Abstract
Background
Antimicrobial resistance (AMR) is a major challenge in the treatment of infections caused by Pseudomonas aeruginosa. Highly drug-resistant infections are disproportionally caused by a small subset of globally distributed P. aeruginosa sequence types (STs), termed “high-risk clones.” We noted that clonal complex (CC) 446 (which includes STs 298 and 446) isolates were repeatedly cultured at 1 medical center and asked whether this lineage might constitute an emerging high-risk clone.
Methods
We searched P. aeruginosa genomes from collections available from several institutions and from a public database for the presence of CC446 isolates. We determined antibacterial susceptibility using microbroth dilution and examined genome sequences to characterize the population structure of CC446 and investigate the genetic basis of AMR.
Results
CC446 was globally distributed over 5 continents. CC446 isolates demonstrated high rates of AMR, with 51.9% (28/54) being multidrug-resistant (MDR) and 53.6% of these (15/28) being extensively drug-resistant (XDR). Phylogenetic analysis revealed that most MDR/XDR isolates belonged to a subclade of ST298 (designated ST298*) of which 100% (21/21) were MDR and 61.9% (13/21) were XDR. XDR ST298* was identified repeatedly and consistently at a single academic medical center from 2001 through 2017. These isolates harbored a large plasmid that carries a novel antibiotic resistance integron.
Conclusions
CC446 isolates are globally distributed with multiple occurrences of high AMR. The subclade ST298* is responsible for a prolonged epidemic (≥16 years) of XDR infections at an academic medical center. These findings indicate that CC446 is an emerging high-risk clone deserving further surveillance.
Keywords: Pseudomonas aeruginosa, antimicrobial resistance, plasmid, phylogenetics, high-risk clone
We describe a prolonged epidemic of an extensively drug-resistant subclade of Pseudomonas aeruginosa CC446 at an academic center. CC446 (containing ST298) is globally distributed with multiple reports of high antimicrobial resistance and should be considered an emerging high-risk clone.
Pseudomonas aeruginosa is a major cause of serious nosocomial infections. Antimicrobial resistance (AMR) in P. aeruginosa is frequent and limits treatment options, which has led the Infectious Diseases Society of America [1] and the World Health Organization [2] to list this bacterium as a priority pathogen for the development of new antimicrobials. Surveillance of highly drug-resistant P. aeruginosa is critical to better understand its epidemiology and limit its spread.
Multilocus sequence typing (MLST) has identified distinct patterns in the epidemiology of multidrug-resistant (MDR) and extensively drug-resistant (XDR) P. aeruginosa infections. While sporadic isolates may demonstrate high AMR, a large proportion of MDR/XDR infections are caused by a relatively small number of globally distributed sequence types (STs) termed “high-risk clones” [3–8]. Known high-risk clones such as ST235, ST111, and ST175 may possess a variety of resistance determinants, both horizontally acquired and mutational [3, 9, 10], suggesting that these clones’ high potential for acquiring AMR plays a role in their survival and spread in human populations [4]. While these STs are relatively common, other high-risk clones also contribute to drug-resistant infections worldwide [3–5, 11], and it is likely that additional high-risk clones have yet to be described.
In this study, we investigated clonal complex (CC) 446, containing major STs 446 and 298, as a potential emerging high-risk clone. We describe the global distribution of this lineage and the presence of highly resistant isolates at both our institution and others. In doing so, we identified the persistence of an XDR ST298 subclade (ST298*) possessing a large novel AMR plasmid at 1 academic medical center for at least 16 years.
METHODS
Bacterial Isolates and Antimicrobial Resistance Testing
Several collections of P. aeruginosa isolates were evaluated (Table 1). These include 3 cohorts of isolates collected from patients and clinical settings at Northwestern Memorial Hospital (NMH) in Chicago: 100 bloodstream isolates collected 1999–2003 (PABL) [12], 301 isolates from clinical specimens and hospital environments collected 2002–2009 (MolEpi), and 99 isolates from patient samples collected 2013–2018 (PA-NM). Other patient isolates screened included 601 bloodstream isolates collected from 10 public hospitals in Spain between 2008 and 2009 (PASP) [13] and 100 isolates from patient samples collected at Brigham and Women’s Hospital in Boston between 2015 and 2016 (BWH). Also included were 58 P. aeruginosa isolates collected from multiple healthcare facility environments (eg, sinks) in the Chicago metropolitan area between 2017 and 2018 (Hosp_Env). While these isolates were not collected from patients, they are healthcare-associated and could be of human origin or serve as a reservoir for potential infections. CC446 isolates were identified from these collections through postsequencing in silico MLST using allele sequences and MLST profiles listed in the PubMLST database [14].
Table 1.
