Significance
The thermogenic function of adipocytes relies on the activation of the thermogenic program that is coordinately modulated by an array of unique transcriptional and epigenetic regulators. Histone methylations represent an essential epigenetic mode of regulation in the establishment of tissue-specific gene expression. However, the role of H4K20 methylation in thermogenic adipocytes is not completely elucidated. Our paper revealed Kmt5c is involved in regulating the thermogenic program in adipose tissues. Adipocyte-specific Kmt5c KO mice demonstrate decreased expression of thermogenic genes and are prone to diet-induced obesity. The findings indicate that activation of methyltransferase activity of KMT5c might be a potential strategy for metabolic diseases.
Keywords: KMT5c, H4K20me3, adaptive thermogenesis, brown adipocyte, beige adipocyte
Abstract
Brown and beige adipocytes harbor the thermogenic capacity to adapt to environmental thermal or nutritional changes. Histone methylation is an essential epigenetic modification involved in the modulation of nonshivering thermogenesis in adipocytes. Here, we describe a molecular network leading by KMT5c, a H4K20 methyltransferase, that regulates adipocyte thermogenesis and systemic energy expenditure. The expression of Kmt5c is dramatically induced by a β3-adrenergic signaling cascade in both brown and beige fat cells. Depleting Kmt5c in adipocytes in vivo leads to a decreased expression of thermogenic genes in both brown and subcutaneous (s.c.) fat tissues. These mice are prone to high-fat-diet-induced obesity and develop glucose intolerance. Enhanced transformation related protein 53 (Trp53) expression in Kmt5c knockout (KO) mice, that is due to the decreased repressive mark H4K20me3 on its proximal promoter, is responsible for the metabolic phenotypes. Together, these findings reveal the physiological role for KMT5c-mediated H4K20 methylation in the maintenance and activation of the thermogenic program in adipocytes.
Brown and beige adipocytes have the distinct capacity of nonshivering thermogenesis by engaging in futile cycling of metabolites (1, 2). The best-characterized mechanism of adipose thermogenesis is via expression of uncoupling protein 1 (UCP1). UCP1 uncouples oxidative respiration from mitochondrial ATP synthesis, resulting in energy derived from fatty acid and glucose oxidation that is dissipated as heat. In addition, alternative UCP1-independent thermogenesis has also been uncovered in recent years (3–5). Brown adipose tissue (BAT) exists in discrete depots in rodents and humans. By contrast, beige adipocytes appear in white adipose tissue (WAT) in response to chronic cold exposure, exercise, and treatment with ligands of peroxisome proliferator activated receptor γ (PPARγ) in a process referred to as browning of white fat (6). The recruitment of new beige cells in adult humans leads to an increase in energy expenditure and an improvement in insulin sensitivity (7, 8). Recent studies further demonstrated that human BAT is metabolically active even in warm conditions and is involved in diet-induced thermogenesis and fat utilization (9). All these findings support the potential significance of thermogenic adipocytes in the treatment of obesity and type 2 diabetes.
The thermogenesis of brown and beige adipose cells relies on the activation of the thermogenic adipose program that is coordinately modulated by an array of unique transcriptional and epigenetic regulators (10). Histone methylations represent an essential epigenetic mode of regulation in the establishment of tissue-specific gene expression. Methylation of histone mainly occurs on histone H3 lysine 4, histone H3 lysine 9 (H3K9), histone H3 lysine 27 (H3K27), histone H3 lysine 36 (H3K36), histone H3 lysine 79 (H3K79), histone H4 lysine 20 (H4K20), and specific arginine residues. Some of them have been reported to be involved in the regulation of adipocyte identity, adipogenesis, or thermogenesis. Notable among these are the dynamics of H3K9 methylation catalyzed by histone methyltransferases and demethylases. H3K9 methyltransferase euchromatic histone lysine methyltransferase 1 (EHMT1) controls brown adipocyte versus muscle cell lineage by depositing H3K9 dimethylation (H3K9me2) and trimethylation (H3K9me3) at muscle-selective gene promoters (11). In mature brown adipocytes, EHMT1 represses the expression of white-fat-selective genes (12). JMJD1a, a H3K9 demethylase, erases H3K9me2 modification on beige-selective gene promoters in s.c. WAT to provide long-term activation of thermogenic genes in response to chronic cold exposure (13). Lysine-specific histone demethylase 1 (LSD1) promotes the expression of BAT-selective genes and the development of beige adipocytes, which demethylates H3K9 to transcriptionally activate the thermogenic adipose program through its interaction with zinc finger protein 516 (ZFP516) (14). A subset of BAT-selective genes distinguishes from common fat genes partly by H3K27me3 marks on promoters in brown preadipocytes. Demethylation of H3K27me3 by Jumonji domain-containing protein 3 (JMJD3) or UTX is required for BAT gene expression and browning of WAT (15, 16). Depletion of histone H3K36 methylation in adipocytes in vivo leads to whitening of BAT (17). However, little is known about the role of H4K20 methylation in thermogenic fat cells.
H4K20 can accept up to three methyl groups, forming H4K20me1, H4K20me2, and H4K20me3 derivatives. Methylated H4K20 is associated with diverse biological processes from the cell cycle, DNA damage repair, and chromatin compaction to transcriptional regulation (18). Three distinct lysine methyltransferase enzymes, KMT5a (also known as SETD8, SET8, and PR-Set7), KMT5b (SUV420H1), and KMT5c (SUV420H2), are responsible for the generation of the methylated H4K20. KMT5a converts unmodified H4K20 to H4K20me1, whereas KMT5b and KMT5c mediate successive methylations of H4K20me1 to H4K20me2 and H4K20me3. Kmt5a is a Pparγ target gene during adipogenesis and positively regulates the expression of PPARγ through H4K20me1 modification and adipocyte differentiation in 3T3-L1 cells (19). Knockdown of Kmt5c suppresses brown adipocyte differentiation in vitro (20). Moreover, double-KO (DKO) Kmt5b and Kmt5c mice display enhanced BAT metabolic activity and increased browning of WAT, thus, counteracting diet-induced obesity (21). The DKO mice in this study constitutively lack Kmt5c and KO Kmt5b in the Myf5 lineage, which expresses in early mesenchymal precursor cells (22) to delete Kmt5b in brown preadipocytes and muscle cells. The activation of Pparγ transcription upon Kmt5b and Kmt5c deletion accounts for the increased energy expenditure in vivo (21). Although these progress, the role of individual H4K20 methyltransferase in committed adipocytes needs to be further explored.
Here, we demonstrate that KMT5c but not KMT5a or KMT5b plays an important role in thermogenic gene expression in fat cells. KO of Kmt5c in adipocytes driven by Adipoq-Cre suppresses the expression of Ucp1 in BAT and inguinal WAT (iWAT). Kmt5c KO mice are susceptible to obesity when fed a high-fat diet (HFD). Mechanically, increased expression of Trp53 provides molecular insight into how Kmt5c deletion can inhibit the thermogenic gene program.
Results
Knockdown of Kmt5c Inhibits the Thermogenic Gene Program in Brown Adipocytes.
