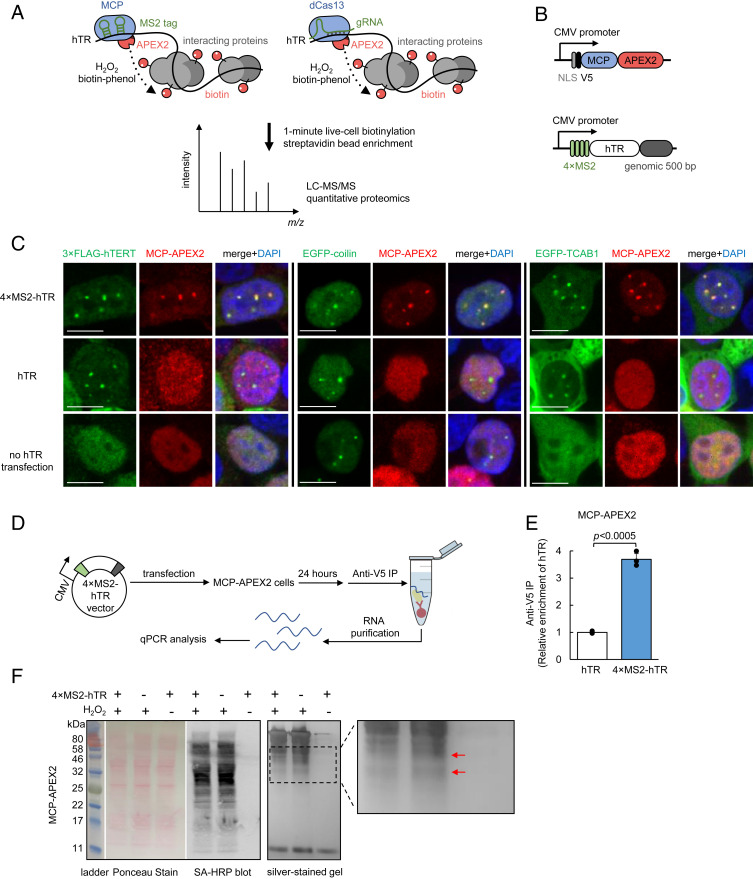
We utilized two complementary approaches to deliver APEX to the site of hTR for proximity labeling (Fig. 1A). The first entails conjugating hTR to the bacteriophage MS2 RNA stem loop, which can specifically bind an MS2 coat protein-fused APEX2 (MCP-APEX2) with high affinity (Kd < 1 nM) (34). In the second approach, a catalytically inactive Cas13-APEX2 fusion (dCas13-APEX2) is programmed using a guide RNA (gRNA) to target unmodified hTR, but with a lower binding affinity (Kd ∼ 10 nM) (35).
Fig. 1.
Targeting APEX2 to human telomerase RNA with MS2-MCP. (A) Scheme of APEX2-mediated proximity labeling of human telomerase RNA (hTR) interacting proteins. (Left) APEX2 is targeted to MS2 stem-loop-tagged hTR via fusion to MS2 coat protein (MCP). (Right) APEX2 is targeted to hTR via fusion to catalytically dead Cas13 protein (dCas13) and a guide RNA (gRNA). To initiate labeling, H2O2 was added for 1 min to cells preloaded with biotin-phenol, which is oxidized by APEX2 into a phenoxyl radical to covalently tag proximal endogenous proteins. Biotinylated proteins are enriched with streptavidin beads, then identified by LC-MS/MS. (B) Design of MCP-APEX2 and 4×MS2-hTR expression constructs. Genomic 500 bp is the 500-bp genomic sequence at the 3′ end of endogenous hTR. (C) Fluorescence imaging of MCP-APEX2 localization to hTR foci. HEK293T cells are transfected with either 4×MS2-hTR (Top), untagged hTR (Middle), or no hTR (Bottom). MCP-APEX2 is visualized by anti-V5 staining (Alexa Fluor 647, Middle column). hTR foci are visualized with either cotransfected 3×FLAG-hTERT (anti-FLAG staining, Alexa Fluor 488, Left image set), cotransfected EGFP-coilin (Middle image set), or cotransfected EGFP-TCAB1 (Right image set). Nuclei are stained with DAPI (Scale bars, 10 µm.) (D) Scheme of RNA immunoprecipitation for validation of MCP-APEX2 targeting to hTR. HEK293T cells stably expressing MCP-APEX2 are transfected with 4×MS2-hTR or untagged hTR. RNA bound to APEX2 is immunoprecipitated with anti-V5 antibody followed by RT-qPCR quantitation. (E) RT-qPCR of RNA immunoprecipitation following experiment in D. The fold enrichment of hTR in cells transfected with 4×MS2-hTR is normalized against the untagged hTR control. Data are analyzed using a one-tailed Student’s t test (n = 3). (F) Biochemical analysis of biotinylated proteins from HEK293T cells stably expressing MCP-APEX2. Lysates were run on SDS/PAGE and analyzed by Ponceau stain (Left) or streptavidin blotting (Middle). Total eluted protein after streptavidin bead enrichment is visualized by silver staining (Right). A magnified view of the boxed region is shown on the right. Red arrows point to proteins differentially biotinylated by targeted vs. nontargeted APEX2.
To validate the MS2-MCP strategy, four tandem MS2 stem loops were inserted at the 5′ end of hTR (4×MS2-hTR, Fig. 1B), which is known to be flexible and single stranded (28), in contrast to hTR’s 3′ end. Furthermore, the 5′ end of hTR (but not 3′ end) is not processed when expressed from a Pol II promoter (e.g., CMV promoter), so that the MS2 tags remain intact in the mature transcript (36). Of note, a recent study (37) showed that fusing three or four MS2 stem loops to the 5′ end of hTR, identical to our approach here, does not impair hTR function in a telomerase activity assay, while 10 to 20 stem-loop fusions are perturbative and result in a loss of signal.
The mature hTR sequence is followed by 500 bp of downstream genomic sequence to ensure proper processing of the 3′ end (36). Because fully processed hTR accumulates in the evolutionarily conserved subnuclear compartment Cajal body (CB) (38, 39), we used fluorescence imaging with CB markers to check for both functional hTR and recruitment of MCP-APEX2 to hTR foci. We coexpressed 4×MS2-hTR with nuclear-localized MCP-APEX2 (Fig. 1B) in HEK293T cells and performed immunostaining (Fig. 1C). We found that MCP-APEX2 specifically colocalizes with the core CB component coilin (40) when MS2-tagged hTR is coexpressed, but not in controls with untagged hTR or no hTR (Fig. 1C). In addition, colocalization of MCP-APEX2 to hTR foci was confirmed by costaining against the telomerase proteins hTERT (human telomerase reverse transcriptase) (41) and TCAB1 (telomerase Cajal body protein 1) (42) (Fig. 1C).
As a separate readout of MCP-APEX2 targeting to 4×MS2-hTR, we performed RIP followed by quantitative RT-PCR (Fig. 1D). Consistent with the imaging data, 3.7-fold greater hTR transcripts were enriched by MCP-APEX2 anti-V5 pulldown in cells expressing 4×MS2-hTR compared to cells expressing the untagged hTR control (Fig. 1E). Together, the imaging and RIP experiments demonstrate the efficient targeting of MCP-APEX2 to hTR.
We assessed the promiscuous labeling activity of MCP-APEX2, targeted to hTR, by Western blot. Fig. 1F shows H2O2-dependent biotinylation in HEK293T cells. Interestingly, biotinylated proteins enriched from cells expressing 4×MS2-hTR vs. untagged hTR showed subtle differences in banding patterns, or “fingerprints,” on the silver-stained gel, consistent with the expectation that hTR-targeted APEX2 tags a different cohort of proteins than untargeted MCP-APEX2, which distributes throughout the nucleus.
To probe the generality of MS2/MCP-based APEX2 targeting, we tested two additional RNA targets. The mRNAs ATP5B and ACTB were each tagged with 2×MS2 stem loops on their 3′ UTRs. These RNAs are known to rapidly localize to stress granules upon 30-min treatment with NaAsO2 (43). SI Appendix, Fig. S1 shows that MCP-APEX2 colocalizes with the stress granule marker G3BP1 (44) specifically in the presence of the MS2-tagged ATP5B but not in omit transcript or unstressed controls. Similar targeting was achieved for ACTB (SI Appendix, Fig. S2), indicating that the MS2/MCP approach is also potentially useful for the study of mRNA interaction partners by APEX2 proximity labeling.