Sequenced CC446 Isolates and Collection Source
| Collection | No. of Isolates | ST298 (No.) | ST446 (No.) | Total CC446 (No.) | Location | Source | Years |
|---|---|---|---|---|---|---|---|
| PABL | 100 | 9 | 2 | 11 | NMH | Blood cultures | 1999–2003 |
| MolEpi | 301 | 10 | 4 | 14 | NMH | Microbiology and molecular epidemiology laboratory | 2002–2009 |
| PA-NM | 99 | 4 | 1 | 5 | NMH | Patient samples | 2013–2018 |
| Hosp_Env | 58 | 1 | 6 | 7 | Chicago metropolitan area | Healthcare facility environments (eg, sinks) | 2017–2018 |
| BWH | 100 | 2 | 1 | 3 | Boston | Patient samples | 2015–2016 |
| PASP | 601 | 2 | 12 | 14 | Spain | Blood cultures | 2008–2009 |
| National Center for Biotechnology Information | 2483 | 21 | 17 | 38 | Variousa | Publicly available genome sequences | … |
Abbreviation: BWH, Brigham and Women’s Hospital; Hosp_Env, Hospital Environment; MolEpi, Molecular Epidemiology; NMH, Northwestern Memorial Hospital, Chicago, Illinois; PABL, Pseudomonas aeruginosa Bloodstream; PA-NM, Pseudomonas aeruginosa Northwestern Memorial Hospital; PASP, Pseudomonas aeruginosa Spain.
aUnited States, Argentina, Belgium, Canada, Columbia, France, Germany, Netherlands, Pakistan, Portugal, and Spain.
Complete methods for antimicrobial resistance testing are described in the Supplementary Methods. Briefly, minimum inhibitory concentrations (MICs) against 8 antibacterial agents from 7 antipseudomonal classes were determined for each isolate using microbroth dilution [15]. Isolates were designated as multidrug-resistant (MDR) if they were nonsusceptible to an antibiotic from ≥3 classes and extensively drug-resistant if they were susceptible to antibiotics from ≤2 classes tested following standardized criteria [16].
Whole-genome Sequencing and Sequence Analysis
In addition to the isolates described above, 2483 P. aeruginosa genomes previously deposited in the National Center for Biotechnology Information (NCBI) database (accessed 26 October 2017, deposited 2006–2017) were screened using in silico MLST to identify CC446 isolates. The PacBio platform was used to sequence isolate PABL048 to generate a complete CC446 genome. De novo assembly yielded the complete circularized genome and 1 circular plasmid (pPABL048). To determine core genome single-nucleotide variants (SNVs), all CC446 genomes were aligned to the PABL048 chromosome. Phylogenetic relationships within CC446 were examined using core genome alignments to construct a maximum likelihood phylogenetic tree, which was then corrected for the impact of recombination. ST298* genomes with known isolation dates were then extracted from a recombination-filtered core genome alignment and the recombination-corrected tree and used to construct a Bayesian time-scaled phylogeny of this subclade. Complete methods for bioinformatic analyses are described in the Supplementary Methods.
RESULTS
Geographic Distribution of CC446 Isolates
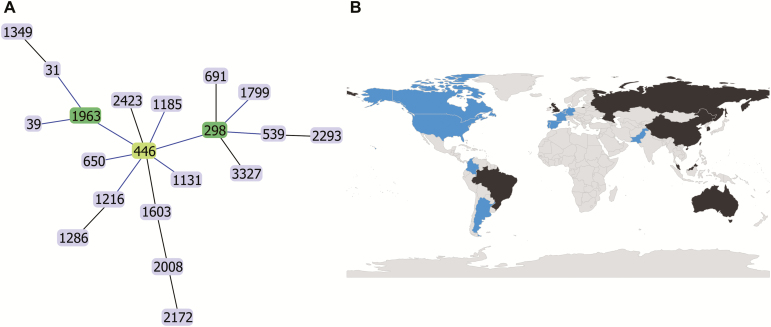
In the process of investigating P. aeruginosa strains from collections obtained at a single medical center (NMH), we noted an unusually large representation of isolates with the closely related ST298 and ST446 genotypes. BURST analysis identified these STs, which are single locus variants, as central members of a larger CC consisting of 20 STs. This CC was termed CC446 after the likely group founder (Figure 1A).
Figure 1.
Clonal complex definition and global distribution of CC446. A, Global optimal eBURST diagram showing sequence types in CC446. The likely founder of the clonal complex (ST446) is indicated in light green, and subgroup founders (ST298 and ST1963) are indicated in dark green. B, World map indicating countries where CC446 isolates have been detected. Countries with at least 1 isolate associated with a genome analyzed in this study are shaded blue. Countries in which a CC446 isolate has been reported but no genome was available are shaded in black.