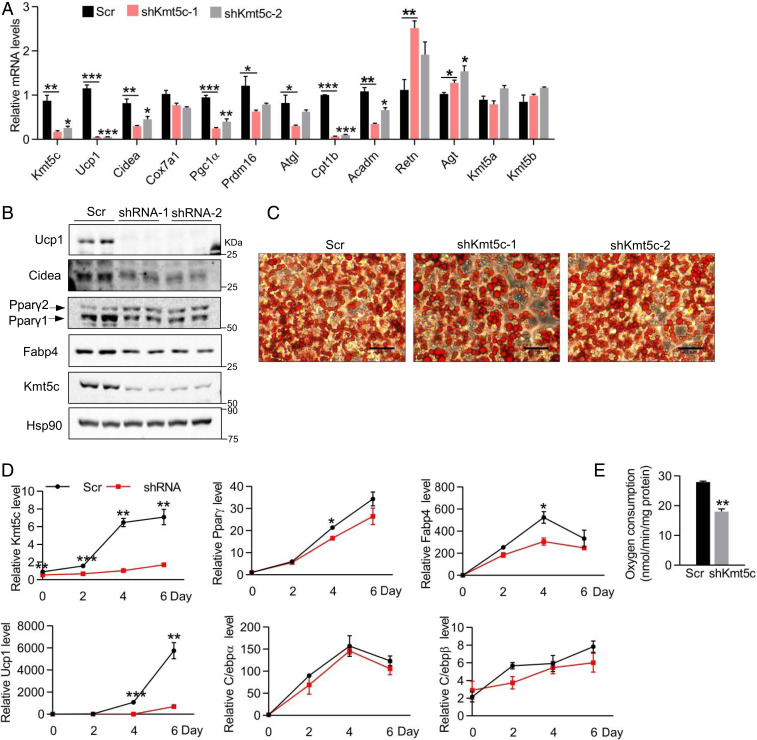
Epigenetic and transcriptional regulators coordinate to modulate the adipose thermogenic program. To test whether the histone H4K20 methylation mark might be involved in the regulation of brown adipose thermogenic gene expression, we first used quantitative RT-PCR to profile the expression of H4K20 methyltransferases, including Kmt5a, Kmt5b, and Kmt5c. All these three methyltransferases express in adipose tissues (SI Appendix, Fig. S1A), and their messenger RNA (mRNA) levels increase during brown adipogenesis (SI Appendix, Fig. S1B). A-196 is a potent and substrate-competitive inhibitor of KMT5b and KMT5c (23). Treating the mature brown adipocytes by A-196 for 24 h remarkably decreased Ucp1, Cox7a1, Cpt1b, and Pparγ2 mRNA levels (SI Appendix, Fig. S1C). To distinguish whether KMT5b, KMT5c, or both of them are responsible for the repression of these gene expressions, we then knocked down Kmt5a, Kmt5b, or Kmt5c with lentiviral short hairpin RNAs (shRNAs) in immortalized brown preadipocytes individually following differentiating them into mature brown adipocytes. The cells with reduced expression of Kmt5a, Kmt5b, or Kmt5c showed regular adipogenesis. Knockdown of Kmt5a or Kmt5b had marginal effects on Ucp1 expression (SI Appendix, Fig. S1 E and F). However, deletion of Kmt5c dramatically decreased both basal and forskolin-induced Ucp1 mRNA expression without eliciting changes in the expression of Kmt5a and Kmt5b (Fig. 1A and SI Appendix, Fig. S1D). Different from the previous study (20), we did not see much defect in cell differentiation when knocking down Kmt5c (Fig. 1C), which may be due to the different growth medium formulation used for the preadipocytes' culture. Strikingly, knockdown of Kmt5c also led to a broad loss of brown-fat-selective gene expression, including reduced levels of Cidea, Pgc1α, Cpt1b, and Acadm (Mcad) (Fig. 1A). Reduction of UCP1 and the CIDEA protein was further confirmed by Western blot analysis (Fig. 1B). On the contrary, the expression of white fat-selective genes Retn and Agt was increased in Kmt5c-knockdown cells (Fig. 1A).
Fig. 1.
Kmt5c regulates the thermogenesis gene program in brown adipocytes. (A) Immortalized brown preadipocytes were infected with shKmt5c or scrambled knockdown lentiviruses and differentiated. Gene expression in mature brown fat cells was analyzed. n = 3. (B) Representative Western blot analysis of UCP1, CIDEA, PPARγ, FABP4, and KMT5c in adipocytes generated as in A. Values represent the results from three experimental replicates. (C) Representative oil-red O staining of mature adipocytes generated as in A. (Scale bar, 40 μm.) (D) Kmt5c was knocked down in brown preadipocytes. Relative mRNA expression of indicated genes during brown adipogenesis. n = 3. (E) Oxygen consumption rate was measured with a Clark-type electrode in brown adipocytes generated as in A. n = 3. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Kmt5c depletion decreased protein levels of general adipocyte markers FABP4 and PPARγ (Fig. 1B), although oil-red O staining showed similar lipid droplet accumulations in Kmt5c-knockdown adipocytes relative to the control cells (Fig. 1C). Then, we asked why PPARγ reduction did not interrupt adipogenic differentiation in Kmt5c-knockdown cells. To address this question, Kmt5c, Pparγ, Cebpα/β, Fabp4, and Ucp1 mRNA levels were further explored during the course of brown adipogenesis. The induction of Kmt5c mRNA during adipogenic differentiation was completely abolished in knockdown cells (Fig. 1D). The retardation of the increase in Pparγ, Fabp4, and Ucp1 was observed on day 4 in knockdown cells (Fig. 1D). There was no significant difference in the expression of C/ebpα and C/ebpβ, another two principal adipogenic transcription factors, between control and knockdown cells (Fig. 1D). PPARγ binds to the Fabp4 promoter at the early differentiation stage (day 2). By contrast, it attaches to brown adipocyte-selective gene promoters and activates their expression until day 4 (24, 25). Transduction of lentiviral Pparγ shRNA to brown fat cells on day 3 did not disturb general adipocyte differentiation as revealed by comparable C/ebpα/β and Fabp4 levels but significantly decreased Ucp1 expression (SI Appendix, Fig. S1G). To minimize the potential effect of defective adipogenesis on brown fat-selective gene expression in Kmt5c depletion cells, we induced brown adipocyte differentiation first following knocking down Kmt5c on differentiation day 3 by shRNAs. The Kmt5c mRNA level was slightly but significantly decreased. The expression of Ucp1, Cidea, and Cox7a1 still displayed remarkable reduction, although with a comparable level of Pparγ2 in Kmt5c-knockdown cells (SI Appendix, Fig. S1H). These data together show the decreased Ucp1 expression is not always linked to the reduced PPARγ level, suggesting that down-regulation of the thermogenic program caused by deletion of Kmt5c is, at least, partly independent of PPARγ.
