Significance
Precision medicine aims to cure cancer without affecting normal tissues. PIWI proteins provide a promising opportunity for precision medicine because they are normally expressed only in the testis for male fertility but gain expression in diverse types of cancers. Thus, inhibiting PIWI expression may stop cancer development (and sperm production) without affecting normal body function. To establish causality between PIWI and cancer, we show here that the expression of PIWIL1, a human PIWI protein, promotes gastric cancer. Surprisingly, this oncogenic function does not require piRNA, the expected partner of PIWI proteins, but involves the nonsense-mediated mRNA decay mechanism. These findings reveal a function and action mechanism of PIWI proteins in oncogenesis, guiding the identification of PIWI inhibitors to cure cancer.
Keywords: gastric cancer, PIWI, piRNA, human, nonsense-mediated mRNA decay
Abstract
Targeted cancer therapy aims to achieve specific elimination of cancerous but not normal cells. Recently, PIWI proteins, a subfamily of the PAZ-PIWI domain (PPD) protein family, have emerged as promising candidates for targeted cancer therapy. PPD proteins are essential for small noncoding RNA pathways. The Argonaute subfamily partners with microRNA and small interfering RNA, whereas the PIWI subfamily partners with PIWI-interacting RNA (piRNA). Both PIWI proteins and piRNA are mostly expressed in the germline and best known for their function in transposon silencing, with no detectable function in mammalian somatic tissues. However, PIWI proteins become aberrantly expressed in multiple types of somatic cancers, thus gaining interest in targeted therapy. Despite this, little is known about the regulatory mechanism of PIWI proteins in cancer. Here we report that one of the four PIWI proteins in humans, PIWIL1, is highly expressed in gastric cancer tissues and cell lines. Knocking out the PIWIL1 gene (PIWIL1-KO) drastically reduces gastric cancer cell proliferation, migration, metastasis, and tumorigenesis. RNA deep sequencing of gastric cancer cell line SNU-1 reveals that KO significantly changes the transcriptome, causing the up-regulation of most of its associated transcripts. Surprisingly, few bona fide piRNAs exist in gastric cancer cells. Furthermore, abolishing the piRNA-binding activity of PIWIL1 does not affect its oncogenic function. Thus, PIWIL1 function in gastric cancer cells is independent of piRNA. This piRNA-independent regulation involves interaction with the UPF1-mediated nonsense-mediated mRNA decay (NMD) mechanism. Altogether, our findings reveal a piRNA-independent function of PIWIL1 in promoting gastric cancer.
Cancer is a malignant disease with a tremendous impact on global health. Surgery, chemotherapy, and radiation therapy are the main types of cancer treatment. Although the overall survival rate of these treatments has improved significantly over the decades, the total therapeutic efficacy is still not ideal, and certain cancers still lack effective therapy. Thus, new treatments, such as immunotherapy, targeted therapies, hormone therapy, and cryoablation, have been developed in recent years for cancer patients (1–4). Among these new types of treatment, targeted therapy has unique strength and potential (2).
Gastric cancer is the fourth most common cancer and the second leading cause of cancer death worldwide (5, 6). Presently, there are only three FDA-approved drugs that target gastric cancer: trastuzumab (7, 8), ramucirumab (9–11), and pembrolizumab (12). These drugs have great efficacy in some cancer types such as breast cancer or colon cancer but are much less effective in treating gastric cancer, especially advanced gastric cancer. Therefore, there is a pressing need for new therapy for gastric cancer treatment.
PIWI proteins have emerged as a new opportunity for targeted cancer therapy. These proteins belong to the PAZ-PIWI domain (PPD) family of RNA-binding proteins, which is composed of the Argonaute and PIWI subfamilies. The PIWI proteins were first discovered for their evolutionarily conserved functions in germline stem cell self-renewal (13, 14). They bind to a class of noncoding small RNAs called PIWI-interacting RNAs (piRNAs) that are generally 24 to 32 nucleotides in length (15–18). PIWI proteins and piRNAs are mostly expressed in the germline. They form the PIWI-piRNA complex that plays essential roles in germline development, stem cell self-renewal, transposon silencing, and gametogenesis in diverse organisms (19–28). However, the somatic function of PIWI has only been reported in lower eukaryotes (13, 14, 28–30), with the expression and function of PIWI proteins in mammalian somatic tissues remaining unclear. In mice, complete knockout of all three PIWI subfamily genes causes no detectable defects in development and growth (31). In humans, there are four human PIWI genes: PIWIL1 (PIWI-Like 1, also known as [aka] HIWI), PIWIL2 (aka HILI), PIWIL3 (aka HIWI3), and PIWIL4 (aka HIWI2) (32). PIWIL1 was first reported to be drastically overexpressed in seminoma, a testicular germ cell tumor (33). Since then, multiple studies have documented the ectopic expression of PIWI proteins in a wide variety of human cancers (34–45). Most of these findings are correlative. Recently, a causative role of PIWIL1 and PIWIL4 genes was demonstrated in pancreatic cancer cells and breast cancer cells, respectively (45, 46). In the pancreatic cancer study, the piRNA expression was not detectable, which led the authors to propose that the function of PIWIL1 is piRNA independent. This proposal is similar to an earlier proposition based on the observation that PIWIL1 did not detectably associate with piRNA in a colon cancer cell line (COLO205) (47). However, some recent studies reported the existence and function of specific piRNAs in cancer, mostly based on correlative evidence such as genetic association studies (48, 49). These contrasting proposals await definitive demonstration of piRNA independence of PIWI proteins for their function in cancer.
Here we demonstrate a piRNA-independent function of PIWIL1 in gastric cancer. Furthermore, we show that PIWIL1 achieves this function by associating with the UPF1-mediated nonsense-mediated mRNA decay (NMD) pathway to regulate mRNA expression. These findings reveal a critical oncogenic mechanism mediated by a PIWI protein that is distinct from the well-established PIWI-piRNA pathway during normal development.
Results
PIWIL1 Is Highly Expressed in Gastric Cancer Patient Samples and Gastric Cancer Cell Lines.
To explore the function of PIWI proteins in gastric cancer, we first examined the expression of all four PIWI genes in six different well-studied gastric cancer cell lines, including adhesion and suspension types of cell lines. Only PIWIL1 was highly expressed in all six gastric cancer cell lines at both mRNA and protein levels, above its expression in the normal gastric epithelial cell line GES-1 (Fig. 1 A and B and SI Appendix, Fig. S1 A and B).
Fig. 1.
