Abstract
The coronavirus disease 2019 (COVID-19) pandemic, which started in China, has created a panic among the general public and health care/laboratory workers. Thus far, there is no medication or vaccine to prevent and control the spread of COVID-19. As the virus is airborne and transmitted through droplets, there has been significant demand for face masks and other personal protective equipment to prevent the spread of infection. Health care and laboratory workers who come in close contact with infected people or material are at a high risk of infection. Therefore, robust biosafety measures are required at hospitals and laboratories to prevent the spread of COVID-19. Various diagnostic platforms including of serological, molecular and other advanced tools and techniques have been designed and developed for rapid detection of SARS-CoV-2 and each has its own merits and demerits. Molecular assays such as real-time reverse transcriptase polymerase chain reaction (rRT-PCR) has been used worldwide for diagnosis of COVID-19. Samples such as nasal swabs or oropharyngeal swabs are used for rRT-PCR. Laboratory acquired infection has been a significant problem worldwide, which has gained importance during the current pandemic as the samples for rRT-PCR may contain intact virus with serious threat. COVID-19 can spread to workers during the sampling, transportation, processing, and disposal of tested samples. Here, we present an overview on advances in diagnosis of COVID-19 and details the issues associated with biosafety procedures and potential safety precautions to be followed during collection, transportation, and processing of COVID-19 samples for laboratory diagnosis so as to avoid virus infection.
Key Words: COVID-19, SARS-CoV-2, Biosafety, Laboratory safety, Diagnosis, Laboratory workers, Laboratory acquired infection
Introduction
Coronavirus disease 2019 (COVID-19) has spread rapidly worldwide, placing the countries around the world on lockdown while posing a huge global threat and panic among mass population (1). As of June 03, 2020, there were 6,287,771 confirmed cases of COVID-19 and 379,941 deaths worldwide (2). Initially, the newly identified coronavirus was named 2019-nCoV, but was later renamed as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) because of its similarities to severe acute respiratory syndrome coronavirus (SARS-CoV) (3). This is the third coronavirus outbreak in two decades, and because of its spread worldwide, a pandemic was declared by the World Health Organization (WHO) on March 11, 2020 (2,4). In just 6 months (as on June 3, 2020), 18, 693 COVID-19-related articles have been published on PubMed, showing the intensity of the situation worldwide.
Presently, there are no antiviral drugs or vaccines available to combat COVID-19. High efforts are being made by researchers of several countries to design and develop effective vaccines, drugs and therapeutics to tackle this pandemic disease (5). Numerous molecular assays and serological assays are available for diagnosis of COVID-19 with varying sensitivity and specificity. The diagnosis of SARS-CoV-2 infected persons/COVID-19 suspected and affected patients need to be carried out accurately and rapidly so as to provide timely treatment and supportive care as well as follow appropriate isolation and quarantine procedures to separate them from naïve population so as to prevent SARS-CoV-2 spread to other individuals. COVID-19 is tested using real-time reverse transcriptase polymerase chain reaction (rRT-PCR), and infected patients are treated symptomatically. Globally, health care workers are under significant pressure not only to curtail the disease and save lives but also to protect themselves from becoming infected. Therefore, health care workers may experience stress because of the panic caused by this pandemic (6). According to the Chinese Centers for Disease Control and Prevention, 1,716 health care workers in China were infected with COVID-19 from a total of 44,672 confirmed cases (3.8%). Contrastingly, other reports assert that more than 3000 health care workers in China were infected with COVID-19 (7, 8, 9). Italy, reports higher morbidity of 10.7% COVID-19 cases among health care workers (10). The mortality rate associated with COVID-19 among health care workers was 0.35% (6 out of 1700 cases), while that associated with severe acute respiratory syndrome (SARS) in 2002 was 1.45% (5 out of 343 cases) (9,11). These data show the importance of having robust biosafety procedures not only for patient care givers like doctors and nurses but also for persons processing the sample for the diagnosis of COVID-19. Health care and laboratory workers are at a high risk of infection because of being in close contact with infected patients and material and a lack of personal protective equipment (PPE) (6). For health care and laboratory workers involved in COVID-19 testing, PPE is the only defense available to avoid being infected (12).
The safety of laboratory workers involved in the diagnosis of COVID-19 is of increased concern as there is no clear picture of infections in laboratory settings. Laboratory-associated infection may occur due to a lack of PPE, improper microbiological techniques, lack of training, and inadequate decontamination protocols (13). A graduate student in Singapore in 2003 and a researcher in Taiwan in 2004 had been infected by SARS in the laboratory setting (13). Thus, there is similar chance of COVID-19 transmission to laboratory workers. Hence, it is important to consider biosafety issues concerning laboratory procedures during the current COVID-19 pandemic. Here, we aimed to address the safety issues faced by laboratory workers involved in the processing COVID-19 samples and detail potential safety precautions that can be implemented to avoid virus infection and prevent the spread of COVID-19. A brief overview on the recent advances in diagnosis of SARS-CoV-2/COVID-19 has also been presented. Biosafety issues during the collection, transportation, and processing of COVID-19 samples for laboratory diagnosis are addressed in detail.
