Significance
Glucagon action in liver is a central response to fasting and type 2 diabetes. Glucagon action has been delineated through regulatory mechanisms involving signaling, transcription factor/coactivator-based gluconeogenic gene expression, and metabolic enzyme activity. Understanding the molecular mechanisms whereby glucagon controls energy metabolism will define new strategies and potential therapies to treat metabolic diseases. Here, we have identified a regulatory mechanism whereby PGC-1α, a known transcriptional regulator of glucagon action, binds RNAs linked to glucose energy metabolism. PGC-1α represents a class of RNA-binding proteins that act as a transcriptional coactivator through transcription factor binding, but also binds to RNA sequences to control specific mRNA transcripts encoding for metabolic and bioenergetic genes.
Keywords: PGC-1α, RNA binding, glucagon, liver, mitochondria
Abstract
The peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is a transcriptional coactivator that controls expression of metabolic/energetic genes, programming cellular responses to nutrient and environmental adaptations such as fasting, cold, or exercise. Unlike other coactivators, PGC-1α contains protein domains involved in RNA regulation such as serine/arginine (SR) and RNA recognition motifs (RRMs). However, the RNA targets of PGC-1α and how they pertain to metabolism are unknown. To address this, we performed enhanced ultraviolet (UV) cross-linking and immunoprecipitation followed by sequencing (eCLIP-seq) in primary hepatocytes induced with glucagon. A large fraction of RNAs bound to PGC-1α were intronic sequences of genes involved in transcriptional, signaling, or metabolic function linked to glucagon and fasting responses, but were not the canonical direct transcriptional PGC-1α targets such as OXPHOS or gluconeogenic genes. Among the top-scoring RNA sequences bound to PGC-1α were Foxo1, Camk1δ, Per1, Klf15, Pln4, Cluh, Trpc5, Gfra1, and Slc25a25. PGC-1α depletion decreased a fraction of these glucagon-induced messenger RNA (mRNA) transcript levels. Importantly, knockdown of several of these genes affected glucagon-dependent glucose production, a PGC-1α–regulated metabolic pathway. These studies show that PGC-1α binds to intronic RNA sequences, some of them controlling transcript levels associated with glucagon action.
The transcriptional peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is a canonical regulatory component of physiological processes such as cold, fasting, and exercise (1–4). PGC-1α is itself regulated at multiple levels including transcription, translation, and posttranslation (5–9). As a transcriptional coactivator, PGC-1α increases expression of genes associated with energy metabolism and mitochondrial biogenesis through binding to transcription factors (10–12). Once bound to a transcription factor it engages additional chromatin-remodeling proteins and the basal transcriptional initiation machinery to increase expression of targeted genes (13, 14). Most of the biological functions of PGC-1α have been focused on recruitment to promoters and enhancers through physical interaction with transcription factors (15). For example, PGC-1α binds to ERRα, NRFs, and YY1 to activate transcription of a large number of nuclear genes encoding for mitochondrial proteins (10, 11, 16, 17), or to HNF4α, FOXO1, and GR to augment transcription of gluconeogenic genes (2, 18–20). Interestingly, PGC-1α is one of the few transcriptional coactivators that contains serine/arginine (SR) and RNA recognition motif (RRM) domains at its C terminus, similar to splicing factors or other RNA-binding proteins (13). Some studies have reported that PGC-1α binds to components of the initial elongation machinery (21, 22). In addition, the RNA methyltransferase NSUN7 has been shown to promote PGC-1α–mediated transcription and corresponds to enrichment of a specific set of enhancer-associated transcripts (23). The C terminus of PGC-1α also binds to cap-binding protein 80 (CBP80) and both proteins appear to associate with the 5′ cap of target transcripts of promyogenic genes (24). However, none of these studies have broadly determined what type of RNAs are bound to endogenous PGC-1α in a metabolic or energetic process.
Glucagon action is a central fasting and diabetic response that controls metabolism and energy balance (25–30). Glucagon uses the ancient and canonical cyclic adenosine monophosphate pathway to control part of the fasting metabolic action response, including hepatic glucose production. Glucagon regulatory metabolic function occurs at different levels, including direct metabolic enzyme activity and fluxes and transcriptional proteins such as CREB-dependent complex assembly with the CBP/p300 and TORC2 coactivators (2, 3, 31–34). One of the CREB targets is PGC-1α that maintains gluconeogenic and fatty acid oxidation gene expression (3, 35, 36). Although PGC-1α has been shown to mediate part of the glucagon and fasting response (37–40), the complete mechanisms and targets, in particular the binding to RNAs, are not entirely understood.
Here, we have performed a transcriptome-wide analysis of PGC-1α target RNAs in glucagon-treated primary hepatocytes. Immunoprecipitated glucagon-induced endogenous PGC-1α was found in complexes that contain specific RNAs. A large fraction of these RNAs were mapped to intronic sequences of transcripts. We carried out specific analysis of a highly glucagon-induced and PGC-1α–bound RNA, the mitochondrial adenosine triphosphate (ATP) transporter SLC25A25, that is necessary for glucagon-dependent glucose production and mitochondrial energetics. These studies identify a mechanism and specific targets whereby the transcriptional coactivator PGC-1α controls glucagon action in hepatocytes.
Results
PGC-1α Binds to RNAs and Forms Protein–RNA Complexes.