Next, we used in silico MLST to screen 6 P. aeruginosa patient and healthcare environmental strain collections from Chicago, Boston, and Spain (a total of 1259 isolates) for CC446 isolates and identified 54 (Table 1). Additionally, we screened 2483 P. aeruginosa genomes previously deposited in the NCBI database to identify another 38 CC446 isolates (Table 1). In total, we identified 92 CC446 isolates (49 ST298 and 43 ST446; Supplementary Table 1). All CC446 isolates in this study were either ST298 or ST446, suggesting that these are the dominant clinical STs in this clonal complex. Whole-genome sequences were available for each of these isolates, and several had been previously published [17–21]. We also found multiple instances of CC446 strains mentioned in the literature and the PubMLST database for which whole-genome sequences were not available [7, 14, 22–26]. These CC446 isolates were cultured from North America, South America, Europe, Asia, and Oceania, indicating that CC446 is globally distributed (Figure 1B).
Antimicrobial Resistance of CC446 Isolates
We had access to the 54 CC446 isolates from the Chicago, Boston, and Spain collections and performed microbroth-dilution antibiotic susceptibility testing on them (Supplementary Table 2). Overall, 51.9% (28/54) of the isolates were MDR, of which 53.6% (15/28) were XDR. AMR was most prevalent in isolates collected at NMH, with 76.7% (23/30) of isolates MDR and 60.9% (14/23) of those XDR. In particular, almost all NMH ST298 isolates (21/23, 91.3%) were MDR, of which many (13/21, 61.9%) were XDR. Of the 14 Spanish CC446 isolates, 4 ST446 isolates were MDR, 1 of which was XDR. One of 3 CC446 isolates from Boston was MDR. In contrast to isolates collected from patient samples, none of the 7 healthcare environmental CC446 isolates were MDR. Additionally, 4 XDR ST298 strains from NMH showed nonsusceptibility to the recently developed β-lactam/β-lactamase inhibitor combinations ceftazidime-avibactam and ceftolozane-tazobactam on disk diffusion testing (Supplementary Table 2). The high prevalence of MDR/XDR isolates in this study, coupled with the global distribution of CC446 (Figure 1B) and previous reports of AMR in this clonal complex [7, 18, 20, 21, 25], support the classification of CC446 as an emerging high-risk clone.
Identification of AMR Integron in1697 in NMH ST298 Isolates
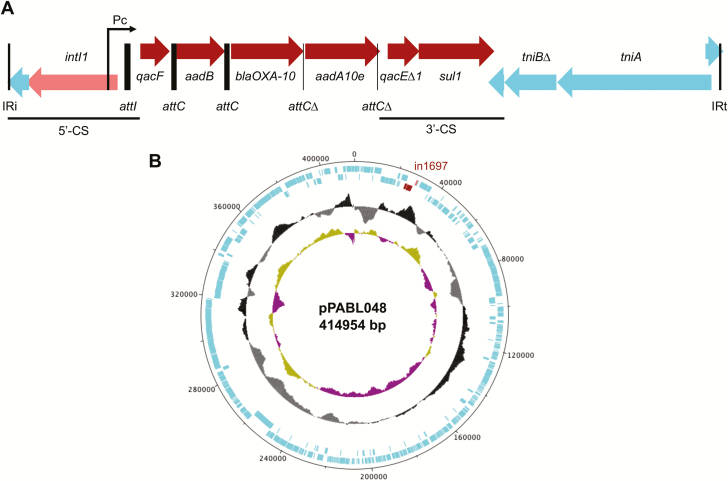
To determine the genetic basis for the high rates of AMR in ST298 isolates from NMH, we identified resistance genes from their whole-genome sequences using the ResFinder database [27]. We identified a locus containing multiple AMR genes present in 16 MDR ST298 isolates from NMH. This locus was present in 76.2% (16/21) of MDR ST298 isolates from NMH, and 81.3% (13/16) of these isolates were XDR (Supplementary Table 2). Characterization of this locus revealed it to be a novel class 1 integron designated in1697 (Figure 2A). As is common for class 1 integrons, it consists of a 5’ conserved segment (5’-CS) containing the intl1 integrase gene and a promoter driving cassette expression, several resistance gene cassettes, and a 3’ conserved segment (3’-CS) containing sul1 (sulphonamide resistance) and qacE∆1 (quaternary ammonium compound [QAC] resistance) [28]. Gene cassettes in in1697 include the β-lactamase blaOXA-10, aminoglycoside resistance genes aadB and aadA10e, and QAC resistance gene qacF. Isolates with in1697 showed high levels of gentamicin resistance (>128 µg/mL) not seen among other CC446 isolates tested. These findings suggest that a novel integron, in1697, contributes to the antibiotic resistance of some CC446 isolates.
Figure 2.
