Significance
Throughout evolutionary history, organisms have evolved features that allow them to interact with their environment in novel ways. When such features lead to increased rates of speciation in a lineage, we call them key innovations. Understanding the genetic and developmental changes involved in the origin of key innovations is of particular interest. Here we identify a gene, POPOVICH, that is crucial to the development of a key innovation, floral nectar spurs, in the columbine genus Aquilegia. While the function of POPOVICH orthologs in other plant taxa suggests an ancestral function of the lineage in leaf development, in Aquilegia, POPOVICH also functions to promote cell division in petals, a key cellular step in the development of nectar spurs in the genus.
Keywords: Aquilegia, petal development, nectar spur, key innovation, mitosis
Abstract
The evolution of novel features, such as eyes or wings, that allow organisms to exploit their environment in new ways can lead to increased diversification rates. Therefore, understanding the genetic and developmental mechanisms involved in the origin of these key innovations has long been of interest to evolutionary biologists. In flowering plants, floral nectar spurs are a prime example of a key innovation, with the independent evolution of spurs associated with increased diversification rates in multiple angiosperm lineages due to their ability to promote reproductive isolation via pollinator specialization. As none of the traditional plant model taxa have nectar spurs, little is known about the genetic and developmental basis of this trait. Nectar spurs are a defining feature of the columbine genus Aquilegia (Ranunculaceae), a lineage that has experienced a relatively recent and rapid radiation. We use a combination of genetic mapping, gene expression analyses, and functional assays to identify a gene crucial for nectar spur development, POPOVICH (POP), which encodes a C2H2 zinc-finger transcription factor. POP plays a central role in regulating cell proliferation in the Aquilegia petal during the early phase (phase I) of spur development and also appears to be necessary for the subsequent development of nectaries. The identification of POP opens up numerous avenues for continued scientific exploration, including further elucidating of the genetic pathway of which it is a part, determining its role in the initial evolution of the Aquilegia nectar spur, and examining its potential role in the subsequent evolution of diverse spur morphologies across the genus.
The pace of species diversification varies across the tree of life, with some lineages exhibiting increased rates of speciation relative to others. The evolution of key innovations, that is, traits thought to promote the process of diversification by increasing ecological opportunities, have often been used to explain particularly species-rich clades (1–3). Floral nectar spurs are considered to be a classic example of a key innovation (4). Nectar spurs are tubular structures, generally formed by floral tissue, that produce a nectar reward for animal pollinators. Such spurs have evolved independently many times in flowering plants, and, in nearly all cases, lineages with nectar spurs are more speciose than their sister lineages that lack them (4–6). Spurs are hypothesized to increase speciation rates because changes in spur morphology can lead to pollinator specialization—either through the differential placement of pollen on the body of a pollinator or visitation by a different animal pollinator altogether—resulting in increased reproductive isolation between plants with different spur morphologies (5). A textbook example of a radiation following the evolution of nectar spurs is the genus Aquilegia (7). Floral nectar spurs evolved as outgrowths of petals in the Aquilegia ancestor 5 million to 7 million years ago (8), after which modifications to spur morphology and other floral features, such as color and orientation, allowed populations to adapt to different animal pollinators (9–12). Although the evolution of novel traits, such as the powered flight of insects, birds, and bats, or the pharyngeal jaws of cichlid fish, is recognized as contributing to lineage diversification (13–15), discovering the genetic and developmental mechanisms that led to their evolution is often difficult, in part, because many of these traits involve complex developmental mechanisms that arose deep in evolutionary history. Given that the Aquilegia nectar spur evolved relatively recently and is formed by modifications to a single floral organ, it provides a unique opportunity to begin to dissect the developmental and genetic basis of a key innovation, which, in turn, will provide insight into its origin.
The development of the spurred petal in Aquilegia is relatively simple, facilitating the identification of key features. The Aquilegia petal is composed of two components, the laminar blade at the distal end of the petal, and the spur, which forms adjacent to the attachment point as a tubular outgrowth with a nectary at the tip (SI Appendix, Fig. S1A). Previous studies of the Aquilegia nectar spur identified key cellular processes involved in its development, which can be broken down into two developmental phases. During phase I, cell divisions that are initially dispersed throughout the petal become localized to the developing spur cup, where they continue until the spur is 7 mm to 10 mm in length (16, 17). As petal development proceeds, the spur enters phase II, in which mitotic activity ceases and the differentiating spur cells elongate anisotropically (16). Cell anisotropy is a major contributor to final spur length, and variation in the degree of anisotropy largely explains the differences in spur length between species (16). However, little is known about the genetic control of persistent mitosis in the spur cup during phase I of development, or the transition to anisotropic cell expansion during phase II of development. An initial RNA sequencing (RNAseq) experiment identified genes that are strongly differentially expressed (DE) between the developing spur and blade tissue during phase I of petal development in Aquilegia coerulea ‘Origami,’ including the TEOSINTE BRANCHED/CYCLOIDEA/PCF gene AqTCP4, which acts to restrain cell proliferation in the distal compartment of the spur, and the AqSTYLISH genes, a small family of transcription factors (TFs) that were subsequently found to be responsible for nectary development (17, 18). While the set of DE genes from that study indicates that cell proliferation pathways are involved, none of the candidates functionally explored so far have revealed potential master regulators of spur development.
One species of Aquilegia native to montane regions of central China, Aquilegia ecalcarata, is the only known species of Aquilegia with petals that do not produce spurs or nectaries (19). While once thought to have diverged from the lineage prior to the evolution of nectar spurs, phylogenetic analyses clearly indicate that the spurless phenotype of A. ecalcarata represents a secondary loss (8). Nonetheless, understanding spur loss in A. ecalcarata has been suggested as key in helping to unravel the genetic and developmental basis of spur development (4). Although the A. ecalcarata petal no longer produces a nectar spur, other aspects of its development remain intact, and the morphology of the A. ecalcarata petal is quite similar to the primitively spurless petals of the Aquilegia sister genus, Semiaquilegia (20). We previously used A. ecalcarata in an effort to further narrow in on key components of the genetic network involved in early spur development using comparative gene expression. A set of genes consistently DE between the developing petals of spurless A. ecalcarata and three phylogenetically divergent spurred taxa was identified (21). Cross-referencing these genes with those identified as DE between blade and spur tissue in A. coerulea ‘Origami’ revealed a list of only 35 genes consistently DE between petal samples containing spur tissue and petal samples lacking spur tissue (21). Interestingly, a genetic cross conducted by W. Prazmo (22) in the 1960s between A. ecalcarata and the spurred species A. vulgaris showed that 25% of the F2 individuals did not produce a spur, while the 75% of the spurred F2 individuals showed continuous variation in spur length. These findings suggest that spur loss in A. ecalcarata is caused by a single locus that is epistatic to other loci affecting morphological traits such as spur length and curvature (22). This genetic cross, combined with the developmental similarities between the spurless Semiaquilegia and A. ecalcarata petals, has been interpreted as an indication that there may be a single genetic factor that plays a crucial role in early spur development (20). Identifying this element would provide a critical component of the genetic network involved in the development of this key innovation, an important step in understanding how this novel feature evolved. Therefore, we made a similar genetic cross to that of Prazmo (22) and made use of tools available in the modern genomics era to identify a gene encoding a C2H2 zinc-finger TF that is crucial to regulating mitosis during phase I in the development of the Aquilegia nectar spur.
Results
Quantitative Trait Locus Mapping and RNAseq Identify a Candidate Gene for Spur Development.
