Significance
Protein synthesis is a fundamental cellular process that occurs from bacteria to humans. Highly accurate protein synthesis has been shown to be critical for the fitness of the cell and is controlled by sophisticated molecular mechanisms. One such mechanism is the release of mature proteins at stop codons to prevent readthrough. Here we identify an environmental stress that substantially increases the level of stop-codon readthrough during protein synthesis in bacteria and provide insights into the underlying mechanism. Intriguingly, the level of readthrough fluctuates extensively among single cells that are genetically identical and grown under the same stress condition. Cells with different levels of readthrough vary in phenotypes, and individual cells with high readthrough recover better from the acid stress.
Abstract
Accurate protein synthesis is a tightly controlled biological process with multiple quality control steps safeguarded by aminoacyl-transfer RNA (tRNA) synthetases and the ribosome. Reduced translational accuracy leads to various physiological changes in both prokaryotes and eukaryotes. Termination of translation is signaled by stop codons and catalyzed by release factors. Occasionally, stop codons can be suppressed by near-cognate aminoacyl-tRNAs, resulting in protein variants with extended C termini. We have recently shown that stop-codon readthrough is heterogeneous among single bacterial cells. However, little is known about how environmental factors affect the level and heterogeneity of stop-codon readthrough. In this study, we have combined dual-fluorescence reporters, mass spectrometry, mathematical modeling, and single-cell approaches to demonstrate that a metabolic stress caused by excess carbon substantially increases both the level and heterogeneity of stop-codon readthrough. Excess carbon leads to accumulation of acid metabolites, which lower the pH and the activity of release factors to promote readthrough. Furthermore, our time-lapse microscopy experiments show that single cells with high readthrough levels are more adapted to severe acid stress conditions and are more sensitive to an aminoglycoside antibiotic. Our work thus reveals a metabolic stress that promotes translational heterogeneity and phenotypic diversity.
Faithful gene expression demands high fidelity during DNA replication, transcription, and translation (1–7). A critical step to maintain such accuracy is proper termination of protein synthesis at UAA, UAG, and UGA stop codons, which is catalyzed by release factors (RFs) (8, 9). In bacteria, RF1 catalyzes release of peptides from the peptidyl-transfer RNA (tRNA) at UAA and UAG codons, whereas RF2 releases growing peptides at UAA and UGA codons. In eukaryotes, a single class I release factor, eRF1, terminates translation at all three stop codons on the ribosome. In nature and in the laboratory, stop codons can be read through with high efficiency by suppressor tRNAs, which carry mutations in the anticodon to match the stop codons (10). This approach is widely used to site-specifically insert noncanonical amino acids into proteins of interest (11, 12). Even in cells lacking cognate suppressor tRNA, readthrough of stop codons is still prevalent, albeit at lower rates. A global analysis of translation termination in Escherichia coli with ribosome profiling reveals that ribosomes occupy the messenger RNA (mRNA) region following the stop codons of hundreds of genes (13). Proteomic analyses also demonstrate that near-cognate aminoacyl-tRNAs are able to read stop codons (14, 15). For example, glutamine and tryptophan suppress UAG and UGA codons, respectively. In addition, readthrough of stop codons are programmed events in multiple mammalian and viral genes (16, 17).
Stop-codon readthrough adds an extended amino acid sequence to the C terminus and has been shown to be critical for expression of functional protein variants. For example, readthrough of a UAG stop codon in gag is critical for expression of a protein variant and assembly of retrovirus (17). Stop-codon readthrough also yields a functional peroxisomal lactate dehydrogenase in mammals (18) and alters protein localization in fungi and animals (19, 20). Furthermore, targeting readthrough of premature stop codons has been actively explored as a therapeutic strategy to treat genetic diseases (21, 22). In bacteria, readthrough of stop codons regulates the expression levels of RF2 and amino acid biosynthesis genes (13, 23, 24). Despite these well-established examples of specific functional readthrough products, how global stop-codon readthrough alters during environmental shifts and affects cell fitness remains largely unknown.
We have previously developed dual-fluorescence reporters to conveniently quantitate stop-codon readthrough levels in the population and single bacterial cells (15). In a screen for growth conditions that affect stop-codon readthrough using these reporters in E. coli, we have uncovered that excess carbon in media substantially increases the readthrough levels of UAA, UAG, and UGA codons. We further show that such an increase depends on a drop of pH during bacterial growth in excess carbon. Interestingly, excess carbon and low pH not only increase the average level of readthrough, but also enhance its fluctuation among single bacterial cells. The individual cells with low and high readthrough levels are better prepared for distinct stress conditions, suggesting that the metabolic stress caused by excess carbon promotes heterogeneity of stop-codon readthrough and phenotypic diversity.
