Significance
The growth and stabilization of dendritic spines is thought to be essential for strengthening the connections between neurons and thereby memories. Actin cytoskeleton remodeling in spines is the basis of this growth and stabilization. A functional prion that binds mRNA, CPEB3, which interacts with actin, has been reported to be involved in the maintenance of long-term memory by forming stable aggregates. Here we study the interaction between actin and CPEB3 and propose a molecular model for the complex structure of CPEB3 bound to an actin filament. Our model gives insights into the actin/CPEB3 positive feedback loop underlying long-term memory which involves CPEB3’s binding to actin, its aggregation, and its regulation by SUMOylation.
Keywords: long-term memory, functional prion, dendritic spine
Abstract
Dendritic spines are tiny membranous protrusions on the dendrites of neurons. Dendritic spines change shape in response to input signals, thereby strengthening the connections between neurons. The growth and stabilization of dendritic spines is thought to be essential for maintaining long-term memory. Actin cytoskeleton remodeling in spines is a key element of their formation and growth. More speculatively, the aggregation of CPEB3, a functional prion that binds RNA, has been reported to be involved in the maintenance of long-term memory. Here we study the interaction between actin and CPEB3 and propose a molecular model for the complex structure of CPEB3 and an actin filament (F-actin). The results of our computational modeling, including both energetic and structural analyses, are compared with novel data from peptide array experiments. Our model of the CPEB3/F-actin interaction suggests that F-actin potentially triggers the aggregation-prone structural transition of a short CPEB3 sequence by zipping it into a beta-hairpin form. We also propose that the CPEB3/F-actin interaction might be regulated by the SUMOylation of CPEB3, based on bioinformatic searches for potential SUMOylation sites as well as SUMO interacting motifs in CPEB3. On the basis of these results and the existing literature, we put forward a possible molecular mechanism underlying long-term memory that involves CPEB3’s binding to actin, its aggregation, and its regulation by SUMOylation.
Structural change of dendritic spines is associated with the formation and maintenance of long-term memory (1–3). A synaptic tag is required to mark growing spines and regulate local translation of synaptic proteins there. Rapid biomolecular turnover (4), however, raises the question of how synaptic structural change is maintained so as to be long-lasting. The mammalian cytoplasmic polyadenylation element-binding protein 3 (CPEB3), a functional prion that binds to RNA (5), is involved in long-term potentiation (LTP) and has emerged as a potentially important factor in the persistence of long-term memory. Homologs of CPEB3 in invertebrates, ApCPEB in Aplysia (6, 7) and Orb2 in Drosophila (8), have also been described as functional prions that regulate local protein synthesis in active synapses by binding to RNA. Similarly, in higher organisms, CPEB3, with its proline- and glutamine-rich (P/Q-rich) N-terminal prion domain (PRD) and C-terminal RNA binding domain (RBD), can form stable aggregates that are thought to consolidate synaptic strength and regulate the local translation of target mRNAs upon synaptic stimulation (5). Numerous structural and cellular experiments on proteins in the CPEB family have been performed. The Cryo-EM structure of Orb2 amyloid core (9) and the NMR structure of the CPEB RBD (10) have been solved. The structures and functions of the remainder of CPEB3, including the structures of monomeric and amyloid PRD as well as the mechanism by which it aggregates, are still unknown. Here we combine bioinformatics studies, molecular simulations, and peptide array experiments to refine the boundaries of an Actin Binding Domain (ABD) in CPEB3 [initially identified by Stephan et al. (5)] and then to model its structure in complex with F-actin.
Actin cytoskeleton remodeling is involved in the structural change of dendritic spines, which is associated with the strengthening of synapses and, therefore, also with the storage of memories (11, 12). Stephan et al. (5) observed that CPEB3 aggregates colocalize with F-actin in neurons and that the depolymerization of F-actin prevents CPEB3 from aggregating in yeast. In previously published theoretical work (13) on the CPEB3 homolog from Aplysia (ApCPEB), it was suggested that the CPEB/F-actin interaction could provide the mechanical force necessary to induce a structural transition of CPEB oligomers from a coiled-coil form into a beta-sheet–containing amyloid-like fiber. Moreover, actin mRNA has been identified as a target of CPEB3, implying then that there may exist a positive feedback loop involving CPEB3 and F-actin (5) wherein F-actin facilitates the aggregation of CPEB3 and then CPEB3 aggregates promote the translation of actin, ultimately influencing spine structure.