In order to identify the genetic region controlling spur loss in A. ecalcarata, we crossed a spurred species, Aquilegia sibirica, to A. ecalcarata (Fig. 1A). A single individual from the F1 generation, whose petals developed nectar spurs (SI Appendix, Fig. S1B), was selfed to create an F2 generation. Various petal phenotypes segregated in the F2 generation, including plants whose petals bore a small pocket but no tubular spur or nectary, as well as plants with clearly developed tubular spurs and nectaries (Fig. 1B). In order to evaluate the previous observations of Prazmo (22), we measured the length of the petal pocket or spur from the attachment point to the apex or nectary, respectively, on the proximal side of the petals for 92 F2 individuals. Plotting these length measurements produced a bimodal histogram confirming that phenotypes could be differentiated into spurred and spurless individuals (SI Appendix, Fig. S1C); 334 F2 individuals were phenotyped using this metric to guide binning of individuals into spurred and spurless classes. Phenotype counts were consistent with expectations for a recessive allele at a single locus causing spur loss (SI Appendix, Fig. S1D; n = 334, 2 = 0.44, degree of freedom = 1, P = 0.51). At the same time, these data indicate that multiple genes contribute to spur length variation downstream of this single locus responsible for spur presence/absence. In this study, we have chosen to focus first on the quantitative trait locus (QTL) for spur loss.
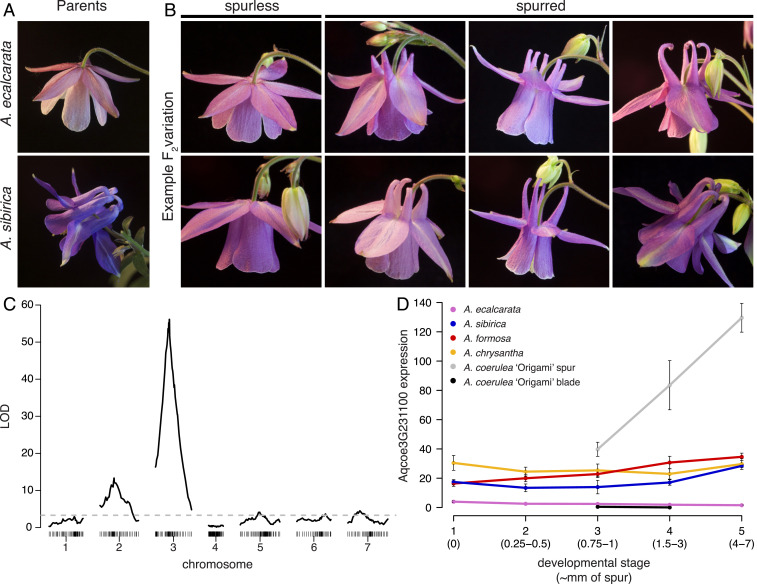
Fig. 1.
Identifying a candidate locus for spur loss in A. ecalcarata. (A) Flowers from the cross parents, spurless A. ecalcarata (Top) and A. sibirica with curved spurs (Bottom). (B) Examples of variation in petal morphology segregating in the F2 generation of the cross between A. ecalcarata and A. sibirica, including plants that do not produce spurs at all (column 1) and plants exhibiting various spur morphologies (columns 2 to 4). (C) Genome-wide association of genotype with the presence/absence of spurs in the F2 generation. Logarithm of the odds (LOD) scores (y axis) for the presence or absence of spurs indicate a major QTL on chromosome 3 (x axis, genomic marker bin position by chromosome in centimorgans). The dashed line represents the 5% LOD false discovery rate. The association on chromosome 2 appears to be a result of a genomic incompatibility (SI Appendix, Table S1). (D) The expression of Aqcoe3G231100, plotted as mean normalized reads of multiple biological replicates (±SE), across several stages spanning phase I of petal development in various petal samples, including some containing spur tissue (spur +) and some lacking spur tissue (spur -). Aqcoe3G231100 is expressed at higher levels in spur + samples. Data for A. ecalcarata (spur–), A. sibirica (spur+), A. formosa (spur+), and A. chrysantha (spur+) are from ref. 21 where RNA was isolated from entire petals (n = 3 for each data point). Data for A. coerulea ‘Origami’ are from ref. 17 where RNA was isolated from petals dissected into blades (spur–) and spur cups (spur+; n = 4 for each data point). Developmental stage is represented numerically 1 to 5 as in ref. 21, and approximate spur lengths in millimeters corresponding to these stages are provided.
Using whole-genome skim sequencing, 286 individuals were genotyped, and 276 of those individuals were phenotyped and used to conduct QTL mapping of spur loss (phenotypes were not recorded for 10 sequenced individuals). As predicted by the phenotype counts, a single major locus associated with spur loss was identified on chromosome 3 (Fig. 1C). An apparent second locus of moderate effect was detected on chromosome 2, as well as several loci of small effect on chromosomes 5, 6, and 7. Closer examination of individual genotypes at the loci on chromosomes 2 and 3 revealed that the association between genotype and phenotype on chromosome 2 is likely caused by a deleterious interaction between A. ecalcarata and A. sibirica alleles at these loci (SI Appendix, Table S1). For example, individuals with an A. sibirica allele (homozygous SS or heterozygous ES) at the chromosome 3 locus were never homozygous for the A. ecalcarata allele (EE) at the chromosome 2 locus, and only one individual homozygous for A. sibirica (SS) at the chromosome 3 locus had an A. ecalcarata allele (ES) at the chromosome 2 locus. Indeed, treating the genotype at the chromosome 3 QTL as a covariate completely eliminates the association between genotype and phenotype at the chromosome 2 locus; however, when treating the genotype on chromosome 2 as a covariate, the significant QTL on chromosome 3 remains (SI Appendix, Fig. S2). Therefore, in order to narrow in on the major genetic element controlling spur development in this cross, we focus on the locus with the highest LOD score, the locus on chromosome 3, which we refer to as POPOVICH (POP).
Genotype at the POP QTL is highly predictive of phenotype—all SS and SE individuals at the QTL produce spurs, whereas 71 of 79 EE individuals lack nectar spurs (SI Appendix, Table S2). Using recombination events that resulted in a shift in genotype between ES and EE that were associated with phenotype, we narrowed our QTL region of interest to a <2-cM region in the middle of chromosome 3. This region has a low recombination rate and encompasses 20 Mbp of sequence containing 1,100 annotated genes. Although the spur loss phenotype could be a result of mutations affecting the coding sequence of one or more proteins in the QTL, we first determined whether any of the 35 genes previously identified as consistently DE between Aquilegia phase I petal samples containing spur tissue (spur +) versus those lacking spur tissue (spur –; ref. 21) are in the QTL for spur loss. Only one of the expression candidates, Aqcoe3G231100, occurs in the POP QTL. This candidate is expressed at much higher levels in the spur tissue relative to the blade tissue of A. coerulea ‘Origami’ petals and is consistently expressed at higher levels in the petals of the spurred taxa, A. sibirica, Aquilegia formosa, and Aquilegia chrysantha, than in spurless A. ecalcarata petals, where it is essentially only expressed at background levels, during phase I of development (Fig. 1D; data from refs. 17 and 21).
Aqcoe3G231100 Encodes a C2H2 Zinc-Finger Transcription Factor.
Aqcoe3G231100 is annotated as a C2H2 zinc-finger TF, one of the largest TF families in plants (23). The subclade of C2H2 TFs containing Aqcoe3G231100, part of the C1-1i clade (23), includes a number of genes with important roles in floral development (SI Appendix, Fig. S3). These genes contain LxLxL-type EAR transcriptional repressor motifs, and related homologs from Arabidopsis have been found to function in regulating the transition between mitotic growth and cellular differentiation in various plant developmental processes by repressing either genes responsible for cellular differentiation or genes involved in the maintenance of mitosis (23). There is no functional information for the Arabidopsis ortholog of Aqcoe3G231100, AT4G17810, although it is expressed in leaves and cotyledons (24), and the ortholog in Medicago truncatula, PALMATE-LIKE PENTAFOLIATA1 (PALM1), functions in the regulation of compound leaf development (25, 26).