Advances in Diagnosis and Mapping of COVID-19
Real time PCR is the globally used in vitro diagnostic assay for the confirmation of SARS-CoV-2 in clinical samples. Availability of whole genome of SARS-CoV-2 in the public domain led to the development of several RT-PCR methods using different target genes (14). Genes like envelope (E), RNA-dependent RNA polymerase (RdRp), spike (S), nuceloprotein (N), open reading frame (ORF) 1a, ORF 1b and non structural protein (NSP) 2 are employed as targets for rRT-PCR detection of the virus (15, 16, 17). Variations and up gradation of the existing assays are increasing day by day to detect SARS-CoV-2 with improved sensitivity of the assays. RdRp/helicase (Hel) gene targeted rRT-PCR was reported to be more sensitive and has limit of detection of 1.80 TCID50/mL using genomic RNA (18,19). Triplex rRT-PCR targeting N and E gene of SARS-CoV-2 along with an internal control has been developed which had reduced sensitivity as compared with uniplex rRT-PCR (20). Occasional issues of false negative results of rRT-PCR have been reported from various countries (21). As per the report of Foundation for Innovative New Diagnostics (FIND), a WHO Collaborating Centre for Laboratory Strengthening and Diagnostic Technology Evaluation, 313 different molecular assay platforms are available for diagnosis of COVID-19.
The rRT-PCR assay requires sophisticated instruments and consumes time. Hence isothermal amplification assays like loop mediated isothermal amplification assays (LAMP) can be employed as a bed-side test (22,23). This assay can minimize the time required for declaring a result and it is a highly sensitive assay. Recently, LAMP assay targeting SARS-CoV-2 ORF1ab and S gene was developed with 100% specificity and sensitivity (24). Similarly, N gene-based LAMP assay has also been developed with 100% sensitivity but its specificity with SARS and MERS were not tested (25). Combination of Recombinase Polymerase Amplification (RPA) with LAMP assay in a single tube termed as Penn-RAMP has been developed which had higher sensitivity than LAMP assay (26). Other platforms like CRISPR based point-of-care Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) and DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) assays have also been developed for SARS-CoV-2 (27). This assay employs both RPA with CRISPR-cas technology and report shows that CRISPR based assay requires lesser time than rRT-PCR but are less sensitive (28). Metagenomic approaches are used for sequencing the SARS-CoV-2 samples thus enabling identification of the novel virus (29). Hence, next generation sequencing (NGS) platforms can be used for efficient diagnosis of emerging or novel diseases. Due to the immediate need for diagnosing COVID-19 affected individuals, diagnostic potential of the few of the developed assays for detection of SARS-CoV-2 has not been properly elucidated, which needs necessary validation for confirmatory and practical use of the advanced diagnostics during COVID-19 pandemic (30).
An array of antibody-based diagnostic assays has been developed to aid in point-of-care testing but they have the inherent problem of reduced sensitivity and specificity (31). Assays like enzyme-linked immunosorbent assay (ELISA) and lateral flow assay (LFA) were developed for diagnosis of SARS-CoV-2. IgG and IgM assays based on recombinant S and N protein showed results comparable to rRT-PCR (32). Reports show that IgA levels can be a marker for diagnosis of SARS-CoV-2 (33). Other platforms like microfluidics and lab-on-chip assays are under development stages for point-of-care diagnosis of SARS-CoV-2.
The real-time COVID-19 tracking and mapping could be performed by using geographical information systems (GIS) (GIS softwareKosmo® 3.1), web-based real-time dashboards, and advanced information technology platforms for designing useful apps, which would aid in checking the spread of the disease by adopting appropriate and timely prevention and control measures (34, 35, 36, 37).
