Significance
Depletion of the neural stem cell (NSC) pool is a major driver of age-related regenerative decline in the hippocampus. While increased quiescence is a major contributor to this decline, NSCs can also undergo terminal differentiation into astrocytes, thus restricting the stem cell pool. The mechanisms underlying this fate switch and their relation to age-related regenerative decline have not yet been fully elucidated. In this study, we report an age-related decline in NSC O-GlcNAcylation, coincident with reduced neurogenesis and increased gliogenesis. We identify loss of O-GlcNAcylation at STAT3 T717 in the hippocampus with age, and demonstrate that O-GlcNAcylation of this site is a critical determinant of NSC fate. Our work expands our understanding of how posttranslational modifications influence the aging brain.
Keywords: neural stem cells, aging, O-GlcNAcylation, neurogenesis, gliogenesis
Abstract
Increased neural stem cell (NSC) quiescence is a major determinant of age-related regenerative decline in the adult hippocampus. However, a coextensive model has been proposed in which division-coupled conversion of NSCs into differentiated astrocytes restrict the stem cell pool with age. Here we report that age-related loss of the posttranslational modification, O-linked β-N-acetylglucosamine (O-GlcNAc), in NSCs promotes a glial fate switch. We detect an age-dependent decrease in NSC O-GlcNAc levels coincident with decreased neurogenesis and increased gliogenesis in the mature hippocampus. Mimicking an age-related loss of NSC O-GlcNAcylation in young mice reduces neurogenesis, increases astrocyte differentiation, and impairs associated cognitive function. Using RNA-sequencing of primary NSCs following decreased O-GlcNAcylation, we detected changes in the STAT3 signaling pathway indicative of glial differentiation. Moreover, using O-GlcNAc–specific mass spectrometry analysis of the aging hippocampus, together with an in vitro site-directed mutagenesis approach, we identify loss of STAT3 O-GlcNAc at Threonine 717 as a driver of astrocyte differentiation. Our data identify the posttranslational modification, O-GlcNAc, as a key molecular regulator of regenerative decline underlying an age-related NSC fate switch.
Within the dentate gyrus (DG) region of the adult mammalian hippocampus, neural stem cells (NSCs) undergo rounds of division and maturation, in a process termed adult neurogenesis, ultimately giving rise to functional neurons that integrate into existing hippocampal circuitry and regulate cognition (1, 2). While much less understood, adult NSCs can alternatively undertake a gliogenic program in which they terminally differentiate into astrocytes (3–6). During aging, neurogenesis precipitously declines in the adult hippocampus (7–11). While adult NSCs in the aging hippocampus remain somewhat controversial in humans (12), the most recent studies report adult hippocampal neurogenesis through the ninth decade of life, with age-related decline exacerbated in Alzheimer’s disease patients (13) and correlating with cognitive dysfunction (14). It is therefore becoming evident that adult NSCs provide a significant source of cellular and cognitive plasticity in the adult brain. However, mechanisms governing age-related neurogenic decline remain to be fully elucidated.
The posttranslational modification O-linked β-N-acetylglucosamine (O-GlcNAc), a dynamic form of intracellular protein glycosylation, is quickly emerging as a potent regulator of brain aging (15–17). Recently, we identified age-related changes in O-GlcNAcylation associated with neuronal dysfunction in old mice (17). Moreover, the catalytic enzymes responsible for the addition and removal of O-GlcNAc, O-GlcNAc transferase (Ogt), and O-GlcNAcase (Oga), have been implicated in Alzheimer’s disease (15, 18, 19), Parkinson’s disease (20), and even lifespan regulation (21, 22). In the context of stem cell biology, previous studies have identified a critical role for O-GlcNAc in embryonic stem cell pluripotency (23) and proper organismal development (24). Additionally, aberrant neural development has been observed following abrogation of either Ogt or Oga (25, 26). Despite involvement in both aging and development, a role for O-GlcNAcylation in regulating age-related regenerative decline in any tissue, including the brain, has yet to be explored.
In this study, we used pharmacological and RNA interference (RNAi) in vitro approaches, in conjunction with RNA-sequencing, to demonstrate that decreasing NSC O-GlcNAcylation promotes a neuron to glia fate switch. Functionally, we utilize genetic knockout (KO) models to demonstrate that decreasing NSC O-GlcNAcylation in young animals mirrors age-related changes in adult neurogenesis, cognitive function, and gliogenesis. Mechanistically, we employ mass spectrometry to identify an age-related loss of STAT3 O-GlcNAcylation at Threonine 717 (T717) in the hippocampus that drives a gliogenic fate switch in NSCs.
Results
NSC O-GlcNAC Levels Decline with Age and Are Coincident with Decreased Neurogenesis and Increased Gliogenesis in the Mature Hippocampus.
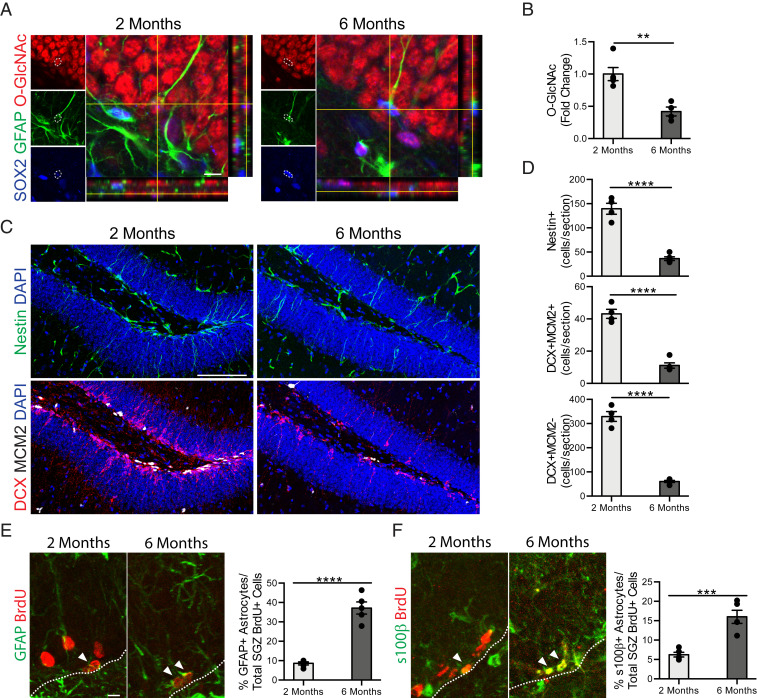
We first compared age-related temporal kinetics of O-GlcNAc expression in adult NSCs with changes in adult neurogenesis and gliogenesis in the hippocampus of mice at young (2 mo) and mature (6 mo) ages by immunohistochemical analysis. We detected a sharp decrease in O-GlcNAc levels in Sox2/GFAP+ NSCs in mature compared to young mice (Fig. 1 A and B). Concurrently, this decline was paralleled by a decrease in the number of Nestin+ NSCs, doublecortin (Dcx)/MCM2+ neuroblasts, and Dcx+/MCM2− newly born neurons in the mature hippocampus (Fig. 1 C and D), consistent with previous reports (7, 10). While glial differentiation has been reported at old age (3, 27), whether the decline in adult neurogenesis in the mature hippocampus is concurrent with an increase in gliogenesis is unknown. Therefore, we assessed astrocyte differentiation using a long-term 5-bromo-2′-deoxyuridine (BrdU) incorporation paradigm, in which differentiated astrocytes express both BrdU and the glial markers GFAP or S100β. We observed an age-dependent increase in the number of long-term retaining BrdU/GFAP+ and BrdU/S100β+ astrocytes in the mature compared to the young hippocampus (Fig. 1 E and F), in a timeframe consistent with decreased neurogenesis. These data raise the possibility that decreased NSC O-GlcNAcylation in the mature hippocampus is associated with age-related regenerative decline.
Fig. 1.
Age-related decreased NSC O-GlcNAc levels is coincident with a decline in neurogenesis and an increase in gliogenesis in the mature hippocampus. (A) Representative single z-planes and orthogonal projections of a confocal image z-stack of 2- or 6-mo-old mouse hippocampus, labeled with anti-Sox2, anti-GFAP, anti–O-GlcNAc, and DAPI. (Scale bar, 10 µm.) (B) Quantification of the fold-change of the average O-GlcNAc intensity normalized to area measured in each GFAP+/Sox2+ NSC across all z-stacks per animal (n = 4 animals per group). (C) Representative maximum intensity projections of a confocal image z-stack of 2- or 6-mo-old mouse hippocampus, labeled with DAPI and anti-Nestin (Upper) or DAPI, anti-Dcx, and anti-Mcm2 (Lower). (Scale bar, 100 µm.) (D) Quantification of Nestin+ radial NSCs (Top), Dcx+/MCM2+ proliferating neuroblasts (Middle), and Dcx+/MCM2− immature neurons (Bottom) in 2- or 6-mo-old mice (n = 4 to 5 animals per group). (E) Representative maximum intensity projections of a confocal image z-stack of 2- or 6-mo-old mouse hippocampus, labeled with anti-GFAP and anti-BrdU. Mice were given six daily intraperitoneal injections of BrdU (50 mg/kg) 30 d before killing. Quantification of the percentage of GFAP+/BrdU+ astrocytes out of total BrdU+ cells in the subgranulzar zone (SGZ) of the DG (n = 4 to 5 animals per group; arrowheads, astrocytes; white dashed line, SGZ). (Scale bar, 10 µm; also applies to F.) (F) Representative maximum-intensity projections of a confocal image z-stack of 2- or 6-mo-old mouse hippocampus, labeled with anti-s100β and anti-BrdU (Left). Mice were treated as in E. Quantification of the percentage of s100β+/BrdU+ astrocytes out of total BrdU+ cells in the SGZ of the DG (n = 4 to 5 animals per group; arrowheads, astrocytes; white dashed line SGZ). Data are represented as mean ± SEM; **P < 0.01, ***P < 0.001, ****P < 0.0001; t test.
