Significance
Brown adipose tissue (BAT) burns fat to maintain body temperature through thermogenesis in mammals. Because heat production requires metabolizing large amounts of calories, increasing the activity of the tissue could treat obesity and related metabolic disorders. Recently, thioesterase superfamily member 1 (Them1) was identified to suppress thermogenesis, whereas deletion of Them1 enhanced the ability of BAT to burn fat and prevented diet-induced obesity. In this study, we show Them1 contains a lipid sensor that binds to fatty acids and lysophosphatidylcholine, which inversely regulate the activity of Them1. This lipid sensor allows for allosteric control of Them1 activity to regulate the burning of fat. Additionally, this study affirms a promising drug target to treat obesity and related disorders.
Keywords: thermogenesis, fatty acids, lysophosphatidylcholine, START domain, STARD14
Abstract
Nonshivering thermogenesis occurs in brown adipose tissue to generate heat in response to cold ambient temperatures. Thioesterase superfamily member 1 (Them1) is transcriptionally up-regulated in brown adipose tissue upon exposure to the cold and suppresses thermogenesis in order to conserve energy reserves. It hydrolyzes long-chain fatty acyl-CoAs that are derived from lipid droplets, preventing their use as fuel for thermogenesis. In addition to its enzymatic domains, Them1 contains a C-terminal StAR-related lipid transfer (START) domain with unknown ligand or function. By complementary biophysical approaches, we show that the START domain binds to long-chain fatty acids, products of Them1’s enzymatic reaction, as well as lysophosphatidylcholine (LPC), lipids shown to activate thermogenesis in brown adipocytes. Certain fatty acids stabilize the START domain and allosterically enhance Them1 catalysis of acyl-CoA, whereas 18:1 LPC destabilizes and inhibits activity, which we verify in cell culture. Additionally, we demonstrate that the START domain functions to localize Them1 near lipid droplets. These findings define the role of the START domain as a lipid sensor that allosterically regulates Them1 activity and spatially localizes it in proximity to the lipid droplet.
Brown adipose tissue (BAT) mediates nonshivering thermogenesis in both mice (1) and humans (1, 2). A key function of nonshivering thermogenesis is to maintain core body temperature upon exposure to cold ambient temperatures. Because high rates of caloric consumption are required to generate heat, pharmacologic approaches to increasing BAT mass and activity are viewed as promising objectives in the management of obesity and related metabolic disorders (3).
In addition to activating thermogenesis in BAT, cold ambient temperatures lead to the transcriptional up-regulation of genes that regulate energy expenditure, including thioesterase superfamily member 1 (Them1) (synonyms: brown fat inducible thioesterase [BFIT], steroidogenic acute regulatory lipid transfer-related domain 14 [STARD14], and acyl-CoA thioesterase 11 [ACOT11]) (4, 5). Expression is induced upon cold exposure, and this was originally believed to contribute to the thermogenic output of BAT (5). However, rather than promoting energy use, Them1 proved to suppress thermogenesis, thereby reducing the energy output of mice (6). Mechanistically, Them1 hydrolyzes long-chain fatty acyl-CoAs that are derived from endogenous lipid droplets within brown adipocytes, preventing their use as fuel for thermogenesis (7, 8). The genetic ablation of Them1 enhances the thermogenic output of mice and protects against diet-induced obesity and metabolic disorders (6).
Them1 comprises two N-terminal thioesterase domains that hydrolyze acyl-CoA and a C-terminal START domain. The Them1 START domain is a member of the larger START domain family which is characterized by a highly conserved ∼210 amino acid sequence found in both plant and animal proteins (9–11). Lipid ligands and functional roles have been proposed for several of the START domains. For instance, cholesterol, 25-hydroxycholesterol, testosterone, phosphatidylcholine, phosphatidylethanolamine, and ceramides bind to STARD1/STARD3/STARD4/STARD5 (12, 13), STARD5 (14), STARD6 (15), STARD2/STARD7/STARD10 (16–18), STARD10 (18), and STARD11 (19), respectively. STARD1 and STARD3 bind and transport cholesterol to the mitochondria for steroidogenesis (12, 20, 21). STARD2, STARD7, and STARD10 bind phosphatidylcholine and influence membrane lipid compositions (12, 17, 18, 22). In the case of Them1, it is not known whether the START domain binds to lipid or whether lipid recognition plays a role in regulating thermogenesis.
We have shown that the Them1 START domain is necessary for full catalytic activity of the acyl-CoA thioesterase domains in the hydrolysis of long-chain fatty acyl-CoAs; purified recombinant acyl-CoA thioesterase domains alone exhibited significantly attenuated catalytic activity, and this was restored upon the addition of the purified recombinant START domain (7). This phenomenon is not restricted to Them1, evidenced by increased activity of the long-chain fatty acyl-CoA thioesterase, Them2, in the presence of STARD2 (17, 23, 24). This study examines the effect of the Them1 START domain in brown adipocytes and explores the mechanism by which the START domain regulates acyl-CoA thioesterase (Acot) activity. We identify long-chain fatty acids and lysophosphatidylcholines (LPCs) as ligands for the Them1 START domain. Certain fatty acids allosterically enhance, whereas 18:1 LPC inhibits Them1 activity in a START domain-dependent manner. We further verify that 18:1 LPC relieves suppression of fatty acid oxidation by Them1 in brown adipocytes in an immortalized brown adipose cell line, in keeping with allosteric inhibition of Them1. Additionally, we discover the START domain is necessary for localizing Them1 to the lipid droplet.
Results
Them1 START Domain Binds Long-Chain Fatty Acids.
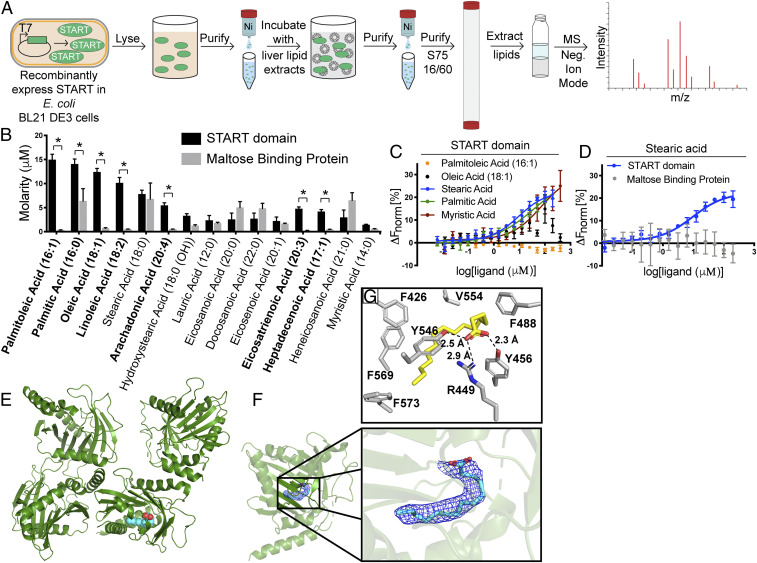
To identify possible ligands for the Them1 START domain, we used affinity purification coupled with mass spectrometry (MS) following exposure of recombinantly expressed 6×His tagged Them1 START domain to mixed-lipid liposomes (Fig. 1A). Because fatty acids are generated as a product of Them1’s enzymatic reaction, we reasoned that these products may bind to the START domain to facilitate product release. Our initial strategy therefore involved a targeted quantitative free fatty acid assay (25). This identified 15 fatty acid species that copurified with the Them1 START domain (Fig. 1B). Seven of the identified fatty acids were significantly enriched within the START domain samples over our negative control (maltose binding protein), namely, palmitoleic acid (16:1), oleic acid (18:1), linoleic acid (18:2), palmitic acid (16:0), arachidonic acid (20:4), eicosatrienoic acid (20:3), and heptadecenoic acid (17:1) (Fig. 1B). The START domain showed a preference for unsaturated fatty acids with a tail length from 16 to 20 carbons, but also bound to saturated fatty acids, such as palmitic acid (16:0), and to a lesser degree myristic acid.