Analyzing the sequence of the A. ecalcarata and A. sibirica Aqcoe3G231100 alleles segregating in the cross and from nine additional phylogenetically distributed Aquilegia species shows that, overall, the predicted protein is highly conserved across the genus, except for in a region of glutamine (Q) repeats near the N-terminus of the predicted protein (SI Appendix, Fig. S4). The nucleotide and predicted protein sequence between the A. ecalcarata and A. sibirica alleles segregating in the F2 generation are nearly identical (both greater than 99% shared sequence identity) with the only amino acid variants occurring in this Q-repeat region (SI Appendix, Fig. S4). Comparing the A. ecalcarata allele to the A. sibirica allele, there is a single nucleotide polymorphism causing an amino acid difference (E26Q) as well as three additional Q residues in the A. ecalcarata allele. It is unlikely that this amino acid variation would result in the spur loss seen in A. ecalcarata, as the A. ecalcarata allele has a predicted amino acid sequence identical to that found in Aquilegia japonica, which has nectar spurs (SI Appendix, Fig. S4).
Cis-Regulatory Changes Contribute to Differential Expression of Aqcoe3G231100 in A. ecalcarata.
Given that there are no predicted amino acid differences that would cause functional effects in the protein and that this gene was first identified in part by transcriptional differences between spur+ and spur– petal tissue, we predicted that the functional difference in the A. ecalcarata allele would be due to cis-regulatory differences in gene expression (while possible, it is less likely that a trans-regulator would be encoded in the same QTL). To test this, we explored allele-specific expression patterns of our gene using RNAseq on F2 individuals with different genotypes at the POP QTL. Gene expression was assayed in developing petals of 20 F2 hybrids: 5 homozygous A. sibirica (SS), 5 homozygous A. ecalcarata (EE), and 10 heterozygous (ES) at the POP QTL. All F2 plants assayed had spur presence/absence phenotypes consistent with their POP QTL genotypes—the EE individuals were spurless, and the ES and SS individuals had spurs. Normalized expression patterns of Aqcoe3G231100 in the F2s significantly differed by their QTL genotype (ANOVA, F = 31.41, P = 1.96e-6; Dunnett modified Tukey–Kramer test results in SI Appendix, Table S3). As expected, expression of Aqcoe3G231100 was lowest in A. ecalcarata homozygotes, highest in A. sibirica homozygotes, and intermediate in the heterozygotes (Fig. 2A). Aside from the correlation between genotype at the POP QTL and spur presence (i.e., EE are spurless and ES and SS are spurred), there does not appear to be a strong correlation between Aqcoe3G231100 expression level and spur length in the spurred F2 individuals that were sampled (R2 = 0.21, P = 0.058, n = 14; SI Appendix, Fig. S5), consistent with the observation that spur length is controlled by multiple loci.
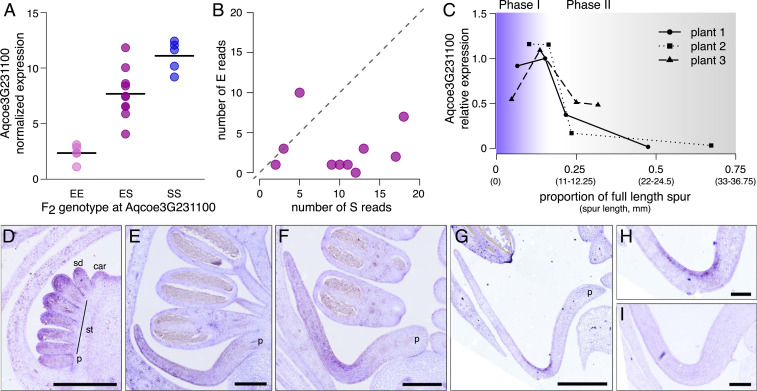
Fig. 2.
Expression of Aqcoe3G231100 in F2 hybrids and in A. coerulea ‘Origami.’ (A) Expression of Aqcoe3G231100 in petals (developmental stage 3 to 4) of F2 individuals that are either homozygous A. ecalcarata (EE, n = 5), homozygous A. sibirica (SS, n = 5), or heterozygous (ES, n = 10) at the spur loss QTL. Each point represents normalized reads as assessed by RNAseq of a single F2 individual, with the black bar indicating mean expression of all individuals for each genotype. (B) The number of Aqcoe3G231100 assignable reads transcribed from the A. ecalcarata allele (E reads) vs. the A. sibirica allele (S reads) for each of the 10 heterozygous F2s in A. The dashed line represents the 1:1 ratio of E and S reads. (C) Relative expression of Aqcoe3G231100 in spur tissue of A. coerulea ‘Origami’ across the transition from mitotic growth (phase I, purple) to cell differentiation (phase II, gray) as assayed at a variety of developmental stages in three different plants. Developmental staging of petals is presented as the sample spur length relative to the spur length of a fully developed petal for each plant (i.e., proportion of full spur length). For various proportions, the range of spur lengths across the three plants assessed is also provided, in millimeters. (D–H) Spatial expression of Aqcoe3G231100 in petals at various developmental stages using in situ hybridization. (D) In a stage 7 floral meristem, Aqcoe3G231100 is expressed throughout early differentiating petal (p), stamen (st), staminode (sd), and carplel (car) primordia (floral stages based on ref. 27; see SI Appendix, Table S4 for stage comparison with RNAseq stage). (E) Aqcoe3G231100 is broadly expressed in the early petal of a stage 9 flower. (F) As development progresses, Aqcoe3G231100 expression contracts to the tip of the developing spur in an early stage 10 flower. (G) Late in stage 10, Aqcoe3G231100 is restricted to the adaxial layer of cells at the spur tip. (H) Magnification of the spur tip in G. (I) The sense probe showed no signal. (Scale bars: D, H, and I, 100 m; E, F, and G, 200 m.)
Focusing on the 10 heterozygotes, the small number of coding sequence nucleotide variants between the A. sibirica and A. ecalcarata alleles were used to assign reads spanning these positions as having been transcribed off of each parental allele. This analysis showed a strong pattern in which, for nearly every F2 heterozygote assessed, a greater number of reads was produced by the A. sibirica allele (directional Wilcoxon signed rank test, P = 0.009), further suggesting that cis-regulatory differences between these alleles significantly contribute to the difference in expression seen both between the parental species and in the F2s with different POP QTL genotypes and otherwise variable genetic backgrounds (Fig. 2B). Comparing the upstream sequence between A. ecalcarata, A. sibirica, and several other spurred species indicates that that there are few variants in the 1.5 kb directly upstream of the transcription start site; however, there is a region containing a number of single nucleotide and indel variants 2 kb upstream of the transcription start site (Chr03:27,452,100 to 27,452,400; SI Appendix, Fig. S6). This region may be a point of focus for future research into cis-regulatory elements related to differences in allele expression.
Temporal and Spatial Expression of Aqcoe3G231100 in A. coerulea ‘Origami’ Is Broadly Consistent with Mitotic Activity during Spur Development.
In order to further examine the temporal and spatial expression patterns of Aqcoe3G231100, we used qRT-PCR and in situ hybridization to assess expression patterns in the horticultural variety A. coerulea ‘Origami.’ As prior RNAseq assays were conducted only on petals during phase I of development (Fig. 1D and ref. 21), we used qRT-PCR to track the expression profile of Aqcoe3G231100 in spurs as petal development transitioned from mitotic growth (phase I) into cell differentiation and anisotropic cell elongation (phase II; Fig. 2C). In each plant assessed, the expression of Aqcoe3G231100 in nectar spurs showed a similar pattern. Expression increased through phase I but dramatically dropped as petals transitioned from mitotic growth to the cell elongation phase (this phase transition was previously identified in ref. 16 as occurring when A. coerulea ‘Origami’ petals are 8 mm to 10 mm in length; Fig. 2C).