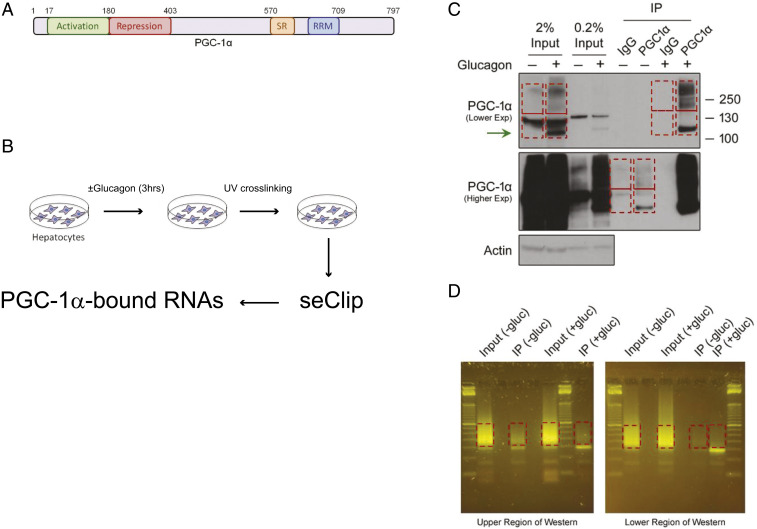
PGC-1α functions as a transcriptional coactivator and contains SR and RRM domains at the C terminus that are predicted to interact with RNA (Fig. 1A). To evaluate if endogenous PGC-1α indeed binds RNA, we used primary hepatocytes treated with vehicle (phosphate-buffered saline; PBS) or glucagon for 3 h (Fig. 1B). Cells were subjected to ultraviolet (UV) irradiation (254 nm at 400 mJ/cm2) to cross-link protein to nucleic acids, followed by immunoprecipitation with a validated commercial antibody specific to PGC-1α. Cross-linked samples were analyzed by Western blot showing that PGC-1α is strongly induced by glucagon, and it appeared in different molecular mass bands, both in whole-cell lysates as well as immunoprecipitates (Fig. 1C and SI Appendix, Fig. S1A). The higher molecular mass PGC-1α bands, greater than 130 kDa, were not detected in non–cross-linked samples. We arbitrarily grouped the different PGC-1α molecular mass bands, from the input (whole-cell lysate) and immunoprecipitates, into upper and lower regions (indicated by the red boxes in Fig. 1C and SI Appendix, Fig. S1A), in both the glucagon-stimulated and unstimulated hepatocyte samples. Complementary DNA (cDNA) libraries were generated from the respective RNAs cross-linked to PGC-1α in accordance with the published eCLIP (enhanced UV cross-linking and immunoprecipitation) protocol (41). Orange G staining indicates the generation of suitable sequencing libraries from the different samples for sequence analysis (Fig. 1D and SI Appendix, Fig. S1B). These experiments indicate that PGC-1α interacts with RNAs. We generated biologically duplicate libraries that were sequenced to an average depth of 13 million reads each (Fig. 2A) (42). In parallel, we also prepared and sequenced paired size-matched input control libraries (30 million reads each). Reads were mapped to the reference mouse genome and irreproducible discovery rate [IDR; P ≤ 0.001; log2(fold change) ≥ 3] analysis (41) was performed to identify reproducible and enriched binding sites within each condition (lower vs. upper band; vehicle vs. glucagon treatment). IDR analysis identified 2,206 and 3,692 reproducible and enriched PGC-1α binding sites within the control samples for the lower and upper regions, respectively. The IDR peaks were distributed among 659 and 986 genes within the lower and upper region samples, respectively. Similarly, 413 and 2,167 sites were identified to be distributed among 118 and 716 genes within the glucagon treatment samples for the lower and upper regions, respectively. Evaluation of the fraction of binding sites within genic regions (proximal introns, distal introns, 5′ splice sites, and noncoding exonic regions) indicated that PGC-1α is largely represented in proximal and distal introns (Fig. 2B), in agreement with its nuclear localization.
Fig. 1.
Profiling of PGC-1α–bound RNA using seCLIP. (A) PGC-1α protein contains an RNA recognition motif at its C terminus. (B) Pipeline for detecting RNAs bound to PGC-1α in hepatocytes following glucagon stimulation and using single-end cross-linking and immunoprecipitation. (C) Immunoblots depicting the upper and lower PGC-1α complexes formed following UV cross-linking and immunoprecipitation. The fragments of the IP and input that were used for RNA sequencing are marked in red. (D) Orange G staining indicates generation of suitable libraries for sequence analysis.
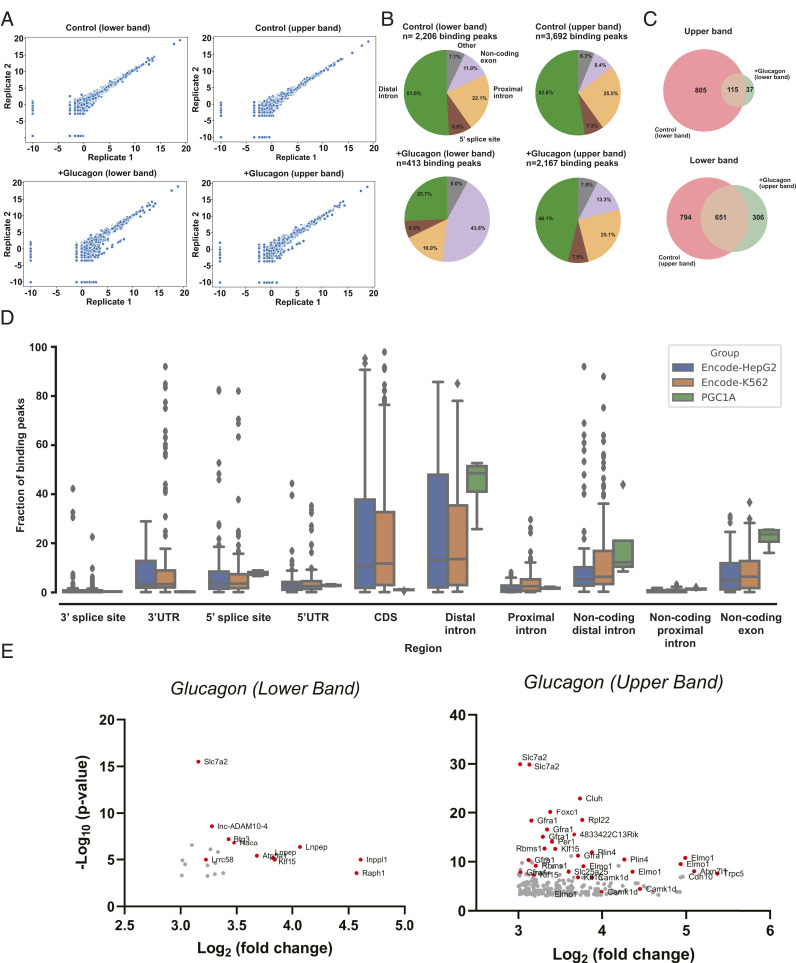
Fig. 2.