Fig. 1.
START domain of Them1 binds to long-chain fatty acids. (A) Schematic of affinity purification mass spectrometry protocol. (B) Concentration of 15 identified fatty acids (FAs) bound to 1 mg of Them1 START domain (black) or maltose binding protein (gray) as determined by negative ion mode mass spectrometry through normalization to deuterated fatty acid standard. Bars indicate average of three technical replicates. Error bars display SE of mean. Statistical analyses were conducted using two-way ANOVA with Sidak’s multiple comparisons test. *P < 0.01. (C and D) Fatty acid binding assay using microscale thermophoresis (MST). (C) Them1 START domain labeled with Monolith RED-Tris-NTA was kept constant at 50 nM, while the concentration of stearic acid (blue), palmitic acid (green), myristic acid (red), oleic acid (black), and palmitoleic acid (orange) were varied between 6.1 nM and 400 μM. Following an overnight incubation at 4 °C, START domain-FA solutions were loaded in standard Monolith NT.115 Capillaries (NanoTemper Technologies) and measured using a Monolith NT.115 instrument (NanoTemper Technologies). An MST on time of 5 s was used for analysis, and baseline-corrected normalized fluorescence values (ΔFnorm[%]) were plotted against fatty acid concentration. Curves were fit with a nonlinear regression model and Kds are reported in SI Appendix, Table S1 (n = 3 independent measurements, error bars represent the SEM). (D) MST binding assay via titration of stearic acid (6.1 to 200 μM) into Monolith RED-Tris-NTA labeled Them1 START domain (blue) or maltose binding protein (gray) held constant at 50 nM. Procedures and analysis were conducted the same as described in C. (E) Asymmetric unit cell of 3.09-Å structure of Them1 START domain contains four monomers (green), with one monomer bound to myristic acid (cyan). (F) Zoomed-in view of the lipid binding pocket of the START domain where myristic acid (cyan) is modeled into a 2Fo-Fc map (blue mesh) contoured to σ = 1.0. (G) Amino acids in close proximity surrounding palmitic acid. Distance between polar amino acids and carboxyl head group of fatty acid is displayed as black dashed line.
To determine the fatty acid binding affinity, we attempted to use traditional fatty acid binding assays that rely on competitive displacement of a fluorescent probe such as 1-anilinonaphthalene-8-sulfonic acid (1,8-ANS) (26, 27); however, the START domain did not bind to any of the probes tested. Therefore, we developed a fatty acid binding assay using microscale thermophoresis, which detects alterations in fluorescence along a temperature gradient induced by ligand binding to fluorophore-labeled protein (28, 29). We tested the top three fatty acids that copurified most abundantly with the START domain from lipid extracts, namely, palmitoleic acid (16:1), palmitic acid (16:0), and oleic acid (18:1). In contrast to expectations, only palmitic acid generated a binding curve (Fig. 1C). Since palmitic acid is a saturated fatty acid, we also tested myristic acid (14:0) and stearic acid (18:0): two saturated fatty acids that also copurified with the START domain in our MS analysis. Both generated binding curves (Fig. 1C). Binding for fatty acid species occurred in the micromolar (µM) range (SI Appendix, Table S1). To ensure these alterations in thermophoresis were not due to micelle formation at high fatty acid concentrations, we titrated stearic acid (critical micellar concentration ∼300 μM) (30) which is most prone to form micelles, into fluorescently labeled maltose binding protein and observed no changes in thermophoresis (Fig. 1D). These results suggest specific binding of saturated long-chain fatty acid to the Them1 START domain.
There were some discrepancies in the fatty acids identified to bind to the START domain by affinity purification-MS and by microscale thermophoresis techniques (Fig. 1 C and D). This may be explained, at least in part, by differences in presentation of lipids to the START domain depending on the technique: the START domain was exposed to liposomes in our affinity purification-MS experiment, whereas a single fatty acid was titrated into the START domain in the microscale thermophoresis experiment. Since unsaturated fatty acids copurified with the START domain and were detected by MS but did not bind in the microscale thermophoresis experiment, Them1 may have only accessed unsaturated fatty acids in the context of a membrane bilayer. Alternatively, unsaturated fatty acid binding may require the presence of other lipids to serve as intermediate ligands prior to lipid exchange.
Fatty Acids Bind within the Hydrophobic Pocket of Them1’s START Domain.
To visualize the START domain–fatty acid complex, we attempted to generate crystals in the presence of a range of fatty acids; however, only myristic acid yielded crystals that diffracted. The myristic acid–START domain structure was solved in the P 1 21 1 space group to a resolution of 3.09 Å, with the asymmetric unit containing four START domain monomers (Fig. 1E). Refinement and model statistics are summarized in SI Appendix, Table S2.
One subunit contained continuous electron density within the interior of the domain that fit myristic acid (Fig. 1 E and F). A polder map displayed clear density at a sigma level of 3.0, strongly supporting the presence of myristic acid binding at this location (SI Appendix, Fig. S1A) (31). The other three subunits did not contain this continuous electron density; therefore, we suspect either no fatty acid was bound or that fatty acid was bound with low occupancy or high conformational mobility. In this connection, myristic acid exhibited a higher B factor than the average B factor for the protein, suggesting the ligand had some mobility in the pocket (SI Appendix, Table S2). Because we crystallized two different states of the START domain (with and without myristic acid) in one crystal, we compared the structures of these two states using ProSMART analysis (32). The root-mean-square deviation (rmsd) between the apo monomers and the myristic acid-bound monomer were mapped onto the structure of the START domain complexed with fatty acid (SI Appendix, Fig. S1B). There were no major conformational differences between the structures, suggesting fatty acid does not induce an appreciable conformational change; however, our data do not discern whether the apo domains are truly devoid of fatty acid. There were some dissimilarities in the N-terminal α-helix, but these changes are likely an artifact of crystal packing.
We next compared our structure with a structure of the Them1 START domain that was solved at higher resolution and in a different space group (33). Our structure exhibits the same overall conformation with an average rmsd of 0.4 Å over aligned atoms (33). The prior structure contained long, tubular electron density in the same location where we modeled a myristic acid. A buffer component (polyethylene glycol [PEG] molecule) was modeled into this density because no ligand was identified in the crystal structure (SI Appendix, Fig. S1C) (33). We observed a branched electron density characteristic of a fatty acid carboxyl-head group; therefore, we modeled palmitic acid, a highly abundant Escherichia coli fatty acid that copurifies with the START domain, into this density (SI Appendix, Fig. S1D) (34). Palmitic acid fit well within the density, providing strong support for placement of this fortuitously copurified ligand. The palmitic acid carboxyl head group is contacted by polar residues arginine 449, tyrosine 456, and tyrosine 546 (Fig. 1G). The curved fatty acyl chain is enclosed by bulky, nonpolar amino acids, including phenylalanine 426, phenylalanine 488, valine 554, phenylalanine 569, and phenylalanine 573, which fully protect it from solvent (Fig. 1G).