In this study, we present a structure of the F-actin/CPEB3-ABD complex found through computational modeling using the RaptorX complex contact prediction webserver in concert with the Associative Memory, Water-Mediated, Structure and Energy Model with Evolutionary Restraints (AWSEM-ER) (14, 15). AWSEM-ER is a coarse-grained force field that has been optimized using energy landscape theory which can be bolstered by predicted contacts, in this case from the RaptorX webserver. Integrating structure prediction and energetic analyses with experimental peptide array data, we conclude that electrostatic interactions are essential to the stability of the bound complex and that a hydrophobic interaction cooperates with the electrostatics to guide the CPEB3-ABD to a binding pocket on the surface of F-actin. This binding pocket is shared by most other actin binding proteins, such as cofilin and Arp 2/3 (16).
By clustering the F-actin/CPEB3-ABD complex structures from the simulation, we found a chameleon sequence in the N-terminal end of the ABD, which can adopt two different secondary structures. When CPEB3 approaches the surface of actin, the electrostatic attraction between F-actin and the two ends of this chameleon sequence facilitates the formation of a beta-hairpin. This beta-hairpin acts as a catalyst for forming intramolecular beta-sheets and could thereby help trigger the aggregation of CPEB3.
Actin-induced beta-sheet formation in CPEB3, however, presents a potential problem if aggregation were unregulated inside neurons. SUMOylation is known to regulate CPEB3 aggregation (17), and we propose that this reversible posttranslational modification may be an important control to the CPEB3/F-actin interaction. We performed bioinformatic searches for both SUMOylation sites as well as SUMO interacting motifs in CPEB3 and found several of both including a SUMO interacting motif in a region of CPEB3 that is crucial for binding to actin in our predicted complex structure. We propose that SUMOylation of CPEB3 in its basal state might repress the CPEB3/F-actin interaction and that deSUMOylation in stimulated synapses then disinhibits this interaction so that the binding, aggregation, and regulatory activity of CPEB3 can be controlled ultimately by synaptic stimulation. This hypothesis is consistent with the experimental observations of Drisaldi et al. (17). As a bridge between synaptic stimulation and CPEB3 aggregation/activation, interaction with F-actin appears to be an essential step in the CPEB3 functional cascade.
Results
Identifying the Actin Binding Domain (ABD) in CPEB3.
The first 431 residues of CPEB3 make up a largely disordered domain, shown in Fig. 1A, which is followed by an RNA binding domain of known structure. Stephan et al. (5) proposed a tripartite model for the unstructured domain of mouse CPEB3. The middle subdomain, a location-mediating domain (LMD; M217-P284), is thought to be the domain that interacts with actin filaments (5). Human CPEB3 and mouse CPEB3 have a high sequence similarity, 94.6%. The schematic diagram of human CPEB3 subdomains based on the definition given by Stephan et al. (5) is shown in Fig. 1A. We conducted peptide array experiments to evaluate the actin-binding activity of peptides taken from human CPEB3 (Fig. 1B). Based on the peptide array data, we have extended the boundaries of the ABD to include residues Q164-T325. This larger range of residues completely covers the two actin-binding intensity peaks found in the peptide array data that overlap with the LMD. The boundaries of the ABD divide the unstructured domain into three subdomains: the N-terminal subdomain (the prion domain [PRD]) is Q/P rich and is required for CPEB3’s prion state to be transmitted from mother to daughter cells in yeast (5); the C-terminal subdomain contains a nuclear export signal (18) and was identified by Stephan et al. (5) as a second prion domain (PRD2; Fig. 1A). This C-terminal subdomain is enriched with charged residues but is not enriched with the types of residues that are typically associated with prion domains (like Q/N/P).
Fig. 1.
CPEB3 subdomain definitions and experimental peptide array data showing per-residue actin binding intensity to peptides derived from the sequence of human CPEB3. (A) Definitions of CPEB3 subdomains used by Stephan et al. (5) (Upper) and in this work (Lower). (B) Experimental residue-level actin-binding intensity along the sequence of human CPEB3. The dashed blue lines enclose the ABD identified in this work (Q164 to T325).
We first ran AWSEM simulations of the entire unstructured domain (residues 1 to 431; Fig. 1A). The resulting contact maps support the tripartite model of Stephan et al. (5) as well as the slightly expanded definition of the actin interacting subdomain (SI Appendix, Fig. S1). The actin cytoskeleton depolymerization experiments by Stephan et al. (5) show that F-actin is required for foci formation of a truncated mouse CPEB3 construct made up of PRD2 and RBD, while foci formation for the RBD alone was not obviously influenced by F-actin depolymerization (5). This result suggests that at least part of PRD2 interacts with F-actin. In summary, we have chosen to extend the original definition of the LMD into a larger actin interacting domain (ABD; Fig. 1A) on the basis of three observations: peaks of actin binding intensity in the peptide array experiments, subdomain structure found in the contact maps generated by folding the unstructured domain of CPEB3 with AWSEM, and experimental evidence from Stephan et al. (5) showing that the localization of a truncated CPEB3 construct including PRD2 and the RBD is influenced by actin depolymerization.