Results
Excess Carbon Promotes Stop-Codon Readthrough.
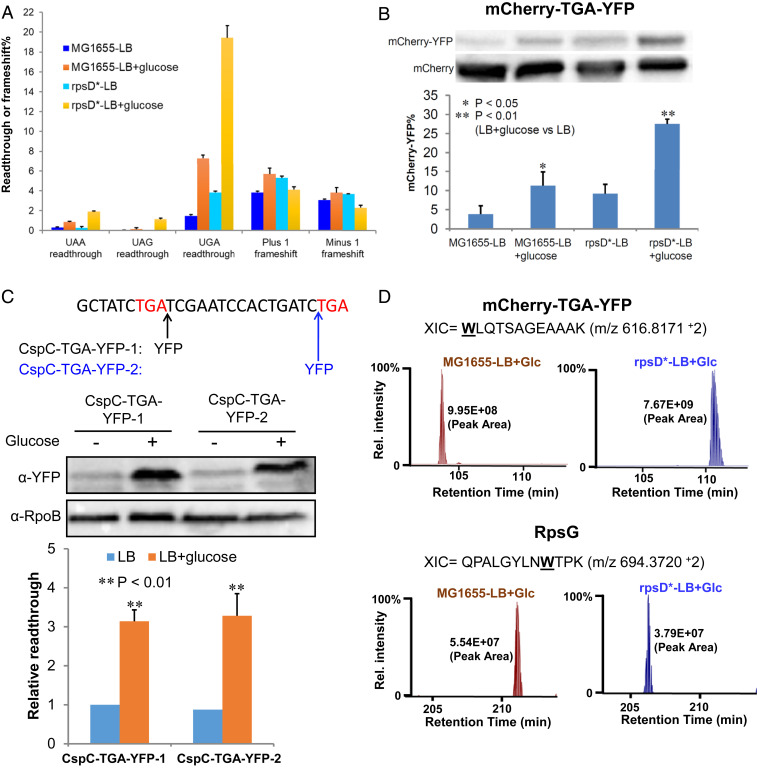
To date, little is known about how environmental factors affect stop-codon readthrough. To address this question, we used our previously developed dual-fluorescence reporter system (15) to screen the UGA readthrough level of wild-type (WT) E. coli (MG1655) grown in various media (SI Appendix, Fig. S1). Previous studies have indicated that carbon starvation increases the level of stop-codon readthrough (25), although the underlying mechanism remains unclear. Unexpectedly, we found that adding extra carbon sources (e.g., glucose) into the rich medium Luria–Bertani broth (LB) also significantly increased the level of UGA readthrough (SI Appendix, Fig. S1). We next tested the effect of excess glucose on stop-codon readthrough and frameshift in both the WT and error-prone (rpsD*) strains. The rpsD* strain carries an I199N mutation in the ribosomal small subunit rpsD gene and displays increased errors during ribosomal decoding (26, 27). Addition of 1% glucose in LB increased the level of UGA and UAA readthrough three- to seven-fold in both MG1655 and rpsD* (Fig. 1A). Although UAG readthrough is not detected with the dual-fluorescence reporter in MG1655, excess glucose clearly increased UAG readthrough in the rpsD* mutant. In contrast, little change was observed for frameshift upon addition of glucose, suggesting that the effect of excess carbon on ribosomal fidelity is specific for stop-codon readthrough.
Fig. 1.
Excess carbon increases the level of stop-codon readthrough. E. coli cells were grown in LB or LB with 1% glucose for 24 h before testing. (A) Dual-fluorescence reporters were used to determine the levels of stop-codon readthrough and frameshift as described previously (15). Fluorescence was quantitated with a plate reader. (B) Western blotting of cell extracts from MG1655 or rpsD* carrying the mCherry-TGA-YFP reporter using an antibody against mCherry. Readthrough of the UGA codon following mCherry yielded the mCherry-YFP fusion protein. (C) An in-frame YFP tag was inserted immediately after the first TGA codon of the native cspC gene (CspC-TGA-YFP-1) or before the second stop codon (CspC-TGA-YFP-2) in E. coli MG1655. The nucleotide sequence of the 3′-end of the cspC gene is shown. Red letters indicate the first and second stop codons. Western blotting shows that readthrough of the cspC UGA codon significantly increased after addition of glucose. Another chromosomal constitutively expressed protein, RpoB, was used as a loading control during Western blotting. (D) Label-free quantitative mass spectrometry analysis of UGA readthrough in the dual-fluorescence reporter protein (mCherry-TGA-YFP) or the native RpsG protein. Extracted ion chromatograms (XIC) for the reporter peptides are shown from MG1655 and rpsD* grown in LB glucose. “W” marks the UGA readthrough site in the peptide. XIC peak area analysis is shown. Error bars represent one SD (n ≥ 3). P values were determined using unpaired t test. *P < 0.05; **P < 0.01.