To explain Them1’s unexpected preference for fatty acids, which are smaller-sized lipids than typically bind START domains, we analyzed the lipid binding pockets of all START domains of known structure. These share a long, continuous C-terminal α-helix that packs across the mouth of a U-shaped incomplete β-barrel, forming the empty interior (SI Appendix, Fig. S1E). The conformation of this helix is radically different in Them1, whereby a kink, enabled by a highly conserved glycine 564 and a steric clash from α-helix α0 (connecting the thioesterase domain and the START domain), constricts the lipid binding pocket (33). SI Appendix, Table S3 displays the surface area (Å2) and volume (Å3) of each lipid binding pocket, which are also shown graphically (SI Appendix, Fig. S1F). Them1 possesses a smaller interior cavity than STARD2 and STARD11, which bind to phosphatidylcholine (PC) and ceramide, respectively. All other START domain proteins contain an interior pocket with similar area and volume, although a different shape, when compared with the Them1 START domain. These other START domain proteins have resisted efforts at cocrystallization with their cholesterol and sterol-like ligands (33, 35, 36). Thus, the calculated size of the pocket size may not accurately reflect the ligand-bound state.
We superposed the Them1 START domain with STARD2 bound to palmitoyl-linoleoyl phosphatidylcholine (Protein Data Bank [PDB] code: 1LN3) (37) and STARD11 bound to C16-ceramide (PDB code: 2E3P) (37, 38). In both instances, the interior cavity of the Them1 START domain was unable to accommodate the same ligands (SI Appendix, Fig. S1G). The Them1 START domain contains some equivalent structural features that enable STARD2 and STARD11 to bind to their respective ligands, such as arginine 449 (STARD2 R78 and STARD11 R442), which electrostatically interacts with the phosphate of phospholipids in STARD2 (37) and a water-mediated hydrogen bond with a ceramide hydroxyl in STARD11 (37, 38). Additionally, these START domains contain an acidic residue (Them1 D453, STARD2 D82, STARD11 E446) that participates in a salt bridge with the conserved arginine and engages in hydrogen bonding with the amide-nitrogen and hydroxyl of ceramide in STARD11 (38). However, Them1 lacks the aromatic cage found in STARD2 that consists of tryptophan 101, tyrosine 114, and tyrosine 155, which together engage in cation–π interactions with the quaternary amine of choline (37). Them1 only contains one structurally analogous aromatic residue (F488), although it also contains tyrosine 456 that could rotate and potentially occupy the same space as tryptophan 101 in STARD2 (SI Appendix, Fig. S1H). Additionally, Them1 does not conserve residues found in STARD11 (Y482, Q467, and N504) that participate in hydrogen bonding with ceramide (38). Them1 lacks some necessary residues for recognition of the larger lipids present in STARD2 and STARD11, while coopting residues such as R449 and D453 to enable fatty acid binding.
Them1 START Domain Binds to Lysophosphatidylcholine.
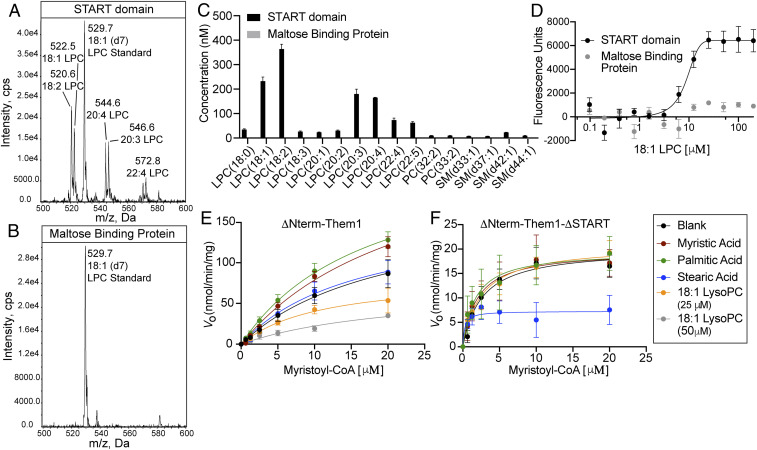
To test whether long-chain fatty acids were the only ligands for the START domain of Them1, we repeated our affinity purification-mass spectrometry experiment using a shotgun MS approach that examined all major phospholipid classes. Several LPC species, including 18:2, 18:1, 20:4, 20:3, and 22:4, were highly enriched in our START domain samples, but absent in our negative control samples (Fig. 2 A–C). Them1 START domain samples were not enriched for any phosphatidylethanolamine, phosphatidylserine, or phosphatidylinositol species. Some sphingomyelin and phosphatidylcholine species were enriched in the START domain samples at low levels.
Fig. 2.
Fatty acids enhance while lysophosphatidylcholine (LPC) inhibits Them1 activity in a START domain-dependent manner. (A and B) Mass spectra intensity counts per second (cps) (precursor ion scan of m/z 184) of lipids containing a phosphatidylcholine head group that copurified with the Them1 START domain (A) and maltose binding protein (B). LPC 18:1 (d7) standard was added to each sample for quantification of lipid concentrations. (C) Graphical analysis of identified LPC species. Bars are an average of three technical replicates. LPC species were not detected in maltose binding protein samples. (D) 18:1 LPC binding assay measuring ligand-induced photo-enhancement of Cy5-labeled Them1 START domain fluorescence (black). Them1 START domain (black) and maltose binding protein (gray) covalently labeled with a Cy5 fluophore were kept constant at 100 nM, while the concentration of 18:1 LPC was varied between 97 nM and 200 μM. Fluorescence was measured using a Dianthus NT.23 Pico (NanoTemper Technologies). Fluorescence values were baseline corrected and plotted against 18:1 LPC concentration. Curve for Them1 START domain was fit with a nonlinear regression model. Kd is reported in SI Appendix, Table S1 (n = 3 independent measurements, error bars represent the SEM). (E and F) Thioesterase activity of ΔNterm-Them1 (E) and Δ Nterm-Them1_ΔSTART (F) (1 μM) to hydrolyze myristoyl-CoA after incubation with buffer (black) or 25 μM stearic acid (blue), palmitic acid (green), myristic acid (red), 18:1 LPC (orange), and 50 μM 18:1 LPC (gray) for 30 min at 37 °C. Saturation curves of V0 plotted against increasing myrstoyl-CoA with solid lines indicating nonlinear analysis of the data. Each point corresponds to the average of a minimum of three replicates. Error bars represent SEM.
To probe the affinity of the START domain for LPC, we titrated 18:1 LPC into a constant amount of fluorescently labeled Them1 START domain and measured ligand-induced alterations in fluorescence. The 18:1 LPC binding to the START domain greatly enhances its fluorescence, presumably through LPC shielding the fluorophore from quenching in the local environment (39), allowing us to track ligand binding (Fig. 2D). The START domain bound to 18:1 LPC with an affinity of 8.5 μM, similar to its affinity to stearic acid (Fig. 2D and SI Appendix, Table S1). To ensure this alteration in fluorescence was due to specific binding, we titrated 18:1 LPC into fluorescently labeled maltose binding protein as a negative control and observed no ligand-induced changes in fluorescence (Fig. 2D).