Development of a CRISPR-Cas13 Strategy to Target APEX to Untagged hTR.
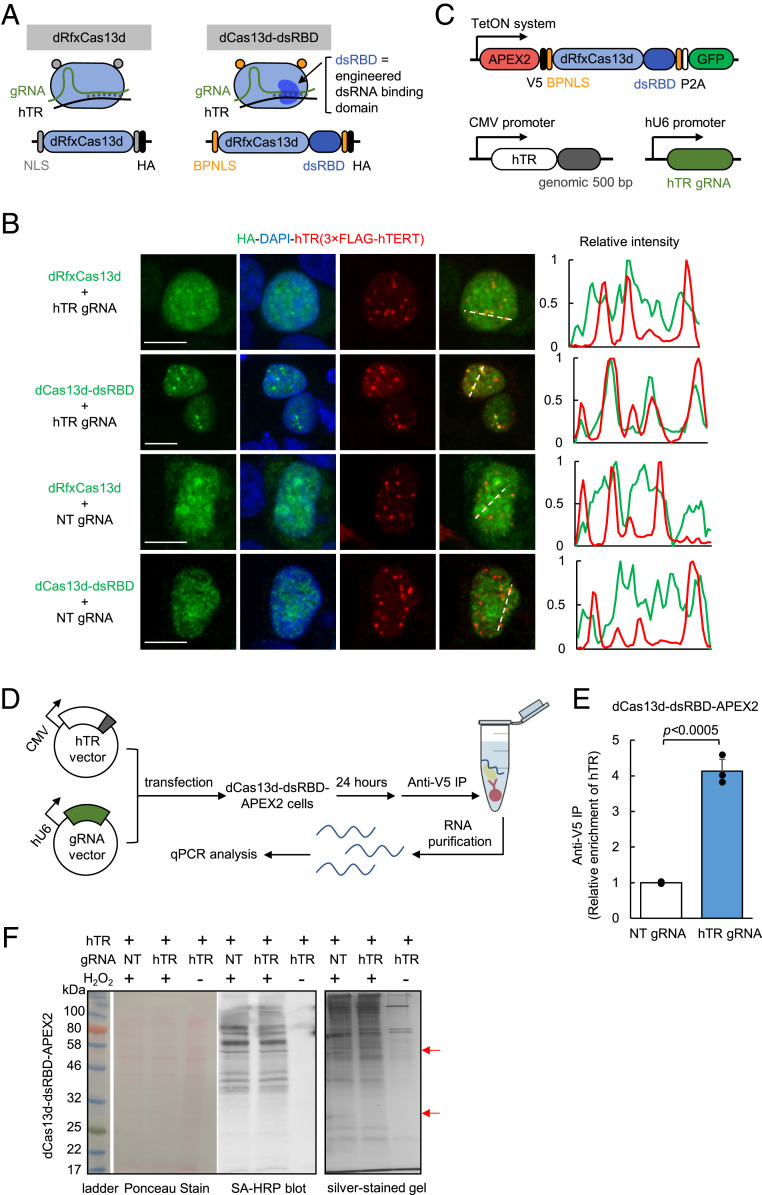
The discovery of RNA-directed CRISPR systems (35, 45) offers the exciting opportunity to target unmodified, native cellular RNAs, in contrast to MS2-based approaches. Thus, we next explored the use of Cas13 to deliver APEX to untagged telomerase RNA. In comparison to the small size of MCP (14 kDa), the bulk of Cas13 enzymes (∼120 kDa) poses a potential steric challenge to identifying accessible sites on the highly structured hTR RNA. Previous studies have established several Cas13 orthologs that are catalytically active inside mammalian cells, including LwaCas13a (139 kDa) (17), PspCas13b (128 kDa) (46), and RfxCas13d (112 kDa) (18). We selected the smallest and most active of these, RfxCas13d (18), for testing. We first designed a total of six gRNAs (gRNA_1-6, SI Appendix, Fig. S3) targeting different regions of hTR and validated their targeting by knockdown with an active RfxCas13d. All six gRNAs significantly reduced hTR levels in HEK293T cells, with efficiency ranging from 20 to 75% (SI Appendix, Fig. S4). However, coexpressing the gRNAs with a catalytically dead RfxCas13d (dRfxCas13d, Fig. 2A) did not result in observable enrichment of dRfxCas13d at hTR foci by imaging (Fig. 2B and SI Appendix, Fig. S5), consistent with observations in literature for other target RNAs (47, 48). A similar test with the catalytically dead PspCas13b (46, 48) using position-matched gRNAs also failed to give enrichment at hTR foci (SI Appendix, Fig. S6). Based on these negative results, we hypothesized that dRfxCas13d binding to hTR may require further optimization to enhance its stability and targeting.
Fig. 2.
Targeting APEX2 to human telomerase RNA with CRISPR-Cas13. (A) Design of dCas13d-dsRBD for improved hTR targeting. A sequence-independent double-stranded RNA binding domain (dsRBD) from the human protein kinase R (PKR) is fused to the C terminus of dRfxCas13d protein to further stabilize the hTR-gRNA RNA duplex. A bipartite nuclear localization sequence (BPNLS) is used in place of the SV40 NLS for improved nuclear localization. SI Appendix, Fig. S5 shows details of the optimization process. (B) Fluorescence imaging of dCas13d-dsRBD localization. HEK293T cells expressing the constructs shown in A along with hTR marker 3×FLAG-hTERT were fixed and stained with anti-HA antibody to visualize dCas13d (Alexa Fluor 488, green), anti-FLAG antibody to visualize hTR foci (Alexa Fluor 647, red), and DAPI (nuclei). Pixel intensity plots of the dashed lines are shown at Right (Scale bar, 10 µm.) SI Appendix, Fig. S8 shows additional fields of view. (C) Design of dCas13d-dsRBD-APEX2, hTR, and gRNA expression constructs used in this study. (D) Scheme of RNA immunoprecipitation for validation of dCas13d-dsRBD-APEX2 targeting to hTR. HEK293T cells stably expressing dCas13d-dsRBD-APEX2 are transfected with hTR and the indicated gRNA. RNA bound to APEX2 is immunoprecipitated with anti-V5 antibody followed by RT-qPCR quantitation. (E) RT-qPCR results from the experiment in D. The fold enrichment of hTR in cells transfected with hTR gRNA is normalized against the nontargeting (NT) gRNA control. Data are analyzed using a one-tailed Student’s t test (n = 3). (F) Biochemical analysis of biotinylated proteins from HEK293T cells stably expressing dCas13d-dsRBD-APEX2. hTR was overexpressed by transfection in all samples. Lysates were run on SDS/PAGE and analyzed by Ponceau stain (Left) or streptavidin blotting (Middle). Total eluted protein after streptavidin bead enrichment is visualized by silver staining (Right). Red arrows point to proteins differentially biotinylated by targeted vs. nontargeted APEX2.
We speculated that the interaction between dRfxCas13d and target RNA might be specifically enhanced by introducing a sequence-independent double-stranded RNA binding domain (dsRBD) (49) to recognize the gRNA-target RNA duplex (Fig. 2A) that forms only when the cognate target is bound. Such an interaction might help to cooperatively stabilize the dRfxCas13d-gRNA-target RNA ternary complex, whose structure (50) features a solvent-exposed dsRNA backbone accessible for additional binding. To test this strategy, we selected the dsRBD from human protein kinase R (PKR), which binds indiscriminately to dsRNA ≥ 16 bp (51–53) during viral infection, and fused it to dRfxCas13d to stabilize the 22-bp gRNA-target duplex. We found that, indeed, the addition of the dsRBD produced detectable enrichment of dRfxCas13d at nuclear foci by imaging (SI Appendix, Fig. S5). We then optimized the site of dsRBD fusion, nuclear localization sequence (NLS), and length of the linker before arriving at the final design (dCas13d-dsRBD, SI Appendix, Fig. S5). Remarkably, clear localization of dCas13d-dsRBD to the expected hTR foci was only observed with gRNA_5 (SI Appendix, Fig. S7), which targets nucleotides 148 to 169 in the single-stranded J2a/3 region of hTR. This region is highly variable in length and sequence across vertebrates (54), suggesting that it might play less of an important structural role than other conserved regions and serve as an accessible binding site for dCas13d. In a side-by-side comparison with the starting template dRfxCas13d (18), the optimized dCas13d-dsRBD formed nuclear foci that tightly overlapped with hTR foci marked by hTERT in the presence of gRNA_5, whereas nontargeting gRNA or original dRfxCas13d only yielded diffuse nuclear signal (Fig. 2B). The greater binding capability of dCas13d-dsRBD was also shown by RIP using another noncoding RNA, MALAT1, as target (SI Appendix, Fig. S9). dCas13d-dsRBD, but not original dCas13d, enriched endogenous MALAT1 transcript in the presence of a matched gRNA. Finally, we tested an endogenous mRNA target, ACTB, which encodes the beta-actin protein, but failed to observe enrichment in stress granules by imaging (SI Appendix, Fig. S10). Further optimization of binding affinity, dCas13d localization (e.g., different NLS), and reduction of background pools of untargeted dCas13 may be necessary to achieve specific targeting to cytosolic mRNAs.