General Recommendations for Laboratory Workers
Social distancing among laboratory workers is vital as an asymptomatic colleague can infect others. In addition, robust hand hygiene should be followed. The health status of all laboratory workers should be regularly monitored for symptoms, and suspected laboratory workers should be quarantined. PPE, such as a lab coat or gown, face mask, face shield, gloves, goggles, and head cap, should be used during all laboratory procedures (10). Aerosols or droplets are also generated while talking, and therefore, a mask should be worn even outside the laboratory (38). Under experimental conditions, SARS-CoV-2 has been found to be viable in aerosols for 3 h. Thus, PPE should always be worn inside the laboratory to prevent infection (39). Although N95 face masks are recommended for all laboratory work that can generate aerosol droplets, there is a considerable shortage of N95 masks worldwide. If an N95 mask is not available, a surgical mask should be used. All workers should be trained on donning and doffing of PPE and decontamination procedures in case a spill occurs. Extra care should be taken while doffing PPE as there is a high chance of contamination during this procedure. Procedures such as centrifugation and vortexing during nucleic acid extraction can generate aerosols; hence, sealed rotor caps should be used while centrifugation and care should be taken to prevent contamination. Lids should be tightly closed during centrifugation, the speed limit should not be exceeded, plastic centrifuge tubes should be used as glass tubes can break, and centrifuges should be used inside a biosafety cabinet (BSC) class II if possible (10). Before processing COVID-19 samples, each laboratory should conduct a risk assessment, identify the potential hazards, prepare a standard operating procedure, and train laboratory workers to prevent the spread of COVID-19 (40).
Biosafety Concerns During Sample Collection
rRT-PCR has been used extensively worldwide to confirm COVID-19 cases. The CDC recommends using upper respiratory samples such as oropharyngeal or nasopharyngeal samples for confirmation of SARS-CoV-2 using rRT-PCR (41,42). However, acquiring oropharyngeal or nasopharyngeal samples require close contact with suspected patients. SARS-CoV-2 is transmitted through respiratory droplets. Therefore, care should be taken during sample collection (43). In the absence of respirators, eye protection goggles, gloves, and full-sleeved gown, N95 respirators, higher-level respirators, or face shield should be used (44). Increased use of face masks by the general public has led to a shortage of masks for health care/laboratory workers (45,46). Health care workers collecting COVID-19 samples may be at risk of infection if the PPE, especially masks or respirators, are not available or not used properly. Other options for sample collection because of PPE shortage may be the self-collection of saliva samples (early-morning coughed-up sample) (47). Saliva samples have been used for detecting several respiratory viruses, including SARS-CoV (48,49). Walk-in sample collection centers are used in India and South Korea. These centers have a barrier between patients and health care workers, and swabs are taken with a gloved hand (50). Although the effectiveness of these centers has not been validated, the transmission of COVID-19 to the health care worker may be reduced because of the barrier. Movement or unwanted entry inside the sample collection area should be prevented since this can increase the risk of further spread.
Biosafety Concerns During Transport
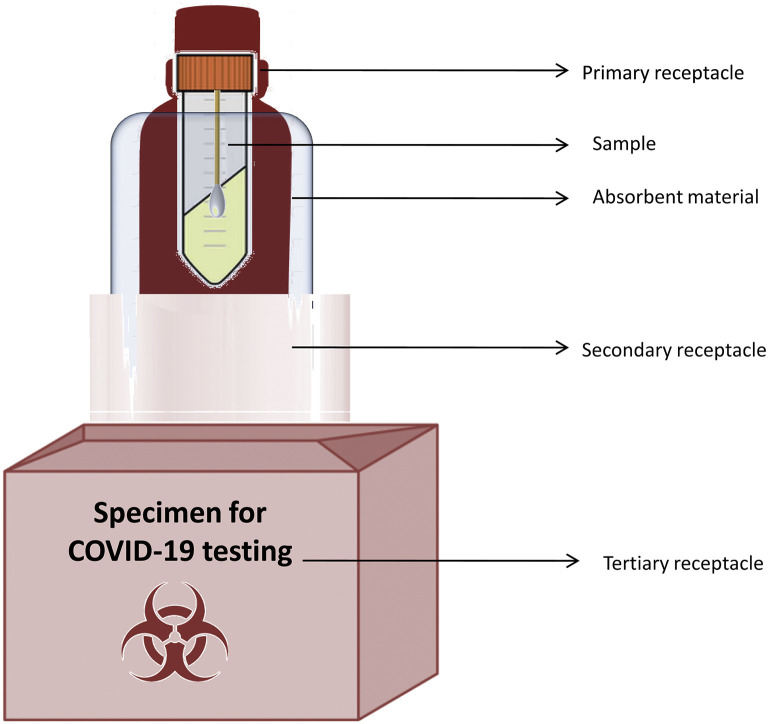
The collected swabs should be transported in a viral transport medium to testing laboratories under cold conditions or at room temperature as per the recommendations (51). If the specimen is being transported a short distance, it should be sealed in a biohazard zip-lock bag or container within a leak-proof cryobox. The outer layer of the box should be disinfected, the labeling of samples should be clear, and a biohazard symbol should be present on the box. All the samples should be labeled properly so that there is no problem for result communication. If the specimens are being transported to another city or country for further processing, triple-layered packaging should be used (10,52). Triple-layered packaging consists of a primary leak-proof receptacle holding the specimen, which is wrapped with a material that can absorb the fluids in case of damage to the receptacle. The primary receptacle should be positioned inside a durable, leak-proof secondary receptacle. More than one primary receptacle can be placed inside the second receptacle with proper absorbent cushioning material in between. The third layer should be a rigid outer packing material that can protect the samples (53). The outer layer should have a label that clearly states “Specimen for COVID-19 testing” and should have details of the transportation company and the receiver (Figure 1 ). People transporting the samples should be aware of the hazardous nature of the sample and should be trained in handling risk group pathogens. In addition, they should be aware of the emergency decontamination procedures in case a spill occurs. Any spill event should be reported to the relevant authorities. The laboratory receiving the sample should be informed in advance regarding the transport of the sample and the number of samples being transported (10).