To assess spatial expression patterns of Aqcoe3G231100, we used in situ hybridization across early developmental stages of A. coerulea ‘Origami’ flowers. During floral development stages 3 to 7 (floral staging as in ref. 27; see SI Appendix, Table S4 for developmental stage comparisons with RNAseq and qRT-PCR data), the gene is broadly expressed in the floral meristem and in early differentiating petals, stamens, staminodes, and carpels (SI Appendix, Fig. S7 A and B and Fig. 2D). Although the expression of Aqcoe3G231100 wanes through development in most floral organs, it is maintained in later stages of petal development (Fig. 2 E–H and SI Appendix, Fig. S7 A–C). The locus is initially expressed broadly in petals (Fig. 2E) but gradually becomes restricted to the spur cup (Fig. 2F and SI Appendix, Fig. S7 C and E). Toward the end of phase I, expression is eventually limited to the adaxial surface of the petal spur cup (Fig. 2 G and H). This pattern of progressive spatial restriction to the spur tip mirrors the contraction of mitotically active cells as indicated by HISTONE4 (HIS4) expression (16, 17), with the notable exception of the adaxial localization of Aqcoe3G231100 late in phase I, which is not seen in cell division markers. In addition to expression in petals, Aqcoe3G231100 expression was also detected in the placenta, ovules, and young leaves (SI Appendix, Fig. S7 A, D, F, and G), all of which are presumably mitotically active during the phase in which expression was assayed (this has been confirmed for young leaves in ref. 17). No signal was detected using a sense probe (Fig. 2I). Thus, both temporally and spatially, Aqcoe3G231100 expression is generally associated with mitotically active tissues.
Gene Knockdown Results in Dramatically Altered Spur Development Caused by a Reduction in Cell Number.
We used Virus-Induced Gene Silencing (VIGS) to transiently knock down the expression of Aqcoe3G231100 in A. coerulea ’Origami.’ As the quantitative, spatial, and temporal degree of target gene knockdown (KD) is highly variable with VIGS, constructs were designed to also target a reporter gene, ANTHOCYANIDIN SYNTHASE (AqANS), which is required to produce the red floral pigments in A. coerulea ‘Origami’ sepals and petals and can be used as a general, although not exact, marker of tissue experiencing target KD versus wild-type (WT) tissue. A range of spur phenotypes were present in petals that showed evidence of VIGS response when Aqcoe3G231100 was targeted. The strongest spur phenotypes resulted in the near-complete loss of the nectar spur, including the nectary, while less severe phenotypes included shorter spurs, some of which still produce nectaries (Fig. 3 A–F). As Aqcoe3G231100 is expressed only early in petal development, largely before pigment is produced, petal samples used to verify and assess the degree of gene KD were identified based on morphology at earlier developmental stages (Fig. 3 E and F). In paired samples (WT vs. KD petals from the same flower), KD petals had, on average, 36.1% of the expression of Aqcoe3G231100 in comparison with WT petals (n = 13, SE 14.4%; SI Appendix, Fig. S8A).
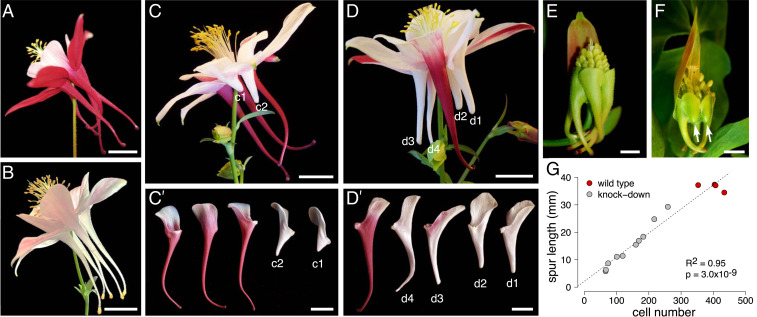
Fig. 3.
Phenotypes of VIGS of Aqcoe3G231100 in A. coerulea ‘Origami.’ (A–D) Flowers at anthesis. (A) WT. (B) AqANS silenced VIGS control. (C) An Aqcoe3G231100-AqANS VIGS treated plant with two KD petals showing greatly reduced nectars spurs. () Petals dissected from the flower in C with the two KD petals indicated (c1 and c2). (D) An Aqcoe3G231100-AqANS VIGS treated plant with four KD petals with variably reduced spurs. () Petals dissected from the flower in D with the four KD petals indicated (d1 to d4). (E and F) Flowers at a developmental stage when Aqcoe3G231100 is normally expressed. (E) A WT flower with multiple sepals removed to see developing petal nectar spurs that have not begun producing anthocyanin pigments. (F) Example of an Aqcoe3G231100-AqANS VIGS treated plant used to verify gene KD, showing a strong reduction in spur development in two petals (indicated by arrows) relative to the other three. (G) Spur length plotted against cell number of WT (red) and Aqcoe3G231100-AqANS KD (gray) petals and the line of best fit, indicating a strong positive correlation between cell number and spur length (R2 = 0.95, P = 3.0e-9, n = 14). (Scale bars: A, B, C, , D, and , 1 cm; E and F, 1 mm.)
Developmental studies of several Aquilegia species have shown that interspecific variation in spur length can largely be attributed to differences in anisotropic cell elongation, rather than differences in overall cell number (16). In order to assess whether the differences in spur length seen in the VIGS KD petals are due to differences in cell elongation, such as those seen with interspecific variation, or in cell number, which would suggest that Aqcoe3G231100 plays a role in regulating mitosis, we counted and measured spur cells and assessed spur length from both WT and KD petals. These comparisons show that variation in spur length is not correlated with cell length (R2 = 0.02, P = 0.60, n = 14; SI Appendix, Fig. S8B), but is highly correlated with cell number (R2 = 0.95, P = 3.0e-9, n = 14; Fig. 3G), indicating that the reduction of spur length in KD spurs is primarily due to the production of fewer cells. Despite the fact that in situ hybridization indicates expression of Aqcoe3G231100 in other floral organs, no other obvious phenotypes were noted in floral tissue; however, many plants exhibiting KD in floral tissue also exhibited a leaf phenotype (SI Appendix, Fig. S9A). In these leaves, the aspect ratio of leaflet shape was altered from rounded in control leaves to elongated and lanceolate in presumably silenced leaves (SI Appendix, Fig. S9 B and C). In more-extreme cases, dissections between leaflet lobes were exaggerated such that the overall leaf architecture shifted from ternately compound to biternately compound leaves (SI Appendix, Fig. S9C).
Discussion
Here we present multiple lines of evidence that Aqcoe3G231100 is critical to the development of nectar spurs in Aquilegia. As prior studies have shown, the gene is expressed in the petals of spurred Aquilegia taxa but not in the petals of A. ecalcarata, which has secondarily lost nectar spurs (21). The gene is located under the POP QTL, which is strongly associated with the presence or absence of spurs. Spatial and temporal expression patterns reveal highly localized expression in developing spurs during phase I, the mitotic phase of spur development. Moreover, although we have not pinpointed the genetic variants responsible for differential expression of Aqcoe3G231100 between the species, allele-specific expression patterns in heterozygotes indicate that cis-regulatory differences contribute to the lack of Aqcoe3G231100 expression in A. ecalcarata. Finally, knocking down expression of the gene in the long spurred variety A. coerulea ‘Origami’ causes a dramatic reduction in the number of spur cells produced, resulting in shortened spurs and, in some cases, the near-complete loss of the spur, including the nectary—traits consistent with the A. ecalcarata phenotype. Given these multiple lines of evidence, we are designating Aqcoe3G231100 as POPOVICH (POP).