Building upon the successful targeting of dCas13d-dsRBD to untagged hTR, we then generated an APEX2 fusion (dCas13d-dsRBD-APEX2, Fig. 2C) for proximity labeling. hTR binding to dCas13d-dsRBD-APEX2 was quantified by RIP-qPCR (Fig. 2D). We found that pulldown of dCas13d-dsRBD-APEX2 with target gRNA resulted in more than fourfold greater enrichment of hTR compared to a control using nontarget gRNA (Fig. 2E). The expected targeting of dCas13d-dsRBD-APEX2 fusion to foci was additionally confirmed by fluorescence microscopy (SI Appendix, Fig. S11).
We then performed a 1-min proximity labeling reaction in HEK293T cells expressing dCas13d-dsRBD-APEX2, followed by streptavidin bead enrichment of biotinylated proteins. The streptavidin-HRP blot in Fig. 2F shows that dCas13d-dsRBD-APEX2 exhibits robust H2O2-dependent biotinylation activity. In addition, the silver-stained gel of biotinylated proteins enriched from hTR-overexpressing cells with hTR gRNA vs. nontarget gRNA shows noticeable differences in banding patterns, consistent with the expectation that APEX2 is targeted to distinct neighborhoods using the two different gRNAs.
Proteomic Mapping of hTR Interacting Partners via APEX Proximity Labeling.
Previous studies (31–33) have suggested that hTR may have roles beyond serving as the RNA template for telomerase. However, the hTR interactome has not been systematically examined due to the shortage of methods for RNA-centered interactome mapping. Therefore, we performed a proteomic analysis of hTR-interacting proteins in living HEK293T cells, using the APEX RNA-targeting strategies developed above. For both strategies, we overexpressed hTR by ∼30-fold compared to endogenous levels of ∼750 hTR molecules per HEK293T cell (29) (SI Appendix, Fig. S12). Even with this overexpression, our hTR copy number of ∼2 × 104/cell is much lower than the endogenous levels of many other well-characterized noncoding RNAs such as U1, U2 (∼106/cell), and 7SK, RNaseP (∼105/cell) (29). Because our hTR is present in stoichiometric excess over telomerase protein components, our experiment would be expected to yield an interactome that goes beyond just the assembled telomerase complex and possibly identifies factors involved in the biogenesis, processing, modification, function, and degradation of hTR.
We designed a 10-plex tandem mass tag (TMT)-based quantitative proteomic experiment, in which half the samples used the MS2-MCP-based targeting strategy, and the other half used CRISPR-Cas13 to target APEX to hTR. Within each sample set, we performed two biological replicates of targeted APEX labeling (Fig. 3A, red/blue) and two biological replicates of the negative control with untargeted APEX (Fig. 3A, black). We also included negative control samples (Fig. 3A, gray) that omitted H2O2 in order to identify endogenously biotinylated proteins and nonspecific binders to the streptavidin beads. Clonal HEK293T cells stably expressing the indicated APEX fusion constructs were incubated with the biotin-phenol probe for 30 min prior to the 1-min H2O2 labeling reaction. All 10 samples were independently lysed, and their biotinylated proteomes enriched using streptavidin magnetic beads. Proteins were digested on-bead to peptides with trypsin, chemically labeled with TMT reagents, pooled, and then analyzed by liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS) (55).
Fig. 3.
Proteomic analysis of hTR interactome via proximity labeling. (A) Design of the proteomic experiment. HEK293T cells stably expressing the indicated APEX2 constructs are transfected with corresponding hTR and gRNA and then labeled with biotin-phenol as in Fig. 1A. Tandem mass tags (TMTs) are used for quantitative proteomic analysis. Each experimental set contains two replicates and negative controls with nontargeted APEX2 or H2O2 omitted. The different TMT channels are represented as 128C, 127C, 129N, 128N, 126C, 130C, 129C, 131N, 130N, and 127N. (B) Filtering of the proteomic data to identify enriched hTR interacting proteins. The table shows the number of proteins remaining after each filtering step. hTR specificity is calculated from interaction partners of known hTR binding proteins (SI Appendix, Table S1) in the BioGRID database (see Methods). Scatterplots in SI Appendix, Fig. S13 show a correlation between TMT ratios across replicates. (C) Gene ontology analysis of proteins enriched by MCP-APEX2 (Left) and dCas13d-dsRBD-APEX2 (Right ). Only a subset of significantly enriched GO terms related to hTR is shown. Node size scales with −log10 (P value). The number of proteins associated with each term is indicated inside each node. (D) hTR specificity analysis, calculated as in B, for the total human proteome (20,996 proteins), nuclear proteome (7,530 proteins), proteins enriched in three or more of our proteomic datasets (49 proteins), and proteins enriched in all four datasets (11 proteins). (E) Protein–protein interaction (PPI) map of proteins enriched in three or more of our proteomic datasets. Known RNA binding proteins are marked with blue stars. Proteins enriched in all four proteomic datasets are marked with black stars. Node size scales with protein biotinylation extent and node color scales with the ratio of enrichment in targeting vs. nontargeting control. Markov clustering was performed with PPI scores from the STRING database (see Methods). L2FC, log2 fold change.
The proteomic experiment identified more than 2,600 proteins with two or more unique peptides (Fig. 3B). Both MCP-APEX2 and dCas13d-dsRBD-APEX2 samples exhibited a high correlation between biological replicates (SI Appendix, Fig. S13, R2 > 0.9 for all). To remove potential contaminants and identify enriched proteins, we adopted a two-step, ratiometric filtering strategy used in our previous APEX studies (23, 56, 57). Because our APEX2 constructs are localized to the nucleus, we first filtered the data by using prior nuclear annotation (possible true positives) or prior mitochondrial or endoplasmic reticulum annotation (possible false positives) as a guide (Fig. 3B, filter 1). Histograms of detected proteins plotted by 128C/126C or 130C/127N TMT ratio show preferential biotinylation of nuclear proteins over false positives (SI Appendix, Fig. S14). We applied a false discovery rate (FDR) cutoff of 2% to remove potential contaminants. The remaining proteins were then filtered on the basis of their preferential biotinylation by targeted vs. untargeted APEX (Fig. 3B, filter 2). Bona fide hTR-interacting proteins should show greater enrichment by hTR-targeted APEX2 compared to nontargeted control (log2 fold change > 0), as seen for known hTR-binding partners DKC1 (58) and DGCR8 (59) (SI Appendix, Fig. S14). This resulted in the enrichment of 218 and 290 proteins for the two MCP-APEX2 datasets, and 153 and 181 proteins for the two dCas13d-dsRBD-APEX2 datasets. We further intersected the enriched proteins from the two biological replicates in each experiment to minimize contaminants and obtained final lists of 129 and 77 proteins in the MCP and dCas13d experiments, respectively (Fig. 3B and SI Appendix, Fig. S15). A total of 49 proteins were enriched in more than three datasets and 11 proteins were identified in all four datasets.