Figure 1.
Triple-layered packaging for sending infectious samples.
When sending samples on a flight for international transport, UN3373 biological substance, category B regulations should be followed (52). After packing and transporting the sample, the surface should be disinfected with agents that are known to be effective against SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) (Table 1 ). Cultures or isolates of SARS-CoV-2 should be transported as a UN2814 biological substance, category A and labeled as an infectious material affecting humans.
Table 1.
Disinfectants effective against SARS-CoV that can be used against SARS-CoV-2
| S. No. | Disinfectant | Concentration | Contact time | References |
|---|---|---|---|---|
| 1 | Hydrogen peroxide | 0.5% | 1 min | (54) |
| 2 | Sodium hypochlorite | 0.1% | 1 min | (55) |
| 3 | Formaldehyde | 0.7–1% | 2 min | (56) |
| 4 | Glutardialdehyde | 2.5% | 5 min | (57,58) |
| 5 | Ethanol | 78–95% | 30 sec | (56,58,59) |
| 6 | 2- propanol | 70–100% | 30 sec | (56,59) |
| 7 | Povidone iodine | 7.5%,0.23% | 15 sec | (60) |
Once the sample is received by the testing laboratory, the outer container needs to be decontaminated and then sent to a biomedical waste management facility.
Biosafety Concerns During the Processing of Samples
The laboratory should have three separate rooms for extraction of nucleic acid from sample, making a reaction mixture, and amplification. Samples should flow in a single direction to prevent contamination and false-positive amplification. Similarly, separate pipettes should be used for sample preparation, reaction mix preparation, and post-amplification procedures. Filtered tips may be used to prevent sample contamination (61). A separate team of people may work on extraction and amplification procedures. The outer surface of the package should be disinfected with 70% ethanol or 0.1% sodium hypochloride before opening the package (62). The specimens intended to undergo a nucleic acid amplification test should only be opened in a BSC class II A1 or A2 or higher-level biosafety cabinets such as BSC class III (63,64). Based on availability, samples can be handled in biosafety level 3 or 4 containment facilities. The required biosafety levels for various laboratory activities during the processing of SARS-CoV-2 samples, as recommended by the CDC, are presented in Table 2 .
Table 2.
Biosafety level requirements for processing SARS-CoV-2-suspected samples
| S. No. | Laboratory activities | Laboratory facility required |
|---|---|---|
| 1 |
|
Biosafety laboratory level 2 facility |
| 2 |
|
Biosafety laboratory level 2 facility with biosafety laboratory level 3 practices |
| 3 |
|
Biosafety laboratory level 3 facility |
PPE, including respirators or masks, face shields or goggles, gloves, aprons or coveralls, and shoe covers, is required when handling samples as they are not deactivated. All staff working on processing SARS-CoV-2-suspected samples should be trained on biosafety practices and the safe donning and doffing of PPE. Nucleic acid extraction should be performed inside a BSC class II as procedures such as centrifuging and vortexing can generate aerosol droplets. If a centrifuge cannot be accommodated inside a BSC, extra care must be taken while handling the samples. A centrifuge with a closed lid should be used to prevent the spread of aerosol droplets. Samples opened for processing should immediately be transferred to a sample lysis buffer containing a guanidinium-based agent and a non-denaturing detergent that can inactivate the virus (65,66). The addition of lysis buffer not only inactivates the virus but also preserves the RNA from degradation. Alternative methods of inactivation include heating the virus at 56°C for 30 min. However, studies have shown that this not only inactivates the virus but also affects the nucleic acid amplification, producing false-negative results (67). An automatic nucleic acid extractor can be used outside a BSC class II after the inactivation of the sample using a lysis buffer.