To validate the results of fluorescence assays, we tested readthrough of the reporter and native proteins using both Western blotting and quantitative mass spectrometry. Using an antibody against mCherry, we detected the full-length readthrough product of the mCherry-TGA-YFP (yellow fluorescence protein) reporter (Fig. 1B). The quantitative result of the readthrough level with and without glucose by Western blotting was very consistent with the dual-fluorescence assay (Fig. 1 A and B). We further inserted a yfp gene at the 3′ untranslated region downstream of the native UGA stop codon of cspC and found with Western blotting that addition of glucose increased readthrough of the cspC UGA codon by three-fold (Fig. 1C). Next, we performed label-free quantitative mass spectrometry of the total proteins extracted from MG1655 and rpsD* grown in LB glucose. Peptides resulting from readthrough in the mCherry-TGA-YFP reporter as well as native proteins were identified (Fig. 1D and SI Appendix, Table S1 and Dataset S1). Compared with the proteomes of the same strains grown in LB (15), the abundance of readthrough products increased in the presence of glucose as indicated by the peak area (SI Appendix, Fig. S2). This further confirmed our results observed with fluorescence assays.
Increased UGA Readthrough in the Presence of Glucose Depends on the Phosphotransferase System.
In E. coli, uptake of glucose into cells primarily relies on the phosphotransferase (PTS) system (28). We found that deleting the PTS transporter (ptsG) abolished the effect of glucose on increasing UGA readthrough (SI Appendix, Fig. S3), suggesting that such an increase depends on glucose uptake. Glucose PTS is involved in catabolite repression of other sugars as well as network regulation through the cAMP/CRP (cAMP receptor protein) pathway (28). Deleting the adenylate cyclase (producing cAMP) gene cyaA or crp did not prevent glucose-induced UGA readthrough (SI Appendix, Fig. S3). These results suggest that excess glucose likely promotes stop-codon readthrough by altering metabolism, rather than by perturbing regulation of the cAMP/CRP pathway.
Glucose Stimulates Expression of RF2 Protein.
Readthrough of UGA codon is directly affected by the competition between RF2 and the EF-Tu:Trp-tRNATrp:GTP complex. Using acidic gel Northern blotting, we found that the level of Trp-tRNATrp was not significantly altered in LB glucose compared with LB (SI Appendix, Fig. S4 A and B). This is in contrast to an increased Trp-tRNATrp level in the presence of the ribosomal inhibitor chloramphenicol, suggesting that, unlike chloramphenicol treatment, addition of glucose does not increase UGA readthrough via inhibiting protein synthesis and activating the feedback loop to produce more ribosomal RNAs and tRNAs (15). We also used Western blotting to determine the relative abundance of EF-Tu and RF2. Due to the lack of an efficient antibody against RF2, we used an MG1655-derived strain with a FLAG tag inserted at the C terminus of RF2 in the chromosome (15). Our result revealed that, whereas the level of EF-Tu remained the same upon glucose addition, the protein level of RF2 increased three-fold in LB glucose compared with LB (SI Appendix, Fig. S4 C and D). The increase in RF2 protein level was not due to an increase in prfB mRNA or protein stability (SI Appendix, Fig. S4 E and F). It is known that translation of RF2 is self-regulated with a negative feedback loop. The prfB gene (encoding RF2) contains a CUU_UGA +1 frameshifting codon, and reducing the RF2 activity and/or level promotes frameshift and translation of RF2 (23, 29). To test the frequency of prfB frameshift, we inserted its frameshifting site between mCherry and YFP. The result shows that addition of glucose increases the prfB frameshift level (SI Appendix, Fig. S4G). An increase in RF2 protein level without reducing its activity is expected to decrease UGA readthrough, in contrast to the increase in readthrough that we observed (Fig. 1). Therefore, our data suggest that increased UGA readthrough in LB glucose is likely due to reduced RF2 activity, which in turn enhances translation and production of RF2 protein.
Glucose Increases UGA Readthrough by Reducing pH.