Decreased NSC O-GlcNAc Levels Impairs Proliferation and Promotes Astrocyte Differentiation In Vitro.
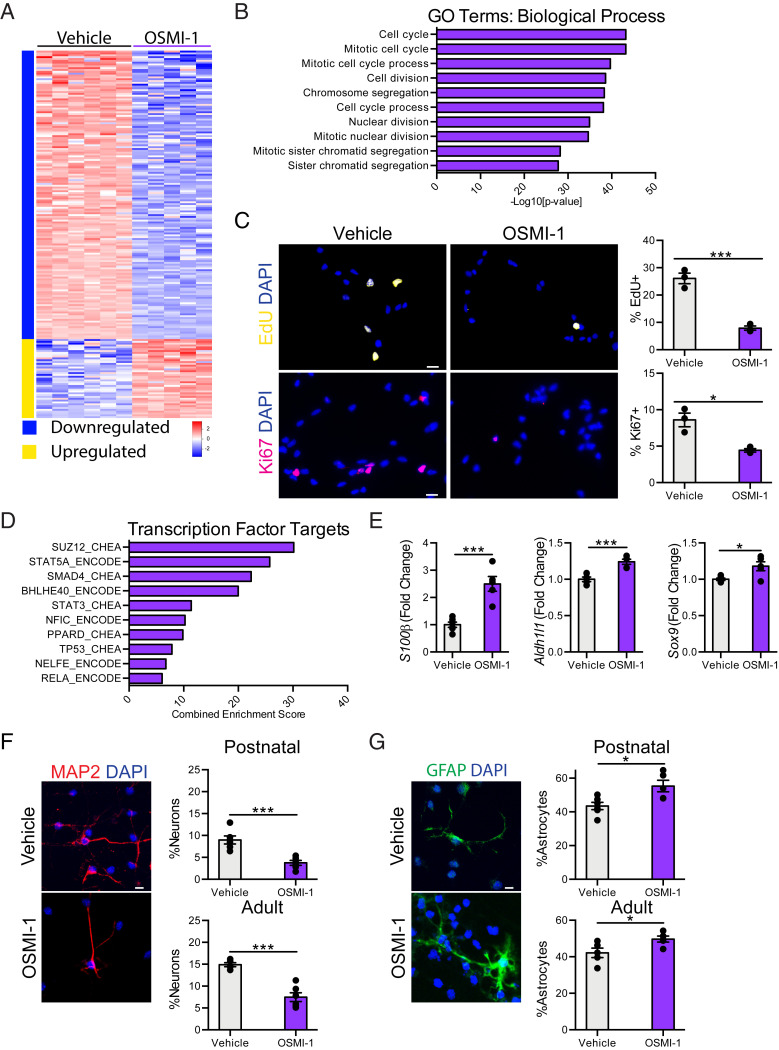
To gain insight into the functional consequence of reduced NSC O-GlcNAcylation, we made use of a primary hippocampal NSC in vitro model. Using RNA-sequencing, we profiled gene expression in primary NSCs following either pharmacological inhibition of Ogt with OSMI-1 treatment (SI Appendix, Fig. S1) or viral-mediated abrogation of Ogt by RNAi (SI Appendix, Fig. S2). We found 845 differentially expressed genes following Ogt inhibition (SI Appendix, Fig. S2) and 741 differentially expressed genes after Ogt abrogation (SI Appendix, Fig. S3A). Gene ontology (GO) analysis of the most differentially down-regulated genes identified biological processes, including cell cycle, mitotic nuclear division, and cell division (Fig. 2B and SI Appendix, Fig. S3B). Functionally, we assessed NSC proliferation in vitro following inhibition or abrogation of Ogt, and observed a significant decrease in the percentage of 5-Ethynyl-2′-deoxyuridine (EdU)+ and Ki67+ proliferating cells compared to control conditions (Fig. 2C and SI Appendix, Fig. S3C). No differences in cell death were detected by immunocytochemical analysis for cleaved caspase-3 (SI Appendix, Fig. S4A). These in vitro data indicate that reducing NSC O-GlcNAcylation negatively regulates NSC proliferation.
Fig. 2.
Decreased NSC O-GlcNAcylation promotes a neuron to glia fate switch in vitro. (A) Heatmap of significantly differentially expressed genes with fold-change > 2 and P < 0.05 in primary hippocampal NSCs following 24-h 12.5 µM OSMI-1 treatment compared to vehicle control in vitro. (B) Biological process GO terms associated with genes down-regulated following OSMI-1 treatment. (C) Representative field and quantification of primary mouse hippocampal NSCs following 24 h of vehicle (Left) or OSMI (Right) treatment, labeled with DAPI and an 8-h EdU-pulse (Upper), or DAPI and anti-Ki67 (Lower) (n = 3 per group). (Scale bars, 10 µm.) (D) Quantification of enriched transcription factor targets as identified by ChEA or ENCODE based on up-regulated genes after 24-h OSMI-1 treatment compared to vehicle control. (E) Fold-change expression (FPKM) of astrocyte-associated genes: S100β, Aldh1l1, and Sox9 in primary hippocampal NSCs following OSMI-1 treatment in vitro (n = 5 to 6 per group). (F) Differentiation was induced in vehicle or OSMI-1 treated primary postnatal (P1) and adult (2-mo-old) hippocampal NSCs by withdrawing growth factors 7 d prior to fixation. Cells were labeled with DAPI and anti-MAP2. MAP2+ neurons were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (Scale bar, 10 µm.) (G) Primary postnatal and adult hippocampal NSCs were treated as in F. Cells were labeled with DAPI and anti-GFAP. GFAP+ astrocytes were quantified as a percentage of total DAPI+ cells (n = 6 per group). (Scale bar, 10 µm.) Data are represented as mean ± SEM; *P < 0.05, ***P < 0.001; t test.
Given the relatively low number of significantly up-regulated genes following Ogt inhibition (Fig. 2A), we performed gene list enrichment analysis using Enrichr for chromatin immunoprecipitation (ChIP) enricher analysis (ChEA) and ENCODE transcription factor targets datasets. We identified transcription factors previously associated with NSC function, including STAT3 (Fig. 2D and SI Appendix, Fig. S3D), a major initiator of astrocyte differentiation (28, 29). Correspondingly, we surveyed expression of known astrocyte genes in NSCs following Ogt inhibition using our RNA-sequencing dataset, and observed an up-regulation of several astrocyte genes, including S100β, Aldh1l1, and Sox9 (Fig. 2E). Next, we examined whether reducing NSC O-GlcNAc levels impacted NSC differentiation in vitro. Primary hippocampal NSCs isolated from either postnatal or adult (2 mo) mice were treated with the Ogt inhibitor OSMI-1 or vehicle control and cultured under differentiation conditions. Ogt inhibition resulted in a concomitant decrease in neuronal (Fig. 2F) and increase in astrocyte (Fig. 2G) differentiation compared to vehicle control conditions. To ensure that these changes in cell fate were not due to increased cell death during differentiation, we analyzed cytotoxicity (SI Appendix, Fig. S4B) and cell viability (SI Appendix, Fig. S4C) throughout the paradigm and observed no difference between Ogt inhibitor-treated and control NSCs. These data suggest that the observed loss of proliferative capacity and neuronal differentiation following decreased NSC O-GlcNAcylation (Fig. 1) may be the result of an altered neuron to glia fate switch.
Mimicking an Age-Related Decline in NSC O-GlcNAcylation Impairs Adult Neurogenesis and Cognitive Function and Increases Gliogenesis in the Young Hippocampus.