PIWIL1 is highly expressed in gastric cancer samples and gastric cancer cell lines. (A) Bar graph showing qRT-PCR of PIWIL1 mRNA expression in a normal gastric epithelial cell line GES-1 and six human gastric cancer cell lines. The mRNA level of GES-1 was normalized to 1. β-Actin was used as an internal standard to normalize the level of PIWIL1 from the same sample. Results are mean ± SD of three independent experiments. (B) Western blot showing the PIWIL1 protein levels in GES-1 and six human gastric cancer cell lines. β-Actin was used as a loading control. (C) qRT-PCR examination of the relative PIWIL1 mRNA levels in gastric cancer samples from 97 patients as compared to their paired normal tissues, using β-actin as an internal control. Bar value represents the difference of PIWIL1 mRNA levels between normal tissue and tumor in log2, so that values >1 and < −1 indicate that PIWIL1 mRNA levels increase and decrease more than twofold in tumors, respectively. Results are mean ± SD of technical triplicates. (D) Representative micrographs of PIWIL1 IHC staining of gastric cancer tissue microarrays. (E) Box plot showing IHC scores of PIWIL1 expression in 104 pairs of paired tumor and normal tissues in the gastric cancer tissue microarrays. Each tissue spot on the tissue microarray was scored by stain strength (range from 0 to 3) and the percentage of PIWIL1-positive cells (range from 0 to 4), respectively. The IHC score = stain strength score × percentage score of PIWIL1-positive cells. *P < 0.05, Student's paired t test. (F) PIWIL1 IHC score of each paired tumor and normal tissue spot in gastric cancer tissue microarrays. T > N, T = N, and T < N denote the number of tissue pairs in which the IHC score in the tumor (T) tissue higher than, equal to, and lower than its paired normal (N) tissue, respectively. The sample number in the T > N group is significantly larger than the sample number in the two other groups. (G) Immunofluorescence staining of PIWIL1 (green) in SNU-1 cells showing the cytoplasmic localization of PIWIL1. The nucleus is stained by DAPI (blue). (H) Bar graphs showing PIWIL1 mRNA levels of 97 paired human gastric cancer samples categorized into different gastric cancer stage, different metastasis state, or different differentiation state. In the Upper graph, I, II, III, and IV denote stages 1 through 4. In the Middle graph, N0: no nodes are involved; N: lymph node metastasis including N1, N2, and N3; M: distant metastasis. In the Lower graph: P: poor differentiation; M: moderate differentiation; H: high differentiation. Error bars represent SE. *P < 0.05, **P < 0.01, ***P < 0.001, Student's unpaired t test.
To further correlate PIWIL1 to gastric cancer, we first conducted a pairwise comparison of its mRNA expression in gastric cancer samples from 97 patients with their normal gastric tissues. In 63 of the 97 patients, the level of PIWIL1 mRNA in the cancer samples was at least twofold higher than their normal samples (Fig. 1C). Among them, 10 patients had PIWIL1 mRNA levels that were at least 100-fold higher than their corresponding normal samples. Consistently, the PIWIL1 mRNA expression data from the Cho gastric dataset of the ONCOMINE database (https://www.oncomine.org/index.jsp) indicate that PIWIL1, but not the three other human PIWI genes, has higher expression in gastric cancer tissues than normal gastric tissues (SI Appendix, Fig. S1 C and D). We then examined the PIWIL1 protein expression in a tissue microarray representing 104 gastric cancer patients by immunohistochemical (IHC) staining. These gastric cancer tissues showed significantly increased levels of the PIWIL1 protein as compared to matched normal tissues (Fig. 1 D–F). Most of the PIWIL1 signal was in the cytoplasm (Fig. 1D). Immunofluorescence staining of the gastric cancer cell line SNU-1 also showed the cytoplasmic localization of PIWIL1 (Fig. 1G), consistent with the known role of PIWIL1 as a cytoplasmic protein in mammalian testes (50).
To further correlate the level of PIWIL1 expression to the severity of gastric cancer, we collected and preprocessed 97 gastric cancer patients’ data by extracting four available clinical factors (stage, metastasis, differentiation, and gender). These data indicated that the up-regulation of PIWIL1 mRNA in gastric cancer samples was significantly correlated with both metastasis and the tumor-node-metastasis (TNM) stage but inversely correlated with the degree of differentiation (Fig. 1H and SI Appendix, Table S1). The percentage of patients with up-regulation of PIWIL1 expression was higher in males than that in females (SI Appendix, Table S1). In addition, we assessed the relationship between PIWIL1 expression and clinical outcome of gastric cancer patients. Kaplan–Meier survival analysis showed that when the level of PIWIL1 mRNA expression was high, patients with poorly differentiated or mixed-classification or 5-fluorouracil (5FU)-based adjuvant therapy had a poor prognosis, especially those with poorly differentiated and high TNM stage (SI Appendix, Fig. S1E). All these data strongly correlate PIWIL1 with the progression of gastric cancer.
PIWIL1 Promotes Gastric Cancer Cell Proliferation, Migration, Tumorigenesis, and Metastasis.
To investigate the effect of aberrant PIWIL1 expression on gastric cancer cells, we chose the gastric cancer cell line SNU-1 as our model. SNU-1 is a poorly differentiated suspension gastric cancer cell line with the highest PIWIL1 expression but low PIWIL2 and PIWIL4 expression among the six gastric cancer cell lines that we examined (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). We deleted a part of the PIWIL1 gene in the SNU-1 cell line by using the CRISPR-Cas9 nickase system (SI Appendix, Fig. S2A). This deletion abolished PIWIL1 protein expression (SI Appendix, Fig. S2B) and significantly inhibited cell proliferation under normal cell culture condition with 10% fetal bovine serum (FBS), as indicated by two independent PIWIL1-knockout (PIWIL1-KO) cell lines (Fig. 2A). Under a malnutritional condition with 5% FBS, the inhibition was even stronger (Fig. 2A). In addition, both PIWIL1-KO cell lines showed a significant increase in the number of cells at G1 phase and a decrease in the number of cells at S phase, but the number of cells at G2/M phase remained mostly unchanged (Fig. 2 B and C). However, PIWIL1-KO did not affect apoptosis (SI Appendix, Fig. S2 C and D). Furthermore, xenografting of SNU-1 cells into nude mice revealed that knocking out PIWIL1 dramatically retarded the tumor growth of gastric cancer cell SNU-1 in vivo (Fig. 2 D and E). Immunostaining of sections from these tumors for two cell proliferation markers, PCNA and ki67, revealed that their expression was significantly reduced in tumors derived from both PIWIL1-KO cell lines (Fig. 2F). Transwell migration assay and tail-vein injection metastasis assay of these cell lines showed that knockout of PIWIL1 significantly inhibited the gastric cancer cell SNU-1 migration ability in vitro (Fig. 2 G and H) and the metastatic potential in vivo (Fig. 2 I and J). These results indicate that PIWIL1 deficiency severely compromised the SNU-1 gastric cancer cell proliferation, migration, tumorigenesis, and metastasis.
Fig. 2.