The Binding Complex of ABD and F-Actin.
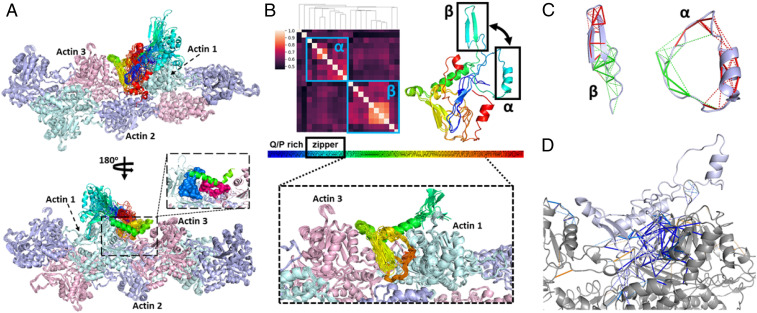
To predict the complex structure of CPEB3’s ABD and F-actin, we ran molecular dynamics simulations using AWSEM-ER. Interface contacts between the ABD and actin were first predicted using the RaptorX ComplexContact webserver (19, 20). RaptorX’s ComplexContact server uses coevolutionary information as well as deep learning techniques to predict interface contacts. The predicted contacts for the ABD-actin interaction are shown in Fig. 2A. We ran AWSEM-ER annealing simulations using only the top 50 RaptorX predicted contacts (those with a reported contact probability >0.3) as input. These high-confidence contacts occur between a repeated VG sequence of the ABD and the D-loop of actin. The final predicted structures, which were obtained by first performing annealing in the presence of the predicted contact restraints and then performing relaxation simulations in the absence of these restraints, show only modest consistency among the 20 predicted structures. We will refer to these structures as the first-generation predictions (Fig. 2B and Movie S1). In the first-generation structures, the contacts between the repeated VG sequence in CPEB3 and actin’s D-loop show strong consistency (Fig. 2C) even after running constraint-free relaxation simulations. When making contact with the D-loop of actin 3 (Fig. 2B), the repeated VG residues also make contact with the hydrophobic cleft of actin 1. These contacts with the hydrophobic cleft, although not included as input to the AWSEM-ER simulations, turn out to be nonetheless consistent with other contacts that were predicted by the RaptorX ComplexContact server but with confidence below our threshold (Fig. 2A).
Fig. 2.
Contact maps and structures for the predicted binding complex of ABD and F-actin in the first-generation AWSEM-ER simulations (after relaxation). (A) Comparison between the RaptorX contact map and the frequency contact maps for 20 AWSEM-ER predicted structures. The x axis is the residue index for three actin monomers, and the y axis is the residue index for ABD, from N terminus to C terminus, blue to red. (Upper) Contact map of three copies of the RaptorX prediction for the ABD/actin complex. Darker color represents higher confidence in the predicted contact. (Lower) Contact map of the sum of contact maps for ABD with three consecutive actin monomers in F-actin in the final structures produced by 20 AWSEM-ER annealing simulations. The white rectangles are used to indicate contacts that are consistent with RaptorX predictions (shown as red and blue rectangles in Upper). The blue rectangle highlights the restraint contacts (top 50 RaptorX contacts) that were included as input to the AWSEM-ER simulations. The red rectangles highlight contacts that were not included as input but are nonetheless consistent with the AWSEM-ER predictions. (B) The overlay of 20 predicted structures. The ABD is shown with a rainbow spectrum (corresponding to the y axis of A). (C) The same as B, but for the ABD, only the repeated VG sequence is shown.
To refine the predicted complex structures, we then conducted second-generation (21) simulations using additional contact restraints based on contacts that were found with high frequency (formed more than 10 times out of 20 simulations) in the first-generation simulations. The second-generation constraint contact map is shown in SI Appendix, Fig. S4. The self-consistency of the second generation complex structures is now quite high: all 20 predicted structures align well with each other. Before running relaxation simulations, the minimal mutual Qcontact for residues M217 to I302 in these 20 structures is 0.812 (Fig. 3 and Movie S2). Relaxation simulations in the absence of constraints confirmed the stability of the second-generation structures (SI Appendix, Fig. S4 and Movie S3). Furthermore, frustration (22–24) analyses performed with the Frustratometer (25) showed that frustration was reduced in F-actin after binding with the ABD (Fig. 3D).
Fig. 3.