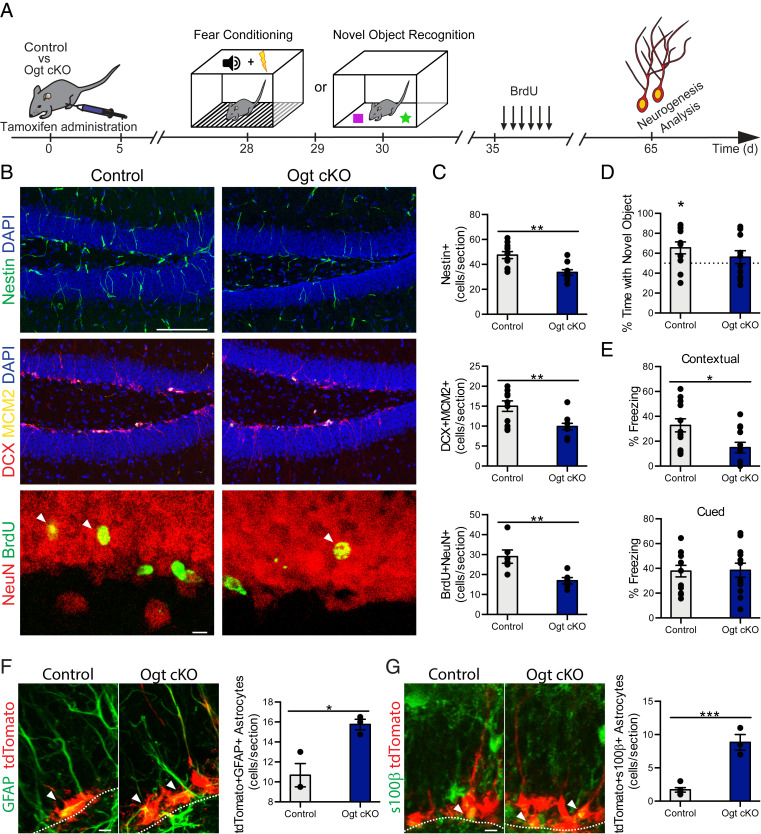
To understand the relationship between O-GlcNAc expression and adult neurogenesis in vivo, we asked whether mimicking an age-related decline in NSC O-GlcNAcylation within the adult hippocampus would impair neurogenesis. To resolve the role of O-GlcNAc in a temporally defined and cell-type–specific manner in the adult brain, we generated male Ogtflox/y mice carrying an inducible NestinCre-ERT2 gene, in which the X-linked Ogt gene is excised specifically in adult NSCs (Ogt cKO) upon tamoxifen administration (Fig. 3A). Young (2 mo) mice were administered tamoxifen and changes in neurogenesis were assessed in young adult (3 mo) Ogt cKO and littermate control (Ogtflox/y) mice by immunohistochemical analysis. Loss of Ogt expression in adult NSCs resulted in a decrease in the number of Nestin+ NSCs and Dcx/MCM2+ neuroblasts in the hippocampus of Ogt cKO compared to littermate control animals (Fig. 3 B and C). We assessed neuronal differentiation and survival using a long-term BrdU incorporation paradigm, in which mature differentiated neurons express both BrdU and the neuronal marker NeuN. We observed a decrease in the number of long-term retaining BrdU/NeuN+ mature differentiated neurons Ogt cKO compared to control animals (Fig. 3 B and C). To ensure that these differences in neurogenesis were not due to increased cell death, we compared the number of cleaved caspase-3+ cells and observed no difference in cell death between Ogt cKO and littermate control mice (SI Appendix, Fig. S5). To corroborate our genetic studies, we abrogated hippocampal Ogt expression in wild-type animals utilizing an in vivo viral-mediated RNAi approach. Young adult (3 mo) animals were stereotaxically injected with high-titer lentivirus encoding Ogt or control shRNA sequences into the DG of contralateral hippocampi (SI Appendix, Fig. S6A). Decreased Ogt in the DG resulted in a significant decrease in the number of Dcx+/EdU+ neuroblasts and long-term retaining BrdU/NeuN+ neurons compared with the control contralateral DG (SI Appendix, Fig. S6B). Collectively, our genetic and in vivo RNAi data indicate that loss of NSC O-GlcNAcylation mirror an aging condition by negatively regulating neurogenesis in the adult hippocampus.
Fig. 3.
Decreased NSC O-GlcNAcylation impairs adult neurogenesis and cognitive function while increasing gliogenesis in the young hippocampus. (A) Schematic of experimental paradigm and cognitive testing timeline. Three-month-old Ogtflox/y (control) or NestinCre-ERT2+/−; Ogtflox/y (Ogt cKO) littermate mice were administered five daily intraperitoneal tamoxifen injections (75 mg/kg). One month later, hippocampal-dependent learning and memory were assessed by NOR or contextual fear-conditioning paradigms. One week after behavioral assays, mice were administered six daily intraperitoneal injections of BrdU (50 mg/kg). Thirty days later mice were killed, and tissue was analyzed. (B) Representative maximum-intensity projections of a confocal image z-stack of control or Ogt cKO hippocampus, labeled with DAPI and anti-Nestin (Top) or DAPI, Dcx, and anti-Mcm2 (Middle), or anti-NeuN and anti BrdU (Bottom). (Scale bar, 100 µm, Top or 10 µm, Bottom.) (C) Quantification of Nestin+ radial NSCs (Top), Dcx+/MCM2+ proliferating neuroblasts (Middle), and NeuN+/BrdU+ newborn mature neurons (Bottom) in control or Ogt cKO littermate mice (n = 10 to 11 animals per group). (D) Object recognition memory was assessed in control and Ogt cKO littermate mice using the NOR task. Quantification of percentage of time spent with the novel object is shown. (n = 11 to 12 per group). (E) Associative fear memory was assessed in Ogt cKO and control littermate mice using contextual fear conditioning. Quantification of percentage freezing in response to cue or context 24 h after training is shown (n = 12 per group). (F) Three-month-old NestinCre-ERT2+/−; Ai14flox/wt (control) or NestinCre-ERT2; Ai14flox/wt; Ogtflox/y (Ogt cKO) littermate mice were administered five daily intraperitoneal tamoxifen injections (75 mg/kg). One month later, mice were killed and tissue was analyzed. Representative maximum intensity projections of a confocal image z-stack of control or Ogt cKO hippocampus, labeled with endogenous TdTomato and anti-GFAP. Quantification of TdTomato+/GFAP+ astrocytes (n = 3 to 5 per group; arrowheads, astrocytes; white dashed line, SGZ). (Scale bar, 10 µm.) (G) Mice were treated as in F. Representative maximum-intensity projections of a confocal image z-stack of control or Ogt cKO hippocampus, labeled with endogenous TdTomato and anti-s100β. Quantification of TdTomato+/s100β + astrocytes (n = 3 to 5 per group; arrowheads, astrocytes; white dashed line, SGZ). (Scale bar, 10 µm.) Data are represented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; t test (C and E–G); one-sample t test versus 50% (D).
To investigate whether mimicking an age-related decline in NSC O-GlcNAcylation also impaired cognitive processes associated with adult neurogenesis, we assessed hippocampal-dependent object recognition and associative fear memory using novel object recognition (NOR) and contextual fear-conditioning paradigms, respectively (Fig. 3A). In the testing portion of the NOR task, control animals exhibited a significant preference for the novel object over the familiar object; however, this preference was lost in Ogt cKO mice (Fig. 3D). During contextual fear-conditioning testing, we observed decreased freezing in Ogt cKO mice compared to controls animals (Fig. 3E). No differences were observed in baseline freezing during fear-conditioning training (SI Appendix, Fig. S7A) or in cued fear-conditioning testing between genotypes (Fig. 3E). As a control, we profiled general health using an open-field paradigm (SI Appendix, Fig. S7B) and observed no differences in overall activity, total distance traveled, or time spent in the periphery or center of the open field, indicative of normal motor and anxiety functions (SI Appendix, Fig. S7 C–E). These behavioral data demonstrate that decreased adult NSC O-GlcNAcylation impairs hippocampal-dependent learning and memory.
Next, to examine the relationship between O-GlcNAcylation and gliogenesis, we asked whether mimicking an age-related decline in NSC O-GlcNAc levels would promote astrocyte differentiation using a genetic lineage-tracing approach. We generated male Ogt cKO and control mice carrying a loxP-flanked STOP cassette preventing transcription of a CAG-promoter driven tdTomato (tdT) reporter inserted into the Gt(Rosa)26Sor locus, in which tdT is expressed in adult NSCs upon tamoxifen administration. Young (2 mo) mice were administered tamoxifen and changes in gliogenesis were assessed in young adult (3 mo) Ogt cKO and littermate control mice by immunohistochemical analysis. Efficacy of our knockout model was confirmed by immunohistochemistry (SI Appendix, Fig. S8). Loss of Ogt expression in adult NSCs resulted in an increase in the number of tdT/GFAP+ and tdT/S100β+ astrocytes in the hippocampus of Ogt cKO compared to control animals (Fig. 3 F and G). Our in vivo lineage tracing analysis identify an increase in adult gliogenesis in young mice following decreased NSC O-GlcNAcylation that reflect age-related changes in the mature hippocampus (Fig. 1 E and F).
O-GlcNAcylation of STAT3 at T717 Is Reduced in the Aging Hippocampus and Promotes a Glial Fate Switch in NSCs.
