Abstract
We assessed the real-world utility of universal broad-range polymerase chain reaction sequencing for pathogen detection. Among 1062 clinical samples, 107/1062 (10.1%) had a clinically significant, positive result, with substantial variation by specimen type. Clinical management was changed in 44/1062 (4.1%). These data can help maximize utility of this emerging diagnostic.
Keywords: broad-range universal PCR, DNA sequencing, molecular diagnostics, pathogen identification
(See the Editorial Commentary by Schuetz on pages 1558–60.)
Molecular diagnostics for pathogen detection are becoming increasingly utilized to guide patient care, especially in complex and difficult-to-diagnose cases. A technique called universal broad-range polymerase chain reaction amplicon sequencing (uPCR) involves the amplification and subsequent sequencing of hypervariable regions of genes that are conserved across classes of pathogens [1]. It allows for the detection of pathogens that do not grow well in standard culture, such as those that are fastidious or present at low levels [2]. While the utility of uPCR has previously been demonstrated for several types of infections [1–8], its performance under routine clinical conditions, including its impact on clinical decision-making, is not well defined.
METHODS
The medical records of patients admitted to University of California, San Francisco, Medical Center or Benioff Children’s Hospital between November 2011 and July 2019 were searched to identify all patients with a clinical specimen sent for uPCR. In all cases, fluid or tissue samples were sent directly for uPCR at the discretion of the primary clinical team, with the involvement of infectious diseases consultants in nearly all cases. The uPCR results were only sent after review by a laboratory provider once routine cultures were negative, with rare exceptions when expedited results were critical for patient care. Formalin-fixed paraffin-embedded (FFPE) specimens were sent only if fresh tissue was not available. No cultured organisms were tested. The University of California, San Francisco, Institutional Review Board approved this study.
The University of Washington Molecular Diagnostics microbiology lab performed uPCR per standardized protocols [3, 9]. Amplification of the following genes was performed: bacterial (16S ribosomal ribonucleic acid [16S rRNA]), fungal (26S rRNA, internal transcribed spacer [ITS] 1 and 2) and/or mycobacterial (16S rRNA, heat shock protein 65 [hsp65], RNA polymerase β subunit [rpoB]). Thus, each clinical specimen had between 1 and 3 associated uPCR results; there was no overlap of bacterial, mycobacterial, or fungal results. Any pathogen-specific results were excluded.
Basic demographic variables were extracted for all patients with a valid uPCR result. For each case with a positive uPCR result, a detailed review was performed to determine: (1) the results of microbiological and pathology studies; (2) a final diagnosis; (3) the clinical significance of a positive result; and (4) whether a positive result changed management. A positive uPCR result was considered clinically significant when the pathogen isolated was compatible with the patient’s clinical syndrome without a better alternative explanation. A result was considered not clinically significant when the organism(s) were a likely contaminant or unlikely to account for the patient’s illness. A positive uPCR result was classified as changing management if it clearly resulted in a change in the patient’s antimicrobial plan (adding or narrowing antimicrobial[s] and/or duration). Tissue biopsies, including fine-needle aspirates, were considered to be tissue specimens, while aspirates of body fluids were classified as fluid specimens. Proportions were compared using Chi-squared tests where appropriate.
RESULTS
We identified 1062 clinical specimens with at least 1 uPCR result from 864 unique patients (median age of 50; interquartile range, 28–64; 15.5% <18 years old; 53.9% male). There were a total of 2280 uPCR results from these specimens: 734 bacterial, 824 fungal, and 722 mycobacterial.
Overall, uPCR was positive 16.5% (95% confidence interval [CI], 14.3–18.8; n = 175/1062) of the time. Results were clinically significant for 10.1% of all samples (95% CI, 8.3–12.0; n = 107/1062) and 61.1% of all positive uPCR results (n = 107/175). Of the 107 clinically significant uPCR results, uPCR was the only positive investigation (including microscopy, culture, pathology, and pathogen-specific PCR) in 38 cases (35.5%). Overall, clinical management was changed by a positive uPCR result 4.1% (95% CI, 3.0–5.5; n = 44/1062) of the time. In the 107 cases where the result was clinically significant, clinical management was changed 41.1% (n = 44/107) of the time. The most common reasons that a clinically significant uPCR result did not change management were (1) appropriate antimicrobial therapy was guided by other positive investigations (n = 31/63, 49.2%); and (2) appropriate antimicrobial therapy had already been started empirically and no changes were indicated (n = 22/63, 34.9%). We identified no instances in which the response to a positive uPCR resulted in inappropriate management.
Bacterial species were the most frequently identified pathogens (n = 100/734; 13.6%), followed by fungal species (n = 58/824; 7.0%) and mycobacterial species (n = 25/722; 3.5%). Although mycobacterial species were infrequently identified, they were almost universally determined to be clinically significant (n = 23/25; 92.0%), compared to 56.0% (n = 56/100) of bacterial and 50.0% (n = 29/58) of fungal results. The uPCR results detected a broad array of true pathogens across a large number of clinical specimens (Supplementary Table 1). Among clinically significant, positive results, the pathogens most frequently detected were Aspergillus fumigatus (n = 12), Mycobacterium tuberculosis complex (n = 8), Staphylococcus aureus (n = 7), Streptococcus pyogenes (n = 7), Mycobacterium avium complex (n = 6), and Propionibacterium acnes (n = 5).