Knockout of PIWIL1 inhibits SNU-1 cell proliferation, migration, G1-S transition, tumorigenesis, and metastasis. (A) Growth curve of SNU-1 cells with or without PIWIL1-KO in 10% FBS or 5% FBS cell culture medium conditions, analyzed by colorimetirc (MTS) assay in biological triplicates. WT: wild-type SNU-1 cells without transfection of CRISPR-Cas9 plasmids; KO-Con: SNU-1 cells cotransfected with PIWIL1-sgRNA plasmid and Cas9 plasmid, but gene editing did not occur in PIWIL1; KO-#20 and KO-#37: two independent SNU-1 cell lines which were cotransfected with PIWIL1-sgRNA plasmid and Cas9 plasmid with gene editing occurring in PIWIL1 to knockout PIWIL1 protein. (B) Flow cytometry analysis of cell cycle of PIWIL1-WT or PIWIL1-KO SNU-1 cells. Results are representatives of biological triplicates. (C) Bar graph showing the percentage of PIWIL1-WT or PIWIL1-KO SNU-1 cells in G1 and S phase (n = 3). **P < 0.01, Student's paired t test. (D) SNU-1-derived tumors isolated from biege/nude/xid (BNX) nude mice 35 d after injection of PIWIL1 WT or KO-control or KO cell clones #20 or #37 SNU-1 cells (5 × 106 cells/mouse). (E) The tumor growth curve of xenograft assay in D (n = 5 animals). (F) Representative IHC staining of cell proliferation markers PCNA and ki67 in the paraffin tumor sections from xenograft assay in D. (G) Transwell assay of SNU-1 cells with or without knockout of PIWIL1. Results are representative of three independent experiments. (H) Bar graph showing the number of migrated cells of each well in the Transwell assay in G (n = 3). **P < 0.01, Student's paired t test. (I) Representative bioluminescent images of BNX nude mice in either the PIWIL1 WT or KO groups at 8 wk after injection of SNU-1 cells, depicting the extent of tumor burden. (J) Chart showing quantitative bioluminescent Flux [P/S] of the BNX nude mice in either the PIWIL1 WT or KO groups at 2, 4, 6, and 8 wk after injection of SNU-1 cells (n = 10 animals).
To assess whether the PIWIL1-KO phenotype we observed in SNU-1 cells reflects a more general function of PIWIL1 in gastric cancer, we performed a similar analysis on another gastric cancer cell line, AGS. After knocking down PIWIL1 in the AGS cell line by three different siRNAs (SI Appendix, Fig. S2E), the G1-S transition and migration of AGS cells were all severely affected (SI Appendix, Fig. S2 F–I). All of the results in this section together demonstrate that PIWIL1 promotes the proliferation, migration, tumorigenesis, and metastasis of gastric cancer cells.
PIWIL1 Directly Targets Many RNAs in Gastric Cancer Cells and Represses the Expression of Most of Its Target RNAs.
To explore the molecular mechanism underlying PIWIL1 function in gastric cancer, we carried out total RNA deep sequencing (RNA-Seq) of two wild-type SNU-1 cell clones (WT) and two PIWIL1-KO SNU-1 cell lines. A total of 1,678 genes are significantly affected in PIWIL1-KO cells (adjusted P value <0.05 and fold change ≥1.5) as assessed by DESeq2 algorithm. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analyses further revealed that 749 down-regulated genes displayed significant enrichment for cell cycle processes (SI Appendix, Fig. S3 A and B), whereas 929 up-regulated genes showed significant enrichment for focal adhesion pathways (SI Appendix, Fig. S3 C and D).
Since DESeq2 algorithm is focused on individual genes with the highest statistical significance, we further analyzed RNA-Seq data from WT and PIWIL1-KO SNU-1 cells using weighted gene coexpression network analysis (WGCNA) that investigates changes in the expression of coexpressed gene clusters in PIWIL1-KO cells, which best reflect the global effect of PIWIL1 on the transcriptome of gastric cancer cells (51, 52). In total, 41 coexpressing modules were identified via the average linkage hierarchical clustering (Fig. 3A). We then investigated the correlation between PIWIL1 and the coexpressed gene modules. Among the 41 modules, only blue and green modules indicate significantly positive correlations with PIWIL1 expression (P = 0.00086 and 0.00014, respectively), whereas the turquoise module shows a significantly negative correlation with PIWIL1 expression (P = 6.3e-06) (51) (Fig. 3 A and B and SI Appendix, Fig. S4A). We assessed the significance of gene coexpression of all 41 modules and found that the blue, green, and turquoise modules were most significant (SI Appendix, Fig. S4B). Especially, genes in blue and turquoise modules are enriched in specific pathways (Fig. 3C; see below), and genes in these two modules have consistent difference in expression patterns among WT and PIWIL1-KO cell lines (SI Appendix, Fig. S4C). Overall, the coexpressed genes in the blue and turquoise modules well represent the difference between WT and PIWIL1-KO cells.
Fig. 3.
PIWIL1 target RNAs and transcriptomic changes in PIWIL1-KO SNU-1 cells. (A) WGCNA analysis of RNA coexpression modules regulated by PIWIL1. Topological overlap dissimilarity measure is clustered by average linkage hierarchical clustering. Module assignments (using a dynamic hybrid algorithm) are denoted in the color bar (Bottom). (B) Heatmap of the correlation between module eigengenes and the trait with or without PIWIL1 expression. Red color represents a positive correlation between a module and the trait, and blue color represents a negative correlation. Each cell contained the corresponding correlation and P value. (C) KEGG pathway analysis of hub genes of the blue or turquoise module. Any gene with correlation with the module eigengenes (kME) ≥0.9 was assigned as a hub gene. GSEA/MSigDB gene sets tool was used for the KEGG pathway analysis. (D) Venn diagram of RNAs positively or negatively regulated by PIWIL1 or bound by PIWIL1. PIWIL1-positively or -negatively regulated RNAs are identified by DESeq2 analysis with P < 0.05 and fold change ≥1.5 cutoff. PIWIL1-bound RNAs are identified with P < 0.05 and fold change ≥1.5 cutoff. (E) Volcano plot of differentially expressed genes (blue dots), including PIWIL1-target genes (orange dots), in PIWIL1-KO SNU-1 cells. Dotted lines represent 1.5 fold change in expression (vertical lines) and P < 0.01 cutoff (horizontal line). (F) Venn diagram of PIWIL1-bound RNAs negatively regulated by PIWIL1 and RNAs of the turquoise module hub genes with kME ≥0.85 and expression fold change ≥1.5. (G) GO biological process analysis of the 171 PIWIL1-target turquoise hub genes that are negatively regulated by PIWIL1. (H) KEGG pathway analysis of the 171 genes in F and G. (I) Quantitative RIP-PCR validation of PIWIL1-enrichment efficiency of six cell migration-related genes (VCL, LAMC3, FLNA, MYO18B, SRCIN1, and TPM2) from the 171 genes in F–H. Results are mean ± SD of biological triplicates. (J) qRT-PCR validation of mRNA expression of the six genes in I. Results are mean ± SD of biological triplicates.
To further reveal the potential hierarchy of gene regulation in blue and turquoise modules by PIWIL1, we identified 808 and 743 hub genes in the blue and turquoise modules, respectively. These hub genes are the most connected in a module and play essential roles in biological processes. The KEGG pathway analysis showed that the cell cycle and DNA replication pathways are highly enriched among the blue module hub genes, whereas the focal adhesion and adherens junction pathways are highly enriched among the turquoise module hub genes (Fig. 3C). The change in the expression of some well-studied oncogenes or tumor suppressor genes in the blue or turquoise module hub genes was validated by quantitative RT-PCR (SI Appendix, Fig. S4D). Thus, consistent with DESeq2-GO analyses, PIWIL1 appears to promote cell proliferation and cell migration mechanisms but inhibit cell adhesion mechanisms.
