Abstract
Bayesian analyses with the arm-based (AB) network meta-analysis (NMA) model require researchers to specify a prior distribution for the covariance matrix of the treatment-specific event rates in a transformed scale, e.g. the treatment-specific log-odds when a logit transformation is used. The commonly-used conjugate prior for the covariance matrix, the inverse-Wishart (IW) distribution, has several limitations. For example, although the IW distribution is often described as non- or weakly-informative, it may in fact provide strong information when some variance components are small (e.g., when the standard deviation of study-specific log-odds of a treatment is smaller than 1/2), as is common in NMAs with binary outcomes. In addition, the IW prior generally leads to underestimation of correlations between treatment-specific log-odds, which are critical for borrowing strength across treatment arms to estimate treatment effects efficiently and to reduce potential bias. Alternatively, several separation strategies (i.e., separate priors on variances and correlations) can be considered. To study the IW prior’s impact on NMA results and compare it with separation strategies, we did simulation studies under different missing-treatment mechanisms. A separation strategy with appropriate priors for the correlation matrix and variances performs better than the IW prior, and should be recommended as the default vague prior in the AB NMA approach. Finally, we re-analyzed three case studies and illustrated the importance, when performing AB-NMA, of sensitivity analyses with different prior specifications on variances.
Keywords: Bayesian inference, covariance matrix, network meta-analysis, prior
1. INTRODUCTION
In recent decades, many systematic reviews and meta-analyses have summarized existing evidence on a given scientific question from multiple independent studies. Traditionally, a meta-analysis of randomized controlled trials compares only two treatments, typically an intervention and a control. With new treatments emerging, network meta-analysis (NMA) has been developed and is increasingly used to synthesize direct and indirect evidence comparing multiple treatments1,2. For instance, if three treatments A, B, and C are available for a certain disease then in NMA, comparing treatments A and C provides direct evidence about A versus C while comparing A versus B plus B versus C offers indirect evidence. Both Bayesian hierarchical approaches3,4,5 and frequentist methods6,7 have been proposed for NMA; this article focuses on Bayesian methods because they have been widely applied.
Broadly, there are two Bayesian approaches for NMA: the arm-based (AB) approach5,8, which models absolute effects, and the contrast-based (CB) approach3,4,9, which models relative effects. Dias and Ades11 and Hong et al.12 thoroughly discussed the differences between these two approaches. The arm-based model has been shown to have several important advantages, e.g., the results produced by the AB-NMA are generally more robust to the choice of treatments to include in the NMA10. Both approaches involve estimating a covariance matrix of random effects and choosing a prior distribution for it, which are generally difficult in Bayesian analysis: the number of parameters in a covariance matrix increases rapidly with the dimension of the matrix, and these parameters are constrained because the matrix must be non-negative definite14. Also, the parameters of the two models are distinct. In the CB approach, if the total number of treatments in the NMA is T, then T − 1 variance parameters need to be estimated for contrasts and (T − 1)(T − 2)/2 parameters for correlations between contrasts but the T − 1 variances are constrained by triangle inequalities4, which complicate prior specification for heterogeneous variances. The AB approach estimates more parameters (i.e., a T-dimensional covariance matrix for variances of absolute effects and correlations between them), but does so without constraints other than positive definiteness. Finally, each study generally includes only a small portion of the NMA’s treatments, generally based on results of previous trials. This selection of treatments produces missing data (treatments excluded from a trial), which affects the AB and CB approaches differently because they use different exchangeability assumptions: CB models require contrasts to be missing at random (MAR)15 while AB models require treatments to be MAR8. Such differences could have distinct effects on estimates of the covariance matrix.
To model the variance structure of contrasts in the CB approach, Lu and Ades4 used an ancillary representation to circumvent the triangle inequalities and compared different prior specifications using case studies. However, in the AB approach little attention has been paid to the choice of priors for covariance matrices and their influence on the results. The obvious choice is the conjugate inverse-Wishart prior, which was used by Zhang et al.5 and Hong et al.8 However, this prior has some problems: the marginal distribution of the variances has low density in the region near zero16 and the prior imposes a dependency between the variances and the correlations17, which may cause the correlations to be underestimated in certain situations. Alternatives have been proposed, such as the scaled inverse Wishart18, a separation strategy14, the Cholesky decomposition19, and the LKJ prior for a correlation matrix20. This article compares, for binary outcomes, the influence of selected priors for the AB model on the estimation of the log odds ratios of treatment comparisons and the correlations between treatment-specific log-odds, under different missing-treatment mechanisms. Based on these comparisons, we aim to recommend appropriate vague priors for the AB model’s covariance matrix.
The rest of this article is organized as follows. Section 2 describes AB approaches for NMA, followed by Section 3 on Bayesian analysis, focusing on priors for the covariance matrix. Section 4 presents simulation studies and results, followed by three case studies in Section 5. Section 6 presents our main conclusions with a brief discussion.