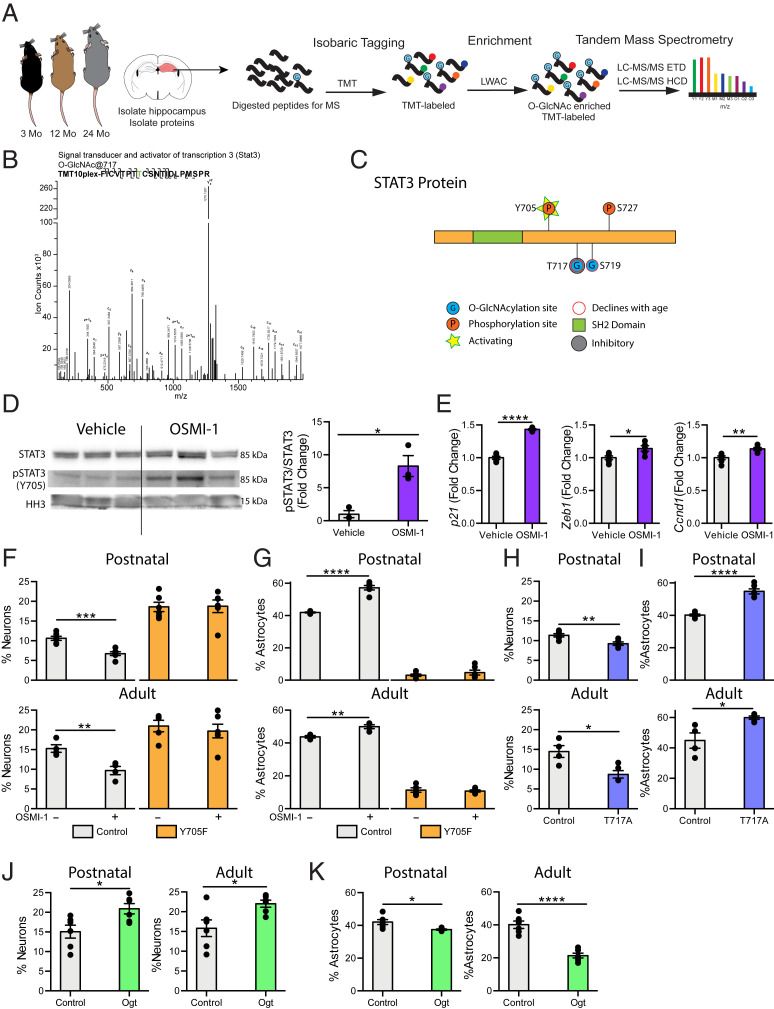
To gain mechanistic insight into site-specific changes in O-GlcNAcylation potentially regulating impaired NSC function during aging, we performed comparative O-GlcNAc–specific proteomics analysis of the hippocampus at ages before and after changes in adult neurogenesis and gliogenesis (Fig. 4A). Due to technical limitations in the number of aged animals necessary to obtain sufficient amounts of primary NSCs to enrich for dynamic posttranslational modifications, we opted to subdissect and pool hippocampi from young adult (3 mo), middle aged (12 mo), and old (24 mo) mice. We isolated total hippocampal protein, trypsin digested, and labeled the resulting peptides with isobaric tandem mass tags (TMT) before enriching for O-GlcNAcylated peptides using lectin weak-affinity chromatography (LWAC). Glycopeptides were analyzed by electron-transfer/higher-energy collision dissociation (EThcD) mass spectrometry. We detected 1,344 O-GlcNAcylation sites across 474 unique proteins (Dataset S1). We next examined mechanistic synergies between decreased sites of protein O-GlcNAcylation in the aging hippocampus (Dataset S1) and cell-type–specific molecular changes elicited in NSCs by decreased O-GlcNAc levels (Fig. 2 and SI Appendix, Fig. S3), and selectively identified STAT3. A representative EThcD spectrum used to resolve an unambiguous O-GlcNAcylation site on a peptide from the STAT3 protein (Fig. 4B), as well as a schematic depicting an age-related decrease in STAT3 O-GlcNAcylation at T717 and Serine 719 (S719) (Fig. 4C) are provided. Collectively, our proteomic analysis of the aging hippocampus, together with RNA-sequencing of primary NSCs, indicate changes in STAT3 signaling downstream of age-related decreased O-GlcNAcylation.
Fig. 4.
STAT3 O-GlcNAcylation at T717 is reduced in the aging hippocampus and mimicking this decline in NSC promotes a gliogenic fate switch. (A) Schematic of proteomic workflow. The hippocampus was isolated from 3-, 12-, or 24-mo-old mice. Hippocampi were pooled to obtain sufficient protein (n = 5 animals per pool). Proteins were isolated from tissue and digested with trypsin before undergoing isobaric tagging. Tagged peptides were enriched for O-GlcNAcylation with LWAC and analyzed via electron transfer dissociation mass spectrometry (ETD-MS). (B) Representative spectra of hippocampal STAT3 O-GlcNAcylation at T717. (C) Diagram of O-GlcNAc sites detected by mass spectrometry in A (T717, S719), and previously identified phosphorylation sites (Y705, S727) on STAT3. (D) STAT3 phosphorylation at Y705 and STAT3 expression in primary hippocampal NSCs following 24-h vehicle or OSMI-1 treatment (n = 3 per group). (E) Expression of STAT3 target genes in primary hippocampal NSCs treated as in D was evaluated using RNA-sequencing. (F) Primary hippocampal postnatal and adult NSCs were infected with a lentiviral construct expressing a nonphosphorylatable STAT3 (Y705F) or wild-type STAT3 (Control). Differentiation was induced in vehicle or OSMI-1 treated NSCs by withdrawing growth factors 7 d prior to fixation. Cells were labeled with DAPI and anti-MAP2. MAP2+ neurons were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (G) Primary postnatal and adult hippocampal NSCs were infected, treated, and differentiated as in F. Cells were labeled with DAPI and anti-GFAP. GFAP+ astrocytes were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (H) Primary postnatal and adult hippocampal NSCs were infected with a lentiviral construct expressing an O-GlcNAcylation–deficient STAT3 (T717A) or wild-type STAT3 (Control). Differentiation was induced by withdrawing growth factors 7 d prior to fixation. Cells were labeled with DAPI and anti-MAP2. MAP2+ neurons were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (I) Primary postnatal and adult hippocampal NSCs were infected, treated, and differentiated as in H. Cells were labeled with DAPI and anti-GFAP. GFAP+ astrocytes were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (J) Primary postnatal and adult hippocampal NSCs were infected with a lentiviral construct expressing Ogt or GFP under the Nestin promoter. Differentiation was by induced withdrawing growth factors 7 d prior to fixation. Cells were labeled with DAPI and anti-MAP2. MAP2+ neurons were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). (K) Primary postnatal and adult hippocampal NSCs were infected, treated, and differentiated as in J. Cells were labeled with DAPI and anti-GFAP. GFAP+ astrocytes were quantified as a percentage of total DAPI+ cells (n = 6 replicates per group). Data are represented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; t test.
Currently, the functional consequences of STAT3 O-GlcNAcylation at S719 are unknown; however, increased STAT3 O-GlcNAcylation at T717 has been previously shown to inhibit STAT3 activation in immune cells by preventing STAT3 phosphorylation at Tyrosine 705 (Y705) (30). We therefore investigated whether decreasing O-GlcNAcylation in primary NSCs could conversely increase STAT3 activation. We assessed NSC STAT3 phosphorylation at Y705 following Ogt inhibition with OSMI-1 treatment in vitro by Western blot (Fig. 4D). While STAT3 expression was not altered, pharmacological inhibition of Ogt significantly increased phosphorylation levels of STAT3 in NSCs (Fig. 4D). Additionally, we made use of our RNA-sequencing dataset to survey expression of known STAT3 target genes in NSCs following Ogt inhibition, and likewise observed an up-regulation of several genes associated with STAT3 activation (31), including p21, Zeb1, and Ccnd1 (Fig. 4E).
To investigate whether blocking STAT3 activation would mitigate the effects of decreasing O-GlcNAcylation on differentiation in postnatal and adult NSCs, we generated viral expression constructs encoding a nonphosphorylatable STAT3 by converting Tyrosine into Phenylalanine at position 705 (Y705F STAT3). As a control, primary NSCs were infected with a virus encoding a wild-type STAT3. When treated with OSMI-1, control NSCs exhibited decreased neuronal (Fig. 4F) and increased astrocyte (Fig. 4G) differentiation, corroborating our previous findings (Fig. 2 F and G). Expression of Y705F STAT3 increased neuronal (Fig. 4F) and decreased astrocyte differentiation (Fig. 4G), consistent with previous reports (28, 29). Moreover, the effects of decreasing NSC O-GlcNAcylation on STAT3 activation (SI Appendix, Fig. S9A) and neuronal differentiation (Fig. 4F) were mitigated by expression of the Y705F STAT3 mutant. Next, we investigated whether mimicking an age-related decline in STAT3 O-GlcNAcylation at T717 altered NSC differentiation. It should be noted that the T717 site is only present on the predominant, full-length STAT3 α-isoform, and not on the shorter β-isoform. We generated viral expression constructs encoding STAT3 with site-directed mutations converting Threonine to Alanine at position 717 (T717A STAT3). Expression of O-GlcNAcylation–deficient T717A STAT3 in NSCs resulted in increased activation of the STAT3 α-isoform (SI Appendix, Fig. S9B), along with decreased neuronal (Fig. 4H) and increased astrocyte (Fig. 4I) differentiation compared to wild-type STAT3 control conditions. These in vitro data indicate that increased STAT3 activity, stemming from reduced NSC O-GlcNAcylation, promotes an astrocyte differentiation program in NSCs.
Increased NSC O-GlcNAc Levels Promote Neuronal and Inhibit Astrocyte Differentiation In Vitro.
Given the progliogenic effects of reducing NSC O-GlcNAc levels, we next investigated whether increasing NSC O-GlcNAc levels would conversely promote neuronal differentiation. We generated a viral expression construct encoding Ogt under the control of the Nestin promoter and infected primary postnatal and adult NSCs with this virus or a control encoding GFP. Ogt overexpression in NSCs resulted in a significant increase in Ogt and O-GlcNAc levels (SI Appendix, Fig. S10). Additionally, we observed an increase in neuronal differentiation (Fig. 4J) and a corresponding decrease in astrocyte differentiation (Fig. 4K) in NSCs overexpressing Ogt compared to a GFP control. These data indicate that increased O-GlcNAcylation is sufficient to promote a neuronal differentiation program and inhibit astrocyte differentiation in NSCs.
Discussion
Cumulatively, our data indicate that age-related loss of NSC protein O-GlcNAcylation regulates a STAT3-mediated neuron to glia fate switch that contributes to a decline in adult hippocampal neurogenesis. While increased NSC quiescence has been highlighted as a major determinant of age-related neurogenic decline in the hippocampus (7, 32), a complementary model has been proposed in which division-coupled conversion of NSCs into mature astrocytes restrict the stem cell pool with age (3). Our data identifying an O-GlcNAc–mediated glial fate switch in the mature hippocampus provide evidence for this coextensive cellular mechanism, in which age-related increased gliogenesis contributes to regenerative decline and associated cognitive impairments.