Profiling of genic regions bound by PGC-1α. (A) Scatterplots showing the correlation of read density, expressed as log2-transformed reads per million [log2(RPM)], of PGC-1α–bound transcripts between the replicates. Each data point represents a bound transcript. R2, Spearman correlation coefficient. (B) The genic regions of the RNA sequences found to bind PGC-1α. (C) Glucagon stimulation in hepatocytes induces binding of specific RNAs to PGC-1α in both upper and lower complexes. (D) Boxplot of binding peak distribution across genic regions for 103 and 120 RBPs from HepG2 (blue) and K562 cells (orange), respectively, compared with PGC-1α (green). Boxes and whiskers indicate quartiles. Outliers are represented by diamonds. (E) Volcano plot of genes that are bound to PGC-1α only in response to glucagon stimulation in lower and upper PGC-1α complexes.
Moderate differences in the fraction of PGC-1α–binding peaks mapping to proximal introns, distal introns, and 5′ splice sites were observed between the upper and lower migrating complexes from glucagon-treated samples. In contrast, the fraction of peaks mapping to noncoding exonic regions was over threefold higher in lower compared with upper complex samples (43.8 vs. 13.3%) (Fig. 2B). Since one gene may be bound across multiple genic regions, we defined a binding target as the genic region bound within a given gene. Using this metric, sequencing analysis identified 842 RNA targets in the lower band, 37 of them specifically enriched by PGC-1α under glucagon treatment; 1,100 targets were bound in the upper band, among them 306 specifically enriched under glucagon treatment (Fig. 2C and SI Appendix, Figs. S2 A–D and S3). Interestingly, comparison of the binding region distribution between RNAs bound by PGC-1α on the one hand, and 223 RBPs (RNA-binding proteins) analyzed by the ENCODE project in HepG2 and K562 human cell lines (43) on the other, showed a selective pattern of RNA binding in genic regions for PGC-1α (Fig. 2D) (44).
Among the most significantly enriched [IDR, P ≤ 0.001; log2(fold change) ≥ 3] glucagon-dependent RNAs, we found that PGC-1α interacts with RNAs encoding for the transcription factors KLF15, FOXO1, and PER1, calcium-related proteins CAMK1δ and TRPC5, proteins involved in translation RPL22 and CLUH, lipid droplet-associated protein PLN4, GCN5 complex subunit ATXN71, and Slc transporters SLC7A2 and SLC25A25 (Fig. 2E and SI Appendix, Fig. S4 A and B). Some of these genes have been previously associated with the liver fasting response or PGC-1α metabolic function, for example, the transcription factors KLF15, FOXO1, and PER1 (18, 45, 46). However, the transcriptional PGC-1α messenger RNA (mRNA) targets, including gluconeogenic genes or OXPHOS genes, were not enriched in the PGC-1α immunoprecipitates (SI Appendix, Fig. S4 C and D). Taken together, these results indicate that endogenous PGC-1α induced by glucagon in hepatocytes binds to RNAs and a large fraction of these RNAs are intronic sequences, suggesting a potential mechanistic function in the metabolic action of this hormone in the liver.
PGC-1α Increases Expression Levels of a Subset of Glucagon-Dependent PGC-1α–Bound RNAs.
In order to determine the regulatory function of the RNA sequences bound to PGC-1α, we measured transcript mRNA expression of top-ranking RNAs that are bound by PGC-1α upon glucagon treatment. A fraction of these transcripts was increased upon glucagon treatment (Fig. 3A), suggesting a regulatory role at the mRNA level. Among the top glucagon-elevated transcripts were Klf15 and the mitochondrial nucleotide transporter Slc25a25. Next, we investigated whether the expression of these glucagon-induced mRNA transcripts was dependent on PGC-1α. Specific small interfering RNAs (siRNAs) were used to deplete PGC-1α in primary hepatocytes (Fig. 3B). Based on their glucagon dependence, we selected 12 RNA transcripts that bind to PGC-1α and quantified them using qPCR upon PGC-1α knockdown (Fig. 3C and SI Appendix, Fig. S5A). Of these 12 transcripts, 6 were decreased when PGC-1α was depleted. Interestingly, consistent with the fact that PGC-1α is highly induced by glucagon, there was a trending pattern with mRNA transcripts that were regulated by glucagon that exhibited PGC-1α dependency on their expression, although there were some exceptions, such as Klf15 that was glucagon-induced but PGC-1α–independent. These results suggest that PGC-1α binds to a fraction of RNAs (most of them at intronic sequences), maintaining their expression levels in response to glucagon.
Fig. 3.
Subset of PGC-1α–bound RNAs are induced by glucagon in hepatocytes. (A) The level of induction by glucagon stimulation of a subset of RNAs that bind PGC-1α. (B) siRNA oligos against Ppargc1a efficiently reduce the expression level of Ppargc1a and the protein level of PGC-1α. (C) qPCR analysis shows that the expression levels of the glucagon-induced genes are controlled by PGC-1α levels. Data were normalized to the siControl of treated (+glucagon) and untreated samples. (D) Depletion of Slc25a25 suppresses glucose release from hepatocytes when either pyruvate/lactate or glycerol are used as substrates for gluconeogenesis. Depletion of Gfra1 increases glucose release from hepatocytes primarily when glycerol is use as a substrate. Gly, glycerol; Pyr/Lac, pyruvate/lactate. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. Error bars represent the standard error of the mean.
A Fraction of Glucagon-Dependent PGC-1α–Binding RNAs Encode for Proteins That Control Hepatic Glucose Production.
We and others have previously shown that PGC-1α controls hepatic glucose production in the fasting and glucagon response through different transcription-dependent mechanisms, including regulation of gluconeogenic gene expression and tricarboxylic acid (TCA) cycle fluxes (2, 3, 31). We therefore tested whether these glucagon-induced RNA transcripts that interact with PGC-1α can control hepatic glucose production. As predicted, previously identified genes involved in this pathway, such as Camk1δ and Klf15, control glucose output (47, 48), although these assays demonstrate a substrate-dependent specificity (SI Appendix, Fig. S5 C–E). Other genes, such as Gfra1, that are mildly induced by glucagon, when depleted, increased glucose production only when glycerol was used as a substrate (Fig. 3D). Importantly, depletion of SLC25A25, a mitochondrial nucleotide transporter (49), strongly suppressed glucose production, particularly upon glucagon treatment, independent of the gluconeogenic substrate used (Fig. 3D). These experiments indicate that PGC-1α, in addition to directly increasing gluconeogenic enzyme gene expression transcription such as Pck1 or G6pc, binds to RNA transcripts that regulate glucagon-dependent hepatic glucose production.