Importantly, the effect of Kmt5c knockdown is functionally relevant as the rate of oxygen consumption in knockdown cells was significantly lower than control cells (Fig. 1E). Collectively, these data demonstrated that KMT5c is required for thermogenic gene expression in brown fat cells in vitro.
KMT5c Is Induced by β3-Adrenergic Signaling.
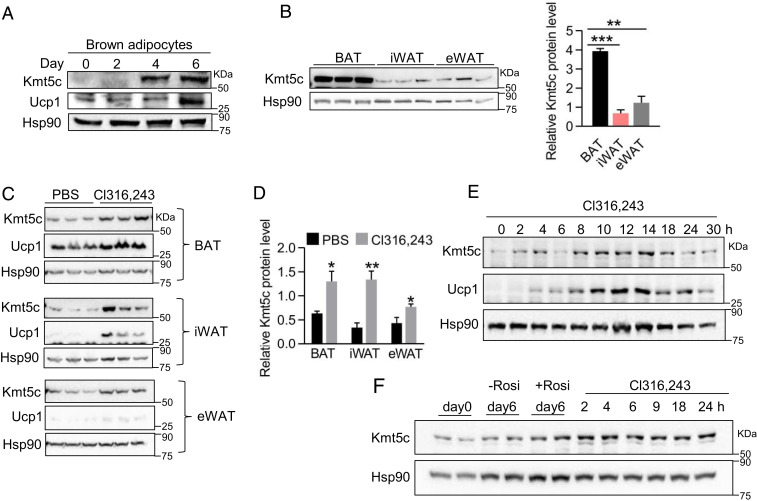
Next, we examine the physiological regulation of Kmt5c expression in adipocytes and adipose tissues. Consisting of its mRNA expression (SI Appendix, Fig. S1B), the KMT5c protein was greatly increased when brown preadipocytes differentiated into mature adipocytes (Fig. 2A). The significantly higher KMT5c protein level in BAT relative to iWAT and epididymal WAT (eWAT) (Fig. 2B) further indicates its potential roles in brown fat tissue thermogenesis in vivo. The thermogenic program in BAT is activated in response to β-adrenergic agonists and cold exposure, and, under these stimuli, recruitable beige cells also develop the ability of adaptive thermogenesis (SI Appendix, Fig.S2A). Although the Kmt5c mRNA level in BAT is reported to be dramatically reduced upon acute cold exposure (21), intraperitoneal (i.p.) injection of the β3-adrenergic receptor agonist Cl316,243 for 5 d significantly increased the KMT5c protein level in brown and white adipose tissues, that mirrored UCP1 expression (Fig. 2 C and D). The expression of the KMT5c protein in adipose tissues was also slightly induced by 2 to 3 d of 4 °C cold challenge (SI Appendix, Fig. S2B). The discrepancy observed in the induction of KMT5c by β-adrenergic signaling in vivo may be due to the different treatment duration. Furthermore, KMT5c induction in adipose tissues by Cl316,243 is cell autonomous since Cl316,243 treatment could strongly promote the expression of KMT5c in both immortalized brown adipocytes (Fig. 2E) and primary iWAT cells (Fig. 2F). Noticeably, the KMT5c protein was marginally increased in rosiglitazone-stimulated beige cells (Fig. 2F, lanes 5 and 6 versus lanes 3 and 4) but was drastically induced by Cl316,243 treatment (Fig. 2F, lanes 7–12), further suggesting β3-adrenergic signaling cascade is involved in the KMT5c induction in beige adipocytes. The amount of functionally active BAT is reversely correlated with body-mass index in human adults and obese (ob/ob) animals (26, 27). Genetically ob/ob mice have lower energy expenditure rates, and BAT depots show impaired thermogenic activity (28, 29). KMT5c expression was twofold increased in BAT of ob/ob mice relative to lean mice (SI Appendix, Fig. S2C), indicating a possible feedback regulation on BAT Kmt5c expression by obesity.
Fig. 2.
KMT5c can be induced by β3-adrenergic signaling. (A) Representative KMT5c protein level during the differentiation of immortalized brown fat cells. Values represent the results from three experimental replicates. (B) Western blot analysis of the KMT5c protein (Left) and its quantification (Right) in BAT, iWAT, and eWAT adipose tissues of 8-wk-old male mice. n = 3. (C) KMT5c and UCP1 protein levels in adipose tissues of 10-wk-old male mice after i.p. injection of Cl316,243 (0.5-mg/kg body weight) or phosphate-buffered saline (n = 3 per group) for 5 d. (D) Quantification of the KMT5c protein in C. (E) Representative Western blot analysis of the KMT5c and UCP1 proteins in brown adipocytes. Mature brown fat cells differentiated from immortalized brown preadipocyte were treated with 10-μM Cl316,243 for the indicated time. (F) Representative KMT5c protein level in beige adipocytes. Stroma vascular fractions (SVFs) isolated from s.c. WAT were induced to beige adipocytes. Rosiglitazone (Rosi, 1 μM) was used during differentiation to promote beige cells formation unless otherwise indicated. Mature beige cells were treated with or without 10-μM Cl316,243 for the indicated time. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Kmt5c Deficiency Abolishes Browning of White Adipocytes.
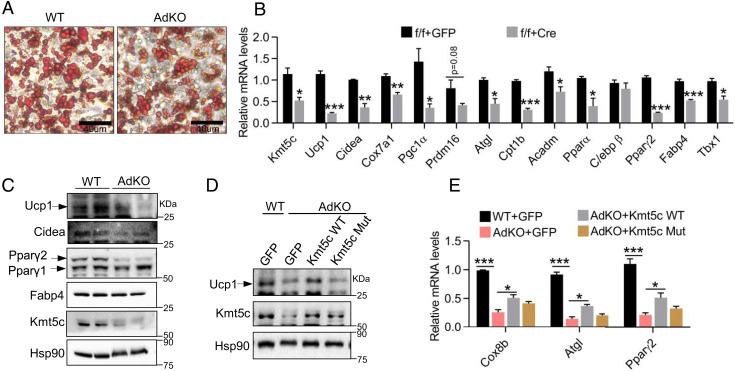
We noted that the KMT5c protein is significantly increased in iWAT and beige cells when the activation of β3-adrenergic receptors occurs (Fig. 2 C and F), suggesting it may be also involved in white fat browning. KO of Kmt5c in beige adipocytes was achieved by infecting Cre recombinase adenovirus to differentiating SVF cells isolated from iWAT of mice carrying a homozygous floxed Kmt5c allele (Kmt5cflox/flox) or isolating SVF cells from iWAT of Kmt5cflox/flox wild type (WT) and Kmt5c KO mice (Kmt5cflox/flox, Adipoq-Cre, and [adipocyte KO] AdKO) (SI Appendix, Fig. S4A). Kmt5c KO adipocytes with little Kmt5c mRNA and protein left (Fig. 3 B and C) appeared to fully differentiate into mature adipocytes (Fig. 3A). At a molecular level, reduction of Kmt5c expression in beige adipocytes led to a systematic decreased expression of adipocyte genes (Fig. 3 B and C), which largely phenocopied the knockdown brown fat cells (Fig. 1A). A decrease in the Pparγ mRNA level was observed from differentiation day 4 to day 6 (SI Appendix, Fig. S3B). The expression of Kmt5c (SI Appendix, Fig. S3A), Ucp1 (SI Appendix, Fig. S3C), and other genes examined, including Cidea, Atgl, Cebpα, Cebpβ, Fabp4, and Cd36, was all reduced in the last 2 d of differentiation (SI Appendix, Fig. S3 D–I). Moreover, Ucp1, Cox8b, Atgl, and Pparγ expressions could be partly restored by reconstitution of Kmt5c in AdKO cells (Fig. 3 D and E), but not by the mutant Kmt5c (G140Y/W174A) that lacks methyltransferase activity (30). Together, Kmt5c deficiency in beige adipocytes reduces the expression of BAT-selective markers, lipid metabolism genes, and common adipogenic markers in the final differentiation stage. The requirement for KMT5c in the beige adipocytes thermogenic program, at least, partly relies on its methyltransferase activities.