Previous reports have observed that the absolute number of newly generated astrocytes in the adult brain remains relatively constant with age (3, 33, 34). Given the well-documented age-related decline of total NSC number, it has been suggested that maintaining this constant level of gliogenesis from a shrinking adult NSC pool requires that an increasing proportion of NSCs adopt a gliogenic fate (3, 33), a phenomenon that we demonstrate in the mature hippocampus. While adult neurogenesis has been shown to be a division-coupled process capable of maintaining the NSC pool (3, 5), gliogenesis is indicative of a terminal cell-cycle exit and leads to a reduction in total NSC number (3). This shift toward terminal astrocyte differentiation with age further exacerbates the age-related decline of the NSC pool. In fact, mathematical modeling of NSC differentiation estimates that maintaining constant levels of gliogenesis accounts for nearly half of the NSCs lost during aging (34). In our study, we demonstrate that this shift in NSC cell fate occurs alongside a decrease in NSC O-GlcNAc. Moreover, we find that O-GlcNAcylation of STAT3 T717 declines in the hippocampus with age, and that loss of this posttranslational modification is sufficient to promote astrocyte differentiation in NSCs. Ultimately the shrinking of the adult NSC pool, which stems in part from terminal astrocyte differentiation, results in decreased neurogenesis during aging, with major functional implications for age-related decreased regenerative capacity and neurodegenerative disease. Indeed, in elderly humans, maintaining higher levels of hippocampal neurogenesis is thought to be neuroprotective, as they are associated with increased cognitive scores and slower disease progression in the context of Alzheimer’s disease (13, 14).
The ability to utilize the neurogenic capacity of adult NSCs is predicated on understanding underlying molecular mechanisms that can be targeted to reverse the effects of aging in the brain. Although posttranslational modification of proteins represents a fundamental mechanism by which information is transmitted, its role in regulating adult NSC function with age has been largely overlooked. Previous work has suggested a role for O-GlcNAc in mediating proper brain development. Specifically, genetic ablation of the O-GlcNAc cleaving enzyme, Oga, in NSCs during development has been shown to increase NSC proliferation and lead to an accumulation of progenitors and immature neurons in the olfactory bulb (26). In our study, we identify O-GlcNAc as a posttranslational regulator of regenerative decline in the adult brain and posit site-specific changes in O-GlcNAcylation as potential targets to counter age-related regenerative dysfunction. In particular, age-related loss of STAT3 O-GlcNAcylation at T717 provides a mechanistic link between increased NSC STAT3 signaling and an aberrant astrocyte differentiation program, pointing to STAT3 activation as a driver of age-related neurogenic decline. Interestingly, inhibition of STAT3 signaling in aged muscle stem cells restores regeneration at old age (35). Moreover, we demonstrate that increasing O-GlcNAcylation in NSCs by overexpressing Ogt is sufficient to promote neuronal differentiation. These data raise the exciting possibility of rejuvenating regenerative capacity in the aging brain by inhibiting STAT3 signaling through an increase in NSC O-GlcNAcylation. Ultimately, our data frame O-GlcNAc as a proneurogenic posttranslational modification, with functional implications for cognitive processes.
From a therapeutic perspective, while decreased O-GlcNAcylation has been implicated in Alzheimer’s disease pathology and neurodegeneration (18, 19), we recently demonstrated that increasing neuronal O-GlcNAcylation ameliorates age-related cognitive decline (17). Similarly, others have shown that inhibiting STAT3 signaling ameliorates cognitive deficits in mouse models of Alzheimer’s disease, in part by preventing astrogliosis (36). This growing body of work positions O-GlcNAcylation, STAT3 signaling and the balance between newly generated neurons and astrocytes as a focal point in combating age-related functional decline in the aging and diseased brain. Of note, the benefits of interventions that ameliorate cognitive-decline during aging and Alzheimer’s disease, such as exercise, have been shown to involve increased adult neurogenesis (37, 38). Given recent observations of the persistence of adult neurogenesis in the aged human brain (13, 14), the promise of restoring cellular and cognitive plasticity in the elderly by targeting adult NSCs is increasingly evident. Here we identify both an age-related NSC fate switch, and the posttranslational modification governing it, as potential cellular and molecular targets by which to maintain regenerative capacity at old age, restore adult neurogenesis and ameliorate cognitive decline.
Methods
Experimental Animal Model and Subject Details.
All mouse handling and use was in accordance with institutional and ethical guidelines approved by the University of California, San Francisco Institutional Animal Care and Use Committee. The following mouse lines were used: C57BL/6J young mice (Jackson Laboratory), B6.129-Ogttm1Gwh/J (The Jackson Laboratory line 004860), C57BL/6-Tg(Nes-cre/ERT2)KEisc/J (Jackson Laboratory line 016261), and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (Jackson Laboratory line 007914). All studies were done in male mice. The numbers of mice used to result in statistically significant differences were calculated using standard power calculations with α = 0.05 and a power of 0.8. We used an online tool (http://www.stat.uiowa.edu/∼rlenth/Power/index.html) to calculate power and sample size based on experience with the respective tests, variability of the assays, and interindividual differences within groups. Mice were housed under specific pathogen-free conditions under a 12-h light/dark cycle. All experiments were randomized and blinded by an independent researcher. Researchers remained blinded throughout histological, molecular, and behavioral assessments. Groups were unblinded at the end of each experiment upon statistical analysis.
Immunohistochemistry.
Tissue processing and immunohistochemistry was performed on free-floating sections following standard published techniques (39). Briefly, mice were anesthetized with a ketamine (100 mg/kg)-xylazine (10 mg/kg) mixture (Patterson Veterinary, Henry Schein) and transcardially perfused with cold PBS. Brains were removed, fixed in phosphate-buffered 4% paraformaldehyde, pH 7.4, at 4 °C for 48 h followed by cryoprotection in 30% sucrose, and coronally sectioned at 40 µm with a cryomicrotome (Leica SM2010 R). Sections were washed three times in Tris-buffered saline with 0.1% Tween 20 (TBST) and incubated in 3% normal donkey serum (Thermo Fisher Scientific) for 1 h. After overnight incubation in primary antibody (mouse anti–O-GlcNAc [RL2; 1:500], mouse anti-Nestin [clone rat-401; 1:200], goat anti-Dcx [C-18; 1:7,500], mouse anti-BM28 [MCM2; 1:250], goat anti-Sox2 [Y-17; 1:200], rabbit anti-GFAP [1:1,000], mouse anti-NeuN [clone A60; 1:1,000], rat anti-BrdU [BU1/75 (ICR1)] [1:1,000], mouse anti-Ki67 [clone B56; 1:500], mouse anti-MAP2 [1:500], rabbit anti-s100β [1:500]) at 4 °C, staining was revealed using fluorescence-conjugated secondary Alexa antibodies (donkey anti-rabbit: Alexa-488 conjugated secondary antibody; donkey anti-rabbit: Alexa-555 conjugated secondary antibody; donkey anti-rat: Alexa-488 conjugated secondary antibody; donkey anti-goat: Alexa-555 conjugated secondary antibody; donkey anti-mouse: Alexa-488 conjugated secondary antibody; donkey anti-mouse: Alexa-555 conjugated secondary antibody; donkey anti-mouse: Alexa-647 conjugated secondary antibody [1:500]). Antigen retrieval for BrdU labeling required incubation in 3M HCl at 37 °C for 30 min before incubation with primary antibody; Nestin labeling required incubation in Citrate Buffer (Sigma-Aldrich) at 95 °C (three times 5 min) prior to incubation with primary antibody. To estimate the total number of immunopositive cells per DG, confocal stacks of coronal sections of the DG (three to six sections per mouse, 40-μm thick, 360-μm apart) were acquired on a Zeiss LSM 800.
In Vivo O-GlcNAc Quantification in NSCs.
Immunohistochemistry and imaging were performed as previously described. To quantify O-GlcNAc levels, NSCs were identified based on GFAP/Sox2+ staining, localization within the subgranulzar zone, and radial morphology. As O-GlcNAc staining is nuclear, DAPI was used to identify the nucleus of the analyzed cells. To quantify O-GlcNAc levels in a single cell, intensity in the O-GlcNAc channel was measured within the region defined by DAPI, and normalized to the area of that region in a single z-plane. This measurement was repeated over every plane in which that nucleus appeared in order to quantify the entirety of the cell. These values were then summed and normalized to the number of planes quantified for that cell. This process was repeated for every neural stem cell which was fully included within the section (i.e., not within terminal z-plane). For each animal, NSCs were analyzed in all sections and averaged to determine a value per animal. Four animals were quantified per group. Fiji was used for quantification.
Isolation of Primary NSCs from the Mouse Hippocampus.
Primary neural stem cell isolation and culture were preformed following previously published techniques (40). Hippocampi were dissected from male postnatal day 1 (P1) or adult (2-mo-old) wild-type C57/BL6 mice and pooled by age for NSC isolation. After removing superficial blood vessels, hippocampi were mechanically dissociated by fine mincing and enzymatically digested for 30 min at 37 °C in DMEM media containing 2.5 U/mL Papain (Worthington Biochemicals), 1 U/mL Dispase II (Boeringher Mannheim), and 250 U/mL DNase I (Worthington Biochemicals). NSCs were purified using a 65% Percoll gradient and cultured (neurobasal A medium supplemented with 2% B27 without vitamin A, 1% glutamax, 1% penicillin streptomycin, 10 ng/mL EGF, 10 ng/mL bFGF) as a monolayer on poly-d-lysine and laminin-coated plates at a density of 105 cells/cm2.
Viral Plasmids and Viruses.