To identify the direct target RNAs of PIWIL1 in gastric cancer, we conducted anti-PIWIL1 RNA coimmunoprecipitation (RIP) followed by deep sequencing on SNU-1 cells. This allowed us to identify 2,194 PIWIL1-bound RNAs, among which 538 displayed altered expression in PIWIL1-KO cells (Fig. 3D and SI Appendix, Fig. S4E). Among these 538 RNAs, 71% were protein-coding mRNAs, 19% were long noncoding RNAs (lncRNAs), and 7% were pseudogene RNAs (SI Appendix, Fig. S4F). Remarkably, 516 of the 538 RNAs were up-regulated in PIWIL1-KO cells, including 365 mRNA and 101 lncRNAs (Fig. 3D). Only 18 mRNAs enriched in metabolic pathways and 3 lncRNAs were down-regulated in PIWIL1-KO cells (SI Appendix, Fig. S4 G and H). Thus, PIWIL1 negatively regulates most of its target RNAs, regardless of whether they are mRNAs or lncRNAs or even pseudogene RNAs (Fig. 3E and SI Appendix, Fig. S4G). Because PIWIL1 is a cytoplasmic protein, it is most likely that PIWIL1 achieves negative regulation by destabilizing its target RNAs instead of repressing their transcription. This posttranscriptional role of PIWIL1 has been demonstrated for mouse PIWIL1 (53). In contrast, 727 indirect target RNAs are positively regulated by PIWIL1, whereas 413 indirect target RNAs are negatively regulated by PIWIL1 (Fig. 3D and SI Appendix, Fig. S4I), indicating a preferentially negative regulatory relationship between the PIWIL1-target RNAs and indirect target RNAs.
Among the 516 genes up-regulated in PIWIL1-KO cells, 171 were turquoise module hub genes (Fig. 3F) enriched in the adhesion process and the focal adhesion pathway, as revealed by KEGG pathway and GO analyses (Fig. 3 G and H). Six of the 171 enriched mRNAs were further confirmed for their binding to, and negative regulation by, PIWIL1 by RIP and qPCR assays (Fig. 3 I and J).
Beyond PIWIL1-target RNAs, the expression of another 1,140 RNAs is changed in PIWIL1-KO cells, including 221 blue module hub genes that are positively regulated by PIWIL1 and are most highly enriched in the cell cycle pathway (SI Appendix, Fig. S4 I and J). These findings corroborate the role of PIWIL1 in promoting cell cycle, proliferation, and tumorigenesis. Taken together, our analyses indicate that PIWIL1 achieves its oncogenic function by directly repressing the expression of many genes involved in cell adhesion and indirectly promoting the expression of many genes involved in cell proliferation.
The PIWIL1 Function in Gastric Cancer Cells Is piRNA Independent.
It has been well demonstrated that PIWI proteins carry out various biological functions by interacting with piRNAs. Our previous work showed that piRNAs derived from transposons and pseudogenes partner with mouse PIWIL1 to degrade many mRNAs and lncRNAs in mouse late spermatocytes (53, 54). We, therefore, investigated whether human PIWIL1 achieves this regulation in gastric cancer via the PIWI-piRNA pathway.
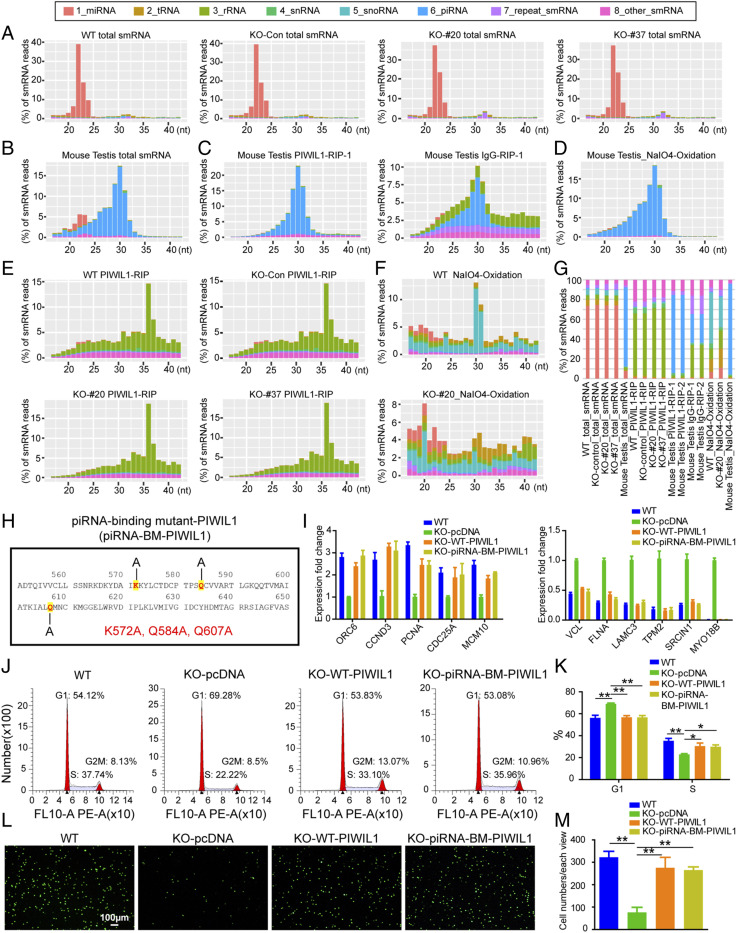
To address this question, we profiled small RNA expression in gastric cancer cells by deep sequencing of the total cellular RNAs and PIWIL1-coimmunoprecipitated RNAs in SNU-1 cells and used the mouse testis as a positive control. The small RNAs in both WT and PIWIL1-KO SNU-1 cells were predominantly miRNAs (22 nucleotides), and there was a barely detectable amount of putative piRNAs (Fig. 4A and SI Appendix, Table S2). Furthermore, the total small RNA size profile in WT SNU-1 cells was not different from that in PIWIL1-KO SNU-1 samples (Fig. 4A), indicating that knocking out PIWIL1 did not affect the expression of miRNAs, as expected. This is contrary to mouse testes, in which most of small RNAs show typical piRNA distributions that peak around 30 nucleotides (Fig. 4B) and are not expressed in PIWIL1-KO mice (53, 54).
Fig. 4.