Our results suggest that POP plays a key morphogenetic role in spur development by promoting cell division in the Aquilegia spur cup. Based solely on what little is known about the function of POP orthologs in other taxa, this locus would seem to be an unlikely candidate gene for controlling spur development. The M. truncatula ortholog of POP, PALM1, regulates the complexity of compound leaf development by suppressing mitotic activity in developing leaves (25). This is achieved through the direct repression of SINGLE LEAFLET1 (SGL1), a LEAFY/FLORICAULA ortholog that functions to maintain indeterminacy during the development of compound leaves in M. truncatula (25, 28). Nothing is known about the function of the Arabidopsis POP/PALM1 ortholog, although it is also expressed in young developing leaves (24), as we similarly saw with POP. The expression in developing leaves of POP and its orthologs in M. truncatula and Arabidopsis suggests that early leaf expression is a common feature of POP orthologs in eudicots, and, although the leaf phenotypes in Medicago and Aquilegia differ considerably, the fact that the perturbation of normal PALM1/POP activity results in mutant leaf phenotypes in both genera may reflect an ancestral function of the gene lineage in leaf development. Further study will be required to determine when the petal expression domain evolved for POP orthologs in the Ranunculaceae and whether this coincides with the evolution of spurs in Aquilegia.
More generally, it is interesting to note that several closely related C2H2 homologs control crucial aspects of plant organ development by repressing genes regulating cell morphogenetic activity. For instance, in Arabidopsis, the closely related homolog RABBIT EARS (RBE) (SI Appendix, Fig. S4) also influences petal shape by controlling the timing of the developmental shift from mitotic growth to differentiation (29). The ultimate role of RBE appears to be more similar to POP in that both loci function to maintain mitosis in petals, which RBE achieves by directly repressing the cell differentiation factor TCP4 (29, 30). Studies of AqTCP4 have confirmed a parallel function in Aquilegia petal spurs, where the gene is responsible for shutting down cell proliferation in what we have termed the distal compartment of the developing spur (17). Although PALM1 and RBE appear to play relatively narrow roles in leaf and petal development, respectively, another more distantly related C2H2, JAGGED (JAG), functions more broadly in the regulation of cell proliferation by directly repressing several cell cycle inhibitors (KRPs; ref. 31). The Aquilegia ortholog, AqJAG, similarly promotes cell proliferation in most lateral organs, including petals. Although not spur specific, the role of AqJAG in promoting cell proliferation and laminar expansion is necessary for spur development, as KD of AqJAG also results in the loss of spur and nectary development (32). What all of these C2H2 TFs have in common is the presence of a repressive EAR domain (23). The previous study of petal transcriptomics that compared A. ecalcarata to three spurred species found that there were generally 50 to 100% more genes up-regulated in A. ecalcarata compared to spurred taxa at equivalent developmental stages (21). That study hypothesized that this was due to a heterochronic shift toward earlier differentiation in A. ecalcarata petals. Loss of expression of a transcriptional repressor (POP) could be the basis for this shift from prolonged cell division in spurred species to rapid differentiation in the spurless A. ecalcarata.
It is notable that POP expression is spatially more restricted than what we have previously observed for cell division markers in the developing petal spur (16, 17). In particular, in petal stages that still show broad AqHIS4 expression, POP expression is localized more narrowly toward the tip of the spur, and, at later developmental stages, it becomes restricted to the adaxial surface inside the developing organ. If POP is actively promoting this comparatively broader domain of cell division, its function must have a non-cell-autonomous component. In terms of the adaxial localization of its expression, there is a parallel observed in the Medicago ortholog PALM1, which is negatively regulated by an ortholog of the abaxial factor AUXIN RESPONSE FACTOR 3 (ARF3) (33). Conserved negative regulation by the abaxial identity program in Aquilegia petals would explain the adaxial localization of POP, but still leaves open the question of how POP promotes cell divisions more broadly in the organ.
Future studies to dissect the genetic interactions among POP, AqTCP4, and AqJAG, as well as identifying additional targets of POP, will provide new insight into the regulatory architecture of Aquilegia spur development. Although POP clearly plays a pivotal role in the earliest phase of spur development, this function appears to be epistatic to continuous morphological variation in traits such as spur length and curvature that is segregating in the spurred individuals of the F2 generation, suggesting that other loci controlling spur morphology vary in this cross. The identification of such loci will allow us to determine whether or not they function through POP to influence spur length. For example, some of the variation in F2 spur length may be directly related to cell number and POP expression levels, either controlled by the cis-regulatory differences at the POP locus itself or through a combination of these and potential variants in trans-regulators. Even though the VIGS KD studies show that variable levels of POP expression can dramatically alter spur length by modulating cell number, the results of the limited analysis comparing POP expression with spur length in the 14 F2s suggest that POP expression is unlikely to be a major determinant of spur length variation in this cross. Alternatively, the spur length differences among the F2s may be caused by changes in the degree of cell anisotropy, which has been shown to be the primary component of interspecific spur length differences in Aquilegia (16). The identification of such loci may also help explain the eight spurred F2s that are homozygous for A. ecalcarata alleles at POP. These individuals all have at least one A. sibirica allele at both of the small-effect QTL on chromosomes 5 and 7. Although most of the F2s with such an allelic combination lack spurs (30/38), under certain circumstances, A. sibirica alleles at these or other loci affecting spur length variation may be able to generate a spur, either by causing POP expression to pass above a threshold level or by affecting cell elongation such that a small spur appears due to changes in cell length, rather than number.
Our studies also indicate that POP is necessary for nectary development, as both F2 plants homozygous for A. ecalcarata alleles and stronger VIGS KD phenotypes lack nectaries. This places it, at least indirectly, up-stream of previously studied AqSTY homologs, which are essential to nectary, but not spur development (18). Interestingly, phenotypes of both the F2 and POP-silenced flowers suggest that there is a relationship between spur length and proper nectary formation. There are several possible explanations for this observation. First, it could be that a higher level of POP expression is required to activate the nectary developmental program. Under this scenario, lower POP expression would promote only limited spur growth and would be insufficient to promote nectary development. Alternatively, the interaction could be more indirect, such that there is some threshold of cell number or duration of expression that is required before the nectary can be initiated. Previous studies have observed that mechanical removal of the nectaries at early developmental stages does not stunt spur development (34), and KD of the essential nectary development loci AqSTY1 and AqSTY2 completely eliminates nectaries without impacting spur length. These results suggest that the nectary itself is not required to promote spur growth. Further study of the function of POP in spur development will help elucidate the relationship between increasing spur length and nectary development.
From an evolutionary perspective, a number of mutational scenarios leading to spur loss in A. ecalcarata can be considered. In one scenario, this cross indicates that the spur loss phenotype in A. ecalcarata could have been largely achieved by mutations at POP alone, rendering additional loci specific to spur morphology moot, at which point they could have secondarily accumulated mutations. On the other hand, spur loss could have happened in a stepwise fashion, with mutations occurring first in these other loci affecting smaller aspects of spur morphology before mutations eventually occurred at POP. Studies focusing on population genetics in A. ecalcarata and its closest relatives may help clarify these scenarios (35). In either case, it is interesting to note that the major component of spur loss in A. ecalcarata appears to be caused by changes in POP expression, rather than function, perhaps because POP has pleiotropic effects on other traits such as leaf development.