LPC contains a single fatty acyl chain typically esterified at the sn-1 position rather than the two fatty acyl chains present in phospholipids. It is an important signaling molecule that is implicated in the pathogenesis of cardiovascular disease, atherosclerosis, diabetes, and neurodegenerative diseases (40). LPC also functions in lipid droplet formation because it is a precursor, along with fatty acyl-CoAs, for phosphatidylcholine molecules that are required to expand the membrane monolayer that coats lipid droplet membranes (41). Upon stimulation of thermogenesis, levels of saturated LPC in brown adipocytes levels dramatically increase in brown adipocytes, which interestingly enhances thermogenesis (42).
Fatty Acids Enhance while 18:1 LPC Inhibits Them1 Acyl-CoA Thioesterase Activity.
Since the START domain was previously shown to alter the enzymatic activity of Them1 (7), we reasoned fatty acids and LPC species may regulate Acot activity through interaction with the START domain. To test this, we monitored myristoyl-CoA hydrolysis in the presence of fatty acid or 18:1 LPC. Incubation with either 25 μM myristic acid or palmitic acid enhanced the maximum enzymatic velocity of ΔNterm-Them1 lacking the intrinsically disordered N terminus (Fig. 2E, red and green). This enhancement in activity is dependent upon the START domain (Fig. 2F). Incubation with stearic acid did not alter the activity of ΔNterm-Them1; however, stearic acid suppressed enzymatic activity of Them1 when the START domain was absent, indicating the START domain relieves the inhibitory effects of stearic acid (Fig. 2 E and F, blue).
Surprisingly, incubation with 25 μM 18:1 LPC greatly inhibited ΔNterm-Them1 activity (Fig. 2E, orange) in a START domain-dependent manner (Fig. 2F). To determine whether this inhibition was dose dependent, we incubated ΔNterm-Them1 with twice the concentration of 18:1 LPC (50 μM) and observed greater inhibition (Fig. 2E, gray).
Them1 Forms Homotrimer Containing a Thioesterase Domain Core Flanked by Mobile START Domains.
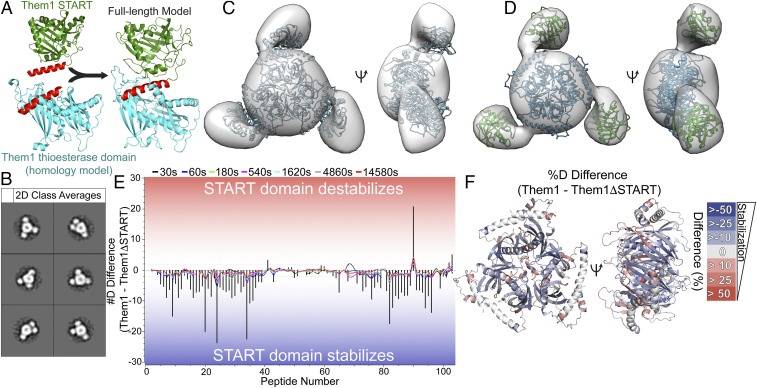
To understand how the START domain interacts with the thioesterase domains to influence catalytic activity, we generated a model of full-length Them1 by joining the START domain structure with a homology model of the Them1 thioesterase domains created using the SWISS MODEL server (43–47) based on the structure of the ACOT12 thioesterase domains (56% sequence identity and 69% sequence similarity) (48). Both structures contained a common α-helix that resides at the C terminus of the thioesterase domain model and N terminus of the START domain structure. We aligned this overlapping α-helix to generate a full-length model of Them1 with similarities to a previously reported model of intact ACOT12 (Fig. 3A) (48).
Fig. 3.
Them1 forms homotrimer with thioesterase domain core and flanking START domains. (A) Generation of full-length Them1 model through structural alignment of linker helix (red) present in crystal structure of Them1 START domain (green) and homology model of Them1 thioesterase domains (blue). (B) Subset of 2D class averages of Them1 using negative stain electron microscopy generated by Relion 3.0. (C) 3D reconstruction of Them1 derived from subset of 2D class averages. Trimeric Them1 model fit into 3D reconstruction using Chimera. (D) Separate START domains (green) and thioesterase domains (blue) modeld into 3D reconstruction using Chimera. (E) Butterfly plot overlayed with a residual plot displaying difference in deuterium uptake between ΔNterm-Them1 and ΔNterm-Them1_ΔSTART. Colored lines depict deuterium uptake difference (y axis) for peptides (x axis) at each time point (black: 30 s, blue: 60 s, green: 180 s, pink: 540 s, gray: 1,620 s, cyan: 4,860 s, and red: 14,580 s). Bars display summed difference in deuterium uptake over all time points for each peptide. Negative values (blue) mean that removal of the START domain increases the incorporation of deuterium for the cooresponding peptide. (F) Percentage difference in deuterium uptake (Them1 – Them1_ΔSTART) at 60 s mapped onto homology model of Them1 thioesterase domains. Negative values (blue) indicate less deuterium exchange (greater protection) in intact Them1, indicating that the START domain stabilizes the thioesterase domains.
We used single-particle negative stain electron microscopy to obtain a low-resolution map of Them1 to fit our structural model. ΔNterm-Them1 was purified as a stable trimer as determined by size exclusion chromatography (SEC) and analytical ultracentrifugation (SI Appendix, Fig. S2 A and B). Negative stain electron microscopy revealed a homogenous distribution of ΔNterm-Them1 trigonal particles with a diameter of 17 nm spread across the grid (SI Appendix, Fig. S2C). We generated two-dimensional (2D) classifications using Relion 3.0 (49) that revealed a trimeric complex consisting of a large spherical body flanked by three protruding lobes representing threefold symmetry (Fig. 3B, all class averages in SI Appendix, Fig. S2D). A three-dimensional (3D) initial model was generated and refined using particles from selected class averages that revealed the trimeric complex (Fig. 3C). The central density of the 3D reconstruction accommodates the core heterotrimeric thioesterase domains; however, fitting the START domain within the peripheral density required relaxing the linker geometry of our model and independently fitting this domain (Fig. 3 C and D).
The negative stain 2D class averages also revealed conformational flexibility between the START domain and thioesterase domains. The main central body of the map that accommodates the thioesterase domains is identical in all 2D class averages; however, the three exterior lobes that accommodate the START domains are not perfectly arranged in a threefold symmetric frame in a few of the 2D class averages as seen in reprojections using C3 symmetry (SI Appendix, Fig. S2E). We postulate this flexibility is the result of a less ordered region lying between the domains (residues 365 to 383, Homo sapiens), which was also disordered in our crystal structure of the START domain.
Them1 START Domain Stabilizes the Thioesterase Domains.