PIWIL1 function in the gastric cancer cell line SNU-1 is piRNA independent. (A) Size profiles of each class of total small RNAs in WT, PIWIL1-KO-Con, PIWIL1-KO-#20, and PIWIL1-KO-#37 samples. WT: wild-type SNU-1 cells without transfection of CRISPR-Cas9 plasmids; KO-Con: SNU-1 cells cotransfected with PIWIL1-sgRNA plasmid and Cas9 plasmid but gene editing did not occur in PIWIL1; KO-#20, KO-#37: two independent SNU-1 cell lines which were cotransfected with PIWIL1-sgRNA plasmid and Cas9 plasmid with gene editing occurring in PIWIL1 to knockout PIWIL1 protein. (B) Size profiles of each class of total small RNAs in mouse testes. (C) Size profiles of each class of small RNAs immunoprecipitated by PIWIL1 antibody in mouse testis; IgG is the IP negative control. (D) Size profiles of each class of the small RNAs in mouse testes after NaIO4 oxidization. (E) Size profiles of each class of small RNAs immunoprecipitated by PIWIL1 antibody in PIWIL1-WT, -KO-Con, -KO-#20, and -KO-#37 samples. (F) Size profiles of each class of small RNAs in PIWIL1-WT and -KO-#20 samples after NaIO4 oxidization. (G) The proportion of each class of small RNAs in A–F. (H) Mutations in the piRNA-binding mutant PIWIL1 (piRNA-BM-PIWIL1). (I) Left graph: qRT-PCR shows that the down-regulation of PIWIL1-target cell cycle mRNAs in PIWIL1-KO cells can be rescued by both WT PIWIL1 (KO-WT-PIWIL1) and piRNA-binding mutant PIWIL1 (KO-piRNA-BM-PIWIL1). Right graph: qRT-PCR shows that the up-regulation of PIWIL1-target cell migration mRNAs in PIWIL1-KO cells can be rescued by both WT-PIWIL1 (KO-WT-PIWIL1) and piRNA-binding mutant-PIWIL1 (KO-piRNA-BM-PIWIL1). Results are mean ± SD of biological triplicates. (J) Flow cytometry analysis of cell cycles of WT and PIWIL1-KO SNU-1 cells transfected with the empty vector (KO-pcDNA), as well as PIWIL1-KO cells transfected with WT or piRNA-binding mutant PIWIL1-expressing plasmids, denoted as KO-WT-PIWIL1 and KO-piRNA-BM-PIWIL1, respectively. Results are representative of three independent experiments. (K) Bar graph shows the percentage of the G1 phase and S phase of WT-, KO-pcDNA, KO-WT-PIWIL1, and KO-piRNA-BM-PIWIL1 SNU-1 cells in J (n = 3 independent experiments). *P < 0.05, **P < 0.01, Student's paired t test. (L) Transwell assay of WT-PIWIL1, KO-pcDNA, KO-WT-PIWIL1, and KO-piRNA-BM-PIWIL1 SNU-1 cells. Results are representative of three independent experiments. (M) Bar graph shows the number of migrated cells in each well in the Transwell assay in L (n = 3 independent experiments). **P < 0.01, Student's paired t test.
To further search for the possible existence of low abundant piRNAs in WT and PIWIL1-KO SNU-1 cells that might not have been detected among other small RNAs, we used an anti-PIWIL1 antibody to enrich PIWIL1-bound piRNAs and NaIO4 oxidation treatment to remove the 2′-O methylation at the 3′-end of piRNAs to facilitate piRNA cloning (55, 56). Mouse testes were used as a positive control. Expectedly, piRNAs were enriched in the mouse testicular small RNA preparations after both PIWIL1 antibody pulldown and NaIO4 treatment but not in the IgG pulldown control (Fig. 4 B–D and SI Appendix, Fig. S5A), indicating the validity of piRNA enrichment by these two methods. However, small RNA preparations from SNU-1 cells showed no piRNA enrichment but the spurious distribution of small RNA categories mainly represented by degraded rRNA fragments in RIP libraries (Fig. 4E and SI Appendix, Tables S2 and S3) and snoRNAs in NaIO4-treated libraries (Fig. 4F). These lines of evidence support the idea that very few bona fide piRNAs, if any, exist in the SNU-1 gastric cancer cell line. In addition, the expression of several key components for piRNA biogenesis was barely detected (SI Appendix, Fig. S5B), further indicating the absence of a functional piRNA biogenesis machinery in SNU-1 cells.
To further demonstrate that PIWIL1 functions independently of piRNAs in SNU-1 cells, we investigated whether mutating piRNA-binding residues in PIWIL1 will compromise its function. We generated a transfection construct containing a piRNA-binding mutant PIWIL1 by introducing K572A, Q584A, and Q607A triple mutations into a WT PIWIL1 cDNA (57, 58) (Fig. 4H). The mutant PIWIL1 was introduced into PIWIL1-KO SNU-1 cells by transfection. The WT PIWIL1 gene and the empty vector were used as a positive and negative control, respectively. The mutant PIWIL1 fully restored the regulation of the PIWIL1 target RNAs in PIWIL1-KO SNU-1 gastric cancer cells, as did the WT PIWIL1 transgene (Fig. 4I). Notably, just like WT PIWIL1, the mutant PIWIL1 blocked the down-regulation of PIWIL1-KO-induced cell cycle-associated mRNAs (CCND3, ORC6, PCNA, CDC25A, and MCM10) and the up-regulation of PIWIL1-KO-induced cell migration-associated mRNAs (VCL, FLNA, LAMC3, TPM2, SRCIN1, and MYO18B) (59–66). Moreover, the mutant PIWIL1 rescued the cell cycle arrest and cell migration defects in PIWIL1-KO SNU-1 cells comparable to WT PIWIL1 (Fig. 4 J–M). Together, these lines of evidence indicate that the function of PIWIL1 in gastric cancer cells is independent of piRNA.
The piRNA-Independent Regulation of PIWIL1 Involves the NMD Mechanism.
To investigate how PIWIL1 carries out piRNA-independent regulation in gastric cancer cells, we performed PIWIL1 coimmunoprecipitation followed by mass spectrometry to identify proteins that interact with PIWIL1 in SNU-1 cells. The PIWIL1 antibody effectively pulled down PIWIL1 protein itself and UPF1 (up-frameshift-1), a core factor of NMD of mRNAs (67, 68) (Fig. 5A). This indicates that PIWIL1 might negatively regulate its direct target RNAs by interacting with the UPF1-mediated NMD complex.
Fig. 5.
PIWIL1 cooperates with UPF1 to negatively regulate PIWIL1-bound RNAs without piRNA. (A) A protein gel of PIWIL1-coimmunoprecipitation identifying PIWIL1 (blue arrow) and UPF1 (red arrow). (B–C) Western blots showing reciprocal coimmunoprecipitated between PIWIL1 and UPF1 (total and phosphorylated form, p-UPF1), between PIWIL1 and UPF2, and between. PIWIL1 and UPF1, p-UPF1, UPF2, and SMG1. (D) PIWIL1 (green) and DCP1A (red) in WT and PIWIL1KO SNU-1 cells. (E) PIWIL1 (green) colocalized with UPF1 (fuchsia) in the P body (red) in SNU-1 cells. (F) Venn diagram of PIWIL1/UPF1-bound RNAs, and PIWIL1-negatively regulated RNAs, P < 0.05, fold change ≥1.5. (G) Volcano plot of PIWIL1- and UPF1-negatively regulated direct targets in SNU-1 cells. Dotted lines represent 1.5-fold change in expression (vertical) and P < 0.01 cutoff (horizontal). (H) qRIP-PCR confirmed that the six indicated genes are bound by UPF1. Mean ± SD; n = 3. (I) KEGG pathway analysis of 203 RNAs targeted by both PIWIL1 and UPF1. (J) Docking model of PIWIL1 (yellow), UPF1 (green), and RNA (orange). Schematic of full-length PIWIL1 and UPF1-interacting domain mutants-PIWIL1. (K) Coimmunoprecipitation mapping of the UPF1-interacting domain of PIWIL1. pcDNA-Flag: empty vector. (L) Western blotting of coimmunoprecipitation showing that Flag-piRNA-BM-PIWIL1 interacts with UPF1 as strongly as WT PIWIL1 (Flag-PIWIL1-FL). (M) qRT-PCR showing that the up-regulation of PIWIL1-UPF1 cotargeted mRNAs in PIWIL1-KO cells can be rescued by WT-PIWIL1 but not ΔNΔC-PIWIL1. Mean ± SD; n = 3. (N) Transwell assays showing that the inhibition of cell migration in PIWIL1-KO cells can be rescued by WT-PIWIL1 but not ΔNΔC-PIWIL1; n = 3. (O) Bar graph shows the numbers of migrated cells of each view in the Transwell assay (n = 3). **P < 0.01, paired t test.