We generated lentiviruses encoding shRNAs targeting Ogt or luciferase using a lentiviral shRNA expression system (pGreenPuro shRNA, System Biosciences) according to the manufacturer's instructions. The targeted sequences were cloned into the pGreenPuro vector (Ogt, 5′-AGGGAACTAGATAACATGCTT-3′; luciferase, 5′- GCCATTCTATCCTCTAGAGGA -3′). Overexpression sequences were generated as follows. The Ogt coding sequence (CDS) and partial 3′ and 5′ UTRs were PCR-amplified form mouse hippocampal cDNA (Ogt forward primer: CACCGTTCAGTATTCTGTGCCGCC; Ogt reverse primer: TAGGGCAATTCTCCTGTGCG) and cloned into the pENTR D-TOPO vector (Thermo Fisher Scientific). The CDS was further amplified and cloned into a lentiviral expression plasmid containing a mouse Nestin promoter using the restriction sites NheI and BamHI. The STAT3 CDS was obtained from the ORFeome 5.1 collection and sequence-verified using Sanger sequencing. STAT3 mutant constructs were generated using the QuikChange Lightning Site-directed Mutagenesis kit (Agilent) in combination with the following primers: GTAGCGCTGCCCCATTCCTGAAGACCAAG (Y705F forward), CTTGGTCTTCAGGAATGGGGCAGCGCTAC (Y705F reverse), CTGTGTGACACCAACGGCCTGCAGCAATACCAT (T717A forward), ATGGTATTGCTGCAGGCCGTTGGTGTCACACAG (T717A reverse). The lentiviral Nestin-promoter plasmid was linearized using the restriction enzymes NheI and SalI and the wild-type and mutant STAT3 expression constructs were generated using Gibson assembly (New England Biolabs). All coding plasmid sequences were verified using Sanger sequencing. Endotoxin-free plasmid maxiprep kits were used for viral plasmid preparation. Lentivirus was generated according to University of California, San Francisco viracore protocols. Briefly Lenti-X 293T cells were transfected with the lentiviral expression plasmid along with Pax2 and VSV-G packaging and envelope plasmids, respectively. Twenty-four hours later, media was supplemented with ViralBoost (Alstem) according to manufacturer’s instructions. Twenty-four hours later, media was removed and filtered through a 0.45-μm filter. The filtered virus-containing media was then concentrated via ultracentrifugation at 24,000 × g. The resulting pellet was resuspended in 100 μL PBS. Viral titer was determined using Lenti-X Go Stix Plus (Takara) according to manufacturer’s instructions.
NSC Proliferation Assay.
NSCs were seeded in 500 μL growth medium (neurobasal A medium supplemented with 2% B27 without vitamin A, 1% glutamax, 1% penicillin streptomycin, 10 ng/mL EGF, 10 ng/mL bFGF) at a density of 10,000 cells per well on PDL/laminin-coated glass coverslips in a 24-well tissue culture plate. Twenty-four hours later, cells were treated with either DMSO or 12.5 μM OSMI-1 (Sigma-Aldrich). Sixteen hours later, cells were treated with 20 μM EdU for 8 h prior to fixing with 4% PFA for 20 min. Acute knockdown of Ogt was induced by treating plated NSCs with shRNA-encoding lentiviruses (10 viral particles per cell) for 3 d prior to EdU treatment as described. Cells were washed three times with PBS after fixation, blocked in 3% donkey serum, and incubated in primary antibody at 4 °C for 16 h. After three washes with PBS, EdU incorporation was revealed using a Click-iT EdU Alexa Fluor Imaging Kit (Thermo Fisher Scientific) and nuclei were counterstained with Hoechst 33342 (1:10,000; Thermo Fisher Scientific). EdU+ cells were counted per field-of-view at three randomly determined locations per coverslip. In vitro experiments were conducted in triplicate for each condition and repeated to ensure reproducibility.
NSC Differentiation Assay.
NSCs were seeded in 500 μL growth medium and treated with OMSI-1 or infected with lentivirus at a ratio of 10 viral particles per cell. Twenty-four hours (OSMI-1) or 72 h (knockdown and overexpression) after treatment, media was replaced with differentiation medium (neurobasal A medium supplemented with 2% B27 without vitamin A, 1% glutamax, 1% penicillin streptomycin). Cells were maintained in differentiation medium for 7 d, with half the media replaced every other day. Cells were fixed with 4% PFA for 20 min. Cells were washed three times with PBS after fixation, blocked in 3% donkey serum, and incubated in primary antibody at 4 °C for 16 h. Staining was revealed using fluorescence-conjugated secondary Alexa antibodies (1:500). MAP2- or GFAP+ cells were counted per field-of-view at three randomly determined locations per coverslip. In vitro experiments were conducted in three to six replicates for each condition and repeated to ensure reproducibility.
Lactate Dehydrogenase Assay.
Primary NSCs were seeded at 3,000 cells per well in a 96-well plate and were treated with OSMI-1 or a vehicle control and differentiated as previously described in NSC Differentiation Assay. At 0, 1, 3, 5, and 7 d after removal of growth factors, cytotoxicity was assessed using the Pierce LDH Cytotoxicity Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Measurements were performed on six individual replicates to ensure reproducibility.
Trypan Blue Assay.
Primary NSCs were treated with OSMI-1 or a vehicle control and differentiated as previously described in NSC Differentiation Assay. At 1, 3, 5, and 7 d after removal of growth factors, cell viability was assessed using Trypan blue. All cells and media were removed from the plate and centrifuged at 400 × g for 5 min. The pellet was resuspended in fresh media and diluted with an equal volume of Trypan blue. Total cells, live cells (Trypan blue excluding), and dead cells (Trypan blue including) were counted using a hemocytometer. Cell viability was determined by dividing the number of lives cells by the total number of cells. Counts were performed on six replicates to ensure reproducibility.
Western Blot Analysis.
Primary NSCs were lysed in RIPA lysis buffer (500 mM Tris, pH 7.4, 150 mM NaCl, 0.5% Na deoxycholate, 1% Nonidet P-40, 0.1% SDS, complete protease inhibitors; Roche, Halt Phosphatase Inhibitor Mixture; Thermo Fisher, and PUGNAc; Sigma-Aldrich). Protein lysates were mixed with 4× NuPage LDS loading buffer (Invitrogen) and loaded on a 4 to 12% SDS polyacrylamide gradient gel (Invitrogen) and subsequently transferred onto a nitrocellulose membrane. The blots were blocked in 5% milk in TBST and incubated with primary antibody (mouse anti–O-GlcNAc [RL2; 1:500] [Abcam, ab2739], rabbit anti-Ogt [1:500; Novus, NBP1-32791], rabbit anti-Histone H3 [1:1,000; Thermo Fisher Scientific, PA5-16183], rabbit anti-STAT3 [1:1,000; Cell Signaling 12640], rabbit anti-pSTAT3 [Y705; 1:1,000] [Cell Signaling, 9131]) at 4 °C for 16 h. Horseradish peroxidase-conjugated secondary antibodies and an ECL kit (GE Healthcare/Bio-Rad) were used to detect protein signals. Blots were imaged with a ChemiDoc (Bio-Rad) and quantified using ImageJ software (v1.8.0_91). Actin or HH3 bands were used for normalization.
PCR Genotyping.
Ogt floxed, Nestin-CreERT2, and TdTomato alleles were genotyped from skin biopsies using PCR. Primers specific for the beta-globin gene were included in the reaction as a control. Ogt floxed forward primer: CATCTCTCCAGCCCCACAAACTG. Ogt floxed reverse primer: GACGAAGCAGGAGGGGAGAGCAC. Primer Cre forward: CACCCTGTTACGTATAGCCG. Primer Cre reverse: GAGTCATCCTTAGCGCCGTA. TdTomato mutant forward primer: CTGTTCCTGTACGGCATGG. TdTomato mutant reverse primer: GGCATTAAAGCAGCGTATCC. TdTomato wild-type forward primer: AAGGGAGCTGCAGTGGAGTA. TdTomato wild-type reverse primer: CCGAAAATCTGTGGGAAGTC. βa-Globin forward primer: CCAATCTGCTCACACAGGATAGAGAGGGCAGG. β-Globin reverse primer: CCTTGAGGCTGTCCAAGTGATTCAGGCCATCG.
Tamoxifen Injections.
Animals were injected via intraperitoneal injection with 75 mg/kg of tamoxifen or vehicle once every 24 h for a total of five injections per animal. Animals were monitored after recovery and 4 wk allowed to pass after the final injection before any analyses were performed.
BrdU Administration and Quantification.
For short-term proliferation studies, mice were intraperitoneally injected with EdU (50-mg/kg body weight, Sigma-Aldrich) daily for 6 d prior to being killed. For study of newborn neuron survival and astrocyte formation, mice were injected with BrdU (50 mg/kg) for 6 d and animals were killed 28 d after administration. To estimate the total number of BrdU+ cells in the brain, we performed fluorescence staining for BrdU on three to six hemibrain sections per mouse (40-μm thick, 360-μm apart). To quantify neuronal fate and maturation of dividing cells, BrdU+ cells across three to six sections per mouse were analyzed by confocal microscopy for coexpression with NeuN. To quantify astrocyte formation, BrdU+ cells across three to six sections per mouse were analyzed by confocal microscopy for coexpression with either GFAP or s100β.
Contextual Fear Conditioning.
Paradigm follows previously published techniques (10). Mice learned to associate the environmental context (fear-conditioning chamber) with an aversive stimulus (mild foot shock; unconditioned stimulus, US) enabling testing for hippocampal-dependent contextual fear conditioning. The mild foot shock was paired with a light and tone cue (conditioned stimulus, CS) in order to also assess amygdala-dependent cued fear conditioning. Conditioned fear was displayed as freezing behavior. Specific training parameters are as follows: Tone duration is 30 s; level is 70 dB, 2 kHz; shock duration is 2 s; intensity is 0.6 mA. On day 1 each mouse was placed in a fear-conditioning chamber and allowed to explore for 2 min before delivery of a 30-s tone (70 dB) ending with a 2-s foot shock (0.6 mA). Two minutes later, a second CS–US pair was delivered. On day 2 each mouse was first place in the fear-conditioning chamber containing the same exact context, but with no administration of a CS or foot shock. Freezing was analyzed for 1 to 3 min. One hour later, the mice were placed in a new context containing a different odor, cleaning solution, floor texture, chamber walls, and shape. Animals were allowed to explore for 2 min before being reexposed to the CS. Freezing was analyzed for 1 to 3 min. Freezing was measured using a FreezeScan video tracking system and software (Clever Sys). All behavior is performed double-blinded.