To determine how the thioesterase and START domains interact, we performed hydrogen-deuterium exchange-MS (HDX-MS). This technique identifies regions of flexibility and rigidity by measuring the rate of exchange of deuterium with amide protons; high deuterium uptake signifies areas of flexibility and high solvent exposure, whereas low deuterium uptake signifies areas of rigidity and low solvent exposure (50). Comparison of deuterium uptake of ΔNterm-Them1 with ΔNterm-Them1-ΔSTART reveals a dramatic stabilizing effect driven by the START domain, as evidenced by a great reduction of deuterium incorporation throughout the thioesterase domains when the START domain is present (Fig. 3 E and F). Heat maps showing identified peptides for each construct are provided in SI Appendix, Fig. S3. Although there is flexibility between the thioesterase and START domains, the START domain significantly stabilizes the thioesterase domains.
Fatty Acids Stabilize while 18:1 LPC Destabilizes START Domain.
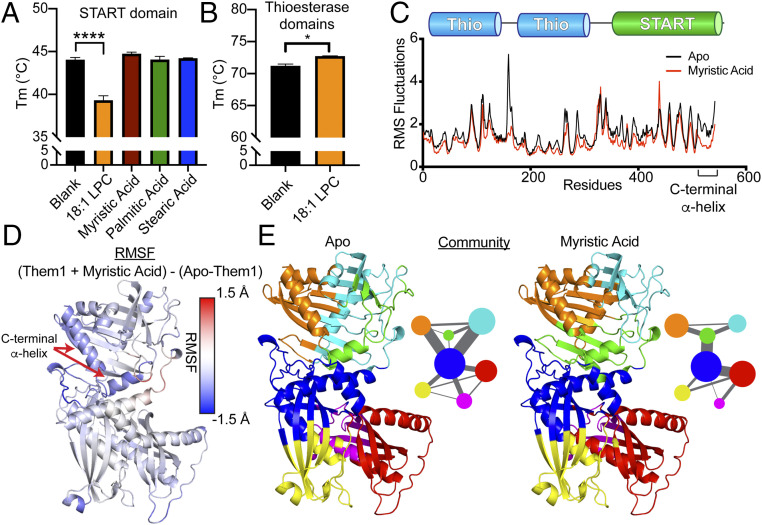
Incubation with myristic acid, palmitic acid, or stearic acid did not alter the thermal melting temperature (Tm) of the START domain, as monitored by differential scanning fluorimetry (DSF); however, this is relative to the START domain that copurifies with fatty acids from E. coli (Fig. 4A). Strikingly, 18:1 LPC destabilized the START domain by nearly 5 °C, which is in line with the reduced catalytic activity driven by this ligand (Fig. 4A). To ensure this destabilizing effect was specific for the START domain, we also tested the effect of 18:1 LPC on the thioesterase domains alone. The thioesterase domains were not destabilized, but slightly stabilized by the presence of 18:1 LPC, verifying LPC specifically destabilizes the START domain (Fig. 4B).
Fig. 4.
Fatty acids stabilize while 18:1 LPC destabilizes the START domain. (A and B) Differential scanning fluorimetry of START domain (A) and thioesterase domains (B) incubated with either buffer (black), 18:1 LPC (orange), myristic acid (red), palmitic acid (green), or stearic acid (blue). LPC 18:1 significantly destabilizes the START domain, while slightly stabilizing the thioesterase domains. Bars depict average of three replicates. Error bars denote SEM. One-way ANOVA and Dunnett’s multiple comparisons test were used to analyze START domain data (****P < 0.0001). Paired t test used to analyze thioesterase domains data (*P < 0.05). (C–E) Molecular dynamics simulation for apo-Them1 and myristic acid-bound Them1 over 500 ns. (C) Root-mean-square fluctuations (rmsfs) across Them1 residues for the apo (black) and myristic acid-bound (red) states. (D) Color coordinated difference in rmsfs between myristic acid-bound and apo states (Them1-MYR-Them1-Apo) mapped onto a full-length model of Them1. Myristic acid stabilizes the C-terminal α-helix; blue color corresponds to Lower rmsfs in myrstic-bound state than apo state. (E) Community analysis that identifies residues that move in coordinated fashion throughout the simulation. Circle size depicts number of residues within community and width of lines corresponds with strength of communication between communities. Myristic acid alters size and connectedness of communities.
To determine how fatty acids enhanced Them1 activity, we performed 500-ns molecular dynamics simulations on Them1 comparing apo vs. lipid-bound Them1. The presence of myristic acid substantially lowered the root-mean-square fluctuations in the START domain and in some parts of the thioesterase domains, suggesting myristic acid generated a more stable complex than apo-Them1 (Fig. 4 C and D). The C-terminal α-helix of the START domain, which plays a role in other START domains binding lipids (51, 52), was significantly stabilized by myristic acid (Fig. 4 C and D). Next, we performed a community analysis, which identifies groups of residues that move in a coordinated manner throughout the simulation. Myristic acid significantly altered the communities within the START domain, changing their size and connectedness (Fig. 4E). In the apo state, multiple communities interface with the blue community of the thioesterase domains, but myristic acid shifts the communities so that only one is connected to the thioesterase domains (Fig. 4E). Although there are fewer connections between the communities in the myristic-bound state, there are stronger connections linking the communities in the START domain (green) with communities within the thioesterase domains where acyl-CoA hydrolysis occurs (blue-red), potentially yielding a more active enzyme (Fig. 4E). Taken together, these data suggest fatty acids allosterically enhance Acot activity through stabilizing the START domain and altering dynamics within the thioesterase domains, while 18:1 LPC inhibits Acot activity through destabilizing the START domain.
LPC 18:1 Reverses Them1-Mediated Suppression of Fatty Acid Oxidation.
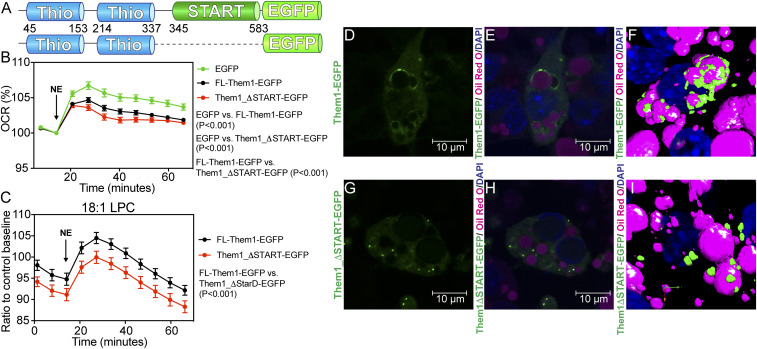
To test the effect of the START domain on Them1’s capacity to suppress thermogenesis in live cells, we measured the oxygen consumption rate (OCR) of mouse-derived immortalized brown adipocytes (iBAs) that were transduced with EGFP alone, EGFP-tagged full-length Them1 (FL-Them1-EGFP), or an EGFP-tagged truncated variant containing only the N-terminal thioesterase domains (Them1_ΔSTART-EGFP) (Fig. 5 A and B). We induced thermogenesis in the iBAs using norepinephrine (NE), which increased the OCR over basal levels (Fig. 5B, green). As expected, Them1 suppressed NE-induced respiration (Fig. 5B, black). However, removal of the START domain did not reduce Them1-mediated suppression of OCR values, but slightly enhanced Them1 activity (Fig. 5B, red). The START domain in part attenuated Them1 activity, suggesting a role for feedback regulation by the START domain.
Fig. 5.