Glucagon-Dependent PGC-1α Binding to Slc25a25 Distal Intronic and Isoform-Specific Near Exon 1 RNA Sequences Increases mRNA and Protein Expression.
Based on the results described above that Slc25a25 gene expression is highly increased by glucagon, Slc25a25 transcript RNA binds to PGC-1α in a glucagon-dependent manner, and SLC25A25 is required for the glucagon action on hepatic glucose production, we decided to investigate the PGC-1α binding to Slc25a25 RNA and how SLC25A25 controls this metabolic pathway. In glucagon-treated hepatocytes, PGC-1α binds to distal intronic regions common to four out of five Slc25a25 isoforms (Fig. 4A). Interestingly, the isoforms of Slc25a25 differ in their exon 1 sequences (Fig. 4A), which encode different numbers of calcium-binding EF hands at the N terminus (49), which potentially provides differential calcium sensitivity to the specific isoforms. Importantly, there was a specific binding of PGC-1α near the exon 1 RNA sequence that is closest to the third Slc25a25 isoform (TV3) that was entirely dependent on glucagon stimulation (Fig. 4A). Interestingly, among the five Slc25a25 isoforms, glucagon specifically promotes the greatest induction of TV3 (Fig. 4B), and depletion of PGC-1α selectively decreased induction of this isoform compared with TV1 and TV2 (Fig. 4C), which translates into diminished Slc25a25 mRNA and protein amounts (Fig. 4 D–F). These results show that PGC-1α controls isoform-specific Slc25a25 gene expression that coincides with specific binding to distal intronic and near exon 1 RNA sequences.
Fig. 4.
Glucagon-dependent PGC-1α binding to Slc25a25 distal intronic and isoform-specific near exon 1 RNA sequences increases mRNA and protein expression. (A) The Slc25a25 gene gives rise to five mRNA transcripts with three different first exons (TV1 to TV3). The specific binding peak to PGC-1α, close to exon 1 of TV3, is indicated with an arrow. (B) Slc25a25 TV3 is the primary variant that is being regulated by glucagon. Data are normalized to the level of induction of total Slc25a25. (C and D) Depletion of Ppargc1a specifically suppresses the expression of Slc25a25 TV3. (E) Western blots showing the effect of siPpargc1a on total PGC-1α and SLC25A25 protein levels. (F) Western blots following mitochondrial isolation show that the protein levels of SLC25A25 are reduced in the mitochondria following siPpargc1a. (G) Depletion of Slc25a25 TV3 using specific siRNA oligos has the strongest effect on suppressing glucagon-induced glucose production. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent the standard error of the mean.
SLC25A25 Regulates ATP and GTP Levels and Controls Hepatic Glucose Production.
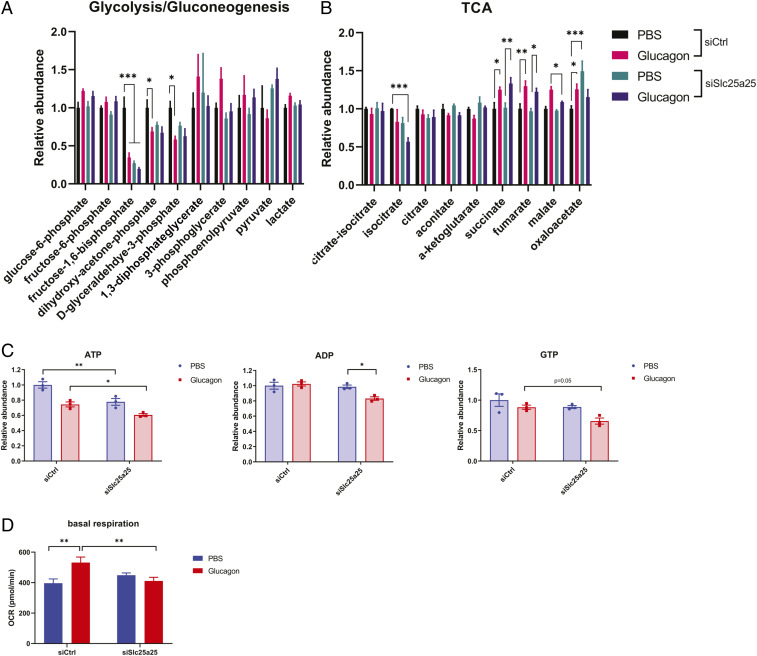
SLC25A25 is a mitochondrial nucleotide transporter that has been shown to transport ATP (49). In order to determine whether SLC25A25 could affect ATP levels in primary hepatocytes, we performed metabolite profile analysis using primary hepatocytes in the presence and absence of SLC25A25 and glucagon treatment. Fig. 5 A–C shows that energetic metabolites such as ATP and guanosine diphosphate were decreased upon SLC25A25 depletion. Moreover, different glycolytic/gluconeogenic metabolites, such as fructose 1,6-phosphate, dihydroxy-acetone-phosphate, and d-glyceraldehyde-3-phosphate, were also decreased (Fig. 5A). In addition, certain TCA metabolites, including isocitrate and malate, were also affected by SLC25A25 depletion (Fig. 5B), indicating that SLC25A25 controls part of the hepatic gluconeogenic metabolite changes in response to glucagon. The decreases in ATP and guanosine triphosphate (GTP) (Fig. 5C) are consistent with a decrease in hepatic glucose production when SLC25A25 is depleted (Fig. 3D). Interestingly, the magnitude of glucose production suppression was higher when the isoform TV3 was depleted compared with TV1 (Fig. 4G). We next evaluated whether these changes in ATP and GTP correlated with altered respiratory activity. Oxygen consumption rates (OCRs) were measured in primary hepatocytes using the Seahorse instrument (Fig. 5D). Glucagon treatment caused an increase in OCR; however, SLC25A25 knockdown significantly blunted this increase, indicating that the activity of this transporter is necessary for the glucagon-dependent oxygen consumption increases. These results suggest that the PGC-1α RNA-binding target SLC25A25 controls part of the glucagon metabolic action, including increased energetics that are necessary to maintain hepatic glucose production.