Conclusion
While many questions regarding the evolution of nectar spurs in Aquilegia still remain, the identification of this key regulator of spur development provides us with a critical point of focus for future research. With the identification of POP, we can now work to elucidate the genetic and developmental pathway that it regulates to better understand its role in spur development. In addition, we can address questions regarding how nectar spurs evolved in Aquilegia by determining how and when POP acquired petal expression in the lineage leading to Aquilegia. The identification of POP will further allow us to explore questions concerning the genetic basis of speciation, as we seek to understand the specific genetic basis for loss of POP expression in A. ecalcarata. More broadly, it will also be of interest to determine whether variation in POP orthologs contributes to other more subtle morphological variation in Aquilegia spur morphology across the genus. Perhaps one of the most notable aspects of this study is that it highlights the power of combining comparative transcriptomics, genetic mapping, and analyses of gene function to discover the identity of a critical locus controlling the development of a key innovation, which pure candidate gene approaches would not have identified.
Materials and Methods
Genetic Cross.
Horticultural lines of A. sibirica and A. ecalcarata were acquired from Suncrest Nurseries. An F1 generation was generated with A. sibirica as the maternal parent and A. ecalcarata as the paternal parent. The F2 generation was generated by selfing a single F1 plant. Approximately 1,000 seeds were sown in Sunshine #4 (Sun Gro Agriculture) soil in individual plug trays and stratified at 4 °C for 4 wk. After stratification, plug trays were moved to the University of California (UC), Santa Barbara greenhouse facilities, and seeds were allowed to germinate under natural light conditions throughout the spring months. In total, approximately 500 seedlings germinated over a span of 3 mo. As vegetative plants matured, they were moved into successively larger pots, eventually ending up in gallon pots. After producing at least 12 vegetative leaves, plants were vernalized at 4 °C for 8 wk in batches of 100, to promote flowering. Each batch of 100 plants put into vernalization was staggered by 1 mo. In total, 400 plants flowered.
Phenotyping.
For nearly every F2 plant that flowered, the first and second flowers to mature were removed from the plant at anthesis. Petals were removed, folded longitudinally, and placed onto the sticky side of clear packing tape that was then affixed to a piece of white paper. Sheets were then scanned at 720 pixels per inch resolution on an Epson Perfection V370 Photo scanner. As a first pass on phenotyping, 92 individuals were measured from the attachment point to the distal extent of the petal pocket or spur (ImageJ; SI Appendix, Fig. S1C). The distribution of these lengths was clearly bimodal such that some F2s formed weakly defined petal pockets with a clear difference in length relative to spurred petals. These petals also differed in shape from spurred petals with short spurs (SI Appendix, Fig. S1 C and D) and always lacked nectaries. Based on this, petals with less than 7-mm pockets were designated “spurless,” while those with greater than 8-mm spurs were designated “spurred.” Spur presence or absence was assessed from the scans and scored as a presence/absence binary trait (SI Appendix, Fig. S1D).
Genotyping.
The F2s were genotyped using a whole-genome skim sequencing method similar to that described in ref. 38, with variations outlined below. For the A. sibirica parent, DNA was extracted from desiccated leaf tissue using Qiagen DNeasy reagents and MagAttract beads (Qiagen Inc.). There was no tissue available for the A. ecalcarata parent or the F1. The sequencing library for the A. sibirica parent was constructed using a half-volume of the Illumina Nextera kit (Illumina Inc.) and sequenced as 100-bp paired end reads to 50 coverage. For the F2s, DNA was extracted from young leaves that were flash frozen using the same protocol as for the A. sibirica parent. Sequencing libraries for the F2s were prepared using 2 ng of DNA as quantified using the Qubit 2.0 fluorometer (ThermoFisher Scientific). Libraries were prepared using Illumina Nextera indices and reagents and a modified protocol allowing for library prep using 1/20th reaction volumes (Illumina Inc.) (36). Given that DNA input amounts were standardized during the library prep, it was initially assumed that library coverage of the F2s would be relatively consistent, and equal volumes of libraries for each of 96 individuals were pooled and sequenced as 100-bp paired end reads in a single lane. In processing the sequence data, it was apparent that there was high variation in coverage per individual. Three additional lanes of sequencing (100 bp paired end) were used to sequence additional F2 libraries, including those from the first lane that had low coverage, however these libraries were quantified using qPCR prior to pooling into groups of 96 individuals. In total, 287 F2s were sequenced aiming for coverage depth of 1x, with those having greater than 0.1x coverage usable for genotyping (n = 286). All sequencing was conducted on a HiSeq3000 at the Vincent J. Coates Genomics Sequencing Laboratory (UC Berkeley).
In order to identify informative sites to determine ancestry in the F2s, the F2 raw sequence reads from a single lane (96 individuals) were merged to reconstitute a mock F1. Reads for the A. sibirica parent, the mock F1, and the F2s were aligned to the A. coerulea ‘Goldsmith’ v3.1 reference genome (https://phytozome.jgi.doe.gov) using the Burrows–Wheeler aligner (37) (details in ref. 38). Informative sites for determining ancestry in the F2 generation were those where the mock F1 is heterozygous and the A. sibirica parent is homozygous. These were identified using SAMtools 0.1.19 (38, 39). Using SAMtools 0.1.19 and custom scripts (available at https://github.com/anjiballerini/POP), the number of reads at informative sites indicative of A. sibirica or A. ecalcarata ancestry were counted for each F2. These read counts were then processed in R (40) to determine the frequency of reads from each parent across nonoverlapping genomic windows, with read frequencies 0.9 or 0.1 indicating homozygosity for either A. sibirica or A. ecalcarata and frequencies between 0.4 and 0.6 indicating heterozygosity. Windows were either 0.5 Mb or 1 Mb in size depending on recombination rate estimates from previous crosses, with larger windows used in low-recombination regions of the genome. Several regions were identified as inconsistent between the A. coerulea ‘Goldsmith’ v3.1 physical map and the genetic map of a previous cross between A. formosa and A. pubescens (38). In these regions, custom bin sizes were created to allow these marker bins to map appropriately in the genetic map.
QTL Mapping.
The F2 genotypes of different marker bins were used to estimate a genetic map using the R package R/qtl v1.35-3 (41). The genetic map was then combined with the spur presence/absence phenotypes and F2 genotypes for 276 individuals to conduct QTL mapping (Dataset S1; gen.w.phen.csv, R/qtl v1.35-3; ref. 41). The spur phenotype was scored as a binary trait (presence/absence), and the R/qtl command scanone was used to calculate logarithm of the odds (LOD) scores for each marker using the expectation-maximization (EM) algorithm. Ten thousand permutations were used to determine the 5% LOD cutoff for significant QTL.
Gene Tree.
The Aqcoe3G231100 predicted protein was queried against the proteomes of Arabidopsis thaliana (AT), M. truncatula (Medtr), and Vitis vinifera (GSVIVG) using the BLAST algorithm in Phytozome 12 (https://phytozome.jgi.doe.gov). Predicted proteins were aligned using the ClustalW algorithm (42) in Geneious (v9.1.16, https://www.geneious.com), adjusted by eye, and trimmed to alignable regions. Initial relationships were estimated using the Neighbor-Joining method (43) implemented in Geneious (v9.1.16, https://www.geneious.com), and the sequence list was trimmed to include sequences in several clades closely related to the one containing Aqcoe3G231100 as well as those related to A. thaliana JAG, which was used as an outgroup. After trimming the sequence list, phylogenetic relationships were estimated using random accelerated maximum likelihood (RAxML, ref. 44) with the Jones–Taylor–Thornton model of amino acid replacement as implemented in the CIPRES web portal (http://www.phylo.org/). We used previously published sequences from nine different species of Aquilegia (A. aurea - SRR405095, A. barnebyi - SRR7965809, A. chrysantha - SRR408559, A. formosa - SRR408554, A. japonica - SRR413499, A. longissima - SRR7965810, A. oxysepala var. oxysepala - SRR413921, A. pubescens - SRR7943924, and A. vulgaris - SRR404349; ref. 38), in addition to the A. sibirica parent (SRR11508011) and an individual A. ecalcarata plant (SRR892115) from the same grower as the parent used in the cross to compare sequence variation in Aqcoe3G231100 across the Aquilegia phylogeny.