To investigate this possibility, we conducted coimmunoprecipitation assays between PIWIL1 and two core factors of the NMD machinery, UPF1 and UPF2. We found that PIWIL1 interacted with both UPF1 and UPF2. Since UPF1 is phosphorylated by SMG1 and then recruits various decay effectors of the NMD degradation machinery to target RNAs, we further explored whether PIWIL1 interacts with SMG1 and/or phosphorylated-UPF1 (p-UPF1) by coimmunoprecipitation of PIWIL1 and SMG1 as well as PIWIL1 and p-UPF1 using SNU-1 cellular extracts. PIWIL1 interacted with SMG1 and p-UPF1 in addition to UPF1 and UPF2 (Fig. 5 B and C). Moreover, the interactions between PIWIL1 and the NMD pathway core factors were also observed in another gastric cancer cell line AGS (SI Appendix, Fig. S6A), confirming that PIWIL1 interacts with the NMD machinery in gastric cancer cells.
To confirm that PIWIL1 is involved in the NMD pathway, we conducted immunofluorescence microscopy to determine whether PIWIL1 and UPF1 colocalize in SNU-1 cells. It was well documented that UPF1 triggers mRNA decay in P bodies, which are large cytoplasmic granules replete with proteins involved in general mRNA decay and related processes (69–71). Indeed, PIWIL1 is colocalized with UPF-1 in P bodies in WT SNU-1 cells, but PIWIL1 signal was not detected in PIWIL1-KO SNU-1 cells (Fig. 5 D and E). In addition, PIWIL1 is localized in P bodies in AGS cells (SI Appendix, Fig. S6B). These observations indicate that PIWIL1 interacts with the NMD pathway components in the P body, and further validate the PIWIL1-UPF1 interaction.
To identify RNA cotargets of PIWIL1 and the NMD pathway, we compared PIWIL1 RIP-Seq data with the UPF1 RIP-Seq data of SNU-1 cells and identified 203 mRNAs that are negatively regulated by PIWIL1 and bound by both PIWIL1 and UPF1 (Fig. 5 F and G). The binding of these PIWIL1-target RNAs by UPF1 was confirmed by UPF1 RIP-qPCR analysis of six representative target hub mRNAs (VCL, FLNA, LAMC3, SRCIN1, TPM2, and MYO18B mRNAs, Fig. 5H). KEGG pathway analysis of these 203 cotarget RNAs shows that they are highly enriched in extracellular matrix (ECM) receptor interaction, focal adhesion, and cell adhesion proteins (Fig. 5I and SI Appendix, Fig. S6C), indicating that PIWIL1 appears to coordinate with UPF1 to regulate these cotargeting mRNAs that may play a negative role in gastric cancer cell migration.
We then mapped specific domains responsible for PIWIL1 interaction with UPF1 and investigated whether such a domain is required for the regulation of PIWIL1-UPF1 cotargeted genes. We developed a UPF1 and PIWIL1 protein structure docking model to guide domain mapping (72–74) (Fig. 5I). Deletion of 251 to 383 amino acid residues and 625 to 758 residues of PIWIL1 abolished its interaction with UPF1, but the piRNA-binding mutant PIWIL1 interacted with UPF1 in the same way as WT PIWIL1 (Fig. 5 J–L). Moreover, the deletion mutant PIWIL1 failed to prevent the up-regulation of all six PIWIL1 and UPF1 cotargeted hub genes that we examined in PIWIL1-KO SNU-1 cells (Fig. 5M). This indicates the importance of PIWIL1-UPF1 interaction for PIWIL1 regulation toward its target genes. In contrast, the piRNA-binding mutant PIWIL1 acted like WT PIWIL1 in preventing the up-regulation of these hub genes caused by PIWIL1-KO (Fig. 4I). In addition, the deletion mutant PIWIL1 failed to rescue the decreased cell migration of PIWIL1-KO cells (Fig. 5 N and O), whereas the piRNA-binding mutant PIWIL1 fully rescued the cell migration defects of PIWIL1-KO as did WT PIWIL1 (Fig. 4 L and M). All these observations indicate that interaction with UPF1 is essential for PIWIL1 function in SNU-1 cells.
Finally, we directly assessed the expression of UPF1 in gastric cancer patient samples. In our 97 paired gastric cancer patient samples, UPF1 mRNA was overexpressed in 74 patients as compared to their corresponding normal samples (SI Appendix, Fig. S7A). Consistent with our results, the Gene Expression Profiling Interactive Analysis (GEPIA) database (gepia.cancer-pku.cn/) indicates that UPF1 mRNA expression in gastric cancer samples is higher than that in normal tissue samples (SI Appendix, Fig. S7B). Finally, the expression of PIWIL1 mRNA is significantly correlated with the expression of UPF1 mRNA in our 97 paired clinical patient samples (Pearson P = 0.0458; SI Appendix, Fig. S7C). Collectively, these results indicate that UPF1 is a crucial partner of PIWIL1 in gastric cancer and that PIWIL1 acts through the NMD mechanism in a piRNA-independent fashion to negatively regulate many of its target RNAs in gastric cancer cells.
Discussion
Although environmental factors such as high-fat diet and Helicobacter pylori are better known to cause gastric cancer, genetic aberrations have been increasingly recognized to play an important role. PIWI proteins have been reported to be ectopically expressed in diverse types of cancer, but their role in gastric cancer or most other types of cancer has not been established. Here we report that PIWIL1 is aberrantly expressed in both gastric cancer tissues and cell lines, which promotes the progression of gastric cancer (Fig. 1). Inhibiting the aberrant PIWIL1 expression reduces cancer cell proliferation, migration, tumorigenesis, and metastasis (Fig. 2 and SI Appendix, Fig. S2). These findings reveal a function of PIWIL1 in gastric cancer and identify PIWIL1 as a potential target for gastric cancer precision therapy. Although human seminoma has been associated with multiplication and overexpression of the PIWIL1 gene (33), such an association has not been reported for gastric cancer. It would be interesting to identify genetic alteration of the PIWIL1 gene that might drive the growth and/or metastasis of gastric cancers.
It has been well documented that PIWIL1 regulates the expression of a range of genes at transcriptional or posttranscriptional levels in the germline by forming a complex with piRNAs. To our knowledge, this study represents a transcriptomic analysis of genes regulated by a PIWI protein in gastric cancer cells. Our work identified 538 RNAs bound and directly regulated by PIWIL1. PIWIL1 regulates most of them negatively (Fig. 3 D and E). These RNAs are enriched in migration-related tumor suppressor genes (Fig. 3 G–J). In addition, PIWIL1 positively but indirectly regulates many cell cycle-related oncogenes (SI Appendix, Fig. S4 I and J). These findings indicate an overall hierarchy of negative regulation by PIWIL1 to its direct targets, such as tumor repressor. These direct targets, in turn, preferentially exert negative regulation toward the indirect targets of PIWILI, including genes that promote cell proliferation and oncogenesis.