Them1 START domain regulates Them1 activity and localization in brown adipocytes. (A) Schematic of adenovirus constructs of full-length Them1 (residues 1 to 594, Top) and Them1_ΔSTART (residues 1 to 344, Bottom) with C-terminal EGFP tags. (B) OCR of iBAs following stimulation with 1 μM norepinephrine (NE). The iBAs were transduced with Ad-FL-Them1-EGFP (black), Ad-Them1_ΔSTART-EGFP (red), and Ad-EGFP (green). OCR values were normalized by the number of live cell nuclei and displayed as a percentage relative to the basal OCR. Graph shows combined data from three independent experiments. Statistical analyses were conducted via two-way ANOVA with Tukey’s correction. P < 0.001, between EGFP versus FL-Them1-EGFP, P < 0.001, between EGFP versus Them1_ΔSTART-EGFP, P < 0.001, between FL-Them1-EGFP versus Them1_ΔSTART-EGFP. (C) OCR of iBAs transduced with Ad-FL-Them1-EGFP (black) or Ad-Them1_ΔSTART-EGFP (red) following stimulation with 1 μM NE. Cells were incubated with 25 μM LPC 18:1 or control buffer for 1 h prior to the start of experiment. OCR values were normalized by the number of live cell nuclei and displayed as ratios relative to the control baseline OCR for each genetic background. Graphs show combined data from three independent experiments. Statistical analyses were conducted via two-way ANOVA with Tukey’s correction. P < 0.0001. (D–I) Confocal fluorescence microscopy of iBAs cells reconstituted with Ad-FL-Them1-EGFP (D–F) and Ad-Them1_ΔSTART-EGFP (G–I). Lipid droplets and nuceli were visualized through staining with Oil Red O and DAPI, respectively. FL-Them1-EGFP localized with lipid droplets, while Them1_ΔSTART-EGFP did not associate with lipid droplets. (F and I) 3D rendering of confocal fluorescence microscopy images.
It was recently reported that LPC levels, specifically 16:0 and 18:0, were elevated in brown adipocytes upon induction of thermogenesis, and that 16:0 LPC enhanced UCP1-mediated respiration (42). Since we identified that 18:1 LPC inhibits Them1 through the START domain (Fig. 2 E and F), we hypothesized that 18:1 LPC regulates thermogenesis in brown adipocytes through interaction with Them1. To test this, we incubated iBAs transduced with FL-Them1-EGFP or Them1_ΔSTART-EGFP with 25 μM 18:1 LPC for 1 h prior to measuring NE-induced respiration. In line with 18:1 LPC inhibition of Them1 through the START domain, respiration of iBAs transduced with FL-Them1-EGFP was enhanced in the presence of 18:1 LPC relative to Them1_ΔSTART-EGFP (Fig. 5C).
Them1 START Domain Is Necessary for Localization to the Lipid Droplet.
To elucidate whether the START domain contributes to Them1 function by altering cellular localization, we visualized Them1 in iBAs stably transduced with the same viral Them1 constructs as above (FL-Them1-EGFP and Them1_ΔSTART-EGFP). FL-Them1-EGFP was primarily localized in puncta near the lipid droplet surface (Fig. 5 D–F), as was previously shown by Li et al. (53). Removal of the START domain disrupted lipid droplet localization and led to puncta dispersed throughout the cytosol (Fig. 5 G–I). Them1 suppresses thermogenesis in this condensed form, but phosphorylation of the N terminus disperses Them1 within the cell, which abrogates Them1-mediated inhibition of thermogenesis (53). To test whether the START domain alters this phosphorylation-mediated dissolution of Them1, we treated cells expressing just the thioesterase domains (Them1_ΔSTART-EGFP) with phorbol 12-myristate 13-acetate (PMA), a PKC activator that leads to Them1 phosphorylation (53). After a 4-h treatment with PMA, Them1 was diffuse, demonstrating that the START domain is not essential for this process (SI Appendix, Fig. S4 A and B). These data reveal a spatiotemporal role for the Them1 START domain, whereby the START domain is necessary for positioning Them1 puncta near the lipid droplet. However, this process is distinct from the phosphorylation-regulated dynamics between puncta and diffuse Them1 that is critical for the suppression of thermogenesis.
Discussion
Them1 suppresses thermogenesis in BAT, limiting its capacity to oxidize endogenous fatty acids (6, 8). Whereas we previously demonstrated that the C-terminal START domain is necessary for full catalytic activity of the enzyme (7), the current study elucidates the multifunctional role of the START domain to act as a lipid sensor to allosterically regulate Them1 activity and spatially localize Them1 near the lipid droplet.
We identified that both long-chain fatty acids and 18:1 LPC bind to the START domain and inversely alter Them1 stability and activity, establishing the START domain as a sensor that has evolved to bind specific lipids to tune enzymatic activity. The allosteric enhancement of activity by myristic and palmitic acids through a feedforward mechanism could drive Them1’s preference to hydrolyze myristoyl-CoA and palmitoyl-CoA (7), distinguishing it from other thioesterases. When taken together with phosphorylation-dependent cellular dispersion of Them1 (53), the observation that 18:1 LPC is an allosteric inhibitor suggests multiple mechanisms have evolved to suppress Them1 activity in order to enhance thermogenesis. One study showed that induction of thermogenesis in brown adipocytes increased levels of 16:0 LPC, but decreased levels of 18:1 LPC (42). However, another study showed that 18:1 LPC levels were increased in browning white adipose tissue of mice treated with a β3-adrenergic agonist (54). Them1 potentially senses the nutritional state of these cells through the START domain and regulates its activity to conserve or oxidize lipids. Additionally, 18:1 LPC may spatiotemporally regulate Them1 activity to control lipid droplet membrane development. Since Them1 is localized to the lipid droplet surface, it could interfere with lipid droplet monolayer formation by hydrolyzing fatty acyl-CoAs, which are the substrates used by LPCAT2 to generate phosphatidylcholines for lipid droplet expansion (41). However, this could be prevented through inhibition by LPC, which would be localized at the lipid droplet and is utilized by LPCAT2 to produce phosphatidylcholines.
Our data suggest the mechanism by which fatty acids enhance and 18:1 LPC inhibits Them1 activity is by differential stabilization of the START domain. The C-terminal α-helix of the START domain is a gate for ligand binding, remaining unfolded in the apo state, and folding and encapsulating the pocket once ligand binds (51, 52). Our molecular dynamic simulations showed myristic acid stabilized this helix, which is apposed to the thioesterase domains. We expect that only specific ligands (myristic acid and palmitic acid) stabilize this helix to enhance activity, while larger ligands, such as stearic acid and 18:1 LPC, prevent this stabilization through steric clash, leading to no enhancement of activity or significant inhibition of activity, respectively. Phospholipids are imperfect pharmacological tools due to their poor pharmacokinetics; however, these findings should aid in the development of small molecule allosteric inhibitors that enhance metabolism. Screening for compounds that destabilize the START domain and in turn inhibit Them1 activity could yield Them1-selective pharmacological tools to treat obesity and related metabolic disorders.