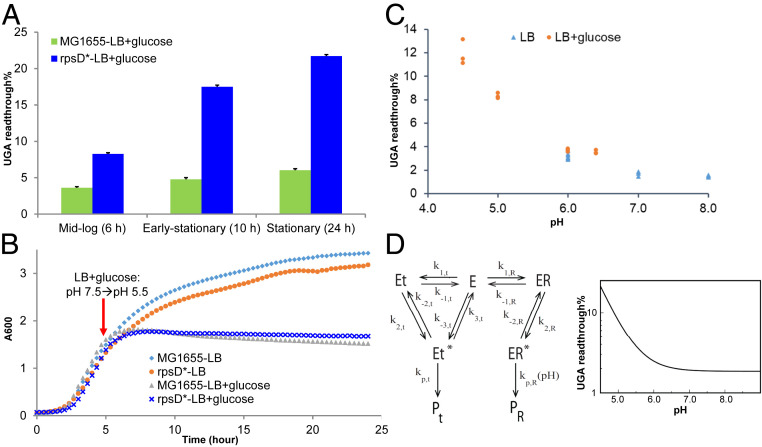
Bacteria growing in excess glucose often produce acidic metabolic products, such as acetate (30). Previous in vitro biochemical analyses have shown that reducing pH impairs the hydrolytic activity of both class I release factors (RF1 and RF2) on the ribosome (31–33), prompting us to hypothesize that increased stop-codon readthrough in the presence of glucose is due to a lower pH. Indeed, we observed that E. coli cultures grown in LB with 1% glucose reduced the pH to below 5 in mid- to late-log phase (Fig. 2 and SI Appendix, Fig. S1B). In contrast, cultures in LB and M9 with glucose remained neutral (SI Appendix, Fig. S1B). We also found that the intracellular pH of E. coli grown in LB + glucose was acidic, whereas cells grown in LB exhibited neutral pH (SI Appendix, Fig. S1C). Consistently, the substantial increase in UGA readthrough in LB glucose occurred in the stationary phase following the pH shift (Fig. 2A). We further adjusted the pH of LB and LB glucose media and found that increasing pH in LB glucose decreased UGA readthrough; reducing pH in LB conversely increased UGA readthrough (Fig. 2C and SI Appendix, Fig. S1D). Further mathematical modeling using published in vitro kinetic data revealed that reduction of pH to below 6, as observed in the presence of excess glucose, substantially increases the level of UGA readthrough (SI Appendix, Table S2 and Fig. 2D).
Fig. 2.
Excess carbon increases UGA readthrough by lowering pH. (A) E. coli cells carrying the mCherry-TGA-YFP reporter were grown in LB or LB with 1% glucose. At 6, 10, and 24 h, fluorescence was determined using a plate reader, and the UGA readthrough levels were calculated. Error bars represent one SD (n ≥ 3). (B) Growth curve of E. coli in LB or LB with 1% glucose. A pH shift from 7.5 to 5.5 was observed at 5 h in the LB glucose media. Excess glucose caused growth arrest at 7 h, correlating with a substantial increase in the UGA readthrough level. (C) E. coli MG1655 was grown in buffered LB or LB glucose for 24 h, and the UGA readthrough level was determined as in A. The pH of the media was determined at 24 h. (D) Mathematical modeling of the pH effect on UGA readthrough. R stands for release factor (RF2); the near-cognate aminoacyl-tRNA (t) is the wrong substrate that suppresses UGA; PR is the hydrolyzed peptide, and Pt represents the elongated peptide. Modeling results suggest that when pH drops to below 6, the UGA readthrough error substantially increases. The kinetic parameters used for modeling are from references (33, 34). See SI Appendix for details of modeling.
Excess Glucose and Low pH Promote Fluctuation of UGA Readthrough among Single Cells.