Fig. 3.
Kmt5c knockdown decreases BAT-selective gene expression in beige cells. (A) Primary iWAT preadipocytes were isolated from WT and Kmt5c AdKO mice and differentiated into beige cells. On day 6, oil-red O staining was performed. (Scale bar, 40 μm.) (B) Gene mRNA expression in primary iWAT adipocytes (n = 3). On differentiation day 2, Cre adenovirus infected cultured iWAT adipocytes isolated from Kmt5cflox/flox mice to the KO Kmt5c allele. A green fluorescent protein virus was used as the control. (C) Representative Western blot analysis in primary mature iWAT adipocytes generated as in A. Values represent the results from three experimental replicates. (D) Representative Western blot analysis of the UCP1 and KMT5c protein expressions in beige cells. Kmt5c KO iWAT adipocytes were infected with WT or a methyltransferase-dead mutant (Mut) Kmt5c adenovirus for 24 h. Values represent the results from three experimental replicates. (E) The mRNA levels of indicated genes were analyzed in cells generated as in D (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Kmt5c Regulates Ucp1 Expression In Vivo.
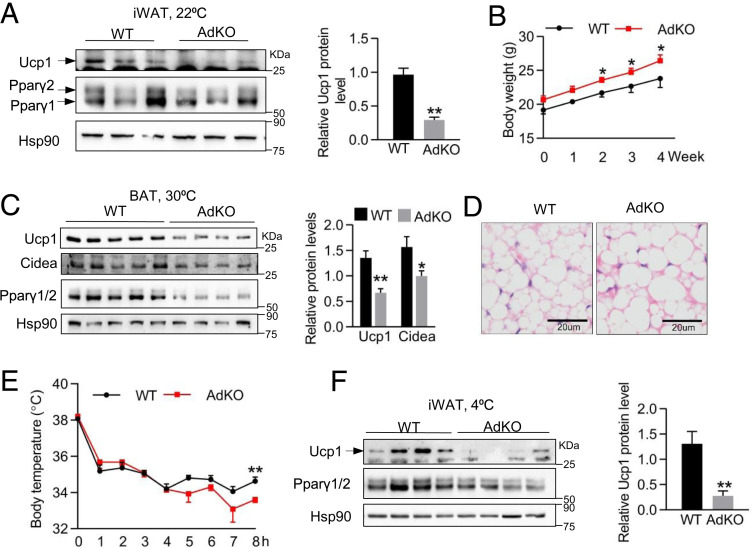
To determine whether KMT5c is genetically required for the thermogenic adipose program in vivo, we generated mice lacking Kmt5c specifically in adipose tissues by crossing Kmt5cflox/flox mice to Adipoq-Cre lines (SI Appendix, Fig. S4A). AdKO mice are apparently indistinguishable from WT littermates at birth. Then, they were raised at room temperature with a chow diet until 2-mo old. Gene expression analysis demonstrated that Adipoq-Cre recombinase decreased Kmt5c mRNA expression specifically in brown and white adipose tissues but did not affect the Kmt5c level in other detected tissues (SI Appendix, Fig. S4B). Kmt5c deletion did not cause a compensatory increase in the expression of Kmt5b in adipose tissues, but a slightly decreased Kmt5a was seen in BAT of AdKO mice (SI Appendix, Fig. S4 C and D). Chow diet-fed AdKO mice had similar body weight, comparable food intake, equal cell size, and cell numbers in adipose tissues as indicated by the unchanged genomic DNA content as well as similar amounts of fat mass as WT mice (SI Appendix, Fig. S4 E–I). AdKO mice had significantly less Ucp1 levels in iWAT compared to WT mice, and the expression of other thermogenic genes and Pparγ2 in both BAT and iWAT is similar (Fig. 4A and SI Appendix, Fig. S4 J and K)
Fig. 4.
Kmt5c deficiency blunts the expression of Ucp1 in adipose tissues. (A) UCP1 and PPARγ protein levels in iWAT tissues of mice housing at 22 °C. Right shows quantification of the UCP1 protein. n = 3 per group. (B) Body weight of mice fed a HFD and housed at 30 °C for 4 wk. n = 4 to 5 per group. (C) The protein levels of UCP1, CIDEA, and PPARγ in BAT of mice in B. Right shows quantification of UCP1 and CIDEA proteins. n = 4 to 5 per group. (D) Representative BAT hematoxylin/eosin (H&E) staining in mice of (B). (E) Rectal temperature of 8-wk-old male WT and AdKO mice at 4 °C. n = 5 to 6 per group. (F) UCP1 and PPARγ protein levels in iWAT of mice in E. Right shows quantification of the UCP1 protein. n = 4 per group. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Room temperature (22 °C) is already below the mice thermoneutral zone, which profoundly impacts BAT metabolism (31). To study the requirement of Kmt5c in adipose thermogenesis at the setting physiologically relevant to humans, WT and AdKO mice were fed a regular or high-fat diet at thermoneutrality (30 °C). A decreased UCP1 protein level was discerned in BAT of AdKO after 3-wk chow feeding at 30 °C (SI Appendix, Fig. S4L). Additionally, 4-wk HFD consumption let AdKOs housed at thermoneutrality gain more body weight (Fig. 4B). Dramatically decreased UCP1, CIDEA, and PPARγ protein levels and increased adipocyte size were observed in BAT of AdKOs (Fig. 4 C and D), implying defects of diet-induced BAT thermogenesis with Kmt5c ablation.
Next, we wonder whether AdKO mice are cold sensitive. WT and AdKO mice were exposed to 4 °C acutely. The core body temperature of AdKO mice was slightly lower than that of WT only at the 8-h time point (Fig. 4E). The expression levels of Ucp1, Atgl, CD36, and PPARγ were remarkably decreased in iWAT of AdKO mice (Fig. 4F and SI Appendix, Fig. S4M) but without significant differences in gene expression in BAT (SI Appendix, Fig. S4N). Alternative mechanisms of adipose adaptive thermogenesis have been discovered in recent years (3, 4). The expression of key genes responsible for creatine-dependent substrate cycling and the calcium-dependent futile cycle did not exhibit any difference in AdKO iWAT after cold challenge (SI Appendix, Fig. S4O). Taken together, KMT5c is important for maintaining BAT Ucp1 and Pparγ expression at thermoneutral settings and is required for the iWAT thermogenic program at ambient temperature and upon acute cold exposure.