Characterization of the plasmid pPABL048. A, Description of the antimicrobial resistance (AMR) class I integron in1697 of pPABL048. in1697 consists of a 5’ conserved segment (5’-CS) with the integrase intI1 and promoter Pc, AMR cassettes, and a 3’ conserved segment (3’-CS) of qacE∆1 and sul1. Complete attC recombination sites were identified downstream of qacF and aadB, and truncated attC sites were identified downstream of blaOXA-10 and aad10Ae. in1697 appears to be part of a transposable-like element that includes a partial tni transposon operon and has as its borders IRi and IRt (25-bp imperfect [92% identity] inverted repeats). B, Diagram of pPABL048 with rings (from in to out) showing guanine-cytosine (GC) skew, GC%, coding sequences, and position in base pairs. The location of in1697 is highlighted in red.
Identification of a Large AMR Plasmid in NMH ST298 Isolates
To investigate the genomic context of in1697, we performed long-read sequencing and complete genome construction for 1 in1697-positive isolate from NMH (PABL048). This yielded a 6 879 622 bp bacterial chromosome and revealed that in1697 is located on a large plasmid (414 954 bp) that we named pPABL048 (Figure 2B). The plasmid pPABL048 contains 496 coding sequences, some of which were predicted to encode for antimicrobial/disinfectant resistance proteins, heavy metal resistance proteins, and chemotaxis proteins (Supplementary Table 3). Screening both the PABL048 chromosome and plasmid against the virulence factor database identified several predicted virulence factors on the plasmid, with 3 related to Type IV pili and 1 potentially related to carbon storage regulation (Supplementary Table 4) [29]. Sequencing reads for all isolates containing in1697 showed substantial alignment to pPABL048 (generally >90% sequence coverage) with few SNVs (≤4; Supplementary Table 5), indicating that these isolates contain very similar plasmids. The exception is PS1875 (57.6% alignment), which is missing a large contiguous portion of the plasmid (data not shown). in1697 was not found outside the context of pPABL048. In summary, pPABL048 is a large ST298-associated plasmid containing a novel AMR class 1 integron.
Two ST298 strains from NMH, PABL036 and PABL067, expressed a heterogeneous pattern of resistance. That is, some colonies from the same culture stock expressed high levels of gentamicin resistance while others did not. Whole-genome sequencing confirmed that this variable pattern of resistance was associated with the presence or absence of the AMR plasmid pPABL048 (Supplementary Table 5). This discrepancy accounts for the lack of in1697 in our initial whole-genome sequence of PABL067. These findings indicate that pPABL048 can be spontaneously lost by some strains, which could potentially cause inaccurate antibiotic susceptibility results.
To explore the function of the plasmid, we generated plasmid-cured variants of PABL048 (PABL048-c1 and PABL048-c2; Supplementary Table 5) and tested the impact of pPABL048 on AMR. Isolates lacking the plasmid showed reduced MICs to gentamicin and piperacillin-tazobactam compared with their isogenic partners possessing the plasmid (Table 2), indicating that pPABL048 encodes for resistance to these antibiotics.
Table 2.
Minimum Inhibitory Concentrations of ST298* Pseudomonas aeruginosa Isolates With and Without pPABL048
| Minimum Inhibitory Concentration (µg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | pPABL048 | Gentamicin | Cefepime | Ceftazidime | Piperacillin-Tazobactam | Meropenem | Aztreonam | Ciprofloxacin | Colistin |
| PABL048 | + | >128–ns | 8 | 2 | 64–ns | 8–ns | 32–ns | 32–ns | 0.5 |
| PABL048-c1 | - | 4 | 4 | 4 | 16 | 4–ns | 16–ns | 16–ns | 0.5 |
| PABL048-c2 | - | 8–ns | 4 | 4 | 16 | 4–ns | 16–ns | 16–ns | 0.5 |
| PABL036-GentR | + | >128–ns | 8 | 4 | 32–ns | 8–ns | 16–ns | 32–ns | 0.5 |
| PABL036-GentS | - | 4 | 4 | 4 | 16 | 8–ns | 16–ns | 32–ns | 0.5 |
| PABL067-GentR | + | >128–ns | 8 | 4 | 64–ns | 8–ns | 32–ns | 32–ns | 0.5 |
| PABL067-GentS | - | 4 | 4 | 4 | 16 | 4–ns | 16–ns | 32–ns | 0.5 |
Abbreviations: +, plasmid pPABL048 is present; —, plasmid pPABL048 is absent; c, experimentally cured of pPABL048; GentR, derived from gentamicin-resistant colony; GentS, derived from gentamicin-sensitive colony; ns, nonsusceptible (intermediate and resistant); as defined by the Clinical Laboratory Standards Institute.