Our confocal fluorescence studies showed the START domain was responsible for localizing Them1 near the lipid droplet in brown adipocytes at a basal state. The START domain of Them1 could directly interact with the lipid droplet membrane or engage in protein–protein interactions at the lipid droplet surface. Previously it was shown that Them1 associated with phosphatidyl inositol-4-phosphate (PIP) through the START domain in a protein–lipid overlay assay, suggesting the START domain of Them1 is capable of directly interacting with a membrane surface (55). There were no PIP species that copurified with the START domain in our affinity purification-MS technique, which would only detect high-affinity ligands that remain bound through several washing steps; therefore, the START domain may engage in a low-affinity interaction with PIP. Recently, it was shown that PIPs are present on the lipid droplet surface, which could potentially explain Them1 localization (56). In our fitted model of Them1 in the low-resolution negative stain map, the START domains are positioned on the exterior of the trimeric complex, where the START domains could cooperatively bind to the membrane surface, anchoring Them1 puncta to the lipid droplet. Although this localization was shown to be dispensable for Them1-mediated suppression of thermogenesis, the START domain of Them1 could perform other functions at the lipid droplet. For instance, many START domains are involved in transporting specific lipids to cellular compartments; however, these possibilities remain to be explored (16, 18, 22).
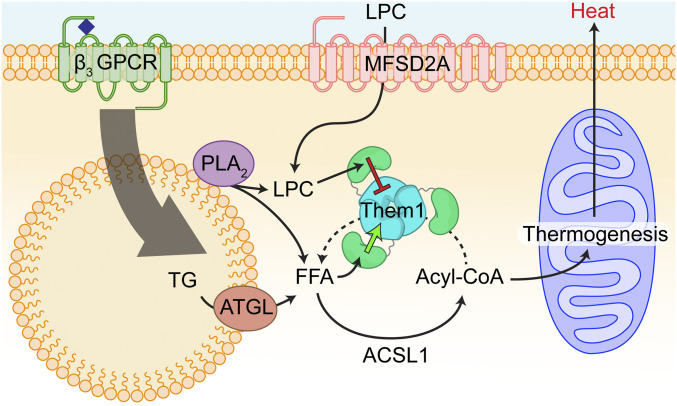
In order to properly traffic acyl-CoAs into specific pathways to drive metabolism, the localization and activity of multiple acyl-CoA thioesterases and synthetases must be controlled (57). The START domain of Them1 allows for fine tuning of Them1 function, regulating both activity and localization (Fig. 6). This control is necessary for correct metabolism in BAT, enabling thermogenesis or preserving resources when needed.
Fig. 6.
Schematic of allosteric regulation of Them1 activity in brown adipocytes. Thermogenesis is activated in brown adipocytes upon adrenergic stimulation of the β3 GPCR. This activates the release of fatty acids from lipid droplets, which are converted into acyl-CoA by ACSL1, and subsequently imported into the mitochondria to fuel thermogenesis. Them1 antagonizes this process by hydrolyzing acyl-CoA into free fatty acids (FFAs) at the surface of the lipid droplet. Select fatty acids (myristic acid and palmitic acid) drive a feed-forward mechanism to enhance Them1 hydrolysis of acyl-CoA through interaction with the START domain (green arrow). LPC levels are elevated in brown adipocytes upon adrenergic stimulation, potentially through the breakdown of phosphatidylcholine by Phospholipase A2 (PLA2) or increased import of LPC from plasma via major facilitator superfamily domain-containing protein 2 (MFSD2A). The 18:1 LPC inhibits Them1 acyl-CoA hydrolysis through interaction with the START domain (red arrow), thus driving forward thermogenesis.
Materials and Methods
Crystallization.
The pure H. sapiens Them1 START domain was incubated with myristic acid in 10-fold excess and concentrated to 10 mg mL−1 in 30 mM Hepes pH 7.5, 300 mM NaCl, 10% glycerol, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP). Crystals of the START domain were grown via hanging drop vapor diffusion at 4 °C from solutions containing 1 μL START domain and 1 μL mother liquor (0.1 M Tris HCl pH 9.4 and 27% PEG 8000).
Negative Stain Electron Microscopy.
Freshly purified Mus musculus ΔNterm-Them1 (residues 43 to 365) after SEC into a buffer containing 30 mM Hepes pH 7.5, 300 mM NaCl, and 0.5 mM TCEP was used for negative staining. Briefly, 4 μL of ΔNterm-Them1 was adsorbed on a carbon coated Cu-400 mesh grid (TedPella) for 1 min and excess liquid was blotted with Whatman filter paper 4, washed twice with a 20-μL drop of water, and stained with 0.75% uranyl formate for 30 s, blotted, and air dried. Negatively stained Them1 was imaged on a Talos 120 C microscope operating at 120 kV with a Lab6 cathode at pixel size of 1.56 Å. Micrographs were recorded at low-dose condition on a Ceta 16M camera (Thermo Fisher).
Acyl-CoA Thioesterase Activity Assay.
Lipids dissolved in ethanol were incubated with purified M. musculus ΔNterm-Them1 or thioesterase domains and 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) or 30 min at 37 °C in assay buffer containing 30 mM Hepes pH 7.5, 150 mM NaCl, and 5% glycerol. Myristoyl-CoA was added to a protein–DTNB–lipid mixture to initiate reaction in a total reaction volume of 200 μL/well, with final protein concentration at 1 μM, DTNB at 300 μM, lipid at 25 μM or 50 μM, and myristoyl-CoA ranging from 0 to 20 μM. Absorbance readings at 412 nm were read every 10 s for 1 h. The initial velocities were plotted against substrate concentrations and fitted with the Michaelis–Menten equation to yield the maximum velocity (Vmax) and Km (the Michaelis constant) using GraphPad Prism 8.0. Each experiment was conducted with two technical replicates for each sample and repeated three times.
Full methods are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Peter E. Prevelige (University of Alabama at Birmingham) for his assistance with HDX-MS, Dr. Pete Lollar and Dr. Anamika Patel (Emory University) for their assistance with analytical ultracentrifugation, and Dr. Bruce Spiegelman (Dana-Farber Cancer Institute at Harvard Medical School) for providing the iBA cells. This work was supported by the NIH (R01 DK 103046 to D.E.C., S.J.H., and E.A.O.). M.C.T. was funded by the T32 GM008602 NIH Pharmacology Training Grant. Crystallographic data were collected at Southeast Regional Collaborative Access Team 22-ID Beamline at the Advanced Photon Source, Argonne National Laboratory and supported by the Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31–109-Eng-38. This study was supported in part by the Emory Integrated Lipidomics Core, which is subsidized by the Emory University School of Medicine, and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical and Translational Science Alliance of the NIH, Award UL1TR002378. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003877117/-/DCSupplemental.
Data Accessibility.
Structure coordinates, diffraction data, and negative stain map have been deposited in the PDB/Electron Microscopy Data Bank (EMDB) under accession codes PDB ID: 6VVQ andEMD ID: 21414.