Fig. 5.
SLC25A25 regulates ATP and GTP levels and bioenergetics in hepatocytes. (A and B) Metabolomics analysis of primary hepatocytes following siSlc25a25. The relative levels of glycolytic/gluconeogenic and TCA cycle intermediates are shown. (C) siSlc25a25 reduces the levels of ATP and GTP in primary hepatocytes. (D) Basal respiration of primary hepatocytes following siSlc25a25 and glucagon stimulation. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent the standard error of the mean.
Discussion
The PGC-1 family agglutinates a series of proteins that function as transcriptional coactivators that respond to nutrient and environmental stresses such as cold, exercise, and fasting (15, 50). All members of this family have an N-terminal transcriptional activation domain and motifs that bind to transcription factors, for example, the LXXLL motifs that bind to the ligand-binding domain of hormone nuclear receptors (1). The canonical mechanisms whereby these proteins activate gene expression are through transcription factor-based recruitment to promoters and engagement of the basal transcriptional machinery (13, 14, 51). Some members including PGC-1α contain a C terminus with SR and RRM domains involved in RNA binding, suggesting that additional mechanisms beyond promoter and transcription factor recruitment are involved (52). In fact, previous studies have implicated some of these mechanisms through binding to transcriptional elongation factors, interaction with the RNA methyltransferase NSUN7 and enhancer-associated transcripts, and CBP80 that binds to 5′ cap transcripts (21–24). Although these are mechanisms that might provide regulation of PGC-1α target genes, the PGC-1α–bound RNAs in a particular biological response were unknown. In this manuscript, we report a transcriptome-wide analysis of PGC-1α–binding RNAs identified in response to glucagon action, a dominant fasting axis, in hepatocytes. A large fraction of these bound regions were intronic sequences, but also included exonic and 5′ splice site regions, suggesting different regulatory mechanisms. This RNA binding site location pattern seems to be unique among the RNA-binding proteins analyzed in the ENCODE project (43). Based on the location pattern of PGC-1α–bound RNAs, it seems difficult to generalize a common mechanism of action integrating the PGC-1α binding with these RNAs and specific regulation. In addition, we cannot conclusively determine at this point whether the RRM domain is required for the PGC-1α–RNA binding or rather this binding is mediated through other domains of PGC-1α or other RNA-binding proteins associated with the PGC-1α complex. Thus, deciphering general principles and additional specific mechanisms underlying PGC-1α binding to RNAs will require further in-depth studies. However, for some RNA sequences, such as the first exons in Slc25a25, PGC-1α binding might provide increases in mRNAs for a specific isoform. This regulation brings an interesting concept based on whether RNA sequences located in proximal regions of gene transcription initiation can provide specific recruitment of transcriptional coactivators to define isoform specificity and mRNA transcript expression levels. In this way, the presence of RNA sequences might function, like transcription factors, as specific recruitment of activators to increase gene expression. It is also conceivable that this mechanism together with transcription factor binding could further accelerate mRNA transcript expression. In fact, this might be the case for the glucagon-inducible gene Slc25a25 that encodes for different N-terminal isoforms, and PGC-1α precisely binds near an exon 1 sequence of an isoform with specific metabolic functions. These studies also indicate that PGC-1α controls genes through different mechanisms of action providing additional points to regulate a metabolic and energetic response. Notably, PGC-1α–binding RNAs identified were not transcripts associated with the known canonical PGC-1α targets that include nuclear genes encoding for proteins linked to mitochondrial biogenesis, OXPHOS, the TCA cycle, fatty acid oxidation, or gluconeogenesis, but some of them were linked to the glucagon and fasting response.
These studies also have implications for how glucagon controls energy metabolism in hepatocytes. Glucagon-induced PGC-1α binds to a series of RNAs that are known to control glucose production, indicating that this hormone uses PGC-1α as an RNA-binding protein to increase specific gene expression. This regulation confers additional mechanisms for dynamic, temporal and spatial, control of glucagon action. PGC-1α is part of a second phase of fasting control after CREB/TORC2 and CBP/p300 activation, reinforcing the fasting response and also providing attenuating feedback mechanisms (20, 34, 38, 39). In addition to the known transcriptional targets, including HNF4α-driven PEPCK and G6Pase (2), PGC-1α uses other modes of action binding RNAs that control the gluconeogenic response, including FOXO1, CAMK1δ, KLF15, and SLC25A25. These multiple mechanisms might function to ensure increased glucose production, a process that is required for survival during fasting. Interestingly, PGC-1α binding to a specific Slc25a25 isoform, a calcium-regulated mitochondrial transporter (49), controls its expression, providing a metabolic and energetic regulation during glucagon action. We propose that glucagon is necessary to initiate transcription of Slc25a25 and that PGC-1α, by binding the RNA of a specific transcript, provides selectivity toward expression of this transcript. The expression of a specific SLC25A25 with different EF-hand calcium binding sites at the N terminus might define the transporter kinetics to control ATP transport between the mitochondria and cytosol in response to calcium fluctuations. Remarkably, this transporter is necessary to maintain cellular ATP and GTP levels, suggesting that it controls energetic mechanisms. In fact, we show that glucagon-induced respiration is defective with SLC25A25 depletion, suggesting that the ATP demand for gluconeogenesis cannot be appropriately met in the absence of SLC25A25, thus compromising the gluconeogenic pathway from different substrates. What points of gluconeogenesis are specifically altered are unclear, but the metabolite analysis suggested that different sites of gluconeogenic substrate entry are altered. A likely point of control might be the mitochondrial pyruvate carboxylase that depends on ATP levels for enzymatic activity (53, 54). In addition, previous studies have suggested that glucagon increases calcium that promotes hepatic glucose production through the inositol-1,4,5-trisphosphate receptor (55) and CAMK2 (56). SLC25A25 is likely to be part of this regulation through direct calcium binding to the EF hands located at the N terminus of the protein. Interestingly, another strong RNA hit was Trpc5, a calcium channel that might provide calcium fluxes to control SLC25A25 during glucagon action. Future studies will define this precise mechanistic regulation in the context of fasting and how it contributes to glucose and energy homeostasis in type 2 diabetes.