Structure of the ABD/F-actin binding complex from the second-generation AWSEM-ER simulations (before relaxation). (A) The overlay of 20 predicted structures shown in two different orientations. The binding interface between the poly(A) helix (S224-N250) in ABD and the hydrophobic pocket in F-actin is zoomed in. The surfaces of the hydrophobic cleft and the D-loop are colored in blue and pink, respectively. (B) Clustering analysis for 20 predicted structures of ABD/F-actin complex using mutual Qcontact (including the intramolecular and intermolecular contacts involving ABD) as the structural similarity metric. Two clusters are labeled by blue rectangles in the mutual Q matrix, and two representative structures (one from each cluster) are aligned and shown on the right (only the ABD is shown here). An obvious difference between these two structures is emphasized by black rectangles, showing two possible secondary structures in this region (R192-K220). This chameleon region (called the “zipper” hereafter) follows the first 25 residues in the ABD, which are Q/P rich. (Lower) Bound structures for the region of the ABD that is structurally most self-consistent across the predictions: M217 to I302. M217-I302 is very well aligned in all 20 structures with a minimal mutual Qcontact of 0.812. (C) Frustration analysis of the zipper sequence in the two clusters. Direct/water-mediated contacts are represented by solid/dashed lines. Minimally frustrated contacts are shown in green, and highly frustrated ones are shown in red. (D) Frustration changes for F-actin (gray) upon binding with ABD (white). Minimally/highly frustrated contacts resulting from ABD binding are shown in blue/orange. Minimal/high frustrations turning neutral after ABD binding are shown in pale orange/pale blue.
The F-Actin/CPEB3-ABD Complex Is Stabilized Primarily by Electrostatic Interactions and Is Supported Cooperatively by Hydrophobic Interactions.
F-actin is highly negatively charged (26) (net negative charges/total residue number = 3.5%, assuming K/R have charges of +1 and D/E have charges of −1), and the percent of positive charges in the ABD is 6.3%, which is very high compared to the unstructured domain as a whole (0.2%). It was recently predicted that Ca2+/calmodulin-dependent kinase II (CaMKII), an actin binding protein involved in the calcium signaling cascade in synapses, binds to F-actin with similarly positively charged surfaces (27).
To study the role of electrostatic interactions in the binding of CPEB3-ABD and actin, we conducted additional peptide array experiments under 3× physiological ionic strength (Fig. 4A). The electrostatic shielding effect in high-salt conditions dramatically reduced the actin-binding intensity for all residues in the ABD, showing that electrostatic interactions are essential for forming this binding complex. Furthermore, the predicted structures show three attractive electrostatic-mediated contact clusters in the ABD/F-actin complex (Fig. 4B).
Fig. 4.
Electrostatic interactions stabilize the ABD/F-actin complex. (A) Residue-level experimental actin-binding intensity for ABD in physiological salt and 3× physiological salt conditions. (B) Electrostatic attractive contact pairs between ABD and F-actin. The ABD is colored in beige. Red residues are negatively charged (D and E), and blue residues are positively charged (K and R). The dashed black circles highlight attractive contact pairs; corresponding residue indexes in ABD/actin are shown in the upper and lower lines of the labels around.
A specific hydrophobic binding pocket, located between actin subdomains 1 and 3, is occupied by hydrophobic helices of many actin-binding proteins (ABPs) (27). This cleft is also a binding target of actin itself in F-actin. The DNase I-binding loop (D-loop) of one actin monomer docks to the cleft of the neighboring monomer and buries part of the binding cleft, such that the exposed parts of the cleft and the D-loop form a hydrophobic pocket that interacts with F-actin binding proteins. For example, CaMKII contains three F-actin–binding domains, and each of these domains has been predicted to bind to the hydrophobic pockets with helical structures (21, 27). In our predicted structure for the CPEB3-ABD/F-actin complex, the hydrophobic binding pocket on the surface of F-actin is also occupied by a polyalanine α-helix of the ABD (Fig. 3A, zoom-in). The peptides containing this alanine-rich α-helix, however, show almost zero actin-binding signal above background in the peptide array experiments. The number of intermolecular contacts for each ABD residue and the residue-level peptide array data are plotted together in Fig. 5A to highlight this apparent discrepancy. We performed peptide-level energy analyses of the predicted complex structure. Starting from the predicted complex structure of F-actin/ABD, the ABD is broken into overlapping peptides corresponding to the peptides that were tethered to the array in the peptide array experiments. The binding energy for each peptide, Ebinding, was calculated with the AWSEM Hamiltonian, as shown in Eq. 1. In Eq. 1, Ecomplex, EF-actin, and Epeptide are the AWSEM energies of the F-actin/peptide complex, the energy of the F-actin by itself, and the energy of the peptide by itself.