Phylogenetic Analysis of CC446
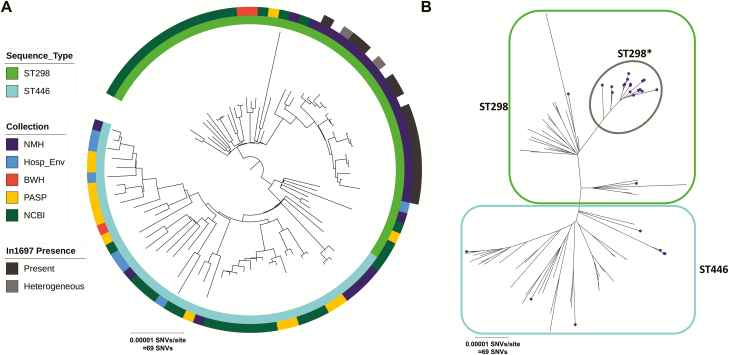
To better understand the relationships between CC446 isolates included in this study, we constructed a recombination-corrected maximum likelihood phylogenetic tree based on core genome alignment to the PABL048 chromosome (Figure 3). We found that while ST298 and ST446 are closely related, they are phylogenetically distinct. The majority (21/30) of CC446 isolates from NMH clustered in a distinct ST298 subclade (designated ST298*), which was not seen in any of the other collections. Both pPABL048 and in1697 are exclusive to this subclade. ST298* isolates were collected between 2000 and 2017 (with pPABL048 first detected in 2001). While ST298* was only detected at NMH, ST298 and ST446 isolates outside of this subclade were also present at NMH. This suggests that a prolonged local epidemic of ST298* had occurred in addition to the general circulation of other CC446 isolates. ST298* isolates showed high levels of AMR, while sporadic CC446 isolates from NMH were largely sensitive to antimicrobials, the exceptions being the ST446 isolates PS1946 (MDR) and PS1948 (XDR; Supplementary Table 2).
Figure 3.
Recombination-corrected maximum likelihood phylogenetic tree of the CC446 isolates included in this study based on core genome alignment to the chromosome of PABL048. A, Midpoint-rooted circular tree annotated (from inner to outer rings) with sequence type, collection of origin, and the presence of in1697. B, Unrooted radial tree with sequence type and subclade indicated by blue (ST446), green (ST298), and gray (ST298*) outlines. Isolates collected from NMH are indicated with purple circles. Abbreviations: BWH, Brigham and Women’s Hospital; Hosp_Env, Hospital Environment; NCBI, National Center for Biotechnology Information; NMH, Northwestern Memorial Hospital; SNV, single-nucleotide variant; PASP, Pseudomonas aeruginosa Spain.
Time-scaled Phylogenetic Analysis of ST298* Subclade
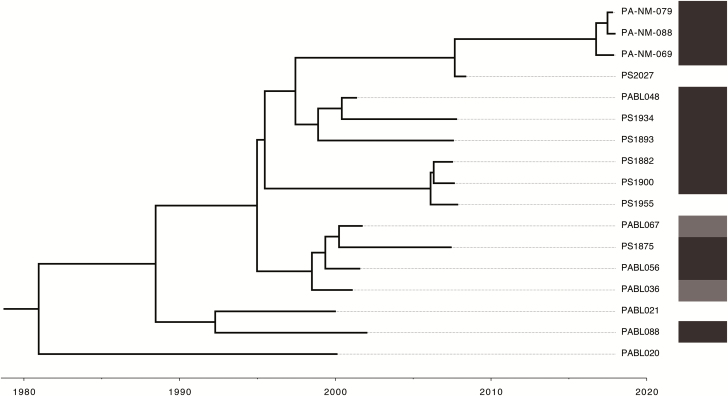
We used Bayesian phylogenetic analysis to construct a time-scaled phylogenetic tree of 17 ST298* isolates with available collection dates (Figure 4). The most recent common ancestor for ST298* is estimated to have arisen in the year 1980 (mean, 1980.9; 95% highest posterior density (HPD) interval, 1973.8–1987.4). Based on this analysis, ST298* has been evolving at a rate of 1.80 (95% HPD interval, 1.32–2.29) core genome SNVs/year. This is comparable to previous estimates in nonhypermutable P. aeruginosa [30, 31].
Figure 4.
Time-scaled phylogenetic tree of ST298* isolates. Tips indicate date of isolation. Root is the estimated last common ancestor of these isolates (mean, 1980.9; 95% highest posterior density interval, 1973.8–1987.4). The presence of in1697 is indicated by shaded bars on the right, with light gray indicating heterogenous presence in only some colonies for a given isolate.