A major surprising finding from this study is the piRNA-independent nature of PIWIL1 function. This is in contrast to the well-established piRNA-dependent function of PIWI proteins both in the germline and in the soma. The gastric cancer cells express very few bona fide piRNAs if any. Although there was a report of the existence of piRNAs in diverse somatic tissues in the mouse and macaque (75) and several reports of piRNA expression in cancers (76–79), tRNA fragments in sperm derived from tRNA-Gly or tRNA-Glu share nearly identical sequences to some of the annotated piRNAs (80–82), so the authenticity and function of some of the reported somatic piRNAs deserve further investigation. Our finding is consistent with two of the latest studies on a colon cancer cell line (COLO205) and pancreatic ductal adenocarcinoma cells, respectively (46, 47). However, unlike the COLO205 cell line in which the knockout of PIWIL1 is inconsequential to the cancer cell transcriptome, we detected significant changes in the transcriptome. Furthermore, we identified a piRNA-independent function of PIWIL1 via interacting with UPF1 and other core components of the NMD mechanism to negatively regulate the expression of its direct target mRNAs (Fig. 5). Specifically, both WT and piRNA-binding-deficient PIWIL1 interact with UPF1 equally effectively with similar regulatory effects and biological impact. Such interaction is essential for PIWIL1 function. The difference between our findings and the COLO205 study could reflect the different requirements of PIWIL1 in different types of cancers. This possibility can be tested by investigating whether PIWIL1 has any oncogenic function in COLO205 cells. In any case, our findings identified a mechanism of PIWIL1 action by interacting with the NMD machinery in a piRNA-independent manner.
The NMD machinery executes mRNA degradation through at least two pathways: 3ʹ-UTR exon junction complex (EJC)-dependent NMD pathway that degrades mRNA containing premature stop codons and 3ʹ-UTR EJC-independent NMD pathway that degrades normal mRNAs (83, 84). Both pathways involve UPF1 and SMG1 kinase, and often UPF2 and UPF3 (84, 85). In addition, in both pathways, UPF1 and SMG1 are recruited to the target mRNA, where SMG1 phosphorylates UPF. This phosphorylation activates UPF1 to further recruit downstream components to degrade the target mRNA. Given that the PIWIL1-UPF1 cotarget mRNAs are presumably normal, it is likely that PIWIL1 moderates the activity of the NMD machinery through the EJC-independent pathway and at least in part through interacting with the phosphorylated form of UPF1. The future systematic analysis will test this possibility and precisely determine the step(s) of the NMD pathway at which PIWIL1 exerts its regulatory function.
All findings in this report converge into an overall model of how PIWIL1 might promote gastric cancer (Fig. 6): PIWIL1 forms a complex with UPF1, UPF2, SMG1, and other components of the NMD machinery in the P body, possibly through the EJC-independent NMD pathway, to degrade its target mRNAs and lncRNAs, especially tumor suppressors mRNAs, This compromises the overall negative regulation of PIWIL1-target genes toward many indirect target mRNAs, including those for cell proliferation and oncogenesis, thus allowing these mRNAs to be well expressed to promote cancer development. Meanwhile, the PIWIL1-NMD pathway also directly degrades some mRNAs for proteins that inhibit migration, such as cell adhesion molecules, which also promotes oncogenic development. Overall, our findings start to reveal a piRNA-independent function of PIWI proteins in partnership with the NMD mechanism, a gastric cancer-promoting mechanism and a potential therapeutic target for precision therapy of gastric cancer and possibly other types of cancers.
Fig. 6.
A piRNA-independent mechanism of PIWIL1 in regulating mRNA turnover in gastric cancer cells (Right), in contrast to the canonical PIWIL1 action mechanism in the germline cytoplasm in which the targeting of PIWIL1 toward a mRNA is guided by the piRNA complementarity with the mRNA. For details, see the main text.
Materials and Methods
Materials and all methods in this paper, including RNA extraction, quantitative RT-PCR, PIWIL1 and -PIWIL1 mutant cDNA cloning, PIWIL1 knockout and knockdown analyses, Western blotting analysis, coimmunoprecipitation, immunohistochemistry, immunofluorescence microscopy, cell proliferation and apoptosis assays, Transwell migration assay, in vivo tumorigenicity and metastasis assays, mass spectrometry, ribonucleoprotein-immunoprecipitation, small RNA-Seq library construction and sequencing, β-elimination by NaIO4 oxidation, and bioinformatic analyses are described in detail in SI Appendix, Supplementary Methods. Experiments involving mice were approved by the ShanghaiTech University Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We are grateful to Dr. Peixiang Ma for doing the PIWIL1-UPF1 docking analysis; Dr. Jiangsha Zhao, Ms. Zhaoran Zhang, Weidong Chen, and Jiawei Zhang for their various technical assistance; and Prof. Bin Shen and Prof. Xingxu Huang for offering us the CRISPR-Cas9 KO system. We thank ShanghaiTech High Performance Computing Platform for providing computation resources and support. We thank Yuedong Huang, Kun-Yong Kim, Nils Neuenkirchen, Yiying Yang, Chen Wang, Zukai Liu, Tingting Lu, and Yuanyuan Gong for comments on the manuscript. Z.-Z.Y. is supported by Yangfan Grant 2018X0301-103-01. This work was supported by ShanghaiTech University.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008724117/-/DCSupplemental.
Data Availability.
All data in this paper are included in the manuscript and SI Appendix. The RNA-Seq and RIP-Seq data have been deposited in the National Center for Biotechnology Information (NCBI) BioProject with accession number PRJNA633677.