| [1] |
The binding-energy profile (Fig. 5B) agrees reasonably well with the experimental data. Particularly for the latter half of the peptide array, both binding energy analyses and peptide array experiment show high signals, which is also consistent with the relatively high probability for the residues in this region to form interface contacts with actin in both the RaptorX and the first-generation AWSEM-ER predictions. The alanine-rich helix itself binds weakly to the hydrophobic pocket from an energetic perspective, consistent with the fact that the corresponding peptides show weak interactions with actin in the peptide array experiments. We also noticed that in the first generation of AWSEM-ER simulations, the binding interface between the alanine-rich helix and the hydrophobic pocket is not formed consistently. In both the RaptorX contact prediction and the first-generation AWSEM-ER structure predictions (Fig. 2A), this α-helix shows a low probability to form intermolecular contacts with actin. Upon using contacts that are formed consistently in the first-generation AWSEM-ER simulations as restraints, which did not involve the alanine-rich sequences, the alanine-rich α–helix structure converges in binding with the hydrophobic pocket in the second-generation AWSEM-ER simulations. In summary, since the hydrophobic interaction between the alanine-rich helix and the binding pocket of F-actin is relatively weak, the hydrophobic interaction of the helix must cooperate with the electrostatic interactions highlighted previously in order to consolidate the ABD’s binding to F-actin.
Fig. 5.
Comparing the predicted ABD/Actin complex structure with experimental peptide array data. (A) Residue-level experimental actin-binding intensity and average number of intermolecular contacts over 20 predicted structures for ABD residues (scaled to a range of 0 to 1). The alanine-rich helix has very low experimental actin-binding signal, although it has some contacts with F-actin in the computational model (shown with green arrow). (B) Peptide-level experimental actin-binding intensity and binding energy analysis of the predicted structure for ABD peptides (scaled to a range of 0 to 1). The blue line represents F-actin binding energy for peptides (averaged over 20 relaxed predicted structures). Peptides including the sequence of the poly-A helix have relatively low binding energy (shown with a green arrow).
Binding with F-Actin Facilitates the Formation of a Potential Aggregation Trigger, the Beta-Hairpin Form of a Chameleon Sequence at the N-Terminal End of the ABD.
Functional prions undergo complicated structural transitions when switching from their inactive soluble states into their active prion states, which are usually beta-sheet–rich. The transition from coiled-coil oligomers to beta-sheet fibers for Q- (or Q/N-) rich prion sequences has been reported in both computational (13) and experimental (28) studies. By clustering our predicted ABD/actin complex structures, we identified a chameleon sequence (R192-K220) in the CPEB3 ABD that alternatively adopts one of two different secondary structures (Fig. 3B). F-actin appears to promote the formation of a beta-hairpin form of this sequence by bringing together the peptide’s two ends, working like a zipper (hence we refer to this motif as the “zipper”). Frustration analysis on this peptide revealed that this sequence is highly frustrated in both of its structural conformations arguing for its polymorphism (Fig. 3C). We subsequently performed a more in-depth study of this peptide by itself to investigate the influence on the preferred secondary structure of bringing together the two ends of the peptide.
At the beginning of the annealing trajectories (Movie S4) of the second-generation ABD/actin complex predictions, at high temperature, the zipper is extended and sometimes helical, but as the system cools down, the two ends of the zipper are captured by F-actin, and sometimes a beta-hairpin is gradually formed, while sometimes the helical form remains. Since the zipper sequence contains multiple alanine residues, which usually promote helical structure, it is not surprising that the zipper is sometimes helical even in high temperature. We also noticed that the attraction between F-actin and the two ends of the zipper results from the electrostatic interactions: the N-terminal end of the zipper has two positive residues (R192 and R193) and the C-terminal end has one (K220). Those three positive residues interact with four negative residues (1D, 2E, 3D, and 4E) of F-actin in the predicted binding complex (Fig. 6A). These electrostatic attractions serve to overcome the electrostatic repulsion that would otherwise occur between the two ends of the zipper, resulting in the zipping up of the sequence into a beta-hairpin. Next, we calculated the free energy profile for the zipper itself (Fig. 6B) and for the zipper with the two ends being attracted to each other by an effective potential that is meant to mimic the effect of the negative residues in actin (Fig. 6C). The free energy analysis showed that the zipper prefers the beta-hairpin structure over the helix structure when the two ends are attracted to each other. In the case of the bare peptide, in the absence of the additional attractive interaction of the two ends, the helical and beta-hairpin forms have roughly equal free energy.
Fig. 6.