RNAseq.
Petal tissue from 20 F2 plants, 5 A. sibirica homozygotes, 5 A. ecalcarata homozygotes, and 10 heterozygotes at the spur loss QTL was collected at the developmental stage equivalent to stage 3 in ref. 21 and flash frozen in liquid nitrogen. Total RNA was extracted using the Qiagen RNeasy Micro kit (Qiagen). For each sample, RNA quantity and quality were assessed prior to library construction as in ref. 21. Libraries were quantified using qPCR, pooled aiming for equal representation across samples, and sequenced in a single lane on the HiSeq4000 (Illumina Inc.) as 50-bp single end reads, at the UC Davis Genome Center. Raw reads were aligned to the A. coerulea ‘Goldsmith’ v3.1 reference transcriptome (https://phytozome.jgi.doe.gov/) and processed to generate read counts per transcript as described in ref. 38. Normalized counts per transcript were estimated using the trimmed mean of M-values method (via the calcNormFactors command) using the R package edgeR (45, 46). Differences in mean normalized expression of Aqcoe3G231100 were assessed using ANOVA as implemented in R, and post hoc testing was done using the Dunnett modified Tukey–Kramer test (for unequal sample sizes) implemented using the DTK package in R (https://CRAN.R-project.org/package=DTK). Spur length for the ES and SS individuals was measured from the attachment point to the nectary on the proximal edge of the spur using ImageJ, and these measurements were used to test for a correlation between Aqcoe3G231100 expression and spur length. We did not have petal scans at anthesis for one ES individual, so the sample size for this test was n = 14. For the 10 heterozygous individuals, reads spanning variant sites between the A. sibirica and A. ecalcarata alleles of Aqcoe3G231100 were counted using the Integrative Genomics Viewer (47), and a one-tailed Wilcoxon signed rank test was used to test for overrepresentation of A. sibirica derived reads.
Developmental qRT-PCR.
Four flowers at various developmental stages were collected from each of three A. coerulea ‘Origami’ plants during a 1-h period. As there was some variability in spur length from plant to plant, a fully developed flower at anthesis was used to get a measure for final spur length for each plant. For each flower collected, spurs were measured, and the ratio of spur length to final spur length was used for developmental staging. Spur tissue was isolated from each petal at the attachment point, and tissue was flash frozen and kept at –80 °C prior to RNA isolation. RNA isolation and genomic DNA removal was done using the RNeasy Plus Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized using the Verso cDNA Synthesis Kit and oligo dT primers (Thermo Fisher). A fragment of Aqcoe3G231100 was amplified and quantified using iTaq Universal SYBR Green Supermix (Bio-Rad) and primers -GACCAATAACCTGATGGCATCCT- and -CGGGGTGGTCTTGATGATCC-. Expression of AqIPP2 (ISOPENTYL PYROPHOSPHATE:DIMETHYLALLYL PHYROPHOSPHATE ISOMERASE2) was used to normalize Aqcoe3G231100 expression (27). The relative expression of Aqcoe3G231100 was calculated using the Ct method with primer efficiency correction.
In Situ Hybridization.
A 250-bp fragment of Aqcoe3G231100, including part of the untranslated region and the beginning of the open reading frame (which is a nonconserved region), was PCR amplified using primers -GTATTCGGAGCGAGGTTCACT- and -AACCCTACCAGGCAAAAC-, and cloned into pCR4-TOPO vector (Thermo Fisher). In situ hybridization steps followed the protocol described by ref. 48. Slides were stained in 1% calcoflour white for 5 min before visualization. Images were taken using the Zeiss AxioImager microscope at the Arnold Arboretum of Harvard University.
VIGS and Phenotypic Assessment.
VIGS targeting Aqcoe3G231100 was conducted following previously published protocols (49, 50). As Aqcoe3G231100 consists of a single exon, a 223-bp sequence of the end was amplified directly from A. coerulea ‘Origami’ DNA using primers -CGGAATTCCATCCTGCCCAACCCTCAAT- and -CGGCTCTAGACGGGGTGGTCTTGATGATCC-, which include sequences to build in EcoR1 and XbaI restriction enzyme sites, respectively. This region does not have similarity with other C2H2 homologs. The fragment was then cloned into a TRV2 construct containing sequence from the Aquilegia ANS gene (49). A. coerulea ‘Origami’ (Goldsmith, Syngenta) plants were grown until they had approximately five to seven true leaves and then vernalized at 4 °C for 4 wk. A total of 331 plants were treated in multiple rounds. Of treated plants, 21.25% exhibited an AqANS phenotype in sepals, 19.34% exhibited an AqANS phenotype in petals, and 17.22% exhibited a spur phenotype (89.1% of those spurs showing an AqANS phenotype). KD of Aqcoe3G231100 was verified in a subset of petal samples by comparing expression of the gene in KD and WT petals from the same flower using qRT-PCR using the same protocol as in Developmental qRT-PCR. Because Aqcoe3G231100 is only expressed early in petal development, largely before petals begin producing anthocyanin pigments, in order to test targeted KD of Aqcoe3G231100, petals with KD phenotype were identified early in development based primarily on shape rather than the lack of pigment production. As we do not know what the final spur length would be and there is variation in KD effect across a single flower, it cannot be ruled out that some petals classified as WT also experienced some KD relative to true WT petals.
Cell count and length measurements were made on fully developed WT (n = 4) and KD (n = 10) petals. WT and KD petals were harvested at anthesis and fixed overnight in FAA (3.7% formaldehyde, 5% glacial acetic acid, 50% ethanol) at 4 °C. The tissue was dehydrated to 95% ethanol and then stained overnight in 1% acid fuchsin. Petals were mounted on microscope slides using Cytoseal 60 (Electron Microscopy Sciences no. 18006) with the proximal side of the spur facing up, and left to dry overnight underneath a coverslip secured by binder clips. Each spur was imaged continuously from the attachment point to nectary at 20× magnification under brightfield illumination using a Zeiss AxioCam 512 mounted on a Zeiss LSM700 confocal microscope. Cells were counted and their lengths measured using the straight line selection tool in FIJI (51, 52).
Supplementary Material
Acknowledgments
We thank J. Wechter, D. Birdseye, and J. Pretorius for contributing to planting, extracting DNA, and conducting qPCR. Members of the S.A.H. and Finkelstein laboratories and M. Shahandeh provided constructive feedback throughout the project. D. Taber and C. Hannah-Bick were invaluable in maintaining the F2 population and VIGS plants. We thank G. Popovich for being an inspirational leader. The NSF (Grant OIA-0963547) provided funds to construct the greenhouses that facilitated plant growth. E.S.B. was supported by the NIH under the Ruth L. Kirschstein National Research Service Award (F32GM103154). Research was funded by the UC Santa Barbara Harvey Karp Discovery award to E.S.B., NSF Grant 1456317 to S.A.H., and NSF Grant 1456217 to E.M.K. The sequencing was carried out by the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01, and the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant. These funding bodies had no role in the design of the study, collection, analysis, and interpretation of data, or in writing the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006912117/-/DCSupplemental.
Data Availability.
Raw sequence data used for genetic mapping and the F2 RNAseq is available in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra) under the following BioProject: PRJNA623619. Scripts for processing mapping data can be found at https://github.com/anjiballerini/POP and the genotype-phenotype file used for mapping is in the SI Appendix (Dataset S1). Otherwise, study data are included in the article and SI Appendix.