Novel Object Recognition.
The NOR task was adapted from a previously described protocol (41). During the habituation phase (day 1), mice could freely explore an empty arena for 10 min. During the training phase (day 2), two identical objects were placed in the habituated arena, and mice could explore the objects for 5 min. For the testing phase (day 3), one object was replaced with a novel object, and mice could explore the objects for 5 min. Time spent exploring each object was quantified using the Smart Video Tracking Software (Panlab; Harvard Apparatus). Two different sets of objects were used. To control for any inherent object preference, half of the mice were exposed to object A as their novel object and half to object B. To control for any potential object-independent location preference, the location of the novel object relative to the trained object was also varied. The objects were chosen based on their ability to capture the animal’s interest, independent of genetic background or age. To determine percent time with novel object, we calculated (time with novel object)/(time with trained object + time with novel object) × 100. In this preference index, 100% indicates full preference for the novel object, and 0% indicates full preference for the trained object. A mouse with a value of 50% would have spent equal time exploring both objects. Mice that did not explore both objects during the training phase were excluded from analysis.
Open Field.
Mice were placed in the center of an open 40-cm × 40-cm square chamber (Kinder Scientific) with no cues or stimuli and allowed to move freely for 10 min. Infrared photobeam breaks were recorded and movement metrics analyzed by MotorMonitor software (Kinder Scientific).
Stereotaxic Injections.
Animals were placed in a stereotaxic frame and anesthetized with 2% isoflurane (2 L/min oxygen flow rate) delivered through an anesthesia nose cone. Ophthalmic eye ointment was applied to the cornea to prevent desiccation during surgery. The area around the incision was trimmed. Solutions were injected bilaterally into the DG of the dorsal hippocampi using the following coordinates: (from bregma) anterior = −2 mm, lateral = 1.5 mm, (from skull surface) height = −2.1 mm. A 2-μL volume was injected stereotaxically over 10 min (injection speed: 0.20 μL/min) using a 5-μL 26s-gauge Hamilton syringe. To limit reflux along the injection track, the needle was maintained in situ for 10 min, slowly pulled out halfway, and kept in position for an additional 5 min. The skin was closed using silk suture. Each mouse was injected subcutaneously with analgesics. Mice were singly housed and monitored during recovery.
Protein Isolation.
Mouse hippocampi were isolated from fresh mouse brain tissue of either 3-, 12-, or 24-mo-old mice after transcardial perfusion with cold PBS (n = 3 replicates per group; n = 3 mice per replicate). Protein was isolated from the hippocampus with TRIzol Reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. Frozen protein pellets were resuspended in 50 mM ammonium bicarbonate containing 6M guanidine hydrochloride, 6X Phosphatase Inhibitor Mixtures II and III (Sigma-Aldrich), and 80 mM PUGNAc (Tocris Bioscience). Protein concentrations were estimated with bicinchoninic acid (BCA) protein assay (ThermoFisher Scientific). The protein lysate (1 mg from each sample) was reduced for 1 h at 56 °C with 2.5 mM Tris(2-carboxyethyl)phosphine hydrochloride and subsequently carbamidomethylated using 5 mM iodoacetamide for 45 min at room temperature in the dark. Lysates were diluted to 1M guanidine hydrochloride with 50 mM ammonium bicarbonate, pH 8.0, and equal amounts of each sample were digested overnight at 37 °C with sequencing grade trypsin (ThermoFisher Scientific) at an enzyme to substrate ratio of 1:50 (wt/wt). Following digestion, samples were acidified with formic acid (FA) (Sigma-Aldrich), desalted using a 360-mg C18 Sep-Pak SPE cartridge (Waters), and dried to completeness using a SpeedVac concentrator (Thermo Electron).
TMT Labeling.
Tryptic peptides were labeled with TMT-10plex according to the manufacturer’s protocol. The TMT labeling was as follows: 3-month-1:TMT127C, 3month-2:TMT127N, 3month-3:TMT128C, 12month-1:TMT128N, 12month-2:TMT129C, 12month-3:TMT129N, 24month-1:TMT130C, 24month-2:TMT130N, 24month-3: TMT-131C. Labeling efficiency was checked on Thermo Scientific Q Exactive Plus Orbitrap. TMT labeled peptides were mixed together, desalted using a 360-mg C18 Sep-Pak SPE cartridge, and dried to completeness using a SpeedVac concentrator before glycopeptide enrichment.
LWAC.
Glycopeptides were enriched as described previously (42, 43). Briefly, desalted TMT-labeled peptides were resuspended in 500 μL LWAC buffer (100 mM Tris pH 7.5, 150 mM NaCl, 10 mM MgCl2, 10 mM CaCl2, 5% acetonitrile) and 100 μL were run over a 2.0 × 250 mm POROS-WGA column at 100 μL/min under isocratic conditions with LWAC buffer and eluted with a 100-μL injection of 40 mM GlcNAc. Glycopeptides were collected inline on a C18 column (Phenomenex). Enriched glycopeptides from five initial rounds of LWAC were eluted with 50% acetonitrile, 0.1% FA in a single 500 μL fraction, dried, and LWAC enrichment was repeated for a total of three enrichment steps.
Glycopeptides were separated on a 1.0 × 100 mm Gemini 3μ C18 column (Phenomenex). Peptides were loaded onto the column in 20 mM NH4OCH3, pH 10 and subjected to a gradient from 1 to 21% 20 mM NH4OCH3, pH10 in 50% acetonitrile over 1.1 mL, up to 62% 20 mM NH4OCH3, pH10 in 50% acetonitrile over 5.4 mL with a flow rate of 80 μL/min while collecting 31 fractions.
Mass Spectrometry Analysis.
Glycopeptides were analyzed on an Orbitrap Fusion Lumos (Thermo Scientific) equipped with a NanoAcquity UPLC (Waters). Peptides were fractionated on a 50-cm × 75-μm ID 2-μm C18 EASY-Spray column using a linear gradient from 3.5 to 30% solvent B over 185 min. Precursor ions were measured from 375 to 1500 m/z in the Orbitrap analyzer (resolution: 120,000; automatic gain control [AGC]: 4.0e5). Each precursor ion (charged 2 to 7+) was isolated in the quadrupole (selection window: 1.6 m/z for EThcD, 0.7 m/z for HCD; dynamic exclusion window: 30 s; MIPS peptide filter-enabled) and underwent two MS2 fragmentation methods. Ions were fragmented by EThcD (maximum injection time: 250 ms, supplemental activation collision energy: 25%) and measured in the Orbitrap (resolution: 30,000; AGC; 5.0e4) for peptide identification. The same selected precursor was also fragmented by HCD (maximum injection time: 86 ms; HCD collision energy: 50%) and measured in the Orbitrap (resolution: 50,000; AGC; 5.0e4; Scan Range: 100 to 400 m/z) for TMT reporter ion intensity measurement. The scan cycle was 18 scans.
Peaklists for EThcD were extracted using Proteome Discoverer 2.2. EThcD peaklists were filtered with MS-Filter and only spectra containing a 204.0867 m/z peak corresponding to the HexNAc oxonium ion were used for database searching. EThcD data were searched against the SwissProt Mus musculus database (downloaded September 6, 2016) (and concatenated with a randomized sequence for each entry) using Protein Prospector (v5.23.0). Cleavage specificity was set as tryptic, allowing for two missed cleavages. Carbamidomethylation of Cys and TMT10plex on lysine and the peptide N terminus were set as constant modifications. The required mass accuracy was 10 ppm for precursor ions and 30 ppm for fragment ions. Variable modifications are listed in Dataset S2. Two modifications per peptide were permitted. Unambiguous posttranslational modifications were determined using a minimum SLIP score of six, which corresponds to a 5% local false localization rate. Modified peptides were identified with a protein and peptide false-discovery rate of 1%. O-GlcNAc and O-GalNAc modifications were differentiated based on known protein subcellular localization and HexNAc oxonium ion fragment ratios (44).
TMT Mass Spectrometry Data Analysis.
Data were filtered to only include O-GlcNAcylated peptides unique to a single protein. Quantitation of TMT data were performed by calculating ratios of reporter ion peak intensities between conditions along with variance for each ratio, and median normalized. Reporter ion peak intensities were obtained from HCD spectra corresponding to peptides identified from EThcD spectra. Peptide abundances were normalized by the median of ratio distributions. The age-dependent changes in O-GlcNAcylation were determined using a normalized median log2 12 mo/3 mo ratio of at least 1.0, corresponding to a 2.0 fold-change with age. Raw data files available under MassIVE submission MSV000084825. All annotated O-GlcNAc-peptide spectra available at MS-Viewer (45) search key qcchi2to8l.
RNA Isolation.
mRNA of NSCs was isolated by lysis with TRIzol Reagent (Thermo Fisher Scientific), separation with chloroform (0.2 mL per mL TRIzol), and precipitated with isopropyl alcohol according to the manufacturer’s instructions.
NSC Library Construction and RNA-Sequencing.