Once inactivated, the samples can undergo nucleic acid extraction. Even though theoretically, the samples have been inactivated after the addition of lysis buffer, care should still be taken, as there may be a chance of droplet spread during centrifugation and other procedures. Samples can be stored at −70°C for extended storage or discarded after proper decontamination procedures. The entire working surface and equipment need to be decontaminated with disinfectants such as bleach, followed by 70% ethanol to remove the bleach residues (Table 1). Regular cross-contamination checks of exposed laboratory surfaces should be performed to prevent erroneous results. The PPE used by laboratory workers and the laboratory medical waste generated should be autoclaved and appropriately packed before disposal to a biomedical waste management facility. Biosafety laboratories level 3 or 4 (BSL 3 or 4) handling virus isolation should store the isolates with clear labeling, and an inventory should be maintained to trace back the isolates stored. During the SARS-CoV outbreak, the WHO recommended the destruction of all unnecessary samples. Similarly, SARS-CoV-2-negative samples may be disposed of after decontamination. SARS-CoV isolates recovered during the outbreak were maintained and regularly audited. Similarly, SARS-CoV-2 isolates should be accounted for and external auditing should be performed to prevent the loss of isolates because of dual-use research concerns (68,69). The biosafety measures to be implemented during COVID-19 testing are shown in Figure 2 .
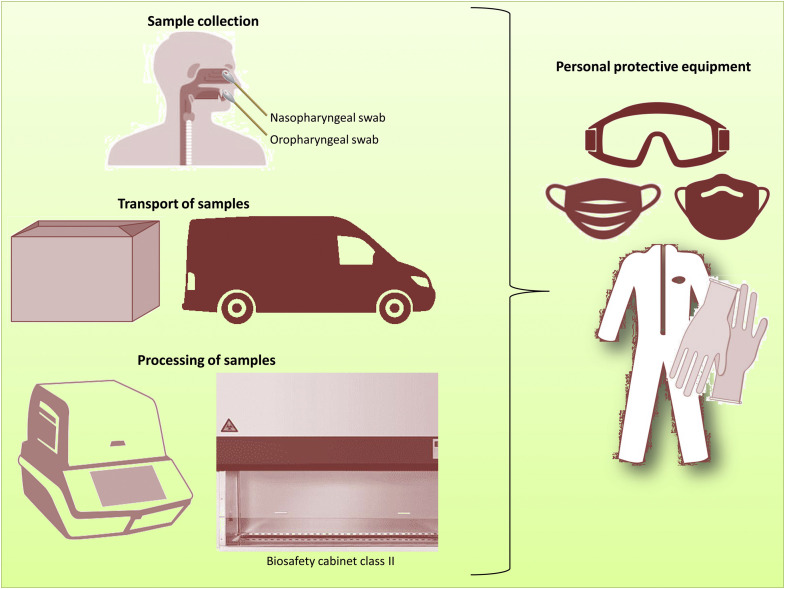
Figure 2.
Personal protective equipment to be used during the sampling, transportation, and processing of COVID-19 samples PPE, such as coveralls or aprons, gloves, masks or respirators, and goggles should be worn at all times from sampling to final processing. During the processing of samples, a biosafety cabinet class II should be used.
Additional Biosafety Concerns
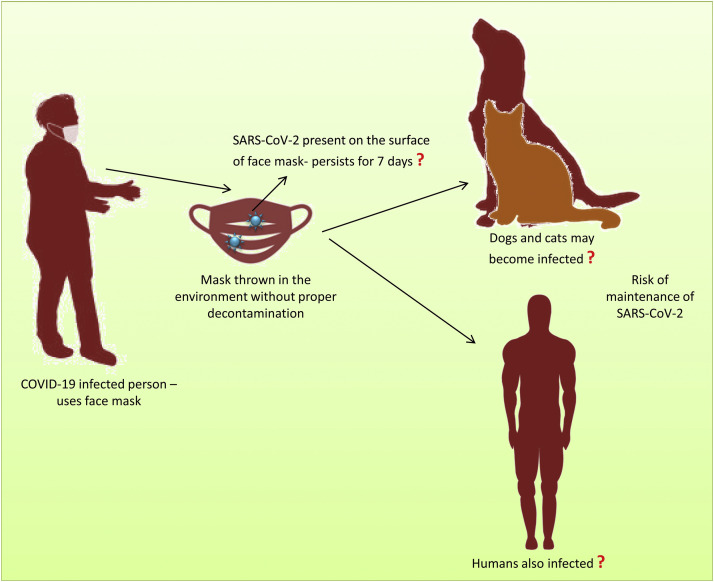
As a general laboratory protocol, all used PPE and laboratory waste generated during the sample processing should be decontaminated before being discarded. Presently masks are used by all people around the world to prevent spread of COVID-19. There is limited awareness among the general public regarding the disposal of masks, and the majority of the masks used are discarded in the environment. It has been found that viruses can be detected at lower levels on the surface of masks, even 7 d after use (63). Hence, face masks should be discarded carefully, and the general public should be informed of the same procedures. Improper disposal of face masks could potentially aid in the spread of COVID-19. Recent reports on the susceptibility of animals to SARS-CoV-2 are also concerning since stray dogs, cats, and other animals may come in contact with these masks, and if this occurs, it will be a significant issue (70) (Figure 3 ).