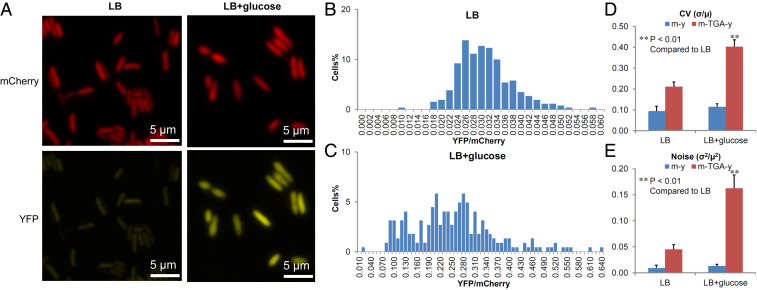
The dual-fluorescence reporter allows us to visualize and quantitate UGA readthrough in individual cells (15). Using fluorescence microscopy, we observed that the average YFP intensity resulting from UGA readthrough was much higher in cells grown in LB glucose compared to LB (Fig. 3A). Flow cytometry of single cells revealed the same trend (SI Appendix, Fig. S5). Such single-cell results were consistent with the population experiments (Fig. 1). Interestingly, the UGA readthrough level (calculated by the ratio of YFP/mCherry) appears to distribute more widely among single cells grown in LB glucose than those grown in LB (Fig. 3 B and C and SI Appendix, Fig. S5). Heterogeneity of gene expression is typically described by coefficient of variation (CV), which is calculated as the ratio of the SD (σ) over the mean (µ), or noise (σ2/µ2) (35–37). E. coli cells cultured in LB glucose displayed high levels of CV and noise (Fig. 3 C and D), suggesting that the metabolic stress induced by glucose promotes heterogeneity of stop-codon readthrough, in addition to elevating the average readthrough level in the population. Likewise, lowering pH also increased the heterogeneity of UGA readthrough (SI Appendix, Figs. S5 and S6).
Fig. 3.
Excess carbon increases the heterogeneity of UGA readthrough among single cells. E. coli cells carrying the mCherry-TGA-YFP reporter were grown in LB or LB with 1% glucose for 24 h and imaged with fluorescence microscopy. Fluorescence levels in single cells were quantitated with Microbe Tracker (38). (A) Fluorescence of mCherry and YFP. (B and C) Distribution of cells based on the relative level of UGA readthrough (YFP/mCherry). Note the difference in the scale of the x-axis in B and C. Cells grown in LB glucose showed a wider distribution than those grown in LB. (D and E) Excess glucose increased the CV and noise of UGA readthrough among single cells. σ, SD of YFP/mCherry; µ, mean of YFP/mCherry among individual cells. Error bars represent one SD (n ≥ 3). P values were determined using unpaired t test. **P < 0.01.
Cells with Different Levels of UGA Readthrough Display Phenotypic Diversity.
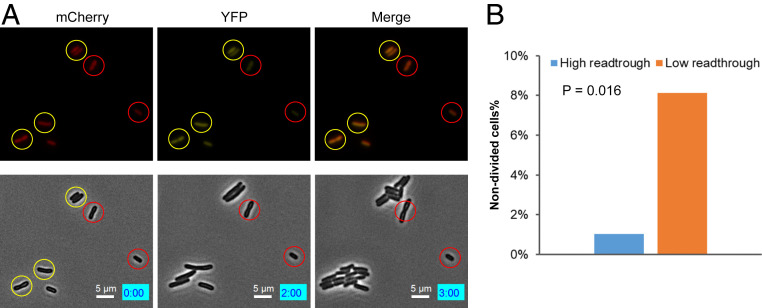
Phenotypic heterogeneity of genetically identical single cells in a microbial population plays critical roles in the population's adaptation to environmental changes (39, 40). To characterize the phenotypes of individual cells with different levels of UGA readthrough, we treated E. coli cells grown in LB glucose with an acid stress (pH 3) or an aminoglycoside antibiotic streptomycin (Str) for 30 min and monitored recovery of growth with time-lapse microscopy. Cells with high levels of UGA readthrough recovered growth better than cells with low readthrough following treatment with low pH (Fig. 4 and Movie S1). However, Str treatment resulted in the opposite trend: i.e., cells with high readthrough appeared to be more sensitive to this antibiotic (SI Appendix, Fig. S7 and Movie S2). A flow cytometry experiment further confirmed that the brief treatment with low pH or Str did not alter the average level or fluctuation of UGA readthrough in cells grown in LB glucose (SI Appendix, Fig. S8). These results support a bet-hedging mechanism whereby cells with different levels of stop-codon readthrough are prepared for challenges from distinct stress conditions.
Fig. 4.
Single cells with high UGA readthrough tolerate acid stress better. MG1655 cells carrying the mCherry-TGA-YFP reporter were grown in LB with 1% glucose for 24 h before treatment with LB at pH 3 for 30 min. Cells were then placed on an LB agar pad for time-lapse microscopy. Fluorescence imaging was taken at time 0. (A) Microscopy images showing individual cells with fluorescence (Top) and phase contrast (Bottom). Cells with high and low UGA readthrough are indicated by yellow and red circles. The numbers in the blue boxes indicate the time in hours and minutes. Note that the cells with low readthrough in these images did not divide or divided after 3 h. (B) Percentage of nondivided cells at 3 h in high and low readthrough groups. The P value was determined using the χ2 test (n = 221).