To separately assess the interactomes identified by MCP-APEX2 and dCas13d-dsRBD-APEX2, we first analyzed the protein lists using Gene Ontology (GO) (Fig. 3C). Both lists showed high enrichment of RNA binding and RNA processing proteins in the nucleus, as expected. Notably, proteins related to the Cajal body and small nuclear RNA binding, which are key in regulating hTR biogenesis and processing, were significantly enriched by both methods. Interestingly, the analysis also revealed protein groups unique to each dataset. For example, telomere maintenance and DNA binding terms were only identified by MCP-APEX2, whereas box H/ACA snoRNA binding and processing terms were selectively enriched by dCas13d-dsRBD-APEX2. Such differences may reflect the unique neighborhoods of the different hTR subregions targeted by each method: the 5′ end of hTR targeted by MCP-APEX2, compared to the J2a/3 region (nt 148 to 169 [22 nt] out of a total 451 nt for hTR) targeted by dCas13d-dsRBD-APEX2. The differences may also reflect the limitations of each method; for instance, the steric bulk of the tandem MS2 tags or dCas13d-dsRBD fusion could block certain interactions but not others. Overall, comparing the two proteomic datasets, we find that the MCP approach is more sensitive, giving a larger number of total interactions (129 vs. 77) and Cajal body proteins (6 vs. 3), while the dCas13d-dsRBD approach is more specific, giving a higher percentage (66% vs. 36%) of known RNA binding proteins.
Because of their respective strengths and tradeoffs, we pooled the data from both MS2-APEX replicates with the data from both dCas13-dsRBD-APEX replicates. A total of 49 proteins were enriched in three or more datasets, while 11 proteins were enriched in all four datasets. The fraction of proteins with prior literature connection (60) to hTR was 33% and 45% for the former and latter groups, respectively (Fig. 3D). In comparison, less than 3% of the human proteome and 7% of the total nuclear proteome exhibit hTR specificity using the same metrics. Each list also contained several proteins with no known connection to hTR (Dataset S1); these could be false positives, or they could be newly discovered hTR interactors. A literature examination suggests that the latter may be true for some of the hits. For example, RPL6, a nucleolar protein enriched in all four datasets (Fig. 3E), binds directly to nucleolin and NOP2 (60), both of which regulate telomerase function (61, 62). Another example is the uncharacterized protein C7ORF50, which was highly enriched by both biotinylation extent and targeted vs. untargeted APEX preferential biotinylation (Fig. 3E). Two telomerase components in yeast, CBF5 and NHP2 (orthologs of human telomerase proteins DKC1 and NHP2), interact genetically (63) and physically (64, 65) with the yeast ortholog of C7ORF50, YCR016W (which is also uncharacterized) (66). Perhaps C7ORF50/YCR016W is an unexplored player in hTR biology that is conserved from yeast to human.
To further analyze the 49 proteins we enriched in three or more datasets, we performed clustering based on prior protein–protein interaction evidence in the STRING database (67) (Fig. 3E). The largest cluster contains almost exclusively RNA binding proteins (15 out of 18), including the key telomerase component DKC1, the hTR degradation complex component DGCR8, and the core Cajal body component COIL. Other smaller groups that closely associate with the major cluster also contain numerous proteins related to hTR, such as the Cajal body component ICE2.
ALKBH5 Binding to hTR Regulates Telomerase Activity through m6A Modification.
We were intrigued by the RNA binding protein ALKBH5 (alkylated DNA repair protein alkB homolog 5), which was consistently enriched in all four proteomic datasets but has no prior connection to hTR biology nor known interaction with other hTR-related proteins that we enriched (Fig. 3E). ALKBH5 was recently identified as an RNA N6-methyladenosine (m6A) demethylase, one of two known to date (68). It catalyzes the removal of m6A from target RNAs and is implicated in the regulation of male fertility (68, 69), glioblastoma (70), and breast cancer (71). m6A is the most abundant internal modification in eukaryotic mRNA (72, 73), known to influence mRNA metabolism, including export, translation, and degradation (74, 75), but its existence and regulatory functions on noncoding RNA are much less explored.
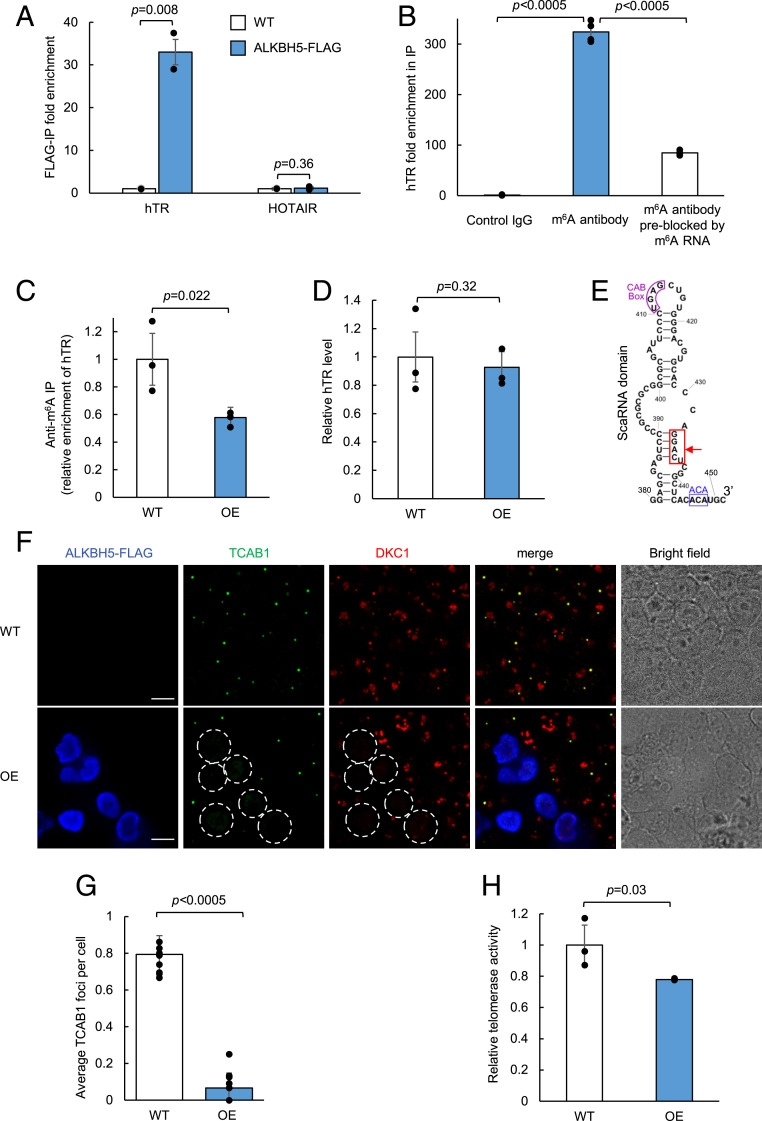
To validate our proteomic identification of ALKBH5 as a possible interaction partner of hTR, we performed an immunoprecipitation experiment. We note that published ALKBH5 CLIP-seq data (76) exist only for polyadenylated RNAs, not noncoding RNAs such as hTR. RT-qPCR analysis in Fig. 4A shows that immunoprecipitation of FLAG-tagged ALKBH5 enriches endogenous hTR but not a different nuclear noncoding RNA, HOTAIR. We next asked whether endogenous hTR is directly modified by m6A. A RIP experiment using anti-m6A antibody gave enrichment of hTR compared to negative controls (Fig. 4B). We further tested if overexpression of ALBKH5 reduces m6A levels on hTR; indeed, anti-m6A RIP enriches less hTR RNA when ALKBH5 is overexpressed, suggesting that ALKBH5 activity can reduce the levels of m6A on hTR (Fig. 4C). A control experiment showed that the total level of hTR was unchanged upon ALKBH5 overexpression (Fig. 4D).
Fig. 4.