Figure 3.
Chance of spreading COVID-19 through improper disposal of face masks in the environment.
Similarly, all laboratory waste generated during the processing of COVID-19 samples should be decontaminated before final disposal to a biomedical waste management facility. A list of disinfectants for SARS-CoV-2 and their manufacturers is available at the United States of America Environmental Protection Agency (https://www.epa.gov/pesticide-registration/list-n-disinfectants-use-against-sars-cov-2).
Conclusion
Since the SARS-CoV-2 is novel, there are several gray areas starting from its pathogenesis to treatment. People around the world are afraid about the fast spreading nature of the disease. Presently testing and isolation is the only known method to prevent the spread of SARS-CoV-2. Early diagnosis of COVID-19 is essential to cut the epidemic curve hence robust diagnostic assays are essential that can detect SARS-CoV-2 accurately and swiftly. Health care and laboratory workers are at risk and hence protective wears play a major role to prevent infection. The proper use of PPE can prevent the spread of SARS-CoV-2 infection among health care and laboratory workers. In addition, biosafety measures such as careful sampling techniques, the transportation of samples in leak-proof containers, the use of biosafety cabinets, good microbiological techniques, decontamination of samples and workspace, and accounting for isolates are essential in preventing laboratory-associated SARS-CoV-2 infection.
Acknowledgments
The authors like to acknowledge and thank their individual institutions.
(ARCMED_2020_797)
Declaration of Competing Interest
All authors declare that there exist no commercial or financial relationships that could, in any way, lead to a potential conflict of interest.
References
- 1.Khosrawipour V., Lau H., Khosrawipour T., et al. Failure in initial stage containment of global COVID-19 epicenters. J Med Virol. 2020;92:863–867. doi: 10.1002/jmv.25883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus disease 2019 (COVID-19) Situation Report – 135. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200603-covid-19-sitrep-135.pdf?sfvrsn=39972feb_2
- 3.Gorbalenya A.E., Baker S.C., Baric R.S., et al. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhama K., Sharun K., Tiwari R., et al. Coronavirus disease 2019 – COVID-19. Preprints. 2020 2020030001. [Google Scholar]
- 5.Dhama K., Sharun K., Tiwari R., et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020;18:1–7. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan S., Nabi G., Han G., et al. Novel coronavirus: how the things are in Wuhan. Clin Microbiol Infect. 2020;26(4):399–400. doi: 10.1016/j.cmi.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. J Am Med Assoc. 2020 doi: 10.1001/jama.2020.2648. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Adams J.G., Walls R.M. Supporting the Health Care Workforce During the COVID-19 Global Epidemic. JAMA. 2020;323:1439–1440. doi: 10.1001/jama.2020.3972. [DOI] [PubMed] [Google Scholar]
- 9.Khan S., Siddique R., Ali A., et al. The spread of novel coronavirus has created an alarming situation worldwide. J Infect Public Health. 2020;13:469–471. doi: 10.1016/j.jiph.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lippi G., Adeli K., Ferrari M., et al. Biosafety measures for preventing infection from COVID-19 in clinical laboratories: IFCC Taskforce Recommendations. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-0633. [DOI] [PubMed] [Google Scholar]
- 11.Xu R.H., He J.F., Evans M.R., et al. Epidemiologic clues to SARS origin in China. Emerg Infect Dis. 2004;10:1030–1037. doi: 10.3201/eid1006.030852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowan N.J., Laffey J.G. Challenges and solutions for addressing critical shortage of supply chain for personal and protective equipment (PPE) arising from Coronavirus disease (COVID19) pandemic - Case study from the Republic of Ireland. Sci Total Environ. 2020;725:138532. doi: 10.1016/j.scitotenv.2020.138532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng H., Bilal M., Iqbal H.M.N. Improved Biosafety and Biosecurity Measures and/or Strategies to Tackle Laboratory-Acquired Infections and Related Risks. Int J Environ Res Public Health. 2018;15 doi: 10.3390/ijerph15122697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udugama B., Kadhiresan P., Kozlowski H.N., et al. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 16.Corman V.M., Landt O., Kaiser M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip C.C., Ho C.C., Chan J.F., et al. Development of a Novel, Genome Subtraction-Derived, SARS-CoV-2-Specific COVID-19-nsp2 Real-Time RT-PCR Assay and Its Evaluation Using Clinical Specimens. Int J Mol Sci. 2020;21:2574. doi: 10.3390/ijms21072574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan J.F., Yip C.C., To K.K., et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol. 2020;58:e00310–e00320. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vashist S.K. In Vitro Diagnostic Assays for COVID-19: Recent Advances and Emerging Trends. Diagnostics (Basel) 2020;10:E202. doi: 10.3390/diagnostics10040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waggoner J.J., Stittleburg V., Pond R., et al. Triplex Real-Time RT-PCR for Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis. 2020;26 doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao A.T., Tong Y.X., Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020 doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu L., Wu S., Hao X., et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse tran-scriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic plat-form: iLACO. medRxiv. 2020 doi: 10.1101/2020.02.20.20025874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek Y.H., Um J., Antigua K.J.C., et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect. 2020;9:998–1007. doi: 10.1080/22221751.2020.1756698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan C., Cui J., Huang L., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R., Wu X., Wan Z., et al. A Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Int J Mol Sci. 2020;21:E2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T., Wang Y.C., Shen C.F., et al. Point-of-Care RNA-Based Diagnostic Device for COVID-19. Diagnostics (Basel) 2020;10:165. doi: 10.3390/diagnostics10030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F., Abudayyeh O.O., Gootenberg J.S. Broad Institute; 2020. A Protocol for Detection of COVID-19 Using CRISPR Diagnostics. [Google Scholar]
- 28.Broughton J.P., Deng X., Yu G., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai J.W., Zhang Y., Zhang H.C., et al. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9(1):597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng M.P., Papenburg J., Desjardins M., et al. Diagnostic Testing for Severe Acute Respiratory Syndrome-Related Coronavirus-2: A Narrative Review. Ann Intern Med. 2020:M20–M1301. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Yi Y., Luo X., et al. Development and Clinical Application of A Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. J Med Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong L., Chuan J., Gong B., et al. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci China Life Sci. 2020;63:777–780. doi: 10.1007/s11427-020-1688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma H., Zeng W., He H., et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by a quantitative and sensitive immunoassay. medRxiv. 2020;2004 2017.20064907. [Google Scholar]
- 34.Arab-Mazar Z., Sah R., Rabaan A.A., et al. Mapping the incidence of the COVID-19 hotspot in Iran - Implications for Travellers. Travel Med Infect Dis. 2020;34:101630. doi: 10.1016/j.tmaid.2020.101630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.KamelBoulos M.N., Geraghty E.M. Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: how 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. Int J Health Geogr. 2020;19(1):8. doi: 10.1186/s12942-020-00202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin C., Braund W.E., Auerbach J., et al. Policy Decisions and Use of Information Technology to Fight 2019 Novel Coronavirus Disease, Taiwan. Emerg Infect Dis. 2020;26(7) doi: 10.3201/eid2607.200574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anfinrud P., Stadnytskyi V., Bax C.E., et al. Visualizing Speech-Generated Oral Fluid Droplets with Laser Light Scattering. N Engl J Med. 2020;382(21):2061–2063. doi: 10.1056/NEJMc2007800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doremalen N., Bushmaker T., Morris D.H., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwen P.C., Stiles K.L., Pentella M.A. Safety Considerations in the Laboratory Testing of Specimens Suspected or Known to Contain the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Lab Med. 2020;51:239–242. doi: 10.1093/labmed/lmaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan P.K., To W.K., Ng K.C., et al. Laboratory diagnosis of SARS. Emerg Infect Dis. 2004;10:825–831. doi: 10.3201/eid1005.030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou L., Ruan F., Huang M., et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bahl P., Doolan C., de Silva C., et al. Airborne or droplet precautions for health workers treating COVID-19? J Infect Dis. 2020 doi: 10.1093/infdis/jiaa189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenco M. CDC updates guidance on PPE for health care personnel; COVID-19 declared a pandemic. 2020. https://www.aappublications.org/news/2020/03/11/coronavirus031120 Available from:
- 45.Wu H.L., Huang J., Zhang C.J.P., et al. Facemask shortage and the novel coronavirus disease (COVID-19) outbreak: Reflections on public health measures. EClinicalMedicine. 2020:100329. doi: 10.1016/j.eclinm.2020.100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shortage W.H.O. of personal protective equipment endangering health workers worldwide. 2020. https://www.who.int/news-room/detail/03-03-2020-shortage-of-personal-protective-equipment-endangering-health-workers-worldwide