Discussion
The impact of environmental stresses on stop-codon readthrough is poorly understood. Previous studies show that carbon starvation induces stop-codon readthrough in E. coli, which in turn destabilizes the proteome and enhances protein oxidation (25). Our work here shows that, surprisingly, excess carbon promotes readthrough of stop codons (Fig. 1 and SI Appendix, Fig. S1). Whereas laboratory media often contain a single carbon source (e.g., amino acids in LB), the carbon sources in natural environments are much more complex. For example, gut microbiota are frequently feasted with excess carbon sources including carbohydrates, proteins, and lipids (41). Excess carbon in bacteria causes an overflow of metabolism and accumulation of metabolic intermediates, such as acetate (30, 42). This leads to a metabolic stress that may inhibit bacterial growth. We demonstrate that excess glucose enhances stop-codon readthrough by lowering pH (Fig. 2), consistent with previous in vitro kinetic studies showing that low pH reduces the catalytic activity of both release factors 1 and 2 (31–33). In contrast, availability of different carbon sources has been shown to alter the frequency of programmed frameshift via activation of several kinases in yeast (43).
With the advancement of single-cell technologies, it has become increasingly clear that not all cells in a genetically identical population exhibit the same phenotypes (39, 44, 45). Such phenotypic heterogeneity allows individual cells to divide labor or bet-hedge against unpredictable environmental shifts (39, 46). One well-studied example of bet-hedging is bacterial persistence, in which a small fraction of dormant cells are capable of surviving high doses of bactericidal antibiotics for a long period of time (47). Phenotypic heterogeneity may arise from stochastic gene expression or fluctuation in the microenvironments (37, 39, 48–52). How heterogeneity of protein synthesis contributes to phenotypic diversity remains poorly understood. We and others have previously developed dual-fluorescence reporters to visualize and quantitate translational errors, including missense incorporation (53), frameshift (15, 54), and stop-codon readthrough (15, 54), in the population and in single cells. Using such reporters, we show that excess carbon and low pH not only increase the average level of readthrough, but also enhance its heterogeneity among single cells (SI Appendix, Figs. S3, S5, and S6). Our previous work suggests that the UGA readthrough level in E. coli cells grown in LB is not sensitive to fluctuation in the RF2 level, presumably due to the relatively high level and activity (15, 55). However, excess carbon and low pH compromise the activity of RF2, which may render cells more sensitive to the stochastic variation of the RF2 level, resulting in a larger noise for UGA readthrough among single cells.
Individual cells grown in excess carbon with different levels of UGA readthrough appear to vary in phenotypes (SI Appendix, Figs. S4 and S7, and Movies S1 and S2). Cells with higher UGA readthrough recover faster following treatment with an acid stress (Fig. 4) yet are more sensitive to treatment with streptomycin. The underlying molecular mechanisms remain unclear. Increased translational errors lead to a higher level of RpoS, the key regulator of the general stress response that protects bacteria against a number of environmental stresses, including the acid stress (27, 56, 57). It is tempting to speculate that individual cells with high readthrough may have a more robust general stress response to enhance acid resistance. On the other hand, streptomycin is an aminoglycoside antibiotic and causes cell death by promoting mistranslation (58, 59). Cells with high readthrough may have already accumulated an elevated level of misfolded proteins and are thus more sensitive to further mistranslation and aggregation of proteins caused by Str (60). Our results therefore highlight previously unknown connections between translational heterogeneity and phenotypic diversity. Further studies to expand the phenotypes and elucidate the underlying molecular mechanisms are warranted in the future.
Methods
Strains, Plasmids, and General Methods.
E. coli K-12 strain MG1655 was used as the wild-type strain in this study, and the error-prone strain rpsD* was constructed as described before (27). The pZ vector (61) carrying the pSC101* replication origin (three to four copies per cell) and a constitutive promoter PLtetO-1 were used for expression of fluorescent reporters as previously described (15).
Unless otherwise noted, E. coli strains were grown in LB with or without 1% glucose at 37 °C. The minimal medium contains 47.8 mM Na2PO4, 22.0 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 4 mM MgSO4, 0.2 mM CaCl2, and 1% carbon source.
Western Blotting and Acidic Gel Northern Blotting.
Western blotting and acid urea gel Northern blotting were performed according to procedures previously described (15).
Fluorescence Microscopy Image Acquisition and Analysis.
Fluorescence microscopy and analysis were performed as described (15) using Olympus IX81-ZDC inverted microscope and MicrobeTracker (38), a MATLAB-based software package.