References
- 1.Simpson G., The Major Features of Evolution (Columbia University Press, New York, NY, 1953). [Google Scholar]
- 2.Cracraft J., “The origin of evolutionary novel pattern and process at different hierarchical levels” in Evolutionary Innovations, Niteki M. H., Ed. (University of Chicago Press, Chicago, IL, 1990), pp. 21–46. [Google Scholar]
- 3.Heard S. B., Hauser D. L., Key evolutionary innovations and their ecological mechanisms. Hist. Biol. 10, 151–173 (1995). [Google Scholar]
- 4.Hodges S. A., Floral nectar spurs and diversification. Int. J. Plant Sci. 158, S81–S88 (1997). [Google Scholar]
- 5.Hodges S. A., Arnold M. L., Spurring plant diversification: Are floral nectar spurs a key innovation? Proc. Biol. Soc. B Biol. Sci. 262, 343–348 (1995). [Google Scholar]
- 6.Kay K. M., et al. , “Floral characters and species diversification” in Ecology and Evolution of Flowers, Harder L., Barrett S. C. H., Eds. (Oxford University Press, Oxford, United Kingdom, 2006), pp. 311–325. [Google Scholar]
- 7.Schluter D., The Ecology of Adaptive Radiation (Oxford University Press, Oxford, United Kingdom, 2000). [Google Scholar]
- 8.Fior S., et al. , Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phytol. 198, 579–592 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Grant V., Isolation and hybridization between Aquilegia formosa and A. pubescens. Aliso A J. Syst. Evol. Bot. 2, 341–360 (1952). [Google Scholar]
- 10.Grant V., Origin of floral isolation between ornithophilous and sphingophilous plant species. Proc. Natl. Acad. Sci. U.S.A. 90, 7729–7733 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulton M., Hodges S. A., Floral isolation between Aquilegia formosa and Aquilegia pubescens. Proc. Biol. Soc. B Biol. Sci. 266, 2247–2252 (1999). [Google Scholar]
- 12.Whittall J. B., Hodges S. A., Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447, 706–709 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Nicholson D. B., Ross A. J., Mayhew P. J., Fossil evidence for key innovations in the evolution of insect diversity. Proc. Biol. Soc. B Biol. Sci. 281, 20141823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peixoto F. P., Braga P. H. P., Mendes P., A synthesis of ecological and evolutionary determinants of bat diversity across spatial scales. BMC Ecol. 18, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galis F., Drucker E. G., Pharyngeal biting mechanics in centrarchid and cichlid fishes: Insights into a key evolutionary innovation. J. Evol. Biol. 9, 641–670 (1996). [Google Scholar]
- 16.Puzey J. R., Gerbode S. J., Hodges S. A., Kramer E. M., Mahadevan L., Evolution of spur-length diversity in Aquilegia petals is achieved solely through cell-shape anisotropy. Proc. Biol. Sci. 279, 1640–1645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yant L., Collani S., Puzey J., Levy C., Kramer E. M., Molecular basis for three-dimensional elaboration of the Aquilegia petal spur. Proc. Biol. Sci. 282, 20142778 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min Y., Bunn J. I., Kramer E. M., Homologs of the STYLISH gene family control nectary development in Aquilegia. New Phytol. 221, 1090–1100 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tang L. L., Yu Q., Sun J. F., Huang S. Q., Floral traits and isolation of three sympatric Aquilegia species in the Qinling Mountains, China. Plant Systemat. Evol. 267, 121–128 (2007). [Google Scholar]
- 20.Tucker S. C., Hodges S. A., Floral ontogeny of Aquilegia, Semiaquilegia, and Enemion (Ranunculaceae). Int. J. Plant Sci. 166, 557–574 (2005). [Google Scholar]
- 21.Ballerini E. S., Kramer E. M., Hodges S. A., Comparative transcriptomics of early petal development across four diverse species of Aquilegia reveal few genes consistently associated with nectar spur development. BMC Genom. 20, 668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prazmo W., Genetic studies on the genus Aquilegia L. I. Crosses between Aquilegia vulgaris L. and Aquilegia ecalcarata Maxim. Acta Soc. Bot. Pol. 29, 423–442 (1960). [Google Scholar]
- 23.Englbrecht C. C., Schoof H., Böhm S., Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genom. 5, 39 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klepikova A. V., Kasianov A. S., Gerasimov E. S., Logacheva M. D., Penin A. A., A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Chen J., et al. , Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 107, 10754–10759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge L., Chen J., Chen R., Palmate-like pentafoliata1 encodes a novel Cys(2)His(2) zinc finger transcription factor essential for compound leaf morphogenesis in Medicago truncatula. Plant Signal. Behav. 5, 1134–1137 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballerini E. S., Kramer E. M., Environmental and molecular analysis of the floral transition in the lower eudicot Aquilegia. EvoDevo 2, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., et al. , Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol. 146, 1759–1772 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J., et al. , RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis. J. Exp. Bot. 67, 6473–6480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nag A., King S., Jack T., miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiessl K., Muiño J. M., Sablowski R., Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111, 2830–2835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Min Y., Kramer E. M., The Aquilegia JAGGED homolog promotes proliferation of adaxial cell types in both leaves and stems. New Phytol. 216, 536–548 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Peng J., Berbel A., Madueño F., Chen R., AUXIN RESPONSE FACTOR3 regulates compound leaf patterning by directly repressing PALMATE-LIKE PENTAFOLIATA1 expression in Medicago truncatula. Front. Plant Sci. 8, 927–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puzey J. R., “Plant microRNA evolution and mechanisms of shape change in plants,” PhD thesis, Harvard University, Cambridge, MA (2012).
- 35.Huang L., et al. , Genetic diversity and evolutionary history of four closely related Aquilegia species revealed by 10 nuclear gene fragments. J. Systemat. Evol. 56, 129–138 (2018). [Google Scholar]
- 36.Baym M., et al. , Inexpensive multiplexed library preparation for megabase-sized genomes. PloS One 10, e0128036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durbin R., Li H., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filiault D. L., et al. , The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife 7, e36426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team , R: A Language and Environment for Statistical Computing, v.3.6.3 (R Foundation for Statistical Computing, Vienna, Austria, 2017). [Google Scholar]
- 41.Broman K. W., Wu H., Sen Ś., Churchill G. A., R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Thompson J. D., Higgins D. G., Gibson T. J., CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou N., Nei M., The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- 44.Stamatakis A., Hoover P., Rougemont J., A rapid bootstrap algorithm for the RAxML web servers. Sys. Bio. 57, 758–771 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson J. T., Thorvaldsdóttir H., Wenger A. M., Zehir A., Mesirov J. P., Variant review with the Integrative Genomics Viewer. Canc. Res. 77, e31–e34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kramer E. M., “Methods for studying the evolution of plant reproductive structures: Comparative gene expression techniques” in Molecular Evolution: Producing the Biochemical Data, Zimmer E. A., Roalson E. H., Eds. (Elsevier Academic, San Diego, CA, 2005), pp. 617–635. [DOI] [PubMed] [Google Scholar]
- 49.Gould B., Kramer E. M., Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3, 6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharma B., Kramer E. M., Virus-induced gene silencing in the rapid cycling columbine Aquilegia coerulea ‘Origami,’ Methods Mol. Biol. 975, 71–81 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data used for genetic mapping and the F2 RNAseq is available in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sra) under the following BioProject: PRJNA623619. Scripts for processing mapping data can be found at https://github.com/anjiballerini/POP and the genotype-phenotype file used for mapping is in the SI Appendix (Dataset S1). Otherwise, study data are included in the article and SI Appendix.