Methods
Primary Hepatocyte Cultures.
Primary hepatocytes from 7- to 10-wk-old male C57BL/6 mice were isolated by perfusion with liver digest medium (Life Technologies; 17703-034), followed by 70-μm mesh filtration. Percoll (Sigma; P7828) gradient centrifugation was then used to separate hepatocytes from debris and other cell types. Isolated hepatocytes were seeded (4 × 105 cells per well, 6-well plates; 2 × 105 cells per well, 12-well plates; 7.2 × 106 cells, 15-cm dish) in plating medium (Dulbecco’s modified Eagle’s medium [DMEM] supplemented with 10% fetal bovine serum, 2 mM sodium pyruvate, 1 μM dexamethasone, 100 nM insulin, and 1% penicillin/streptomycin). Three hours postseeding, the medium was changed to maintenance medium (DMEM supplemented with 0.2% bovine serum albumin [BSA], 2 mM sodium pyruvate, 0.1 μM dexamethasone, 1 nM insulin, and 1% penicillin/streptomycin). Cells were cultured at 37 °C in a humidified incubator containing 5% CO2 and medium was replaced daily with fresh maintenance medium.
siRNA Transfection.
siRNA oligonucleotides against mouse genes were purchased from OriGene Technologies (Ppargc1a, SR427524; Slc25a25, SR412336; Gfra1, SR415185; Klf15, SR413764; Camk1d, SR412965). Hepatocytes were transfected with a 1:1:1 mixture of three unique 27-mer siRNA duplexes (20 µM each, with a final experimental concentration of 10 nM), using Lipofectamine RNAiMAX (Invitrogen) by reverse transfection followed with immediate seeding of hepatocytes, according to the manufacturer’s protocol and as previously described (39). The following morning, the medium was replaced with maintenance medium, accompanied by another round of forward transfection, as described earlier. After 6 h, the medium was replaced again.
Treatment of Cells with Stimuli.
For treatment of primary hepatocytes with glucagon, cells were incubated overnight in starvation medium (DMEM supplemented with 0.2% BSA, 2 mM sodium pyruvate, and 1% penicillin/streptomycin). The following morning, cells were stimulated with 200 nM glucagon for 3 h (unless indicated otherwise) in starvation medium.
Single-End Enhanced Cross-Linking and Immunoprecipitation.
Primary hepatocytes were cultured in 15-cm dishes as described above. Cells were stimulated with or without glucagon for 3 h (three plates for each condition, done in experimental duplicates), following which they were washed twice with ice-cold PBS (pH 7.4). Cells were subjected to UV cross-linking (254 nm at 400 mJ/cm2) to generate protein–nucleic acid cross-links as described (41).
Single-end enhanced cross-linking and immunoprecipitation (seCLIP) was performed as described (41). Briefly, lysates were generated, sonicated, and treated with RNase I to fragment RNA. Two percent of each lysate sample was stored for preparation of a parallel size-matched input (SMInput) library. The remaining lysates were immunoprecipitated. Bound RNA fragments in the immunoprecipitates (IPs) were dephosphorylated and 3′ end-ligated to an RNA adapter. Protein–RNA complexes from SMInputs and IPs were run on a sodium dodecyl sulfate (SDS) polyacrylamide gel and transferred to nitrocellulose membrane. Membrane regions were excised and RNA was released from the complexes with proteinase K. SMInput samples were dephosphorylated and 3′ end-ligated to an RNA adapter. All RNA samples (IPs and SMInputs) were reverse-transcribed with AffinityScript (Agilent). cDNAs were 5′ end-ligated to a DNA adapter. cDNA yields were quantified by qPCR and 100 to 500 fmol of libraries generated with Q5 PCR Master Mix (New England Biolabs). Libraries were sequenced on an Illumina HiSeq 4000 platform.
Sequencing reads were processed as described (41). Briefly, reads were adapter-trimmed using Cutadapt (v1.14) and mapped to human-specific repetitive elements from RepBase (v18.05) by STAR (v2.4.0i) (57). Repeat-mapping reads were removed, and remaining reads were mapped to the mouse genome assembly (mm10) with STAR. PCR duplicate reads were removed using the unique molecular identifier sequences in the 5′ adapter and remaining reads were retained as “usable reads.” Peaks were called on the usable reads by CLIPper (58) and assigned to gene regions annotated in GENCODE (mm10) with the following descending priority order: coding sequence (CDS), 5′ untranslated region (UTR), 3′ UTR, proximal intron, and distal intron. Proximal intron regions are defined as extending up to 500 bp from an exon–intron junction. Each peak was normalized to the SMInput by calculating the fraction of the number of usable reads from immunoprecipitation to that of the usable reads from the SMInput. Peaks were deemed significant at ≥8-fold enrichment and P ≤ 10−3 (χ2 test, or Fisher’s exact test if the observed or expected read number in the eCLIP or SMInput was below 5). Sequencing and mapping statistics are described in Datasets S1 and S2. Code is available on GitHub (https://github.com/YeoLab/eclip).
Cell Lysis and Western Blotting.
Cell lysis.
Cells were washed twice in ice-cold PBS (pH 7.4) (Life Technologies) and lysed in ice-cold buffer. For whole-cell extracts, cells were lysed in buffer B (1× PBS, pH 7.4, 0.5% sodium deoxycholate [wt/vol], 0.1% SDS [wt/vol], 1 mM ethylenediaminetetraacetate [EDTA], 1 mM dithiothreitol [DTT], 1% IGEPAL [vol/vol], 5 mM NaF, 5 mM β-glycerophosphate, 5 mM sodium butyrate, and 20 mM nicotinamide), supplemented with cOmplete EDTA-free Protease Inhibitor Mixture (Roche Diagnostics). To isolate nuclei, cells were harvested in buffer C (10 mM Hepes-KOH, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.25% IGEPAL [vol/vol], 5 mM NaF, 5 mM β-glycerophosphate, 5 mM sodium butyrate, and 20 mM nicotinamide), supplemented with protease inhibitor mixture. Cytoplasmic fractions were separated, and nuclear pellets were lysed in buffer D (20 mM Hepes-KOH, pH 7.9, 125 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% IGEPAL [vol/vol], 10% glycerol [vol/vol], 5 mM NaF, 5 mM β-glycerophosphate, 5 mM sodium butyrate, and 20 mM nicotinamide), supplemented with protease inhibitor mixture.