KMT5c-Deficient Mice Gain More Body Weight on a HFD Feeding.
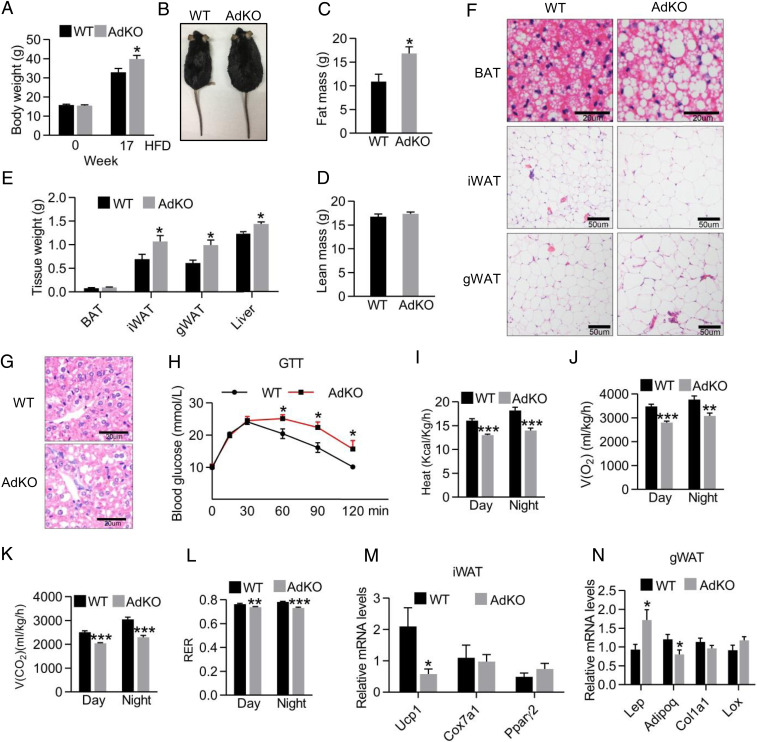
Then, we sought to investigate whether the impaired thermogenic program in iWAT is functionally relevant. We assessed the effects of Kmt5c deficiency by challenging the KO mice with a HFD. The female AdKO mice were susceptible to diet-induced obesity. The average body weight of AdKO mice was 6.9 g more than that of WT after 17 wk of a HFD feeding (Fig. 5A and SI Appendix, Fig. S5A), although they consumed similar amounts of high-fat food (SI Appendix, Fig. S5B). AdKO mice appeared substantially obese (Fig. 5B) because of more fat accumulation as revealed by increased fat mass whereas similar lean mass as WTs (Fig. 5 C and D). Accordingly, weights of inguinal and epididymal adipose depots and livers were considerably more in HFD AdKOs (Fig. 5E and SI Appendix, Fig. S5C). The morphology of adipocytes and hepatocytes was also examined. In HFD AdKOs, brown adipocytes and hepatocytes accumulated substantially more fat, and white adipocytes were more hypertrophic (Fig. 5 F and G). Moreover, a glucose tolerance test found that HFD AdKOs were more glucose intolerant (Fig. 5H). However, an insulin tolerance test and phosphorylated Akt level in tissues revealed that the AdKO mice did not display signs of insulin resistance (SI Appendix, Fig. S5 D and E). Accordingly, WT and AdKO mice had similar serum insulin levels (SI Appendix, Fig. S5F). Metabolic analysis of HFD AdKOs showed reduced heat production, decreased O2 consumption, decreased CO2 production, and respiratory exchange ratio (RER) (Fig. 5 I–L and SI Appendix, Fig. S5 G–J) in a 24-h time period relative to WTs. Importantly, male AdKO mice also gained slightly but significantly more weight than WTs when fed a HFD (SI Appendix, Fig. S5K).
Fig. 5.
Kmt5c deficient mice are predisposed to diet-induced obesity. (A) WT and AdKO female mice body weights before and after a HFD feeding. n = 5 to 6 per group. Values represent the results from two experimental replicates. (B) Mice in A after 17 wk of HFD feeding. (C and D) Fat mass (C) and lean mass (D) of mice in A. n = 5 to 6 per group. (E) Fat pad and liver weight of WT and AdKO mice in A. n = 5 to 6 per group. (F and G) Representative H&E staining of adipose tissues (F) and livers (G) from mice in A. (Scale bars, 20 μm for BAT and liver and 50 μm for iWAT and eWAT.) (H) Glucose tolerance test in WT and AdKO mice in A at 15 wk of a HFD. n = 5 to 6 per group. (I–L) Metabolic cage analyses in WT and AdKO mice at 17 wk of a HFD. Heat production (I), oxygen consumption (J), carbon dioxide production (K), and RER (L) were measured. n = 5 to 6 per group. (M) iWAT mRNA levels of Ucp1, Cox7a1, and Pparγ2 in mice of A. n = 5 to 6 per group. (N) gWAT mRNA levels of Lep, Adipoq, Col1a1, and Lox in mice of A. n = 5 to 6 per group. *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Quantitative PCR revealed thermogenic gene expression in BAT was comparable between AdKO and WT mice (SI Appendix, Fig. S5L). On the other hand, a significant decrease in the Ucp1 mRNA level was seen in iWAT of HFD AdKOs (Fig. 5M). A higher level of Lep, decreased Adipoq, and similar fibrosis marker gene expression were seen in the Kmt5c-deficient gonadal WAT (gWAT) relative to WT mice (Fig. 5N).
Depletion of Kmt5c Enhances Trp53 Expression.