References
- 1.Farkona S., Diamandis E. P., Blasutig I. M., Cancer immunotherapy: The beginning of the end of cancer? BMC Med. 14, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padma V. V., An overview of targeted cancer therapy. Biomedicine (Taipei) 5, 19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo R. A., Hormone-replacement therapy: Current thinking. Nat. Rev. Endocrinol. 13, 220–231 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Erinjeri J. P., Clark T. W., Cryoablation: Mechanism of action and devices. J. Vasc. Interv. Radiol. 21 (8, suppl.), S187–S191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferro A. et al., Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer 50, 1330–1344 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Jemal A. et al., Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Bang Y. J. et al.; ToGA Trial Investigators , Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 376, 687–697 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Gunturu K. S., Woo Y., Beaubier N., Remotti H. E., Saif M. W., Gastric cancer and trastuzumab: First biologic therapy in gastric cancer. Ther. Adv. Med. Oncol. 5, 143–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs C. S. et al.; REGARD Trial Investigators , Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 383, 31–39 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Young K., Smyth E., Chau I., Ramucirumab for advanced gastric cancer or gastro-oesophageal junction adenocarcinoma. Therap. Adv. Gastroenterol. 8, 373–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilke H. et al.; RAINBOW Study Group , Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): A double-blind, randomised phase 3 trial. Lancet Oncol. 15, 1224–1235 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Shitara K. et al.; KEYNOTE-061 investigators , Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): A randomised, open-label, controlled, phase 3 trial. Lancet 392, 123–133 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lin H., Spradling A. C., A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124, 2463–2476 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Cox D. N. et al., A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard A., Sachidanandam R., Hannon G. J., Carmell M. A., A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Aravin A. et al., A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Grivna S. T., Beyret E., Wang Z., Lin H., A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau N. C. et al., Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Houwing S. et al., A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129, 69–82 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Seipel K., Yanze N., Schmid V., The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int. J. Dev. Biol. 48, 1–7 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Alié A. et al., Somatic stem cells express piwi and Vasa genes in an adult ctenophore: Ancient association of “germline genes” with stemness. Dev. Biol. 350, 183–197 (2011). [DOI] [PubMed] [Google Scholar]
- 22.De Mulder K. et al., Stem cells are differentially regulated during development, regeneration and homeostasis in flatworms. Dev. Biol. 334, 198–212 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Palakodeti D., Smielewska M., Lu Y. C., Yeo G. W., Graveley B. R., The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174–1186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szakmary A., Cox D. N., Wang Z., Lin H., Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 15, 171–178 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Unhavaithaya Y. et al., MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507–6519 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smulders-Srinivasan T. K., Szakmary A., Lin H., A Drosophila chromatin factor interacts with the Piwi-interacting RNA mechanism in niche cells to regulate germline stem cell self-renewal. Genetics 186, 573–583 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliano C., Wang J., Lin H., Uniting germline and stem cells: The function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 45, 447–469 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A., SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Funayama N., Nakatsukasa M., Mohri K., Masuda Y., Agata K., Piwi expression in archeocytes and choanocytes in demosponges: Insights into the stem cell system in demosponges. Evol. Dev. 12, 275–287 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Juliano C. E. et al., PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. U.S.A. 111, 337–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng E. C., Kang D., Wang Z., Lin H., PIWI proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells. PLoS One 9, e97821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross R. J., Weiner M. M., Lin H., PIWI proteins and PIWI-interacting RNAs in the soma. Nature 505, 353–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiao D., Zeeman A. M., Deng W., Looijenga L. H., Lin H., Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 21, 3988–3999 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Lee J. H. et al., Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum. Mol. Genet. 15, 201–211 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Liu X. et al., Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int. J. Cancer 118, 1922–1929 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Yang L. J. et al., Effect of betulinic acid on the regulation of Hiwi and cyclin B1 in human gastric adenocarcinoma AGS cells. Acta Pharmacol. Sin. 31, 66–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taubert H. et al., Expression of the stem cell self-renewal gene Hiwi and risk of tumour-related death in patients with soft-tissue sarcoma. Oncogene 26, 1098–1100 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Jiang J., Zhang H., Tang Q., Hao B., Shi R., Expression of HIWI in human hepatocellular carcinoma. Cell Biochem. Biophys. 61, 53–58 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y. M. et al., HIWI is associated with prognosis in patients with hepatocellular carcinoma after curative resection. Cancer 118, 2708–2717 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Xie Y. et al., Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells. Mol. Med. Rep. 11, 1455–1461 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Navarro A. et al., The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget 6, 31544–31556 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan P. et al., Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget 7, 37944–37956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C., Liu J., Xu G., Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark. 13, 315–321 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Wang Y. et al., The PIWI protein acts as a predictive marker for human gastric cancer. Int. J. Clin. Exp. Pathol. 5, 315–325 (2012). [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z., Liu N., Shi S., Liu S., Lin H., The role of PIWIL4, an argonaute family protein, in breast cancer. J. Biol. Chem. 291, 10646–10658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li F. et al., piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat. Cell Biol. 22, 425–438 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Genzor P., Cordts S. C., Bokil N. V., Haase A. D., Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 116, 11111–11112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu A., Jacobs D. I., Hoffman A. E., Zheng T., Zhu Y., PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 36, 1094–1102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs D. I. et al., piRNA-8041 is downregulated in human glioblastoma and suppresses tumor growth in vitro and in vivo. Oncotarget 9, 37616–37626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng W., Lin H., miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Zhang B., Horvath S., A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Dong J., Horvath S., Understanding network concepts in modules. BMC Syst. Biol. 1, 24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe T., Cheng E. C., Zhong M., Lin H., Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 25, 368–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T., Lin H., Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol. Cell 56, 18–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirino Y., Mourelatos Z., Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 14, 347–348 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Ohara T. et al., The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 14, 349–350 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto N. et al., Crystal structure of silkworm PIWI-clade argonaute siwi bound to piRNA. Cell 167, 484–497.e9 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Parker J. S., Roe S. M., Barford D., Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23, 4727–4737 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng X., Cuff L. E., Lawton C. D., DeMali K. A., Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. J. Cell Sci. 123, 567–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao Y. et al., Loss of ERα induces amoeboid-like migration of breast cancer cells by downregulating vinculin. Nat. Commun. 8, 14483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calderwood D. A. et al., Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3, 1060–1068 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Xu Y. et al., Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J. Exp. Med. 207, 2421–2437 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He Y. H. et al., A novel messenger RNA and long noncoding RNA signature associated with the progression of nonmuscle invasive bladder cancer. J. Cell. Biochem., 10.1002/jcb.28089 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Cui J. et al., Epigenetic silencing of TPM2 contributes to colorectal cancer progression upon RhoA activation. Tumour Biol. 37, 12477–12483 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Wang P. et al., SRCIN1 suppressed osteosarcoma cell proliferation and invasion. PLoS One 11, e0155518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Ouderkirk J. L., Krendel M., Non-muscle myosins in tumor progression, cancer cell invasion, and metastasis. Cytoskeleton (Hoboken) 71, 447–463 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lykke-Andersen J., Shu M. D., Steitz J. A., Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 103, 1121–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Lykke-Andersen J., Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22, 8114–8121 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mühlemann O., Lykke-Andersen J., How and where are nonsense mRNAs degraded in mammalian cells? RNA Biol. 7, 28–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brogna S., Ramanathan P., Wen J., UPF1 P-body localization. Biochem. Soc. Trans. 36, 698–700 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Durand S. et al., Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J. Cell Biol. 178, 1145–1160 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vajda S. et al., New additions to the ClusPro server motivated by CAPRI. Proteins 85, 435–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozakov D. et al., The ClusPro web server for protein-protein docking. Nat. Protoc. 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozakov D. et al., How good is automated protein docking? Proteins 81, 2159–2166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan Z. et al., Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39, 6596–6607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng J. et al., piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta 412, 1621–1625 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Assumpção C. B. et al., The role of piRNA and its potential clinical implications in cancer. Epigenomics 7, 975–984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim S. L. et al., Overexpression of piRNA pathway genes in epithelial ovarian cancer. PLoS One 9, e99687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moyano M., Stefani G., piRNA involvement in genome stability and human cancer. J. Hematol. Oncol. 8, 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng H. et al., A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 22, 1609–1612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Q. et al., Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Tosar J. P., Rovira C., Cayota A., Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 1, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karousis E. D., Nasif S., Mühlemann O., Nonsense-mediated mRNA decay: Novel mechanistic insights and biological impact. Wiley Interdiscip. Rev. RNA 7, 661–682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metze S., Herzog V. A., Ruepp M. D., Mühlemann O., Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA 19, 1432–1448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data in this paper are included in the manuscript and SI Appendix. The RNA-Seq and RIP-Seq data have been deposited in the National Center for Biotechnology Information (NCBI) BioProject with accession number PRJNA633677.