Western blot analysis.
Protein samples were resolved by SDS/polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes (EMD Millipore). Membranes were blocked with either 5% BSA or nonfat dry milk in Tris buffered saline with Tween 20 (TBST) for 1 h, and then incubated with primary antibodies at 4 °C overnight, according to the manufacturer’s protocol. Following incubation with the appropriate horseradish peroxidase-conjugated secondary antibody, chemiluminescence detection was performed with ECL Western Blotting Detection Reagents (Thermo Fisher Scientific).
Commercial antibodies.
The following antibodies were purchased from 1) Millipore: anti–PGC-1α (4C1.3) (ST1202), anti–β-tubulin (05-661), anti-actin clone C4 (MAB1501); 2) Proteintech Group: anti-Scl25a25 (21568-1-AP); 3) Santa Cruz Biotechnology: anti–PGC-1α (sc-13067), anti-VDAC1 (sc390996); 4) Abcam: Total OXPHOS Rodent WB Antibody Mixture (ab110413); and 5) Jackson ImmunoResearch: anti-rabbit IgG secondary (711-035-152), anti-mouse IgG secondary (715-035-150).
Gene Expression Analysis.
Total RNA was isolated from cells or homogenized liver using TRIzol (Life Technologies) according to the manufacturer’s protocol. cDNA was synthesized using random primers and a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A gene expression analysis was performed using a CFX384 Real-Time PCR System (Bio-Rad) and Power SYBR Green PCR Master Mix (Applied Biosystems). The ΔΔCT method was used to calculate fold change and the genes 36B4 and Rpl13 and U6 small nuclear RNA were used as normalization controls.
Glucose Production Assays.
Primary hepatocytes were incubated overnight in starvation medium (DMEM supplemented with 0.2% BSA, 2 mM sodium pyruvate, and 1% penicillin/streptomycin). Hepatocytes were then washed twice in warm PBS (pH 7.4). Glucose secretion by primary hepatocytes was measured by incubating the cells for 6 h in glucose-free medium (phenol-red/glucose-free DMEM, 0.2% BSA, and ±2 mM sodium pyruvate and ±20 mM sodium lactate or ±10 mM glycerol), with or without concomitant stimulation with glucagon (200 nM). Medium aliquots were collected, and glucose levels were quantified using an enzyme-based glucose assay, according to the manufacturer’s protocol (Glucose/Glucose Oxidase Assay Kit; Sigma-Aldrich). The glucose concentration was calculated based on a standard curve.
Measurement of Oxygen Consumption.
Oxygen consumption of primary hepatocytes was measured using a Seahorse instrument. Primary hepatocytes were seeded on an XFe24 24-well Seahorse plate and reverse transfection with siRNA oligos was performed before seeding the cells with plating medium (15,000 cells per well). The next morning, medium was changed to maintenance medium and, in the afternoon, medium was changed again to low-glucose maintenance medium (2.5 mM glucose). Following overnight incubation with low-glucose maintenance, medium cells were treated for 3 h with glucagon (200 nM) in no-glucose starvation medium. After 3 h, cells were washed and medium was changed to Seahorse medium (DMEM, 0.2% BSA, 1% penicillin/streptomycin, 4 mM glutamine, 2.5 mM glucose, 2 mM pyruvate, and 150 μM oleic acid) with and without glucagon. Plates were equilibrated for 45 min in a 37 °C incubator without CO2 and oxygen consumption rates were measured using the Seahorse bioanalyzer.
Metabolomics.
For metabolomics analysis, primary hepatocytes were treated in a similar manner to glucose production assays using glucose-free medium containing pyruvate/lactate for the last 4 h. Following a 4-h incubation with glucagon, medium was aspirated, cells were washed with ice-cold PBS, and 800 μL of 80% methanol solution was immediately added to each well. After a 15-min incubation at −80 °C, cells were scraped and debris was cleared (10 min, 9,000 × g). Supernatant was transferred to a new tube and the pellet was resuspended in an additional 100 μL of 80% methanol solution. Resuspended pellets were centrifuged again (5 min, 9,000 × g) and the supernatant was combined with the previous one. The combined supernatant was dried overnight using a SpeedVac and dried pellets were resuspended in 20 μL H2O before being subjected to metabolomics profiling using the AB/SCIEX 5500 QTRAP triple quadrupole instrument.
Statistics.
All data are presented as means ± SEM. One-way ANOVA tests and t tests were conducted, along with corresponding posttests, as indicated; P < 0.05 was considered significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank the members of the P.P. laboratory for the help and discussions on this project. C.D.J.T. was partially funded by a mentor-based American Diabetes Association postdoctoral fellowship, and also by a postdoctoral fellowship from the American Diabetes Association (Grant 1-16-PDF-111). K.S. was partially funded by a postdoctoral fellowship from the American Heart Association (15POST22880002) and Charles King Trust. G.W.Y. was supported by grants from the NIH (HG004659 and HG009889). This work was supported by NIH/National Institute of Diabetes and Digestive and Kidney Diseases funding (R01 DK089883 and DK081418 to P.P.).
Footnotes
G.W.Y. is cofounder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. G.W.Y.’s interests have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. The authors declare no other competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000643117/-/DCSupplemental.
Data Availability.
All sequencing data reported in this paper have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE152303. The RBP binding data of HepG2 and K562 cells was retrieved from www.encodeproject.org, accession code ENCSR456FVU. The code for read mapping and peak calling is available on GitHub, https://github.com/YeoLab/eclip.