- 47.To K.K., Tsang O.T., Chik-Yan Yip C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;12 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W.K., Chen S.Y., Liu I.J., et al. Detection of SARS-associated coronavirus in throat wash and saliva in early diagnosis. Emerg Infect Dis. 2004;10:1213–1219. doi: 10.3201/eid1007.031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goff J., Rowe A., Brownstein J.S., et al. Surveillance of Acute Respiratory Infections Using Community-Submitted Symptoms and Specimens for Molecular Diagnostic Testing. PLoS Curr. 2015;7 doi: 10.1371/currents.outbreaks.0371243baa7f3810ba1279e30b96d3b6. ecurrents.outbreaks.0371243baa7f3810ba1279e30b96d3b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S.I., Lee J.Y. Walk-Through Screening Center for COVID-19: An Accessible and Efficient Screening System in a Pandemic Situation. J Korean Med Sci. 2020;35:e154. doi: 10.3346/jkms.2020.35.e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodino K.G., Espy M.J., Buckwalter S.P., et al. Evaluation of saline, phosphate buffered saline and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol. 2020 doi: 10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CDC Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID-19) 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/biosafety-faqs.html Available from:
- 53.World Health Organization (WHO) 3rd edition. 2004. Laboratory biosafety manual; pp. 94–97. (Accessed May 20, 2020) [Google Scholar]
- 54.Omidbakhsh N., Sattar S.A. Broad-spectrum microbicidal activity, toxicologic assessment, and materials compatibility of a new generation of accelerated hydrogen peroxide-based environmental surface disinfectant. Am J Infect Control. 2006;34:251–257. doi: 10.1016/j.ajic.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kampf G., Todt D., Pfaender S., et al. Persistence of coronaviruses on inanimate surfaces and its inactivation with biocidal agents. J Hosp Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabenau H.F., Cinatl J., Morgenstern B., et al. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol. 2005;194:1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kariwa H., Fujii N., Takashima I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatol (Basel, Switzerland) 2006;212(Suppl 1):119–123. doi: 10.1159/000089211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rabenau H.F., Kampf G., Cinatl J., et al. Efficacy of various disinfectants against SARS coronavirus. J Hosp Infect. 2005;61:107–111. doi: 10.1016/j.jhin.2004.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siddharta A., Pfaender S., Vielle N.J., et al. Virucidal Activity of World Health OrganizationRecommended Formulations Against Enveloped Viruses, Including Zika, Ebola, and Emerging Coronaviruses. J Infect Dis. 2017;215:902–906. doi: 10.1093/infdis/jix046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eggers M., Eickmann M., Zorn J. Rapid and Effective Virucidal Activity of Povidone-Iodine Products Against Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and Modified Vaccinia Virus Ankara (MVA) Infect Dis Ther. 2015;4:491–501. doi: 10.1007/s40121-015-0091-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta P. Why is SARS-CoV-2 testing not possible in every medical laboratory? Indian J Pathol Microbiol. 2020;63:173–174. doi: 10.4103/0377-4929.282722. [DOI] [PubMed] [Google Scholar]
- 62.Hong K.H., Lee S.W., Kim T.S., et al. Guidelines for Laboratory Diagnosis of Coronavirus Disease 2019 (COVID-19) in Korea. Ann Lab Med. 2020;40:351–360. doi: 10.3343/alm.2020.40.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chin A.W.H., Chu J.T.S., Perera M.R.A., et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;8:1391–1399. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu D.K.W., Pan Y., Cheng S.M.S., et al. Molecular Diagnosis of a Novel Coronavirus (2019- nCoV) Causing an Outbreak of Pneumonia. Clin Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blow J.A., Dohm D.J., Negley D.L., et al. Virus inactivation by nucleic acid extraction reagents. J Virol Methods. 2004;119:195–198. doi: 10.1016/j.jviromet.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Burton J.E., Easterbrook L., Pitman J., et al. The effect of a non-denaturing detergent and a guanidinium-based inactivation agent on the viability of Ebola virus in mock clinical serum samples. J Virol Methods. 2017;250:34–40. doi: 10.1016/j.jviromet.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 67.Pan Y., Long L., Zhang D., et al. Potential false-negative nucleic acid testing results for Severe Acute Respiratory Syndrome Coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. 2020 doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim W., Ng K.C., Tsang D.N. Laboratory containment of SARS virus. Ann Acad Med Singapore. 2006;35:354–360. [PubMed] [Google Scholar]
- 69.Ma S., Gao P., Lu D., Wang G. Prudently conduct the engineering and synthesis of the SARS-CoV-2 virus. Synth Syst Biotechnol. 2020;5(2):59–61. doi: 10.1016/j.synbio.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi J., Wen Z., Zhong G., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]