After RNA isolation, RNA-sequencing libraries were constructed using the Smart-Seq2 protocol from (46), with modifications. Briefly, 1 ng high-quality RNA was reverse transcribed using SuperScript II (Life Technologies, 18064-014) with a poly-dT anchored oligonucleotide primer, and a template switching oligonucleotide primer that generated homotypic PCR primer binding sites. The cDNA underwent 10 rounds of PCR amplification using KAPA HiFi Hotstart (Kapa Biosystems, KK2601), followed by Ampure bead (Agencourt) clean-up. The quality of the amplified cDNA was tested using qPCR for GAPDH and nucleic acid quantitation. One nanogram of high-quality amplified cDNA was fragmented with the Tn5 transposase from the Illumina Nextera kit (FC-131-1096) to a median size of ∼500 bp. The fragmented library was amplified with indexed Nextera adapters (FC-131-1002) using 12 rounds of PCR. Final libraries were purified with Ampure beads and quantified using a qPCR Library Quantification Kit (Kapa Biosystems, KK4824). Libraries were pooled for sequencing on an Illumina HiSEq. 2500 (paired reads 2 × 100 bp).
Bioinformatic Analysis of NSC RNA-Sequencing Data.
Alignment of RNA-sequencing reads to the mouse mm10 transcriptome was performed using STAR v2.7.3a (47) following ENCODE standard options, read counts were generated using RSEM v1.3.1, and differential expression analysis was performed in R v3.6.1 using the DESeq2 package v1.38.0 (48) (detailed pipeline v2.0.1 and options available on https://github.com/emc2cube/Bioinformatics/). Significance was determined using a corrected P < 0.05 and a log2 fold-change > 0.5. GO and ChEA/ENCODE transcription factor enrichment analysis was performed with Enrichr (https://amp.pharm.mssm.edu/Enrichr/). Heatmaps were generated using iDEP (http://bioinformatics.sdstate.edu/idep/). All data available under Gene Expression Omnibus accession no. GSE143388.
Data and Statistical Analysis.
Graphed data are expressed as mean ± SEM. Statistical analysis was performed with Prism 6.0 software (GraphPad Software). Unless otherwise noted, means between two groups were compared with two-tailed, unpaired Student’s t test. NOR data were compared with a one sample t test against 50% (expected preference). Comparisons of means from multiple groups with each other or against one control group were analyzed with one-way ANOVA and Tukey’s post hoc test. All histology and behavior experiments conducted were done in a randomized and blinded fashion. For each experiment, the overall size of the experimental groups corresponded to distinct animals or cultures. Unique samples were not measured repeatedly within the same characterization of a given cohort.
Supplementary Material
Acknowledgments
We thank Dr. Barbara Panning for insightful comments; and the Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results. Computing for this project was performed on the Sherlock cluster. This work was funded by the HHMI (J.C.M. and A.L.B.); NIH National Institute of General Medical Sciences 8P41GM103481 (to J.C.M. and A.L.B.); the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (J.C.M. and A.L.B.); NIH Ruth L. Krischstein National Research Service Award predoctoral Fellowship F31-AG050415 (to E.G.W.); National Institute on Aging (R01 AG055797, RF1 AG062357); and a gift from Marc and Lynne Benioff (S.A.V.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007439117/-/DCSupplemental.
Data Availability.
RNA-sequencing data were deposited at GEO (accession no. GSE143388). Mass Spectrometry raw data were deposited at MassIVE (MSV000084825). Mass spectrometry spectra can be viewed at MS-Viewer (qcchi2to8l) (49–51).
References
- 1.Christian K. M., Song H., Ming G. L., Functions and dysfunctions of adult hippocampal neurogenesis. Annu. Rev. Neurosci. 37, 243–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonçalves J. T., Schafer S. T., Gage F. H., Adult neurogenesis in the Hippocampus: From stem cells to behavior. Cell 167, 897–914 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Encinas J. M. et al., Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8, 566–579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steiner B. et al., Differential regulation of gliogenesis in the context of adult hippocampal neurogenesis in mice. Glia 46, 41–52 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Bonaguidi M. A. et al., In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonzano S. et al., Neuron-astroglia cell fate decision in the adult mouse hippocampal neurogenic niche is cell-intrinsically controlled by COUP-TFI in vivo. Cell Rep. 24, 329–341 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Lugert S. et al., Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445–456 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Kuhn H. G., Dickinson-Anson H., Gage F. H., Neurogenesis in the dentate gyrus of the adult rat: Age-related decrease of neuronal progenitor proliferation. J. Neurosci. 16, 2027–2033 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villeda S. A. et al., The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gontier G. et al., Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 22, 1974–1981 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn H. G., Toda T., Gage F. H., Adult hippocampal neurogenesis: A coming-of-age story. J. Neurosci. 38, 10401–10410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorrells S. F. et al., Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno-Jiménez E. P. et al., Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Tobin M. K. et al., Human hippocampal neurogenesis persists in aged adults and Alzheimer’s disease patients. Cell Stem Cell 24, 974–982.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang A. C., Jensen E. H., Rexach J. E., Vinters H. V., Hsieh-Wilson L. C., Loss of O-GlcNAc glycosylation in forebrain excitatory neurons induces neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 113, 15120–15125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagerlöf O., O-GlcNAc cycling in the developing, adult and geriatric brain. J. Bioenerg. Biomembr. 50, 241–261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheatley E. G. et al., Neuronal O-GlcNAcylation improves cognitive function in the aged mouse brain. Curr. Biol. 29, 3359–3369.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y., Shan X., Yuzwa S. A., Vocadlo D. J., The emerging link between O-GlcNAc and Alzheimer disease. J. Biol. Chem. 289, 34472–34481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S. et al., Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer’s disease. J. Pathol. 243, 78–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine P. M. et al., α-Synuclein O-GlcNAcylation alters aggregation and toxicity, revealing certain residues as potential inhibitors of Parkinson’s disease. Proc. Natl. Acad. Sci. U.S.A. 116, 1511–1519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love D. C. et al., Dynamic O-GlcNAc cycling at promoters of Caenorhabditis elegans genes regulating longevity, stress, and immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 7413–7418 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman M. M. et al., Intracellular protein glycosylation modulates insulin mediated lifespan in C. elegans. Aging 2, 678–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang H. et al., O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 11, 62–74 (2012). [DOI] [PubMed] [Google Scholar]
- 24.O’Donnell N., Zachara N. E., Hart G. W., Marth J. D., Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parween S. et al., Higher O-GlcNAc levels are associated with defects in progenitor proliferation and premature neuronal differentiation during in-vitro human embryonic cortical neurogenesis. Front. Cell. Neurosci. 11, 415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivier-Van Stichelen S., Wang P., Comly M., Love D. C., Hanover J. A., Nutrient-driven O-linked N-acetylglucosamine (O-GlcNAc) cycling impacts neurodevelopmental timing and metabolism. J. Biol. Chem. 292, 6076–6085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Praag H., Shubert T., Zhao C., Gage F. H., Exercise enhances learning and hippocampal neurogenesis in aged mice. J. Neurosci. 25, 8680–8685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He F. et al., A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 8, 616–625 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiwari N. et al., Stage-specific transcription factors drive astrogliogenesis by remodeling gene regulatory landscapes. Cell Stem Cell 23, 557–571.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X. et al., Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 214, 1093–1109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter R. L., Lo H. W., STAT3 target genes relevant to human cancers. Cancers (Basel) 6, 897–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalamakis G. et al., Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell 176, 1407–1419.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Beccari S., Valero J., Maletic-Savatic M., Sierra A., A simulation model of neuroprogenitor proliferation dynamics predicts age-related loss of hippocampal neurogenesis but not astrogenesis. Sci. Rep. 7, 16528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziebell F., Dehler S., Martin-Villalba A., Marciniak-Czochra A., Revealing age-related changes of adult hippocampal neurogenesis using mathematical models. Development 145, dev153544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tierney M. T. et al., STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 20, 1182–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichenbach N. et al., Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer’s disease model. EMBO Mol. Med. 11, e9665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horowitz A. M. et al., Blood factors transfer beneficial effects of exercise on neurogenesis and cognition to the aged brain. Science 369, 167–173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi S. H. et al., Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer’s mouse model. Science 361, eaan8821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith L. K. et al., β2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat. Med. 21, 932–937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babu H. et al., A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Front. Neurosci. 5, 89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubal D. B. et al., Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J. Neurosci. 35, 2358–2371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trinidad J. C. et al., Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S. et al., Schwann cell O-GlcNAc glycosylation is required for myelin maintenance and axon integrity. J. Neurosci. 36, 9633–9646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halim A. et al., Assignment of saccharide identities through analysis of oxonium ion fragmentation profiles in LC-MS/MS of glycopeptides. J. Proteome Res. 13, 6024–6032 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Baker P. R., Chalkley R. J., MS-viewer: A web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trombetta J. J. et al., Preparation of single-cell RNA-Seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1-4.22.17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White C. W. III, Couthouis J., Villeda S. A., RNA-Seq Data. Gene Expression Omnibus. (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143388). Deposited 9 January 2020.
- 50.White C. W. III, Maynard J. C., Villeda S. A., Mass spectrometry data: Raw data. MassIVE Database. https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=633978a3da404371acd5cc0b954cac31. Deposited 21 January 2020.
- 51.White C. W. III, Maynard J. C., Villeda S. A., Mass spectrometry data: Spectra. MS-Viewer. http://msviewer.ucsf.edu/prospector/cgi-bin/mssearch.cgi?report_title=MS-Viewer&search_key=qcchi2to8l&search_name=msviewer. Deposited 22 January 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data were deposited at GEO (accession no. GSE143388). Mass Spectrometry raw data were deposited at MassIVE (MSV000084825). Mass spectrometry spectra can be viewed at MS-Viewer (qcchi2to8l) (49–51).