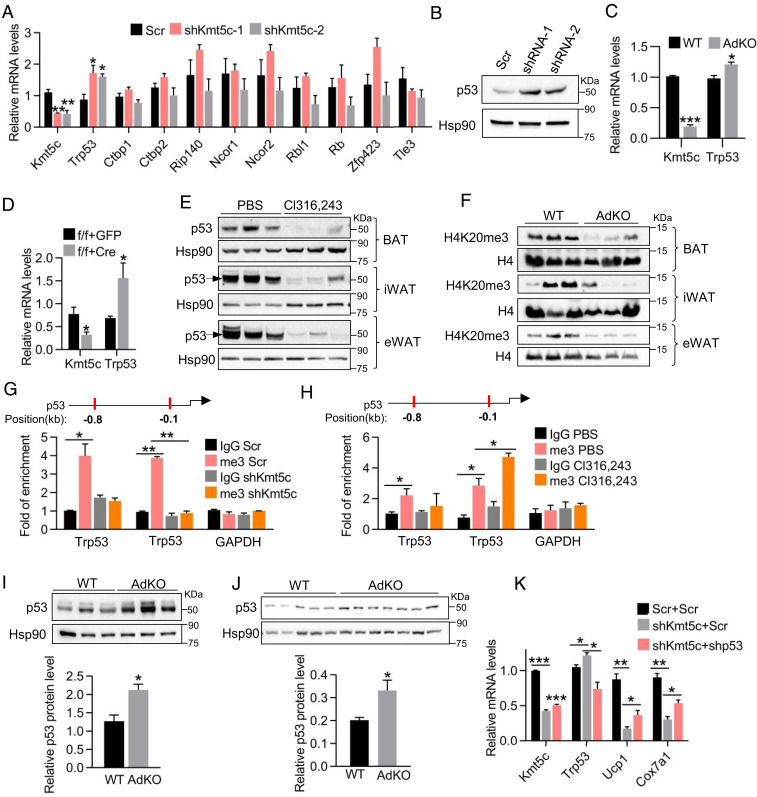
KMT5c is predominantly responsible for trimethylation deposition to histone H4K20. A decrease in H4K20me3 modification is usually associated with transcriptional activation (32). The fact that repressed the adipocyte thermogenic gene program in Kmt5c KO cells in vitro and in vivo leads us predict Ucp1, Cox7a1, or Pparγ genes should not be the direct targets of KMT5c. Therefore, we assume KMT5c represses the expression of a negative regulator of adipose thermogenic machinery. To test this hypothesis, we screened a list of known negative regulators of thermogenesis in Kmt5c-knockdown brown fat cells (10). Trp53 was the only candidate whose expression was significantly increased in Kmt5c-deficient cells both in brown and in beige adipocytes (Fig. 6 A–D). Given the fact that adipose KMT5c is significantly induced by a β3-adrenergic receptor agonist (Fig. 2C), we examined the p53 protein level in adipose tissues after Cl316,243 injection. As expected, the protein level of p53 was considerably decreased by Cl316,243 in brown and white fat tissues (Fig. 6E, the specificity of the p53 antibody was shown in SI Appendix, Fig. S6A by including Trp53 knockdown samples). These data demonstrate that the protein levels of KMT5c and p53 are negatively correlated in adipocytes and adipose tissues.
Fig. 6.
Increased p53 expression in Kmt5c KO adipocytes is responsible for repression of the thermogenic program. (A) Quantitative RT-PCR screening of negative regulators in the thermogenic program in Kmt5c knockdown brown adipocytes. n = 3. (B) Representative Western blot analysis of the p53 protein in Kmt5c knockdown brown adipocytes. Values represent the results from three experimental replicates. (C) Trp53 mRNA level in isolated iWAT adipocytes from WT and AdKO mice (n = 3). (D) Trp53 mRNA level in primary iWAT adipocytes as generated in Fig. 3B (n = 3). (E) p53 protein level in adipose tissues after Cl316,243 injection for 5 d. n = 3 mice per group. (F) Adipose tissue H4K20me3 protein level in WT and AdKO mice. (G) Representative H4K20me3 chromatin immunoprecipitation (ChIP) assay on the Trp53 promoter in Kmt5c-knockdown brown adipocytes (n = 3). Values represent the results from three experimental replicates. (H) Representative H4K20me3 ChIP assay on the Trp53 promoter in s.c. fat tissues. n = 4 per group. (I) p53 protein expression and its quantification in iWAT of 8-wk-old WT and AdKO mice. (J) p53 protein expression and its quantification in iWAT of WT and AdKO mice after 17-wk HFD feeding. (K) Relative mRNA levels in Kmt5c knockdown and scrambled control brown adipocytes. Lentiviral Trp53 shRNA was infected with Kmt5c-knockdown cells on differentiation day 2 (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM.
Elevated Trp53 expression was observed from day 4 to day 6 but not in preadipocytes with knockdown of Kmt5c in brown fat cells (SI Appendix, Fig. S6 B and C). While the levels of well-defined Trp53 target genes, which are involved in cell-cycle arrest, apoptosis, or cell proliferation, were not altered in mature adipocytes (SI Appendix, Fig. S6 D and E). It may be due to mature adipocytes as a terminal differentiated cell type have left the cell cycle and lost the ability to cell divide.
Kmt5c regulates target gene expression through deposition of the H4K20me3 mark on gene promoter regions (33). Next, we examined the H4K20me3 level in Kmt5c-knockdown cells and fat tissues of KO mice. Western blot and immunofluorescence staining revealed Kmt5c depletion reduced the H4K20me3 level in fat tissues (Fig. 6F), immortalized brown adipocytes (SI Appendix, Fig. S6F), and primary iWAT cells (SI Appendix, Fig. S6G). The proximal promoter of Trp53 is enriched in the H4K20me3 mark in both brown and beige fat cells (SI Appendix, Fig. S6 H and I). Knockdown of Kmt5c in brown fat cells erased H4K20me3 on the Trp53 proximal promoter region (Fig. 6G). By contrast, Cl316,243 treatment increased H4K20me3 modification on the Trp53 promoter in s.c. fat tissues (Fig. 6H).
The role of p53 in cellular metabolism is emerging, positioning p53 as a fundamental regulatory hub in adipose tissue metabolism (34). It is reported that elevated p53 in WAT aggravates systemic insulin resistance (35). Whole-body Trp53 KO mice gained less weight and showed increased BAT and beige thermogenic activity (36). To verify whether p53 is responsible for the phenotypes in Kmt5c-deficient mice, p53 expression in iWAT of AdKO mice fed with normal chow or a HFD was detected. Elevated p53 mRNA and protein expressions were seen in iWAT of AdKO mice (Fig. 6 I and J and SI Appendix, Fig. S6 J and K). There was no difference in the Trp53 mRNA level in BAT between WT and AdKO mice housing at 22 °C (SI Appendix, Fig. S6L), which was in agreement with the relatively normal gene expression profile in BAT of AdKOs. In addition, overexpression of Trp53 at the physiological level in primary iWAT SVF cells did not affect adipocyte differentiation (SI Appendix, Fig. S6M) but repressed the expression of Ucp1, Cox7a1, and Cidea (SI Appendix, Fig. S6N). A similar gene expression profile was also observed in brown adipocytes when Trp53 was overexpressed (SI Appendix, Fig. S6O). Knockdown of Trp53 can partly resume Ucp1 and Cox7a1 expressions in Kmt5c-knockdown fat cells (Fig. 6K). Together, these lines of molecular evidence suggest that p53 functions as one of the key mediators downstream of KMT5c in adipocytes.
KMT5c and YBX1 Cooperate to Suppress Trp53 Expression.