Mutational Resistance in ST298*
While pPABL048, likely through in1697, contributes to increased resistance to aminoglycosides and penicillins, acquired AMR genes do not explain the resistance of ST298* isolates to other antibacterials. We investigated whether mutations could explain these resistance patterns. PABL048 harbors a T83I substitution in GyrA and a S87L substitution in ParC that, in combination, confer high fluoroquinolone resistance [32]. PABL048 also possesses a 4 amino acid deletion (residues 12–15) and 3 single amino acid substitutions in NalC compared with the reference strain PAO1. While the impact of these mutations is unknown, inactivation of NalC increases resistance to multiple antibacterials through MexAB-OprM efflux pump overproduction [33]. Mutations impacting the porin OprD can play a role in carbapenem resistance [34]. ST298* isolates, all of which are meropenem nonsusceptible, show multiple amino acid substitutions of unclear significance in OprD compared with PAO1. The ST298* isolates with the highest meropenem resistance (PS2027, PA-NM-069, PA-NM-079, PA-NM-088) show both amino acid deletions from residues 12–54 as well as amino acid substitutions in residues 2–10 (Supplementary Figure 1). Ceftazidime resistance in the ST298* isolates PS1793, PS1796, and PS1797 is likely secondary to a deletion of amino acid residues 2–30 in AmpD, leading to AmpC overproduction [35]. These 3 isolates also share 2-amino-acid substitutions in the plasmid-borne OXA-10, which may confer extended-spectrum β-lactamase activity. These substitutions include the G157D substitution previously seen in the extended spectrum OXA-10 variant OXA-14 [36] as well as a F153S substitution. These findings suggest that ST298* isolates have accumulated mutations that confer antibiotic resistance.
Comparative Genomics of pPABL048
While pPABL048 appears to be exclusive to ST298*, evidence of related plasmids can be seen in other isolates, including LLTF and MPVG from this study (Supplementary Table 5). We identified 16 complete plasmids with substantial sequence alignment (≥70% coverage) to pPABL048 in multiple Pseudomonas species (Supplementary Table 6), suggesting that pPABL048 is part of a family of large Pseudomonas genus plasmids. No similar plasmids were found in available sequences from non-Pseudomonas Gammaproteobacteria. We defined the “plasmid backbone” as sequence positions present in 16/17 of these plasmids (Supplementary Figure 2A). While in1697 is not part of the backbone, other genetic features, such as replication and partitioning genes, a chemotaxis locus, putative pilus locus, and a tellurium resistance locus are common to these plasmids. The “backbone” replication protein gene common to these plasmids has not been characterized. Of note, other cases of integron-mediated AMR have been described in this family of plasmids [37–39]. To identify additional Pseudomonas isolates that may carry pPABL048-like plasmids, we screened publicly available draft genomes and identified 32 with >70% alignment to pPABL048 (Supplementary Table 7). Phylogenetic analysis of all 63 pPABL048-like sequence alignments shows that they do not appear to segregate by species (Supplementary Figure 2B, C). Additionally, ST298* pPABL048 alignments form a distinct group, showing that pPABL048 itself has not been previously reported. These results indicate that pPABL048 is a novel member of a family of large Pseudomonas genus plasmids.
DISCUSSION
In this study, we identified a subclade of CC446, termed ST298*, that is responsible for a prolonged epidemic of XDR P. aeruginosa infections at NMH from at least 2001 through 2017. Extensive antimicrobial resistance in ST298* was due in part to the presence of the large novel AMR plasmid pPABL048, but ST298* isolates lacking this plasmid were still universally MDR. The long-term persistence of ST298* P. aeruginosa is clinically significant, both from the standpoint of infection prevention at this institution and in highlighting the potential risk posed by CC446 at healthcare centers in general. Additionally, with its global distribution and multiple incidents of high AMR both from this study and other reports in the literature [7, 18, 20, 21, 25], we provide evidence that CC446 is an emerging high-risk lineage of P. aeruginosa.
While we were able to identify the XDR subclade ST298* at NMH and show that it has repeatedly caused highly AMR infections, we lack additional epidemiological data to link these cases. However, our findings suggest the existence of a persistent reservoir for ST298* isolates over the last 2 decades. We hypothesize that this reservoir could be within NMH itself, from a common source outside the hospital (eg, a long-term acute care hospital), or more widespread throughout Chicago healthcare settings. It is notable that the estimated date of emergence of the last common ancestor for the ST298* subclade (1980; Figure 4) is nearly 20 years prior to the opening of the current NMH inpatient facility in 1999. It is important to note that only a limited number of isolates from Chicago came from sources outside of NMH, and we were unable to determine the extent to which ST298* has spread throughout the region. It will be critical to assess whether this lineage is unique to NMH or more widespread. As such, future work integrating both microbiological and epidemiological approaches is needed to identify the reservoir and geographic spread of ST298*.
The plasmid pPABL048 containing the AMR integron in1697 has contributed to the XDR phenotypes of ST298* isolates. While pPABL048 is unique to ST298*, it is part of a family of large Pseudomonas genus plasmids. The involvement of both pPABL048 and related plasmids in drug-resistant infections [37–39] highlights the clinical importance of this plasmid family. Further investigation is needed to determine the impact of the pPABL048 family of plasmids on bacterial phenotypes that could contribute to increased persistence or fitness, with a specific focus on predicted virulence factors that may affect adhesion, motility, and carbon storage (Supplementary Table 4).