Regulation of hTR function by m6A demethylase ALKBH5. (A) RT-qPCR of RNA immunoprecipitation for validation of ALKBH5 binding to hTR. Wild-type (WT) HEK293T cells are either mock transfected or transfected with ALKBH5-FLAG. RNA bound to ALKBH5 is immunoprecipitated with anti-FLAG antibody followed by RT-qPCR quantitation. Fold enrichment of the respective RNA in ALKBH5-FLAG-expressing cells is normalized against WT cells. Data are analyzed using a one-tailed Student’s t test (n = 2). (B) RNA immunoprecipitation followed by RT-qPCR of hTR. Total RNA extract from wild-type HEK293T cells is immunoprecipitated with either a control rabbit monoclonal IgG antibody, an anti-m6A rabbit monoclonal IgG antibody, or the m6A antibody preblocked with a synthetic m6A modified luciferase RNA. The immunoprecipitated RNA is quantified by RT-qPCR. Fold enrichment of hTR is normalized against the control IgG pulldown. Data are analyzed using a one-tailed Student’s t test (n = 4). (C) Anti-m6A immunoprecipitation of hTR. Total RNA extract from either WT or ALKBH5-overexpressing (OE) HEK293T cells is immunoprecipitated with an anti-m6A antibody and quantified by RT-qPCR. Fold enrichment of hTR in OE cells is normalized against the WT cells. Data were analyzed using a one-tailed Student’s t test (n = 3). (D) RT-qPCR of hTR expression level in WT and ALKBH5-OE cells. Total RNA extract from either WT or ALKBH5-OE HEK293T cells was quantified by RT-qPCR. hTR level in OE cells was normalized against WT cells. Data were analyzed using a one-tailed Student’s t test (n = 3). (E) Sequence and structure of nucleotides 380 to 451 in hTR. The m6A consensus motif GGACU is boxed in red with an arrow pointing to the putative methylated adenosine. The ACA box and CAB box motifs in the small Cajal body-specific RNA (scaRNA) domain are highlighted in blue and purple, respectively. (F) Fluorescence imaging of telomerase component TCAB1 and DKC1 in WT and ALKBH5-OE cells. WT or ALKBH5-OE HEK293T cells are fixed and immunostained with antibodies against endogenous TCAB1 (Alexa Fluor 488), DKC1 (Alexa Fluor 405), and ALKBH5-FLAG (anti-FLAG phycoerythrin conjugate). ALKBH5-OE cells are circled with white dotted lines (Scale bars, 10 µm.) (G) Quantitation of imaging data in E. The total number of TCAB1 foci is divided by the total number of WT cells or by the total number of ALKBH5-OE (FLAG-positive) cells in each field of view. Data were analyzed using a one-tailed Student’s t test (n = 10). (H) Telomerase activity assay in WT and ALKBH5-OE cells. The 1 × 106 WT or ALKBH5-OE HEK293T cells were analyzed using a commercialized PCR-based telomerase activity assay (TRAP). Telomerase activity in OE cells is normalized against the WT cells. Gel showing the resulting PCR products is shown in SI Appendix, Fig. S17. Data were analyzed using a one-tailed Student’s t test (n = 3).
If hTR is modified by m6A methylation, where are the modification sites? We surveyed previous m6A transcriptome-wide sequencing studies (SI Appendix, Fig. S16). Comparing datasets obtained via m6A antibody enrichment (77, 78) to those obtained via pulldown of various m6A regulators [writers METTL3 and METLL14 (79), eraser FTO (80), nuclear m6A reader YTHDC1 (81–83)], we found evidence for m6A modification in the H/ACA scaRNA domain of hTR. This domain contains a 5-nt GGACU sequence that matches the reported m6A consensus motif (Fig. 4E). Interestingly, the adenosine within this motif (A435) is in a predicted RNA duplex region, whose secondary structure could be affected by m6A modification (84). Previous studies have shown that the duplex structure of the scaRNA domain is important for the assembly of telomerase complex (28). Hence, m6A modification of hTR (regulated by ALKBH5) might be a mechanism to regulate telomerase assembly and hence function.
To explore this hypothesis, we probed two telomerase protein components in the context of HEK293T cells with or without ALKBH5 overexpression (which lowers m6A modification of hTR, Fig. 4C). TCAB1 and dyskerin (DKC1) are both well-characterized telomerase components that directly bind hTR at the H/ACA scaRNA domain, where the postulated m6A modification site is located. Fig. 4F shows that in wild-type HEK293T cells, endogenous TCAB1 forms discrete foci that colocalize with a subset of DKC1. However, when we overexpressed ALKBH5 in HEK293T cells, we observed a >10-fold reduction in both TCAB1 and DKC1 foci specifically in ALKBH5-overexpressing cells (outlined in Fig. 4F and quantified in Fig. 4G). To rule out the possibility that ALKBH5 directly affects the mRNAs encoding telomerase components (i.e., TERT, TCAB1, NHP2, NOP10, GAR1, and DKC1), we analyzed published CLIP-seq and differentially expressed mRNA datasets for ALKBH5 (68, 70, 76) and confirmed that those mRNAs are not targets of ALKBH5. This suggests that ALKBH5 promotes the disassembly of telomerase components, possibly via removal of m6A on hTR. In addition, we performed an assay of telomerase activity using the telomeric repeat amplification protocol (TRAP, see Methods). We found that ALKBH5 overexpression caused a 22% reduction in telomerase activity compared to controls (Fig. 4H), suggesting that ALKBH5 may also play a role in the down-regulation of telomerase activity.
Discussion
Systematic characterization of the protein interactome of specific cellular RNAs can improve our understanding of the function and regulation of diverse RNAs inside living cells. Mass spectrometry-based proteomic identification of RNA interactomes, however, has been hampered by the shortage of methods to efficiently target and enrich specific cellular RNAs along with their associated binding partners. Here, we developed two methods for RNA-centered profiling based on MS2 or CRISPR-Cas13 targeting of APEX for 1-min live-cell proximity labeling. The application of both methods to the noncoding RNA hTR recovered a list of candidate hTR interaction partners. We note that both approaches require careful screening of the MS2 stem-loop fusion site or CRISPR gRNA targeting regions to find accessible sites on the target RNA, and the regions are likely different for each method. In hTR, while the 5′ end is compatible with stem-loop fusions, it is poorly accessible to the 22-nt dRfxCas13d gRNA due to a much shorter (17 nt) single-stranded region.
BioID has previously been targeted to cellular RNAs using MS2 and BoxB hairpin motifs (12, 13). Our APEX-based approaches here complement these tools by enabling RNA interactome profiling on a 1-min time scale. While long labeling times can be beneficial to integrate interactors that may visit an RNA briefly but repeatedly over a long time window, short labeling times enable analysis and deconvolution of dynamic RNA interactomes, such as those that may occur during RNA processing and transport. APEX’s 1-min labeling also minimizes the toxicity associated with 1 mM H2O2 addition to cells. Previous studies (85–87) have shown that cellular stress responses generally occur on a longer time scale (>15 min) and with higher oxidant concentration. For in vivo applications, the requirement of biotin-phenol and H2O2 delivery makes APEX less suitable compared to BioID-based methods. Our study also introduces dCas13d-dsRBD as an alternative method to MS2 for targeting APEX to RNA, with the advantages of no tag on the RNA and higher proteome specificity.
Several recent studies describe similar RNA–protein interaction mapping methods that combine CRISPR-Cas13 with proximity labeling (14, 47, 88). The use of dPspCas13b or dLwCas13a by others, together with dRfxCas13d by us, demonstrates that many orthologs in the Cas13 family can potentially serve as targeting modules for RNA-centered proximity labeling. Notably, both Zhang et al. (14) and Li et al. (47) found that controlling the expression level of the Cas13 protein, via an inducible vector in a clonal cell line that is similar to our approach, is key for optimizing signal-to-noise ratio. In addition, these studies validated the application of different classes of proximity labeling enzymes, including BioID2 (47), APEX2 (88), and PafA (14), for promiscuous tagging of RNA-associated endogenous proteins. While others have chosen to study endogenous RNAs that are highly abundant [e.g., ACTB mRNA (47), total polyA RNAs (88), and the lncRNA NORAD (14)], our work demonstrates the profiling of an overexpressed target of interest in the case of a low-abundance RNA. Because the scope of accessible RNA regions for different Cas13 orthologs are likely to be different, and the labeling radius, kinetics, and chemistry vary widely among proximity labeling enzymes, these studies collectively expand the toolbox for investigating the landscape of a broad range of RNA interactions inside living cells.
Our strategies for targeting APEX to RNA should enable other applications as well, including electron microscopy (19) and RNA–RNA interactome mapping. The latter could be achieved using the recently developed APEX-seq approach (89), whereby APEX catalyzes the direct and covalent biotinylation of proximal RNAs. We anticipate a growing repertoire of RNA-centered proximity labeling tools to further expand our analysis of cellular RNAs.