Electrostatic attraction from F-actin zips a potential aggregation trigger into beta-hairpin configuration. (A) The two ends of the zipper sequence (R192-K220) are positively charged and attracted by four negatively charged residues in F-actin in the predicted complex structures. The zipper sequence is colored from blue to red, N terminus to C terminus, in rainbow spectrum, and the actin contacting the zipper it is colored in cyan. The surface of negative-/positive-charged residues is shown in red/blue. Both the α-helical form (Left) and beta-hairpin form (Right), corresponding to the two clusters found in Fig. 3B, are shown here. These two structures were used as references for calculating Qalpha and Qbeta in the free energy landscape analysis. (B) Free energy landscape for the zipper sequence plotted using Qbeta and Qalpha as the two order parameters. Three basins are labeled, and the representative structures are shown in the right upper corners. (C) Free energy landscape for the zipper sequence with the two ends attracted via a biasing potential. Only the beta-hairpin form is in a low free energy basin when the two ends are attracted together.
Furthermore, the beta-hairpin form of the zipper suggests that it might be able to trigger extensive beta-sheet formation in the N-terminal prion domain, PRD. As shown in Movie S5, when annealing an extended ABD construct that includes the PRD on the N-terminal end of the ABD, residues in the PRD form beta-sheets after they interact with the zipper in its beta-hairpin form. These beta-sheets could, in turn, participate in further intermolecular interactions with free CPEB3 monomers, triggering a cascade of aggregation. These findings suggest a candidate mechanism for F-actin’s involvement in CPEB3 aggregation, consistent with previous experimental studies (5). To further probe the role of the beta-hairpin on the N-terminal end of the ABD in triggering CPEB3 aggregation, we suggest that future experimental studies could use mutagenesis to decrease the repulsion between the positive residues at the ends of the zipper sequence or, more dramatically, generate constructs that link the two ends with a disulfide bond using mutations to cysteine.
SUMOylation of CPEB3 Potentially Inhibits the Interaction between CPEB3 and F-Actin.
The basal form of CPEB3 is soluble and is colocalized with P bodies (29), membraneless cytoplasmic processing bodies. Upon stimulation, CPEB3 aggregates and becomes colocalized with F-actin. The CPEB3/F-actin interaction is apparently required for CPEB3 aggregation in vivo (5). Stimulation therefore ultimately regulates the distribution and activity of CPEB3 in neurons and results in the redistribution of CPEB3 from P bodies to the F-actin network. Multiple factors are likely involved in the modification of the distribution of CPEB3. In particular, after stimulation, in addition to an increase of CPEB3 levels, SUMOylation of CPEB3 is reduced.
SUMOylation is a reversible posttranslational modification involved in numerous crucial cellular regulation mechanisms, including preventing protein aggregation. It has been reported that SUMOylation of Huntingtin protein inhibits its pathogenic aggregation, which results in Huntington’s disease (30). Likewise, the aggregation of alpha-synuclein implicated in Parkinson’s disease declines when alpha-synuclein is SUMOylated (31). Small ubiquitin-like modifier (SUMO) proteins function by covalently attaching to lysine residues of target proteins (a process known as SUMOylation) and then subsequently being catalytically detached from target proteins (deSUMOylation). Drisaldi et al. (17) found that CPEB3 is SUMOylated and soluble in its basal state. After stimulation, CPEB3 is deSUMOylated and aggregates. In its aggregated form, CPEB3 promotes the translation of its target mRNAs.
To further probe the regulatory mechanism of SUMOylation/deSUMOylation for CPEB3, we conducted a SUMOylated site and SUMO-Interacting Motifs (SIMs) bioinformatic search using the CPEB3 sequence. We employed multiple webservers, including SUMOplot (32), JASSA (33), and GPS-SUMO (34). Table 1 summarizes potential SUMO sites of the CPEB3 unstructured domain, including sites with the top three predicted scores in each webserver, along with their corresponding scores. Three SIMs predicted for the full-length CPEB3 sequence are also shown in Table 2. These predictions pointed to two probable SUMOylation sites (K50 and K294) and three SIMs (P484-W489, V273-V277, and V279-V283). Intriguingly, several of the SUMOylation sites and SIMs include key residues in the predicted binding complex of CPEB3 and F-actin. Several possible mechanisms by which SUMOylation could regulate the CPEB3/F-actin interaction are discussed in SI Appendix, Fig. S6. In all of these mechanisms, the actin-binding sites of the ABD are blocked in CPEB3’s basal state and exposed after CPEB3 is deSUMOylated upon stimulation. We therefore propose that SUMOylation of CPEB3 is a potential inhibitor for the CPEB3/F-actin interaction, which in turn regulates the distribution and activity of CPEB3. Further experimental and theoretical work is required to determine which, if any, of these mechanisms is operating in neurons.
Table 1.
Predicted SUMOylated sites in CPEB3 unstructured sequence via sequence analyses in SUMOplot, JASSA, and GPS-SUMO
 |
Red represents region from PRD, and blue represents regions from ABD. Red font represents predicted sites.
Table 2.
Predicted SIMs in full length CPEB3 sequence via sequence analyses in JASSA and GPS-SUMO
 |
Yellow represents regions from RBD, and blue represents regions from ABD. Red font represents predicted sites.