Although recognized high-risk clones such as ST235, ST111, and ST175 are enriched for antibiotic resistance and cause a large proportion of MDR/XDR infections worldwide [3, 5–8], relatively susceptible isolates from these STs also occur [5, 8, 18, 40]. Additionally, the genetic bases for AMR in these STs are diverse [3, 9], suggesting that the propensity to acquire antibiotic resistance is a hallmark of high-risk clones. Our findings show that CC446 has many of these same features. Although our study documents XDR ST298* isolates at only a single institution, CC446 organisms as a whole are responsible for clinically significant infections worldwide (Figure 1B). While not all CC446 isolates tested in this study were MDR/XDR, the frequency of these phenotypes unmasks the high potential for AMR within this clonal complex. Previous findings support this, with multiple geographically distinct reports of MDR CC446 isolates [7, 18, 20, 21, 25]. Mechanisms of resistance among these isolates are varied and include other AMR plasmids [20, 21], extended spectrum β-lactamases (eg, VIM-2) [7, 21–24], and chromosomal mutations [17, 20, 21, 25]. These findings are consistent with the assertion that CC446 represents an emerging high-risk clone with the potential to cause further MDR outbreaks.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. This research was supported in part through the computational resources and staff contributions provided by the Genomics Compute Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose of advancing research in genomics. The authors thank Dr Teresa Zembower and the staff of the Northwestern Memorial Hospital Molecular Epidemiology Laboratory for assisting with strain collection. The authors thank the Spanish Network for Research in Infectious Diseases (REIPI) for providing the Spanish isolates. The authors also thank the staff of the Brigham and Women’s Hospital Clinical Microbiology Laboratory for their help with collection of the Brigham and Women’s Hospital strains.
Financial support. This work was supported by the National Institute of General Medical Sciences (T32 GM008061 and T32 GM008152 to N. B. P.), the American Cancer Society (130602-PF-17-107-01-MPC to K. E. R. B.; MRSG-13-220-01-MPC to E. A. O.), and the National Institute of Allergy and Infectious Diseases (T32 AI007061-36A1 to K. E. R. B. and R01 AI118257, R21 129167, and U19 AI135964 to A. R. H.). A. O. is supported by Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, and Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0004) and is cofinanced by the European Development Regional Fund “A Way to Achieve Europe” and operative program Intelligent Growth 2014‐2020.
Potential conflicts of interest. A. R. H. serves on the scientific advisory board and as a consultant for Microbiotics, Inc. E. A. O. serves on the clinical advisory board for Gladius Pharmaceuticals. N. J. R. has received research support from Hartford Hospital and honoraria from the American Society of Health System Pharmacists and has patent WO2017161296A1 pending. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Talbot GH, Bradley J, Edwards JE Jr, Gilbert D, Scheld M, Bartlett JG; Antimicrobial Availability Task Force of the Infectious Diseases Society of America Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin Infect Dis 2006; 42:657–68. [DOI] [PubMed] [Google Scholar]
- 2. Tacconelli E, Sifakis F, Harbarth S, et al. ; EPI-Net COMBACTE-MAGNET Group Surveillance for control of antimicrobial resistance. Lancet Infect Dis 2018; 18:e99–e106. [DOI] [PubMed] [Google Scholar]
- 3. Oliver A, Mulet X, López-Causapé C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 2015; 21-22:41–59. [DOI] [PubMed] [Google Scholar]
- 4. Woodford N, Turton JF, Livermore DM. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 2011; 35:736–55. [DOI] [PubMed] [Google Scholar]
- 5. Slekovec C, Robert J, van der Mee-Marquet N, et al. . Molecular epidemiology of Pseudomonas aeruginosa isolated from infected ICU patients: a French multicenter 2012–2013 study. Eur J Clin Microbiol Infect Dis 2019; 38:921–6. [DOI] [PubMed] [Google Scholar]
- 6. Peña C, Cabot G, Gómez-Zorrilla S, et al. ; Spanish Network for Research in Infectious Diseases Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 2015; 60:539–48. [DOI] [PubMed] [Google Scholar]
- 7. Correa A, Del Campo R, Perenguez M, et al. . Dissemination of high-risk clones of extensively drug-resistant Pseudomonas aeruginosa in Colombia. Antimicrob Agents Chemother 2015; 59:2421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gomila M, Del Carmen Gallegos M, Fernández-Baca V, et al. . Genetic diversity of clinical Pseudomonas aeruginosa isolates in a public hospital in Spain. BMC Microbiol 2013; 13:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Treepong P, Kos VN, Guyeux C, et al. . Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 2018; 24:258–66. [DOI] [PubMed] [Google Scholar]
- 10. Cabot G, López-Causapé C, Ocampo-Sosa AA, et al. . Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 2016; 60:7415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wright LL, Turton JF, Livermore DM, Hopkins KL, Woodford N. Dominance of international ‘high-risk clones’ among metallo-β-lactamase-producing Pseudomonas aeruginosa in the UK. J Antimicrob Chemother 2015; 70:103–10. [DOI] [PubMed] [Google Scholar]