An important limitation of our approach and other RNA proximity labeling studies (12, 13) is that they have only been demonstrated on overexpressed, and/or highly abundant cellular RNAs. Based on previous studies using MCP-GFP for single-molecule imaging of MS2-tagged mRNAs (90–92), it is likely that the MCP-APEX2 strategy should have sufficient sensitivity to be generally extensible to lower-abundance RNAs, albeit with an increase in the number of MS2 stem loops (to >10 stem loops, compared to the 4 used here). Our CRISPR-Cas13 approach, however, has much lower sensitivity, even with the introduction of the dsRBD domain. In this study, we were only able to observe APEX targeting to untagged hTR overexpressed via transfection, but not to endogenous hTR in HEK293T cells. While a small number of previous studies (48), including by us (17), have used CRISPR-Cas13 for imaging endogenous RNAs, the targets have been highly abundant cellular RNAs, and we have found that these published systems are not as effective as our dCas13d-dsRBD for targeting APEX to hTR specifically (SI Appendix, Fig. S6). Future improvements are needed to develop CRISPR-Cas13 systems that can broadly target endogenous, low-abundance cellular RNAs with high sensitivity.
In terms of proteome identification, the use of TMT labeling for quantitative comparison to reference control samples (untargeted APEX2) increased the spatial specificity of our protein lists. However, this ratiometric analysis workflow increases specificity at the expense of sensitivity, a tradeoff we also noted in our previous APEX studies of open subcellular compartments (23, 56). Dual-localized proteins that bind to hTR but also reside elsewhere in the nucleus tend to be removed by the ratiometric filtering step we performed to increase spatial specificity. For instance, we failed to enrich known hTR-interacting proteins PARN, EXOSC10, and TOE1 (59, 93–95), likely because of their known dual localization. Our analysis enriched the core telomerase protein DKC1 (Fig. 3E), but not TERT, TCAB1, NHP2, or NOP10. Interestingly, these four core telomerase proteins were not detected at all by our mass spectrometer, nor in previous APEX nuclear proteomes (96–98), suggesting they are low abundance in HEK293T cells or lack surface-exposed tyrosines for APEX biotinylation.
The utility of our method is exemplified by the discovery and validation of the hTR interactor ALKBH5. Though our proteomic experiment was performed with overexpressed hTR in stoichiometric excess over endogenous telomerase complex proteins, our resulting list of candidate protein interaction partners is highly enriched for RNA binding function and known hTR interactors. We performed biological followup on one of our most surprising hits—the m6A demethylase ALKBH5—and found that it binds to endogenous hTR to regulate its m6A modification extent. Additional experiments showed that changes in normal ALKBH5 expression level disrupt telomerase complex assembly and function.
Although m6A is predominantly understood as a regulator of mRNA stability and abundance, growing evidence suggests that it also has functions on noncoding RNAs. Recent work on the long noncoding RNA MALAT1 and its binding partner hnRNPC (84) showed that m6A can destabilize RNA duplexes to increase the accessibility of hairpin structures to RNA binding proteins. Interestingly, the putative methylated A (A435) in hTR is also within a duplex region and our data are consistent with methylation facilitating telomerase component binding. This raises the interesting possibility that the mechanism underlying m6A regulation of hTR is similar to that of MALAT1. Further experimentation, including mutagenesis at the putative m6A site of hTR, could shed light on the detailed molecular mechanism of this regulation.
Methods
Methods related to cloning, Western blots, proteomic sample preparation and analysis, additional data analysis, confocal fluorescence imaging, m6A pulldown assay, and telomerase activity assay are detailed in SI Appendix.
Cell Culture, Transfection, and Clonal Cell Line Construction.
HEK293T cells from the ATCC (American Type Culture Collection, passages <25) were cultured in a 1:1 DMEM (Dulbecco's Modified Eagle Medium)/MEM (Minimum Essential Medium Eagle) mixture (Cellgro) supplemented with 10% fetal bovine serum, 50 units/mL penicillin, and 50 mg/mL streptomycin at 37 °C under 5% CO2. For fluorescence microscopy imaging experiments, cells were grown on 7 × 7 mm glass coverslips in 48-well plates. To improve the adherence of HEK293T cells, we pretreated glass slides with 50 mg/mL fibronectin (Millipore) for 20 min at 37 °C before cell plating and washing three times with Dulbecco’s phosphate-buffered saline (DPBS) (pH 7.4). All constructs were transfected using Lipofectamine 2000 (Thermo) according to the manufacturer’s protocol. The amount of plasmids used for each experiment is detailed below. To generate the MCP-APEX2 clonal cells, a stable population was first created by infecting HEK293T cells with lentivirus containing MCP-APEX2 followed by blasticidin selection (8 µg/mL). Single colonies were then picked from the stable population using glass cloning cylinders (Millipore). To generate the dCas13d-dsRBD-APEX2 clonal cells, a stable population was first created by transfecting HEK293T cells with the dCas13d-dsRBD-APEX2 plasmid, followed by puromycin selection (1 µg/mL). To generate clonal cell lines from the drug-selected stable population, single colonies were picked with glass cloning cylinders (Millipore) through serial dilution. Individual clones were expanded and then tested for APEX2 expression level by either V5 antibody Western blotting or imaging of the P2A-GFP fluorescence upon addition of doxycycline (400 ng/mL). Clones with low expression are preferable for subsequent experiments because they have a reduced pool of unbound APEX protein that may contribute to background labeling.
APEX Labeling.
HEK293T cells stably expressing the appropriate APEX2 fusion were plated in T150 flasks at 70 to 80% confluency, 18 to 24 h prior to transfection. For the MCP-APEX2 samples, cells were transfected with 48 µg of the 4×MS2-hTR plasmid for 24 h prior to APEX labeling. For the dCas13d-dsRBD-APEX2 samples, cells were transfected with 48 µg of the hTR plasmid and 48 µg of the guide RNA plasmid, and doxycycline (400 ng/mL) was also added to induce expression for 24 h prior to APEX labeling. Biotin phenol (Iris Biotech) was dissolved in dimethyl sulfoxide as 500 mM stock solution and added directly to cell culture media to a final concentration of 500 µM. After incubating the cells for 30 min at 37 °C, H2O2 was spiked into the cell culture media to a final concentration of 1 mM to induce biotinylation. After 1 min of very gently swirling, the media was decanted as quickly as possible and the cells were washed three times with phosphate-buffered saline (PBS) containing 100 mM sodium azide, 100 mM sodium ascorbate, and 50 mM TROLOX (6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid). Cells were scraped and transferred to 15-mL Falcon tubes with ice-cold PBS, spun at 3,000 × g for 5 min, flash frozen in liquid nitrogen, and stored at −80 °C.
Streptavidin Bead Enrichment of Biotinylated Proteins.
APEX-labeled cell pellets from a T150 flask were used as input for each sample. The pellets were then lysed in RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 1% Triton X-100, protease mixture [Sigma-Aldrich], and 1 mM phenylmethylsulfonyl fluoride) for 5 min at 4 °C. The lysates were cleared by centrifugation at 15,000 × g for 10 min at 4 °C. Streptavidin-coated magnetic beads (Pierce) were washed twice with RIPA buffer, and 8 mg of total protein cell lysate from each sample was separately incubated with 450 µL of magnetic bead slurry with rotation for 1 h at room temperature or overnight at 4 °C. The beads were subsequently washed twice with 1 mL of RIPA lysis buffer, once with 1 mL of 1 M KCl, once with 1 mL of 0.1 M Na2CO3, once with 1 mL of 2 M urea in 10 mM Tris⋅HCl (pH 8.0), and twice with 1 mL RIPA lysis buffer. For streptavidin blot analysis, biotinylated proteins were then eluted by boiling the beads in 75 µL of 3× protein loading buffer supplemented with 20 mM dithiothreitol (DTT) and 2 mM biotin, and run on SDS-polyacrylamide gel electrophorsesis (PAGE) gel.
RNA Immunoprecipitation and Quantitative PCR.