Discussion
A Positive Feedback Loop Involving the F-Actin/CPEB3 Interaction May Support Long-Term Memory.
Stephan et al. proposed that intact F-actin is required for CPEB3 aggregation (5). Our predicted CPEB3-ABD/F-actin complex structure gives insight into the molecular mechanism possibly underlying these cellular-level findings. In this proposed mechanism, upon stimulation, actin monomers polymerize into F-actin to support the growth of dendritic spines. Meanwhile, CPEB3 is deSUMOylated and binds to F-actin. The specific binding between CPEB3 and F-actin increases the local concentration of CPEB3 in dendritic spines, where CPEB3 aggregates to consolidate synaptic strength and regulates the local translation of target mRNAs. F-actin, exhibiting a periodic helix twist structure (35), provides multiple binding pockets for CPEB3 and thereby raises the local level of CPEB3 up to a critical concentration for aggregation. Through electrostatic interactions between F-actin and the CPEB3 actin binding domain, the zipper sequence in CPEB3 is prone to fold into a beta-hairpin structure, serving as a core for the formation of intramolecular beta-sheets, a potential intermediate in the transition toward prion-like aggregates. Previous work (13) has proposed that F-actin exerts mechanical force on CPEB oligomers to facilitate fiber formation. The specific binding of the F-actin/ABD complex in our current predictions supports the possibility that F-actin would be able to supply the pulling force necessary to achieve this transition without detaching from CPEB3. Overall, through its interaction with F-actin, CPEB3 is locally concentrated and goes through a structural transition into a fiber form.
The convergence of multiple ABPs (including cofilin, CPEB3, and CaMKII) onto the same hydrophobic binding pocket of F-actin suggest this binding could serve to regulate the assembly and function of actin networks. For example, cofilin, an ABP responsible for severing F-actin, also binds with F-actin at the same hydrophobic pocket (16). The maintenance of the actin cytoskeleton and synaptic strength then might involve the competition between CPEB3 and cofilin or other ABPs. Actin mRNA is one of the targets of CPEB3 (5), implying that CPEB3 aggregation/activation induces the translation of actin mRNA and, therefore, further polymerization into F-actin.
As a molecular mechanism underlying long-term memory, this positive feedback loop should be controlled by synaptic stimulations. We proposed that SUMOylation, as a known regulator of CPEB3 activity, is a potential trigger of the CPEB3/F-actin interaction, thereby the positive feedback loop between CPEB3 and actin.
In conclusion, our computational structural model of the F-actin/CPEB3-ABD complex gives insight into the molecular mechanisms of CPEB3’s functions: in its basal state, CPEB3 is SUMOylated, and its actin binding domain is buried. After deSUMOylation upon stimulation, the CPEB3-ABD is exposed and is available to interact with F-actin. The interaction between F-actin and CPEB3 triggers the aggregation of CPEB3 and the translation of its target mRNAs, including actin mRNA, ultimately resulting in strengthened synaptic junctions. Future experimental and computational studies are still needed to identify the SUMOylated sites and SIMs in CPEB3. A fiber structure for a CPEB3 homolog was recently solved (9), but we do not yet have a fiber structure for CPEB3 itself. Having a fiber structure for CPEB3 will allow us to study its aggregation pathways in greater detail (36–38) and to understand how it regulates target mRNA translation in its active state.
Supplementary Material
Acknowledgments
This work was supported by the Center for Theoretical Biological Physics sponsored by NSF Grant PHY-1427654. This work was also supported by NSF Grants CHE-1614101 and CHE-1743392. Additional support was provided by the D. R. Bullard-Welch Chair at Rice University, Grant C-0016, and an endowment from the William Wheless III Professorship at University of Texas Health Science Center at Houston.
Footnotes
Competing interest statement: N.P.S. is the founder of Schafer Science, LLC.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012964117/-/DCSupplemental.
Data Availability.
Raw data, input files for simulations, and analysis scripts have been deposited in Xinyu-Gu/CPEB3_Actin at Zenodo, https://zenodo.org/badge/latestdoi/274487374, and GitHub, https://github.com/Xinyu-Gu/CPEB3_Actin) (39).