References
- 1.Puigserver P. et al., A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Yoon J. C. et al., Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Herzig S. et al., CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413, 179–183 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Handschin C., Spiegelman B. M., The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454, 463–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominy J. E. Jr., Lee Y., Gerhart-Hines Z., Puigserver P., Nutrient-dependent regulation of PGC-1alpha’s acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim. Biophys. Acta 1804, 1676–1683 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantó C., Auwerx J., PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 20, 98–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villena J. A., New insights into PGC-1 coactivators: Redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 282, 647–672 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Krämer A. I., Handschin C., How epigenetic modifications drive the expression and mediate the action of PGC-1α in the regulation of metabolism. Int. J. Mol. Sci. 20, 5449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian X. et al., KDM3A senses oxygen availability to regulate PGC-1α-mediated mitochondrial biogenesis. Mol. Cell 76, 885–895.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Wu Z. et al., Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Mootha V. K. et al., Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U.S.A. 101, 6570–6575 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knutti D., Kaul A., Kralli A., A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 20, 2411–2422 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puigserver P. et al., Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286, 1368–1371 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Wallberg A. E., Yamamura S., Malik S., Spiegelman B. M., Roeder R. G., Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Scarpulla R. C., Vega R. B., Kelly D. P., Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol. Metab. 23, 459–466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber S. N. et al., The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 6472–6477 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham J. T. et al., mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Puigserver P. et al., Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Rodgers J. T. et al., Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434, 113–118 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto M., Pocai A., Rossetti L., Depinho R. A., Accili D., Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Monsalve M. et al., Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6, 307–316 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Sano M. et al., Ménage-à-trois 1 is critical for the transcriptional function of PPARgamma coactivator 1. Cell Metab. 5, 129–142 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Aguilo F. et al., Deposition of 5-methylcytosine on enhancer RNAs enables the coactivator function of PGC-1α. Cell Rep. 14, 479–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H. et al., Transcriptional coactivator PGC-1α contains a novel CBP80-binding motif that orchestrates efficient target gene expression. Genes Dev. 32, 555–567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell J. E., Drucker D. J., Islet α cells and glucagon—Critical regulators of energy homeostasis. Nat. Rev. Endocrinol. 11, 329–338 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Petersen M. C., Vatner D. F., Shulman G. I., Regulation of hepatic glucose metabolism in health and disease. Nat. Rev. Endocrinol. 13, 572–587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller R. A., Birnbaum M. J., Glucagon: Acute actions on hepatic metabolism. Diabetologia 59, 1376–1381 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Lee Y. H., Wang M. Y., Yu X. X., Unger R. H., Glucagon is the key factor in the development of diabetes. Diabetologia 59, 1372–1375 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Ramnanan C. J., Edgerton D. S., Kraft G., Cherrington A. D., Physiologic action of glucagon on liver glucose metabolism. Diabetes Obes. Metab. 13 (suppl. 1), 118–125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin H. V., Accili D., Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 14, 9–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess S. C. et al., Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha)-deficient mice. J. Biol. Chem. 281, 19000–19008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai E., Miner J. N., Mitchell J. A., Yamamoto K. R., Granner D. K., Glucocorticoid receptor-cAMP response element-binding protein interaction and the response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids. J. Biol. Chem. 268, 5353–5356 (1993). [PubMed] [Google Scholar]
- 33.Hanson R. W., Reshef L., Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 66, 581–611 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Koo S. H. et al., The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Koo S. H. et al., PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 10, 530–534 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Rodgers J. T., Puigserver P., Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U.S.A. 104, 12861–12866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estall J. L. et al., Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic peroxisome proliferator-activated receptor-gamma coactivator-1alpha expression. Diabetes 58, 1499–1508 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rines A. K., Sharabi K., Tavares C. D., Puigserver P., Targeting hepatic glucose metabolism in the treatment of type 2 diabetes. Nat. Rev. Drug Discov. 15, 786–804 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharabi K. et al., Selective chemical inhibition of PGC-1alpha gluconeogenic activity ameliorates type 2 diabetes. Cell 169, 148–160.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Besse-Patin A. et al., PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proc. Natl. Acad. Sci. U.S.A. 116, 4285–4290 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Nostrand E. L. et al., Robust, cost-effective profiling of RNA binding protein targets with single-end enhanced crosslinking and immunoprecipitation (seCLIP). Methods Mol. Biol. 1648, 177–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavares C. D. J., et al. , Transcriptome-wide analysis of PGC-1α–binding RNAs identifies genes linked to glucagon metabolic action. Gene Expression Omnibus (GEO). http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152303. Deposited 16 August 2020. [DOI] [PMC free article] [PubMed]
- 43.Van Nostrand E. L. et al., A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Nostrand E. L., et al. , Data from “A large-scale binding and functional map of human RNA-binding proteins.” ENCODE. https://www.encodeproject.org/publication-data/ENCSR456FVU/. Deposited 11 February 2020.
- 45.Takashima M. et al., Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes 59, 1608–1615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C., Li S., Liu T., Borjigin J., Lin J. D., Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 447, 477–481 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Haney S. et al., RNAi screening in primary human hepatocytes of genes implicated in genome-wide association studies for roles in type 2 diabetes identifies roles for CAMK1D and CDKAL1, among others, in hepatic glucose regulation. PLoS One 8, e64946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray S. et al., Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 5, 305–312 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofherr A. et al., The mitochondrial transporter SLC25A25 links ciliary TRPP2 signaling and cellular metabolism. PLoS Biol. 16, e2005651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin J. D., Minireview: The PGC-1 coactivator networks: Chromatin-remodeling and mitochondrial energy metabolism. Mol. Endocrinol. 23, 2–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabata M. et al., Cdc2-like kinase 2 suppresses hepatic fatty acid oxidation and ketogenesis through disruption of the PGC-1α and MED1 complex. Diabetes 63, 1519–1532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martínez-Redondo V., Pettersson A. T., Ruas J. L., The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 58, 1969–1977 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Kumashiro N. et al., Targeting pyruvate carboxylase reduces gluconeogenesis and adiposity and improves insulin resistance. Diabetes 62, 2183–2194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cappel D. A. et al., Pyruvate-carboxylase-mediated anaplerosis promotes antioxidant capacity by sustaining TCA cycle and redox metabolism in liver. Cell Metab. 29, 1291–1305.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y. et al., Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature 485, 128–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ozcan L. et al., Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 15, 739–751 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A. et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lovci M. T. et al., Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 20, 1434–1442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data reported in this paper have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE152303. The RBP binding data of HepG2 and K562 cells was retrieved from www.encodeproject.org, accession code ENCSR456FVU. The code for read mapping and peak calling is available on GitHub, https://github.com/YeoLab/eclip.