RNA immunoprecipitation experiments to test the binding between MCP-APEX2 or dCas13d-dsRBD-APEX2 and the target hTR were performed as previously described in refs. 17 and 99 with minor modifications. Briefly, HEK293T cells stably expressing the corresponding APEX2 fusion were plated in six wells at 70 to 80% confluency, 18 to 24 h prior to transfection. For the MCP-APEX2 samples, cells were transfected with 3 µg of the 4×MS2-hTR plasmid. For the dCas13d-dsRBD-APEX2 samples, cells were transfected with 3 µg of the hTR plasmid and 3 µg of the guide RNA plasmid, and doxycycline (400 ng/mL) was also added to induce expression. At 48 h posttransfection, cells were fixed with 0.1% paraformaldehyde (Ricca Chemical) in DPBS for 10 min at room temperature, and then quenched with 125 mM glycine in PBS for 5 min. Cells were lysed in RIPA buffer supplemented with protease inhibitor mixture (Sigma-Aldrich) and RiboLock RNase inhibitor (Thermo) before centrifugation at 16,000 × g for 10 min at 4 °C. After saving 5% of the supernatant as input, the rest of the clarified lysate was then incubated with 25 μL Protein G magnetic beads (Pierce) coupled with 1 µL mouse anti-V5 (Life Technologies) antibody for 2 h at 4 °C with rotation. The beads were then pelleted and washed four times with RIPA buffer supplemented with 0.02% Tween-20 and RiboLock. Enriched RNAs were released from the beads in 100 μL of elution buffer (2% N-lauryl sarcoside, 10 mM EDTA, 5 mM DTT, in PBS supplemented with 200 μg proteinase K [Thermo] and RiboLock) at 42 °C for 1 h, followed by 55 °C for 1 h. Eluted samples were cleaned up using Agencourt RNAClean XP magnetic beads (Beckman Coulter) and eluted into 85 μL H2O. Thereafter, contaminating DNA was removed by digestion with RNase-free DNase I (Promega). Purified RNAs were again cleaned up using Agencourt RNAClean XP beads as above and eluted into 30 μL H2O. The resulting enriched RNA and input RNA is used as the template for cDNA synthesis using SuperScript III (Invitrogen) and quantified by qRT-PCR (Bio-Rad) with Maxima SybrGreen qPCR Master Mix (Thermo) and specific primers (SI Appendix, Table S3). The recovery of specific RNAs is calculated by dividing transcript abundance in the enriched sample by its input.
Proteomic Data Analysis.
LC-MS/MS data were analyzed using Spectrum Mill (Agilent) against a human Uniprot database (12/28/2017) that included 264 common laboratory contaminants. Individual TMT ratios for human proteins identified with at least two peptides were global median normalized prior to the filtering and thresholding described below. Each of the replicates was analyzed independently. To select cutoffs for proteins biotinylated by APEX2 over nonspecific bead binders (filter 1), we first classified three classes of proteins according to their Gene Ontology cellular component (GOCC) annotation (10/02/2019): 1) mitochondrial matrix, 2) endoplasmic reticulum, and 3) nucleus. Proteins that were annotated to be in either class 1 or 2 but not in class 3 were defined as false-positive hits, as they should not be biotinylated by our nucleus localized APEX2. We then ranked the data in each replicate by the enrichment ratio over the unlabeled negative control samples (i.e., 128C/126C, 129N/126C, 130C/127N, and 131N/127N) and calculated the false-positive rate we would obtain if we retained only proteins above that TMT ratio. A false-positive rate of <2% was chosen as cutoff for filter 1, which corresponds to cutoff at 0.4375, 0.3375, 0.6985, 0.622, for ratios 128C/126C, 129N/126C, 130C/127N, and 131N/127N, respectively. To select cutoffs for proteins that were preferentially biotinylated by APEX2 targeted to hTR vs. general nucleus-targeted APEX2 (filter 2), we ranked the data in each replicate by the enrichment ratio over nucleus-targeted APEX2 samples (i.e., 128C/127C, 129N/128N, 130C/129C, and 131N/130N) after the application of filter 1. The enrichment ratio is then normalized by the median of the remaining class 3 proteins to account for differences in total protein amount between samples within the TMT experiment. Proteins with enrichment ratio log2 fold change >0 were retained for further analysis. After applying filter 1 and filter 2 cutoffs, each of the two replicates in the MCP-APEX2 experiment or dCas13d-dsRBD-APEX2 experiment were intersected to produce the final proteomes of 129 and 77 proteins, respectively. To assess the specificity of the enriched proteins, we analyzed the hTR specificity and Gene Ontology enrichment, respectively. We defined hTR-specific proteins using a list of 10 known hTR-binding proteins curated from literature (SI Appendix, Table S1), their direct interaction partners in the BioGrid database (https://thebiogrid.org/), and components of Cajal body according to GOCC (10/02/2019). For Gene Ontology analysis, we uploaded the final proteomes to the Gene Ontology database search portal (geneontology.org/, 10/02/2019) on cellular compartment, biological process, or molecular function to retrieve the plotted terms with their corresponding P values (Fisher’s exact test with no correction). For the protein network analysis, the enriched proteins were clustered by their reported protein–protein interactions and corresponding confidence scores (STRING database, 10/02/2019) using a Markov clustering algorithm (cutoff 0.25, inflation value set at 3) and plotted in Cytoscape (v3.7.1).
ALKBH5 Pulldown Assay.
One 10-cm dish of HEK293T cells was transfected with 8 µg of ALKBH5-FLAG plasmid at 70 to 80% confluency for 24 h. The cells were rinsed with PBS three times, scraped off the plate in 10 mL of PBS, and collected by centrifugation. The cell pellets were lysed with 1 mL lysis buffer (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Nonidet P-40, 1:200 SUPERase•In [Invitrogen], 1:100 protease inhibitor mixtures [Sigma-Aldrich]) on ice for 10 min, then flash frozen with liquid nitrogen and stored at −80 °C. After thawing, the lysate was centrifuged at maximum speed and the supernatant was taken for pulldown. For target RNA pulldown, 20 µL suspension of anti-FLAG M2 magnetic beads (Sigma-Aldrich) were washed with lysis buffer three times and mixed with 500 µL of supernatant lysate. The bead mixture was incubated at 4 °C for 2 h with rotation, washed five times with lysis buffer, and eluted by adding TRIzol directly. The enriched RNA and input total RNA from 5% of lysate were purified using the Direct-zol RNA Purification Kit (Zymo). Target genes in input total RNA and enriched RNA were quantified by qRT-PCR using a SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen). Control group with untransfected HEK cells was processed with the same protocol in parallel.
Bioinformatic Analysis of Published Sequencing Datasets.
Reanalysis of the published datasets in SI Appendix, Fig. S16 was carried out with IGV software. PA-m6A-seq and miCLIP peak data (bed files) were downloaded directly from GEO (accession nos. GSE54921 and GSE63753). The remaining CLIP-seq peak data were acquired using the ENCORI starBase platform (100) (starbase.sysu.edu.cn/download.php). All entries were loaded into the same IGV session and aligned to the hg19 reference genome. The proposed modified region at the 3′ end of hTR was shown in SI Appendix, Fig. S16 (range defined by RefSeq Sequence marked with blue bar and TERC label) with center motif highlighted in the red box.
Supplementary Material
Acknowledgments
We thank Omar Abudayyeh and Jonathan Gootenberg (MIT) for help and advice on Cas13-related experiments and members of the A.Y.T. laboratory for technical support and advice. S.H. is supported by the Stanford Bio-X Bowes Graduate Fellowship. B.S.Z. was a Wu-Tsai Neurosciences Institute interdisciplinary scholar and is a Shurl and Kay Curci Foundation fellow of the Life Sciences Research Foundation. A.Y.T. is an investigator of the Chan Zuckerberg Biohub. C.H. is an investigator of the Howard Hughes Medical Institute. This work was supported by the National Institutes of Health (U24-CA210986 to S.A.C., U01-CA214125 to S.A.C., and R01-DK121409 to S.A.C. and A.Y.T.) and the Wu-Tsai Neurosciences Institute of Stanford.
Footnotes
C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006617117/-/DCSupplemental.
Data Availability.
Proteomics data and all log2 ratio values associated with each protein detected (with two or greater unique peptides) are available in Dataset S1.