References
- 1.Lüscher C., Nicoll R. A., Malenka R. C., Muller D., Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat. Neurosci. 3, 545–550 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Harris K. M., Fiala J. C., Ostroff L., Structural changes at dendritic spine synapses during long-term potentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 745–748 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey C. H., Kandel E. R., Structural changes accompanying memory storage. Annu. Rev. Physiol. 55, 397–426 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Crick F., Memory and molecular turnover. Nature 312, 101 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Stephan J. S. et al., The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Rep. 11, 1772–1785 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Si K., Lindquist S., Kandel E. R., A neuronal isoform of the aplysia CPEB has prion-like properties. Cell 115, 879–891 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Si K. et al., A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115, 893–904 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Majumdar A. et al., Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148, 515–529 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Hervas R. et al., Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 367, 1230–1234 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afroz T. et al., A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev. 28, 1498–1514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakahata Y., Yasuda R., Plasticity of spine structure: Local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matus A., Actin-based plasticity in dendritic spines. Science 290, 754–758 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Chen M., Zheng W., Wolynes P. G., Energy landscapes of a mechanical prion and their implications for the molecular mechanism of long-term memory. Proc. Natl. Acad. Sci. U.S.A. 113, 5006–5011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davtyan A. et al., AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J. Phys. Chem. B 116, 8494–8503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirovetz B. J., Schafer N. P., Wolynes P. G., Protein structure prediction: Making AWSEM AWSEM-ER by adding evolutionary restraints. Proteins 85, 2127–2142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galkin V. E. et al., Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 20568–20572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drisaldi B. et al., SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep. 11, 1694–1702 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao H.-W. et al., NMDAR signaling facilitates the IPO5-mediated nuclear import of CPEB3. Nucleic Acids Res. 40, 8484–8498 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng H. et al., ComplexContact: A web server for inter-protein contact prediction using deep learning. Nucleic Acids Res. 46, W432–W437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T.-m., Wang S., Xu J., Deep learning reveals many more inter-protein residue-residue contacts than direct coupling analysis. bioRxiv:10.1101/240754 (April 28, 2018).
- 21.Prentiss M. C., Hardin C., Eastwood M. P., Zong C., Wolynes P. G., Protein structure prediction: The next generation. J. Chem. Theory Comput. 2, 705–716 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Ferreiro D. U., Komives E. A., Wolynes P. G., Frustration in biomolecules. Q. Rev. Biophys. 47, 285–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreiro D. U., Hegler J. A., Komives E. A., Wolynes P. G., Localizing frustration in native proteins and protein assemblies. Proc. Natl. Acad. Sci. U.S.A. 104, 19819–19824 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreiro D. U., Hegler J. A., Komives E. A., Wolynes P. G., On the role of frustration in the energy landscapes of allosteric proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 3499–3503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra R. G. et al., Protein frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Res. 44, W356–W360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J. X., Janmey P. A., The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. J. Biol. Chem. 271, 8556–8563 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Wang Q. et al., Assemblies of calcium/calmodulin-dependent kinase II with actin and their dynamic regulation by calmodulin in dendritic spines. Proc. Natl. Acad. Sci. U.S.A. 116, 18937–18942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiumara F., Fioriti L., Kandel E. R., Hendrickson W. A., Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell 143, 1121–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford L., Ling E., Kandel E. R., Fioriti L., CPEB3 inhibits translation of mRNA targets by localizing them to P bodies. Proc. Natl. Acad. Sci. U.S.A. 116, 18078–18087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffan J. S. et al., SUMO modification of Huntingtin and Huntington’s disease pathology. Science 304, 100–104 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Krumova P. et al., Sumoylation inhibits α-synuclein aggregation and toxicity. J. Cell Biol. 194, 49–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gramatikoff K., Wu C., Shi X., Fang F., Frontiers of Biotechnology and Pharmaceuticals, (Science Press USA Inc., 2004). [Google Scholar]
- 33.Beauclair G., Bridier-Nahmias A., Zagury J.-F., Saïb A., Zamborlini A., JASSA: A comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Zhao Q. et al., GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujii T., Iwane A. H., Yanagida T., Namba K., Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Zheng W., Tsai M.-Y., Chen M., Wolynes P. G., Exploring the aggregation free energy landscape of the amyloid-β protein (1-40). Proc. Natl. Acad. Sci. U.S.A. 113, 11835–11840 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen M., Schafer N. P., Wolynes P. G., Surveying the energy landscapes of Aβ fibril polymorphism. J. Phys. Chem. B 122, 11414–11430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Chen M., Schafer N. P., Wolynes P. G., Exploring the interplay between fibrillization and amorphous aggregation channels on the energy landscapes of tau repeat isoforms. Proc. Natl. Acad. Sci. U.S.A. 117, 4125–4130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu X., et al. , Modelling F-actin/CPEB3 interaction. GitHub. https://github.com/Xinyu-Gu/CPEB3_Actin. Deposited 23 June 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data, input files for simulations, and analysis scripts have been deposited in Xinyu-Gu/CPEB3_Actin at Zenodo, https://zenodo.org/badge/latestdoi/274487374, and GitHub, https://github.com/Xinyu-Gu/CPEB3_Actin) (39).