Time-Lapse Microscopy.
Overnight cultures were diluted 1:100 in LB or LB glucose and grown for 24 h at 37 °C. Cultures were placed on a 120-μL 1% agarose LB pad. Microscopy was performed using BZ-X800 fluorescence microscope (Keyence). Fluorescent images were taken at the initial time point. Cells were followed for 5 to 6 h at room temperature with phase-contrast images taken at regularly spaced intervals throughout the experiment. Image analysis and editing were performed using BZ-X800 analyzer.
Calculation of CV and Noise.
CV was calculated as the SD (σ) divided by the mean (µ) of the YFP/mCherry ratio of each cell in the same microscopic image frame. Noise was calculated as the ratio of the variance (σ2) over the square of the mean (µ2).
Flow Cytometry.
E. coli cells grown in LB or LB glucose for 24 h were diluted in phosphate buffer containing 25 µg/mL chloramphenicol and subjected to flow cytometry analysis using BD FACSCanto II according to standard procedures.
Protein Preparation and Proteomics.
Protein preparation and proteomics were performed as described previously (15), except that cells were grown in LB with 1% glucose for 24 h.
Supplementary Material
Acknowledgments
This work was funded by National Institute of General Medical Sciences (NIGMS) Grants R01GM115431 and R35GM136213 (to J.L.); NIGMS Grant R01GM117230 (to J.R.); and NSF Grant PHY-1427654 and Welch Foundation Grant C-1995 (to O.A.I.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. B.S.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013543117/-/DCSupplemental.
Data Availability.
All study data discussed in the paper are available in the main text and SI Appendix.
References
- 1.Francklyn C. S., DNA polymerases and aminoacyl-tRNA synthetases: Shared mechanisms for ensuring the fidelity of gene expression. Biochemistry 47, 11695–11703 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley C. C., Gordon A. J. E., Halliday J. A., Herman C., Transcription fidelity: New paradigms in epigenetic inheritance, genome instability and disease. DNA Repair (Amst.) 81, 102652 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohler K., Ibba M., Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2, 17117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaher H. S., Green R., Fidelity at the molecular level: Lessons from protein synthesis. Cell 136, 746–762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffer E. D., Maehigashi T., Fredrick K., Dunham C. M., Ribosomal ambiguity (ram) mutations promote the open (off) to closed (on) transition and thereby increase miscoding. Nucleic Acids Res. 47, 1557–1563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fersht A. R., Kaethner M. M., Mechanism of aminoacylation of tRNA. Proof of the aminoacyl adenylate pathway for the isoleucyl- and tyrosyl-tRNA synthetases from Escherichia coli K12. Biochemistry 15, 818–823 (1976). [DOI] [PubMed] [Google Scholar]
- 7.Eldred E. W., Schimmel P. R., Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247, 2961–2964 (1972). [PubMed] [Google Scholar]
- 8.Youngman E. M., McDonald M. E., Green R., Peptide release on the ribosome: Mechanism and implications for translational control. Annu. Rev. Microbiol. 62, 353–373 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Stoppel R. et al., Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell 23, 2680–2695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggertsson G., Söll D., Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol. Rev. 52, 354–374 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C. C., Schultz P. G., Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010). [DOI] [PubMed] [Google Scholar]
- 12.O’Donoghue P., Ling J., Wang Y. S., Söll D., Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 9, 594–598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baggett N. E., Zhang Y., Gross C. A., Global analysis of translation termination in E. coli. PLoS Genet. 13, e1006676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aerni H. R., Shifman M. A., Rogulina S., O’Donoghue P., Rinehart J., Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 43, e8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y. et al., Heterogeneity of stop codon readthrough in single bacterial cells and implications for population fitness. Mol. Cell 67, 826–836.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loughran G. et al., Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honigman A., Wolf D., Yaish S., Falk H., Panet A., Cis Acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology 183, 313–319 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schueren F. et al., Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. eLife 3, e03640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn J. G., Foo C. K., Belletier N. G., Gavis E. R., Weissman J. S., Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2, e01179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stiebler A. C. et al., Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in fungi and animals. PLoS Genet. 10, e1004685 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dabrowski M., Bukowy-Bieryllo Z., Zietkiewicz E., Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons. Mol. Med. 24, 25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng M. Y. et al., New in vitro assay measuring direct interaction of nonsense suppressors with the eukaryotic protein synthesis machinery. ACS Med. Chem. Lett. 9, 1285–1291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baranov P. V., Gesteland R. F., Atkins J. F., Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 3, 373–377 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelberg-Kulka H., Amiel A., Miller C., Schoulaker-Schwarz R., Studies on the involvement of the UGA readthrough process in the mechanism of attenuation of the tryptophan operon of Escherichia coli. Mol. Gen. Genet. 188, 156–160 (1982). [DOI] [PubMed] [Google Scholar]