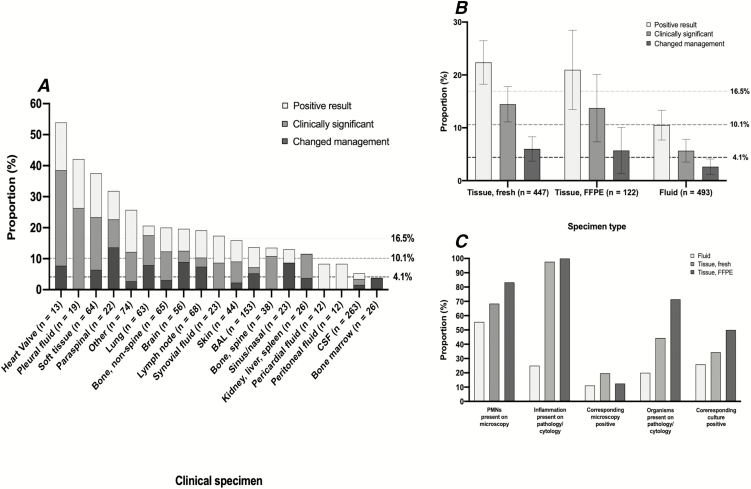
Next, we assessed the performance of uPCR by specimen type (Figure 1A; Supplementary Table 2). There were large differences in the proportion of specimens with any positive result (range, 3.8–53.9%); a clinically significant, positive result (range, 0–38.5%); and a positive result that changed management (range, 0–13.6%). Among frequently tested specimen types (≥20 uPCR tests), the clinical specimens that were most likely to have an associated clinically significant, positive uPCR result that changed management were soft tissue (proportion clinically significant = 23.4%; proportion that changed management = 6.4%), paraspinal (22.7% and 13.6%, respectively), lung (17.5% and 7.9%, respectively), brain (12.5% and 8.9%, respectively), and lymph node (10.3% and 7.4%, respectively) biopsies. Cerebrospinal fluid was the most frequently tested specimen type (n = 263), but only 3.4% (n = 9/263) of samples had a clinically significant, positive result and clinical management was changed in only 1.5% (n = 4/263) of cases. There was little difference in utility with respect to sterile versus nonsterile specimens (Supplementary Table 2).
Figure 1.
A, The clinical utility of universal broad-range polymerase chain reaction amplicon sequencing (uPCR) by clinical specimen type. The bars are overlapping and the 3 horizontal, dashed lines represent the overall proportion of all samples with corresponding positive uPCR results (16.5%), positive uPCR results that were clinically significant (10.1%), and positive uPCR results that changed clinical management (4.1%). B, The clinical utility of uPCR by clinical sample type: fresh tissue, formalin-fixed paraffin-embedded tissue, or fluid sample. C, The proportion of clinical samples, stratified by sample type, that have a corresponding positive microbiological or pathology result. Abbreviations: BAL, bronchoalveolar lavage; CSF, cerebrospinalfluid; FFPE, formalin-fixed paraffin-embedded; PMN, polymorphonuclear neutrophil.
There were no differences between fresh and FFPE tissue specimens in the proportions with either a clinically significant, positive uPCR result (14.3% vs 13.1%, respectively; P = .74) or a positive result that changed management (5.8% vs 4.9%, respectively; P = .70; Figure 1B). The data show that uPCR on fluid specimens was less likely than all tissue specimens to yield a clinically significant, positive result (5.5% vs 14.1%, respectively; P < .001) or change management (2.4% vs 5.6%, respectively; P = .009).
We evaluated the factors associated with a clinically significant, positive uPCR result (n = 107; Figure 1C). Nearly all positive, tissue-based uPCR results had inflammation on corresponding pathology (n = 65/66; 98.5%) in which the inflammatory pattern appropriately corresponded to the underlying pathogen (Supplementary Table 3), and a majority (n = 45/64; 70.3%) also had neutrophils on corresponding microscopy. For FFPE specimens, the majority of patients with a clinically significant, positive uPCR had organisms directly visualized on corresponding pathology (n = 10/14; 73.3%). Fluid specimens had lower concordance between results of routine microbiology/pathology and clinically significant, positive uPCR results (Figure 1C).
Discussion
This study evaluated the clinical utility of uPCR for pathogen detection under real-world conditions on more than 1000 clinical specimens at an academic medical center over nearly 8 years. We found that uPCR was positive and clinically significant in about 10% of cases and that a positive uPCR result changed management in about 4% of cases. Among those cases with a clinically significant, positive result, uPCR was the only positive investigation in 36% and changed the antimicrobial plan in 41%.