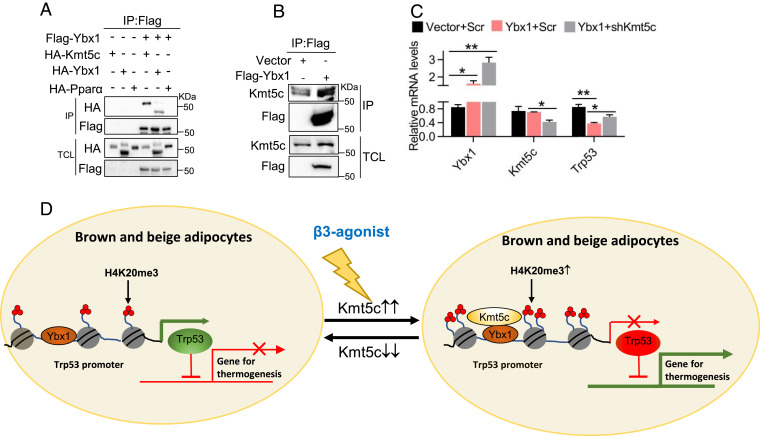
Lastly, we asked the molecular basis for the recruitment of KMT5c to the Trp53 promoter. Transcription factors often orchestrate chromatin-modifying enzymes binding to the promoter of specific genes and result in the activation or silencing of the gene expression. To find out the involved transcription factors, we purified the KMT5c protein complex in brown adipocytes stably expressing a HA-tagged KMT5c protein. Then liquid chromatography coupled with tandem mass spectrometry was performed (SI Appendix, Table S1). Our attention is attracted to the transcription factor YBX1, which is reported to interact with Ewing sarcoma (EWS) to activate the expression of Bmp7 in the early stage of adipocytes differentiation (37). YBX1 is also a potential negative regulator of Trp53 in several tumor cell lines (38), but the underlying mechanism is still unknown. We assume YBX1 may be responsible for recruitment of KMT5c to the Trp53 promoter. An interaction between YBX1 and KMT5c was readily detected when overexpressing Flag-YBX1 and HA-KMT5c in HEK293T cells (Fig. 7A). Lentiviral expression of Flag-YBX1 at the physiological level (Fig. 7C) could associate with endogenous KMT5c in brown adipocytes (Fig. 7B). Overexpressed YBX1 in brown adipocytes inhibited the expression of Trp53 (Fig. 7C). Simultaneous knockdown of Kmt5c partly restored the mRNA level of Trp53 (Fig. 7C). To summarize, our data together uncover a molecular network leading by KMT5c that regulates adipose adaptive thermogenesis and systemic energy expenditure in vivo (Fig. 7D).
Fig. 7.
YBX1 coordinates KMT5c to inhibit p53 expression. (A) Representative coimmunoprecipitation analysis in HEK293T cells cotransfected with Flag-Ybx1 and HA-Kmt5c plasmids. YBX1 can form the homodimer. The interaction between Flag-YBX1 and HA-YBX1 is used as a positive control. The interaction between YBX1 and PPARα is used as a negative control. Values represent the results from two experimental replicates. (B) Detection of KMT5c in Flag-YBX1 immunoprecipitates isolated from brown fat cell extracts. Values represent the results from two experimental replicates. (C) Immortalized brown preadipocytes were infected with Ybx1 overexpression lentivirus and differentiated. Lentiviral Kmt5c shRNA or scrambled control infected the Ybx1-overexpressing adipocytes at differentiation day 2. Gene expression was analyzed at day 6 (n = 3). *P < 0.05; **P < 0.01, Two-tailed unpaired Student’s t test was performed. The results shown are the mean ± SEM. (D) A model for KMT5c action in thermogenic adipocytes. KMT5c modulates the thermogenic program in adipocytes, in part, through downregulating Trp53 expression. YBX1 cooperates with KMT5c to suppress the expression of Trp53.
Discussion
We report, here, that KMT5c is induced upon activation of the β-adrenergic signaling cascade and modulates the adaptive thermogenesis gene expression, specifically in BAT at thermoneutrality and iWAT at ambient and cold challenges. Knockdown of Kmt5c in cultured brown or primary iWAT adipocytes dramatically reduces the expression of a broad panel of thermogenic, mitochondrial, and fatty acid oxidation genes, which indicates that the effects caused by Kmt5c KO in vivo are adipocyte autonomous. Although BAT Kmt5c expression is much higher than WAT, there were no alterations in thermogenic gene expression in BAT of Kmt5c AdKO mice at room temperature. We predict there could be some compensation mechanisms in BAT to maintain regular thermogenesis at the subthermoneural environment (22 °C). PGC1α and PRDM16 are two fundamental transcriptional factors responsible for the adipocyte thermogenic program (39), but intact brown fat thermogenic gene expression has also been reported in Pgc1α and Prdm16 KO mice (40, 41).
KMT5c deposits di- and trimethylation to H4K20, which is associated with transcription repression and chromatin compaction. The proximal region of the Trp53 promoter is remarkably marked by H4K20me3 modification in adipocytes (SI Appendix, Fig. S6 H and I), which is also observed in T98G glioma cell lines (42). There is increasing evidence underscoring that p53 regulates lipid metabolism, adipocyte differentiation, and systemic energy metabolism (34). Overexpression of Trp53 inhibits 3T3-L1 cell adipogenesis, concomitantly, the expression of Pparγ, Fabp4, and Adipoq is largely reduced (43). In our study, the decreased Pparγ and Fabp4 level after knockdown of Kmt5c phenocopied that in 3T3-L1 cells with ectopic expression of Trp53. It is worth noting with the knockdown of Kmt5c, the decreased Pparγ and Fabp4 expressions were only observed in the final differentiation stage (day 4 to day 6) without an obvious defect in adipogenesis per se. It may be because the twofold increase in p53 expression in Kmt5c knockdown cells (Fig. 6 A–D) is not as high as the overexpression of p53 (more than fourfold of the endogenous level) in 3T3-L1 cells (43) to block the adipocyte differentiation. The Trp53-deficient mice exhibit the activated thermogenic program in iWAT and are more resistant to HFD-induced obesity under thermoneutral conditions (36). Overexpression of Trp53 in adipose tissues results in insulin resistance even at normal chow, and, conversely, loss of Trp53 in adipocytes improves insulin resistance on a high-fat high-sucrose diet (35). All these findings show that p53 negatively regulates metabolism in thermogenic adipocytes, supporting our conclusion that the increased p53 protein level elicited by Kmt5c ablation is responsible for the decrease in thermogenic gene expression and inclination to diet-induced obesity.
A recent study took advantage of whole-body Kmt5c−/− and Myf5-driven KMT5b DKO mice to investigate the role of KMT5 in systemic energy metabolism (21). The mice deficient of Kmt5b and Kmt5c demonstrate enhanced mitochondria respiration in BAT, increased WAT browning, and resistance to obesity. Pparγ is a direct target of KMT5b and KMT5c in preadipocytes, and increased PPARγ expression accounts for H4K20me3 metabolic regulation. All these data are completely contrary to our results. We cannot exclude the contribution of Kmt5c expressed in nonadipose cells on metabolism when whole-body Kmt5c KO mice were used there. Systemic loss of Kmt5c may also cause interorgan crosstalk targeting adipocyte metabolic activity. Myf5 is expressed in precursor cells of skeletal muscle and brown adipocyte, and Myf5-Cre is usually used to investigate BAT development and myogenesis. Deletion of Kmt5b in the Myf5 lineage may concomitantly affect adipocyte and skeletal muscle functions since Kmt5b is reported to regulate skeletal muscle stem cell quiescence and muscle regeneration (44). By contrast, in our study, Adipoq-Cre was utilized for KO Kmt5c mainly in mature brown and white adipocytes. Considering that dynamic epigenetic modification is highly tissue and cell specific (45, 46), the H4K20me3 code may modulate distinct target gene sets in precursor and mature adipocytes. In light of all these factors, it is not surprising that loss of H4K20me3 modification at different adipogenic stages results in diverse metabolic consequences.