- 25.Ballesteros M., Fredriksson A., Henriksson J., Nyström T., Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20, 5280–5289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Björkman J., Samuelsson P., Andersson D. I., Hughes D., Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 31, 53–58 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Fan Y. et al., Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 43, 1740–1748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutscher J., Francke C., Postma P. W., How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donly B. C., Edgar C. D., Adamski F. M., Tate W. P., Frameshift autoregulation in the gene for Escherichia coli release factor 2: Partly functional mutants result in frameshift enhancement. Nucleic Acids Res. 18, 6517–6522 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luli G. W., Strohl W. R., Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56, 1004–1011 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhlenkoetter S., Wintermeyer W., Rodnina M. V., Different substrate-dependent transition states in the active site of the ribosome. Nature 476, 351–354 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Shaw J. J., Trobro S., He S. L., Åqvist J., Green R., A Role for the 2′ OH of peptidyl-tRNA substrate in peptide release on the ribosome revealed through RF-mediated rescue. Chem. Biol. 19, 983–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Indrisiunaite G., Pavlov M. Y., Heurgué-Hamard V., Ehrenberg M., On the pH dependence of class-1 RF-dependent termination of mRNA translation. J. Mol. Biol. 427, 1848–1860 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Bieling P., Beringer M., Adio S., Rodnina M. V., Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat. Struct. Mol. Biol. 13 (5), 423–428 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S., Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Blake W. J., Kærn M., Cantor C. R., Collins J. J., Noise in eukaryotic gene expression. Nature 422, 633–637 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi Y. et al., Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sliusarenko O., Heinritz J., Emonet T., Jacobs-Wagner C., High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol. Microbiol. 80, 612–627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann M., A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Evans C. R., Fan Y., Weiss K., Ling J., Errors during gene expression: Single-cell heterogeneity, stress resistance, and microbe-host interactions. MBio 9, e01018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland I. et al., Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 57, 1–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dauner M., Storni T., Sauer U., Bacillus subtilis metabolism and energetics in carbon-limited and excess-carbon chemostat culture. J. Bacteriol. 183, 7308–7317 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Türkel S., Kaplan G., Farabaugh P. J., Glucose signalling pathway controls the programmed ribosomal frameshift efficiency in retroviral-like element Ty3 in Saccharomyces cerevisiae. Yeast 28, 799–808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norman T. M., Lord N. D., Paulsson J., Losick R., Stochastic switching of cell fate in microbes. Annu. Rev. Microbiol. 69, 381–403 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Stuart T., Satija R., Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019). [DOI] [PubMed] [Google Scholar]
- 46.West S. A., Cooper G. A., Division of labour in microorganisms: An evolutionary perspective. Nat. Rev. Microbiol. 14, 716–723 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Balaban N. Q. et al., Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez A., Golding I., Genetic determinants and cellular constraints in noisy gene expression. Science 342, 1188–1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Levine J. H., Lin Y., Elowitz M. B., Functional roles of pulsing in genetic circuits. Science 342, 1193–1200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaern M., Elston T. C., Blake W. J., Collins J. J., Stochasticity in gene expression: From theories to phenotypes. Nat. Rev. Genet. 6, 451–464 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Newman J. R. et al., Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441, 840–846 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Balázsi G., van Oudenaarden A., Collins J. J., Cellular decision making and biological noise: From microbes to mammals. Cell 144, 910–925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su H. W. et al., The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat. Microbiol. 1, 16147 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Rakauskaite R., Liao P. Y., Rhodin M. H., Lee K., Dinman J. D., A rapid, inexpensive yeast-based dual-fluorescence assay of programmed-1 ribosomal frameshifting for high-throughput screening. Nucleic Acids Res. 39, e97 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adamski F. M., McCaughan K. K., Jørgensen F., Kurland C. G., Tate W. P., The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol. 238, 302–308 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Fredriksson A. et al., Decline in ribosomal fidelity contributes to the accumulation and stabilization of the master stress response regulator sigmaS upon carbon starvation. Genes Dev. 21, 862–874 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battesti A., Majdalani N., Gottesman S., The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J., Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis B. D., Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51, 341–350 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ling J. et al., Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48, 713–722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutz R., Bujard H., Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data discussed in the paper are available in the main text and SI Appendix.