References
- 1.Smith R. E., Horwitz B. A., Brown fat and thermogenesis. Physiol. Rev. 49, 330–425 (1969). [DOI] [PubMed] [Google Scholar]
- 2.Cypess A. M. et al., Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cypess A. M., Kahn C. R., Brown fat as a therapy for obesity and diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 17, 143–149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricquier D., Mory G., Bouillaud F., Thibault J., Weissenbach J., Rapid increase of mitochondrial uncoupling protein and its mRNA in stimulated brown adipose tissue. Use of a cDNA probe. FEBS Lett. 178, 240–244 (1984). [DOI] [PubMed] [Google Scholar]
- 5.Adams S. H. et al., BFIT, a unique acyl-CoA thioesterase induced in thermogenic brown adipose tissue: Cloning, organization of the human gene and assessment of a potential link to obesity. Biochem. J. 360, 135–142 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y. et al., Targeted deletion of thioesterase superfamily member 1 promotes energy expenditure and protects against obesity and insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 5417–5422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S., Cohen D. E., Functional characterization of thioesterase superfamily member 1/acyl-CoA thioesterase 11: Implications for metabolic regulation. J. Lipid Res. 53, 2620–2631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada K. et al., Thioesterase superfamily member 1 suppresses cold thermogenesis by limiting the oxidation of lipid droplet-derived fatty acids in brown adipose tissue. Mol. Metab. 5, 340–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schrick K., Nguyen D., Karlowski W. M., Mayer K. F., START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5, R41 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soccio R. E., Breslow J. L., StAR-related lipid transfer (START) proteins: Mediators of intracellular lipid metabolism. J. Biol. Chem. 278, 22183–22186 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Ponting C. P., Aravind L., START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24, 130–132 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Alpy F., Legueux F., Bianchetti L., Tomasetto C., [START domain-containing proteins: A review of their role in lipid transport and exchange]. Med. Sci. (Paris) 25, 181–191 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Reitz J., Gehrig-Burger K., Strauss J. F. 3rd, Gimpl G., Cholesterol interaction with the related steroidogenic acute regulatory lipid-transfer (START) domains of StAR (STARD1) and MLN64 (STARD3). FEBS J. 275, 1790–1802 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Agudo D. et al., Human StarD5, a cytosolic StAR-related lipid binding protein. J. Lipid Res. 46, 1615–1623 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Létourneau D. et al., STARD6 on steroids: Solution structure, multiple timescale backbone dynamics and ligand binding mechanism. Sci. Rep. 6, 28486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horibata Y., Sugimoto H., StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. J. Biol. Chem. 285, 7358–7365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanno K. et al., Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J. Biol. Chem. 282, 30728–30736 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Olayioye M. A. et al., StarD10, a START domain protein overexpressed in breast cancer, functions as a phospholipid transfer protein. J. Biol. Chem. 280, 27436–27442 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Hanada K., Kumagai K., Tomishige N., Kawano M., CERT and intracellular trafficking of ceramide. Biochim. Biophys. Acta 1771, 644–653 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Alpy F. et al., The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein. J. Biol. Chem. 276, 4261–4269 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Lin D. et al., Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science 267, 1828–1831 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Yang L. et al., The phosphatidylcholine transfer protein Stard7 is required for mitochondrial and epithelial cell homeostasis. Sci. Rep. 7, 46416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei J., Kang H. W., Cohen D. E., Thioesterase superfamily member 2 (Them2)/acyl-CoA thioesterase 13 (Acot13): A homotetrameric hotdog fold thioesterase with selectivity for long-chain fatty acyl-CoAs. Biochem. J. 421, 311–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang H. W., Niepel M. W., Han S., Kawano Y., Cohen D. E., Thioesterase superfamily member 2/acyl-CoA thioesterase 13 (Them2/Acot13) regulates hepatic lipid and glucose metabolism. FASEB J. 26, 2209–2221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cajka T., Fiehn O., Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 61, 192–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastukhov A. V., Ropson I. J., Fluorescent dyes as probes to study lipid-binding proteins. Proteins 53, 607–615 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Norris A. W., Spector A. A., Very long chain n-3 and n-6 polyunsaturated fatty acids bind strongly to liver fatty acid-binding protein. J. Lipid Res. 43, 646–653 (2002). [PubMed] [Google Scholar]
- 28.Duhr S., Braun D., Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 103, 19678–19682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seidel S. A. et al., Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukerjee P., Mysels K. J., Critical Micelle Concentrations of Surfactant Systems, (Nat. Stand. Ref. Data Ser., NSRDS-NBS 36, National Bureau of Standards, 1971). [Google Scholar]
- 31.Liebschner D. et al., Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D Struct. Biol. 73, 148–157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholls R. A., Fischer M., McNicholas S., Murshudov G. N., Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D Biol. Crystallogr. 70, 2487–2499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorsell A. G. et al., Comparative structural analysis of lipid binding START domains. PLoS One 6, e19521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw M. K., Ingraham J. L., Fatty acid composition of Escherichia coli as a possible controlling factor of the minimal growth temperature. J. Bacteriol. 90, 141–146 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath M. P. et al., Structure of the lutein-binding domain of human StARD3 at 1.74 Å resolution and model of a complex with lutein. Acta Crystallogr. F Struct. Biol. Commun. 72, 609–618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan L., Tong J., Chun C., Im Y. J., Structural analysis of human sterol transfer protein STARD4. Biochem. Biophys. Res. Commun. 520, 466–472 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Roderick S. L. et al., Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 9, 507–511 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Kudo N. et al., Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. U.S.A. 105, 488–493 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee E., Shim S. H., Cho M., Fluorescence enhancement of a ligand-activated fluorescent protein induced by collective noncovalent interactions. Chem. Sci. (Camb.) 9, 8325–8336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law S. H. et al., An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. 20, E1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cotte A. K. et al., Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat. Commun. 9, 322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer S. et al., The lipidome of primary murine white, brite, and brown adipocytes-Impact of beta-adrenergic stimulation. PLoS Biol. 17, e3000412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waterhouse A. et al., SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bienert S. et al., The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guex N., Peitsch M. C., Schwede T., Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30 (suppl. 1), S162–S173 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Benkert P., Biasini M., Schwede T., Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T., Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 7, 10480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swarbrick C. M. et al., Structural basis for regulation of the human acetyl-CoA thioesterase 12 and interactions with the steroidogenic acute regulatory protein-related lipid transfer (START) domain. J. Biol. Chem. 289, 24263–24274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zivanov J. et al., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chalmers M. J. et al., Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Roostaee A., Barbar E., Lavigne P., LeHoux J. G., The mechanism of specific binding of free cholesterol by the steroidogenic acute regulatory protein: Evidence for a role of the C-terminal alpha-helix in the gating of the binding site. Biosci. Rep. 29, 89–101 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Roostaee A., Barbar E., Lehoux J. G., Lavigne P., Cholesterol binding is a prerequisite for the activity of the steroidogenic acute regulatory protein (StAR). Biochem. J. 412, 553–562 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Li Y., et al. , Thioesterase superfamily member 1 undergoes stimulus-coupled reorganization to regulate metabolism 10.1101/2020.02.18.954818 (19 February 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He P. et al., Lipid profiling reveals browning heterogeneity of white adipose tissue by Β3-Adrenergic stimulation. Biomolecules 9, E444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen D. et al., Human brown fat inducible thioesterase variant 2 cellular localization and catalytic function. Biochemistry 51, 6990–6999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du X. et al., ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 219, e201905162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cooper D. E., Young P. A., Klett E. L., Coleman R. A., Physiological consequences of compartmentalized acyl-CoA metabolism. J. Biol. Chem. 290, 20023–20031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates, diffraction data, and negative stain map have been deposited in the PDB/Electron Microscopy Data Bank (EMDB) under accession codes PDB ID: 6VVQ andEMD ID: 21414.