In conclusion, our analyses of KMT5c ablation mice have revealed that KMT5c plays an important role in regulating thermogenic gene expression in adipose tissues. The fact that KMT5c serves as a key epigenetic regulator in adipocyte thermogenesis may provide a potential drug target aiming at activating its methyltransferase activity for the therapeutics of obesity.
Materials and Methods
Kmt5cflox mice in a C57BL/6 background were prepared by Beijing Biocytogen Co, Ltd. The immortalized BAT preadipocytes and primary SVF cells were induced to differentiate as decribed in the SI Appendix. Cellular respiration was measured with a Clark-type oxygen electrode (Oxygraph+ system, Hansatech). The shRNA target sequences, antibodies used in this study, and the procedures for ChIP, qRT-PCR, and coimmunoprecipitation are described in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Yongxu Wang for providing the immortalized brown fat cell line; Drs. Junli Liu and Xinran Ma for reagents. This work was supported by grants from the National Key R&D Program of China (2018YFA0800401 to Q.-Q.T., 2016YFC1305102 to D.P.), and the National Natural Science Foundation of China (31970710, 31570783 and 31770839 to D.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.-Y.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922548117/-/DCSupplemental.
Data Availability.
All study data, methods, and results of statistical analyses are reported in this paper and associated SI Appendix. We welcome any specific inquiries.
References
- 1.Harms M., Seale P., Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Cohen P., Spiegelman B. M., Brown and beige fat: Molecular parts of a thermogenic machine. Diabetes 64, 2346–2351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazak L. et al., A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikeda K. et al., UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 23, 1454–1465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y. et al., Thermal stress induces glycolytic beige fat formation via a myogenic state. Nature 565, 180–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kajimura S., Spiegelman B. M., Seale P., Brown and beige fat: Physiological roles beyond heat generation. Cell Metab. 22, 546–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee P. et al., Temperature-acclimated brown adipose tissue modulates insulin sensitivity in humans. Diabetes 63, 3686–3698 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chondronikola M. et al., Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 63, 4089–4099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir G. et al., Substantial metabolic activity of human brown adipose tissue during warm conditions and cold-induced lipolysis of local triglycerides. Cell Metab. 27, 1348–1355.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inagaki T., Sakai J., Kajimura S., Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 17, 480–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno H., Shinoda K., Ohyama K., Sharp L. Z., Kajimura S., EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504, 163–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms M. J. et al., Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 19, 593–604 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe Y. et al., Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch. Nat. Commun. 9, 1566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambeat A. et al., LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. 15, 2536–2549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan D. et al., Jmjd3-mediated H3K27me3 dynamics orchestrate brown fat development and regulate white fat plasticity. Dev. Cell 35, 568–583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zha L. et al., The histone demethylase UTX promotes brown adipocyte thermogenic program via coordinated regulation of H3K27 demethylation and acetylation. J. Biol. Chem. 290, 25151–25163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang L. et al., Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat. Commun. 9, 1796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Nuland R., Gozani O., Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol. Cell. Proteomics 15, 755–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakabayashi K. et al., The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 29, 3544–3555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son M. J. et al., Methyltransferase and demethylase profiling studies during brown adipocyte differentiation. BMB Rep. 49, 388–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedrotti S. et al., The Suv420h histone methyltransferases regulate PPAR-γ and energy expenditure in response to environmental stimuli. Sci. Adv. 5, eaav1472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan T. et al., Distinct populations of adipogenic and myogenic Myf5-lineage progenitors in white adipose tissues. J. Lipid Res. 54, 2214–2224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bromberg K. D. et al., The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat. Chem. Biol. 13, 317–324 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Rajakumari S. et al., EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17, 562–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen E. D., MacDougald O. A., Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885–896 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Cypess A. M. et al., Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X. et al., Brown adipose tissue transplantation reverses obesity in Ob/Ob mice. Endocrinology 156, 2461–2469 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Himms-Hagen J., Defective brown adipose tissue thermogenesis in obese mice. Int. J. Obes. 9, 17–24 (1985). [PubMed] [Google Scholar]
- 29.Jung R. T., Shetty P. S., James W. P., Barrand M. A., Callingham B. A., Reduced thermogenesis in obesity. Nature 279, 322–323 (1979). [DOI] [PubMed] [Google Scholar]
- 30.Nicetto D. et al., Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLoS Genet. 9, e1003188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldmann H. M., Golozoubova V., Cannon B., Nedergaard J., UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Balakrishnan L., Milavetz B., Decoding the histone H4 lysine 20 methylation mark. Crit. Rev. Biochem. Mol. Biol. 45, 440–452 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Shinchi Y. et al., SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp. Cell Res. 334, 90–99 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Krstic J., Reinisch I., Schupp M., Schulz T. J., Prokesch A., p53 functions in adipose tissue metabolism and homeostasis. Int. J. Mol. Sci. 19, 2622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minamino T. et al., A crucial role for adipose tissue p53 in the regulation of insulin resistance. Nat. Med. 15, 1082–1087 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Hallenborg P. et al., p53 regulates expression of uncoupling protein 1 through binding and repression of PPARγ coactivator-1α. Am. J. Physiol. Endocrinol. Metab. 310, E116–E128 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Park J. H. et al., A multifunctional protein, EWS, is essential for early brown fat lineage determination. Dev. Cell 26, 393–404 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lasham A. et al., The Y-box-binding protein, YB1, is a potential negative regulator of the p53 tumor suppressor. J. Biol. Chem. 278, 35516–35523 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Seale P., Transcriptional regulatory circuits controlling brown fat development and activation. Diabetes 64, 2369–2375 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleiner S. et al., Development of insulin resistance in mice lacking PGC-1α in adipose tissues. Proc. Natl. Acad. Sci. U.S.A. 109, 9635–9640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen P. et al., Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156, 304–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soto-Reyes E., Recillas-Targa F., Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene 29, 2217–2227 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Huang Q. et al., Role of p53 in preadipocyte differentiation. Cell Biol. Int. 38, 1384–1393 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Boonsanay V. et al., Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. Cell Stem Cell 18, 229–242 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Hayashi-Takanaka Y. et al., Histone modification dynamics as revealed by multicolor immunofluorescence-based single-cell analysis. J. Cell Sci. 133, jcs243444 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamadema N., Burr S., Brewer A. C., Dynamic regulation of epigenetic demethylation by oxygen availability and cellular redox. Free Radic. Biol. Med. 131, 282–298 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data, methods, and results of statistical analyses are reported in this paper and associated SI Appendix. We welcome any specific inquiries.