This study builds upon 2 smaller, retrospective evaluations of uPCR in pediatric (n = 247) [9] and adult (n = 71) [10] patients. In those studies, uPCR detected a clinically significant pathogen in 14% and 27% of cases, respectively, and changed management in 6% and 15% of cases, respectively [10]. Differences in the diagnostic yields between these 3 studies are likely due to many factors, including patient characteristics, specimen types, and institutional practice variations. However, all demonstrate the important clinical utility of uPCR under real-world conditions.
We found that the performance of uPCR differed substantially across clinical specimens, with tissue-based specimens more likely to yield a clinically significant result and change management than fluid samples [10]. For cerebrospinal fluid, uPCR detected a clinically significant pathogen in only 3.4% of cases, reflecting the significant challenge of diagnosing central nervous infections. These data suggest that alternative diagnostic techniques are needed for pathogen detection in fluid-based specimens, and emerging evidence suggests that metagenomic, next-generation sequencing may provide improved diagnostic yields [11, 12]. Notably, nearly all tissue specimens with clinically significant uPCR results had inflammation present on the corresponding pathology specimen, indicating that the presence of inflammation is a useful parameter for selecting those samples likely to yield positive results.
Strengths of this study include the analysis of a large, real-world cohort at a representative tertiary hospital, with the inclusion of a broad array of clinical specimens. We were unable to assess the true sensitivity and specificity of uPCR as compared to a culture-based reference standard, since uPCR is not routinely sent as part of infectious disease investigations. We also did not evaluate how negative uPCR results changed management, as how negative test results change clinical management is rarely documented. Finally, given the retrospective nature of the study, the impact of uPCR results on clinical decision-making may have been misclassified; however, this was likely uncommon, as providers usually documented clearly how uPCR influenced their clinical decisions.
In conclusion, uPCR has important clinical utility for pathogen detection in challenging cases in which infection is suspected. To date, there has been no clear guidance regarding when to send uPCR testing. Our results suggest that the yield of uPCR is likely to be maximized on tissue-based specimens, especially those with inflammation and/or direct visualization of organisms on corresponding pathology.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. A. D. K. and R. L. R. performed the chart review and developed the first manuscript draft. J. M. B. gave input on the manuscript draft. A. D. K. undertook the statistical analyses and developed the figures, with input from J. M. B., R. L. R., and S. M. All authors contributed to interpretation of data and editing of the article and approved the final version of the manuscript before submission.
Acknowledgments. The authors thank their infectious disease colleagues at the University of California, San Francisco, for thoughtful discussion about how to best incorporate broad-range polymerase chain reaction technology into practice at our institution.
Financial support. A.D.K. was supported by the National Institute of Allergy and Infectious Diseases (grant number T32 AI060530). R.L.R. was supported by the National Institute of Allergy and Infectious Diseases (grant number K23AI134327).
Potential conflicts of interest. All authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 2000; 38:3623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rampini SK, Bloemberg GV, Keller PM, et al. . Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis 2011; 53:1245–51. [DOI] [PubMed] [Google Scholar]
- 3. Rakeman JL, Bui U, Lafe K, Chen YC, Honeycutt RJ, Cookson BT. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol 2005; 43:3324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rampini SK, Zbinden A, Speck RF, Bloemberg GV. Similar efficacy of broad-range ITS PCR and conventional fungal culture for diagnosing fungal infections in non-immunocompromised patients. BMC Microbiology 2016; 16: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han XY, Pham AS, Tarrand JJ, Sood PK, Luthra R. Rapid and accurate identification of mycobacteria by sequencing hypervariable regions of the 16S ribosomal RNA gene. Am J Clin Pathol 2002; 118:796–801. [DOI] [PubMed] [Google Scholar]
- 6. Marín M, Muñoz P, Sánchez M, et al. . Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine 2007; 86:195–202. [DOI] [PubMed] [Google Scholar]
- 7. Schuurman T, de Boer RF, Kooistra-Smid AM, van Zwet AA. Prospective study of use of PCR amplification and sequencing of 16S ribosomal DNA from cerebrospinal fluid for diagnosis of bacterial meningitis in a clinical setting. J Clin Microbiol 2004; 42:734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bémer P, Plouzeau C, Tande D, et al. ; Centre de Référence des Infections Ostéo-articulaires du Grand Ouest (CRIOGO) Study Team Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J Clin Microbiol 2014; 52:3583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. University of Washington Department of Laboratory Medicine . Molecular diagnosis microbiology section. Available at: http://depts.washington.edu/molmicdx/mdx/available_tests.shtml. Accessed 19 October 2019. [Google Scholar]
- 10. Basein T, Gardiner BJ, Andujar Vazquez GM, et al. . Microbial identification using DNA target amplification and sequencing: clinical utility and impact on patient management. Open Forum Infect Dis 2018; 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson MR, Sample HA, Zorn KC, et al. . Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 2019; 380:2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol 2019; 14:319–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.