- 12. Scheetz MH, Hoffman M, Bolon MK, et al. . Morbidity associated with Pseudomonas aeruginosa bloodstream infections. Diagn Microbiol Infect Dis 2009; 64:311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peña C, Suarez C, Gozalo M, et al. ; Spanish Network for Research in Infectious Diseases Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob Agents Chemother 2012; 56:1265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008; 3:163–75. [DOI] [PubMed] [Google Scholar]
- 16. Magiorakos AP, Srinivasan A, Carey RB, et al. . Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18:268–81. [DOI] [PubMed] [Google Scholar]
- 17. Kos VN, Déraspe M, McLaughlin RE, et al. . The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 2015; 59:427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Belkum A, Soriaga LB, LaFave MC, et al. . Phylogenetic distribution of CRISPR-Cas systems in antibiotic-resistant Pseudomonas aeruginosa. MBio 2015; 6:e01796–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roach DJ, Burton JN, Lee C, et al. . A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 2015; 11:e1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonnin RA, Bogaerts P, Girlich D, et al. . Molecular characterization of OXA-198 carbapenemase-producing Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2018; 62:e02496–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Zee A, Kraak WB, Burggraaf A, et al. . Spread of carbapenem resistance by transposition and conjugation among Pseudomonas aeruginosa. Front Microbiol 2018; 9:2057. doi:10.3389/fmicb.2018.02057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee JY, Peck KR, Ko KS. Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol 2013; 62:1015–24. [DOI] [PubMed] [Google Scholar]
- 23. Kim MJ, Bae IK, Jeong SH, et al. . Dissemination of metallo-β-lactamase-producing Pseudomonas aeruginosa of sequence type 235 in Asian countries. J Antimicrob Chemother 2013; 68:2820–4. [DOI] [PubMed] [Google Scholar]
- 24. Chen Y, Sun M, Wang M, Lu Y, Yan Z. Dissemination of IMP-6-producing Pseudomonas aeruginosa ST244 in multiple cities in China. Eur J Clin Microbiol Infect Dis 2014; 33:1181–7. [DOI] [PubMed] [Google Scholar]
- 25. Domitrovic TN, Hujer AM, Perez F, et al. . Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis 2016; 3:ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Avetisian LR, Voronina OL, Chernukha M, et al. . Persistence of Pseudomonas aeruginosa strains in patients of Federal Scientific Center of Transplantology and Artificial Organs. Zh Mikrobiol Epidemiol Immunobiol 2012; 4:99–104. [PubMed] [Google Scholar]
- 27. Zankari E, Hasman H, Cosentino S, et al. . Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 2012; 67:2640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 2009; 33:757–84. [DOI] [PubMed] [Google Scholar]
- 29. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 2019; 47:D687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duchêne S, Holt KE, Weill FX, et al. . Genome-scale rates of evolutionary change in bacteria. Microb Genom 2016; 2:e000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marvig RL, Johansen HK, Molin S, Jelsbak L. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet 2013; 9:e1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruchmann S, Dötsch A, Nouri B, Chaberny IF, Häussler S. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 2013; 57:1361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev 2015; 28:337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22:582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bagge N, Ciofu O, Hentzer M, Campbell JI, Givskov M, Høiby N. Constitutive high expression of chromosomal beta-lactamase in Pseudomonas aeruginosa caused by a new insertion sequence (IS1669) located in ampD. Antimicrob Agents Chemother 2002; 46:3406–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Danel F, Hall LM, Gur D, Livermore DM. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 1995; 39:1881–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiong J, Alexander DC, Ma JH, et al. . Complete sequence of pOZ176, a 500-kilobase IncP-2 plasmid encoding IMP-9-mediated carbapenem resistance, from outbreak isolate Pseudomonas aeruginosa 96. Antimicrob Agents Chemother 2013; 57:3775–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yuan M, Chen H, Zhu X, et al. . pSY153-MDR, a p12969-DIM-related mega plasmid carrying blaIMP-45 and armA, from clinical Pseudomonas putida. Oncotarget 2017; 8:68439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun F, Zhou D, Wang Q, et al. . Genetic characterization of a novel blaDIM-2-carrying megaplasmid p12969-DIM from clinical Pseudomonas putida. J Antimicrob Chemother 2016; 71:909–12. [DOI] [PubMed] [Google Scholar]
- 40. Maatallah M, Cheriaa J, Backhrouf A, et al. . Population structure of Pseudomonas aeruginosa from five Mediterranean countries: evidence for frequent recombination and epidemic occurrence of CC235. PLoS One 2011; 6:e25617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.