References
- 1.Müller-McNicoll M., Neugebauer K. M., How cells get the message: Dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14, 275–287 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Lee S. R., Lykke-Andersen J., Emerging roles for ribonucleoprotein modification and remodeling in controlling RNA fate. Trends Cell Biol. 23, 504–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai B. P., Wang X., Huang L., Waterman M. L., Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol. Cell. Proteomics 10, M110.007385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon J. H., Srikantan S., Gorospe M., MS2-TRAP (MS2-tagged RNA affinity purification): Tagging RNA to identify associated miRNAs. Methods 58, 81–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong C., Popp M. W. L., Maquat L. E., Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods 58, 88–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogell B. et al., Specific RNP capture with antisense LNA/DNA mixmers. RNA 23, 1290–1302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu C. et al., Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McHugh C. A. et al., The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mili S., Steitz J. A., Evidence for reassociation of RNA-binding proteins after cell lysis: Implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jazurek M., Ciesiolka A., Starega-Roslan J., Bilinska K., Krzyzosiak W. J., Identifying proteins that bind to specific RNAs–focus on simple repeat expansion diseases. Nucleic Acids Res. 44, 9050–9070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faoro C., Ataide S. F., Ribonomic approaches to study the RNA-binding proteome. FEBS Lett. 588, 3649–3664 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Ramanathan M. et al., RNA-protein interaction detection in living cells. Nat. Methods 15, 207–212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee J. et al., β-Actin mRNA interactome mapping by proximity biotinylation. Proc. Natl. Acad. Sci. U.S.A. 116, 12863–12872 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z. et al., Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res. 48, e52 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han S., Li J., Ting A. Y., Proximity labeling: Spatially resolved proteomic mapping for neurobiology. Curr. Opin. Neurobiol. 50, 17–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roux K. J., Kim D. I., Raida M., Burke B., A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abudayyeh O. O. et al., RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konermann S. et al., Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam S. S. et al., Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee H. W. et al., Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paek J. et al., Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell 169, 338–349.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobingier B. T. et al., An approach to spatiotemporally resolve protein interaction networks in living cells. Cell 169, 350–360.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han S. et al., Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells. Cell Chem. Biol. 24, 404–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J. et al., Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180, 373–386.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branon T. C. et al., Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theimer C. A., Feigon J., Structure and function of telomerase RNA. Curr. Opin. Struct. Biol. 16, 307–318 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Chen J. L., Greider C. W., Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem. Sci. 29, 183–192 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q., Kim N. K., Feigon J., Architecture of human telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20325–20332 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xi L., Cech T. R., Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res. 42, 8565–8577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J. et al., The RNA component of human telomerase. Science 269, 1236–1241 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Ivanyi-Nagy R. et al., The RNA interactome of human telomerase RNA reveals a coding-independent role for a histone mRNA in telomere homeostasis. eLife 7, e40037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazzaniga F. S., Blackburn E. H., An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity. Blood 124, 3675–3684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kedde M. et al., Telomerase-independent regulation of ATR by human telomerase RNA. J. Biol. Chem. 281, 40503–40514 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Johansson H. E. et al., A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proc. Natl. Acad. Sci. U.S.A. 95, 9244–9249 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abudayyeh O. O. et al., C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu D., Collins K., Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell 11, 1361–1372 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Laprade H., et al. , Single-molecule imaging of telomerase RNA reveals a Recruitment – Retention model for telomere elongation. bioRxiv: 10.1101/2020.01.31.929026 (2020). Posted 2 February 2020. [DOI] [PubMed]
- 38.Jády B. E., Bertrand E., Kiss T., Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jády B. E., Richard P., Bertrand E., Kiss T., Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 17, 944–954 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebert M. D., Matera A. G., Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11, 4159–4171 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong F. L. et al., TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmoudi S. et al., WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 8, e1000521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortega Á. D., Willers I. M., Sala S., Cuezva J. M., Human G3BP1 interacts with β-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 123, 2685–2696 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Aulas A. et al., G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 209, 73–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.East-Seletsky A. et al., Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox D. B. T. et al., RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., et al. , CBRPP: A new RNA-centric method to study RNA-protein interactions. Cold Spring Harbor Lab. bioRxiv: 10.1101/2020.04.09.033290 (2020). Posted 9 April 2020. [DOI]
- 48.Yang L. Z. et al., Dynamic imaging of RNA in living cells by CRISPR-cas13 systems. Mol. Cell 76, 981–997.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Masliah G., Barraud P., Allain F. H. T., RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Cell. Mol. Life Sci. 70, 1875–1895 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang C. et al., Structural basis for the RNA-guided ribonuclease activity of CRISPR-cas13d. Cell 175, 212–223.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bevilacqua P. C., Cech T. R., Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35, 9983–9994 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Eguchi A. et al., Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat. Biotechnol. 27, 567–571 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryter J. M., Schultz S. C., Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J. L., Blasco M. A., Greider C. W., Secondary structure of vertebrate telomerase RNA. Cell 100, 503–514 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Thompson A. et al., Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Hung V. et al., Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 55, 332–341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loh K. H. et al., Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166, 1295–1307.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell J. R., Wood E., Collins K., A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999). [DOI] [PubMed] [Google Scholar]
- 59.Macias S., Cordiner R. A., Gautier P., Plass M., Cáceres J. F., DGCR8 acts as an adaptor for the exosome complex to degrade double-stranded structured RNAs. Mol. Cell 60, 873–885 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oughtred R. et al., The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khurts S. et al., Nucleolin interacts with telomerase. J. Biol. Chem. 279, 51508–51515 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Hong J., Lee J. H., Chung I. K., Telomerase activates transcription of cyclin D1 gene through an interaction with NOL1. J. Cell Sci. 129, 1566–1579 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Costanzo M. et al., A global genetic interaction network maps a wiring diagram of cellular function. Science 353, 42–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogan N. J. et al., Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Schwer B., Erdjument-Bromage H., Shuman S., Composition of yeast snRNPs and snoRNPs in the absence of trimethylguanosine caps reveals nuclear cap binding protein as a gained U1 component implicated in the cold-sensitivity of tgs1Δ cells. Nucleic Acids Res. 39, 6715–6728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cherry J. M. et al., Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szklarczyk D. et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng G. et al., ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang C. et al., ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U.S.A. 115, E325–E333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang S. et al., m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang C. et al., Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U.S.A. 113, E2047–E2056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desrosiers R., Friderici K., Rottman F., Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perry R. P., Kelley D. E., Friderici K., Rottman F., The methylated constituents of L cell messenger RNA: Evidence for an unusual cluster at the 5′ terminus. Cell 4, 387–394 (1975). [DOI] [PubMed] [Google Scholar]
- 74.Zhao B. S., Roundtree I. A., He C., Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi H., Wei J., He C., Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baltz A. G. et al., The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Chen K. et al., High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 54, 1587–1590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Linder B. et al., Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J. et al., A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartosovic M. et al., N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu C. et al., Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Xiao W. et al., Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Roundtree I. A. et al., YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu N. et al., N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakamura J., Purvis E. R., Swenberg J. A., Micromolar concentrations of hydrogen peroxide induce oxidative DNA lesions more efficiently than millimolar concentrations in mammalian cells. Nucleic Acids Res. 31, 1790–1795 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Daroui P., Desai S. D., Li T. K., Liu A. A., Liu L. F., Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J. Biol. Chem. 279, 14587–14594 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Sablina A. A. et al., The antioxidant function of the p53 tumor suppressor. Nat. Med. 11, 1306–1313 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin X., Lawrenson K., In vivo analysis of RNA proximity proteomes using RiboPro. bioRxiv: 10.1101/2020.02.28.970442 (2020). Posted 28 February 2020. [DOI]
- 89.Fazal F. M. et al., Atlas of subcellular RNA localization revealed by APEX-seq. Cell 178, 473–490.e26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu B., Buxbaum A. R., Katz Z. B., Yoon Y. J., Singer R. H., Quantifying protein-mRNA interactions in single live cells. Cell 162, 211–220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu B., Eliscovich C., Yoon Y. J., Singer R. H., Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grünwald D., Singer R. H., In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moon D. H. et al., Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet. 47, 1482–1488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shukla S., Schmidt J. C., Goldfarb K. C., Cech T. R., Parker R., Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol. 23, 286–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deng T. et al., TOE1 acts as a 3′ exonuclease for telomerase RNA and regulates telomere maintenance. Nucleic Acids Res. 47, 391–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao X. D. et al., C-BERST: Defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat. Methods 15, 433–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Myers S. A. et al., Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat. Methods 15, 437–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benhalevy D., Anastasakis D. G., Hafner M., Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods 15, 1074–1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaewsapsak P., Shechner D. M., Mallard W., Rinn J. L., Ting A. Y., Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife 6, e29224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J. H., Liu S., Zhou H., Qu L. H., Yang J. H., starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data and all log2 ratio values associated with each protein detected (with two or greater unique peptides) are available in Dataset S1.