2. THE ARM-BASED NETWORK META-ANALYSIS
2.1. Notation
Suppose we have collected K studies comparing a total of T treatments and each study contains only a subset of the T treatments. Let Ak (k = 1, … , K) be the subset of treatments investigated in the kth study. If the number of elements in the set Ak (denoted by |Ak|) is larger than 2, then study k is called multi-armed. Most randomized clinical trials are two-arm studies with |Ak| = 2. Let Dk = {(rkt, nkt), t ∊ Ak} be the data collected in the kth study, where rkt and nkt are the numbers of events and total subjects in the tth treatment group in the kth study. Finally, let pkt be the true probability of an event (i.e., absolute risk) for the tth treatment in the kth study.
2.2. The arm-based approach
Zhang et al.5 proposed the following arm-based NMA model:
| (1) |
where μt represents the overall fixed effect of treatment t and the vector (vk1, … , vkT)′ is a random effect specific to study k, following the multivariate normal distribution with mean 0 and covariance matrix Σ having dimension T. Here, a′ denotes the transpose of the vector a. We used logit instead of probit transformation to estimate both marginal and conditional log odds between treatments. Let δt be the between-study standard deviation of log-odds for treatment t, i.e., the square root of the tth diagonal element in the covariance matrix Σ Here, Σ can also be written as ΔPΔ with Δ a diagonal matrix having standard deviation δt as its tth diagonal element, and correlation matrix P with entries ρij.
The marginal event rate of treatment t is pt = E[pkt|μt, δt]; for the logit link as in Equation (1), pt can be approximated as in Zeger et al.,21
Using these marginal absolute risks of the T treatments, the risk ratio (RR) and risk difference (RD) for each pair of treatments can be estimated accordingly as RRij = pi/pj and RDij = pi − pj. We can also compute two log odds ratio estimands: 1) the marginal log odds ratio between treatments i and j , and 2) the conditional log odds ratio cLORij = μi – μj, which is more commonly used in the meta-analyses literature. Agresti22 (pp. 496–497) provided further details differences between these two LORs. Other link functions could also be used in Equation (1). For example, using the probit link Φ−1(pkt) = μt + vkt as in Zhang et al.5, the marginal absolute risk has an exact from, , where Φ(·) and Φ−1(·) denote the cumulative distribution function of the standard normal distribution and its inverse, respectively. When the logit link is used, both cLOR and mLOR can be estimated; using the probit link, only mLOR can be directly estimated.
3. PRIOR SPECIFICATIONS FOR THE COVARIANCE MATRIX
This section describes specifications of prior distributions for the AB-NMA in Bayesian analysis. Specifically, we place vague N(0, 1002) priors on μt (t = 1, … , T), and discuss prior distributions for the covariance matrix Σ. Two common ways to specify the prior distribution for the covariance matrix Σ are the natural conjugate prior for the multivariate normal likelihood, which treats the covariance matrix as a whole, and the separation strategy proposed by Barnard et al.14, which decomposes the covariance matrix into separate parts as Σ = ΔPΔ and assigns priors to the components Δ and P separately. As above, Δ is a diagonal matrix with standard deviation δi as its ith diagonal element, and P is a correlation matrix with diagonal elements 1 and off-diagonal elements ρij. The following subsections give more details about these two methods and their variations.
3.1. The Inverse-Wishart prior
The inverse-Wishart (IW) distribution for the covariance Σ is a conjugate prior for the multivariate normal likelihood, which can speed up computation compared to other priors. The density function of the IW distribution with degrees of freedom m (> T − 1) and positive definite scale matrix ψ is:
| (2) |
Where | · | and tr(·) denote the determinant and trace of a matrix, respectively. The scale matrix ψ is often selected to be the T × T identity matrix I.
We now give some remarks about the IW prior’s properties and problems. The marginal distribution of each (the ith diagonal element of Σ) is the inverse-gamma distribution for i = 1, … , T, where is the shape parameter and is the scale parameter. Here, ψii is the ith diagonal element of ψ.
The IW prior has limitations. For example, simulation studies by Alvarez et al.17 showed that although the estimated covariances (posterior means) Σij (i ≠ j) are unbiased, the estimated correlations (posterior means) ρij are biased towards zero and the estimated variance (posterior mean) Σii is biased upward when the true variance Σii is small. These biases are caused by the lack of prior density near zero in the marginal prior for the variance and the dependence induced by the IW prior between the correlation matrix P and the variances. Section 4’s simulation studies also illustrate this problem.
Finally, the IW prior does not allow a user to specify different amounts of prior knowledge about different variance components; the single parameter m controls this uncertainty for all diagonal elements. To add more flexibility, the scaled IW prior18 and the hierarchical half-t prior23 have been proposed; however, such flexibility may be limited compared to the separation strategy17.
3.2. The separation strategy
As mentioned, the separation strategy allows more flexibility by decomposing the covariance matrix Σ as ΔPΔ and placing independent prior distributions on the standard deviations δi and the correlation matrix P. Popular priors for δi include the inverse-gamma prior for the variance , the uniform prior between 0 and a certain upper bound, the half-Cauchy16 and the log-normal.14 There is less consensus about choice of a prior for the correlation matrix P, given the difficulty of constraining p to be positive definite. The following subsections elaborate this by discussing four possible choices.
3.2.1. The restricted inverse-Wishart prior and restricted Wishart Prior
We start with the restricted inverse-Wishart (RIW) prior mentioned by Barnard et al.14, where the correlation matrix P follows an IW distribution with the restriction that its diagonal elements are fixed as 1. Specifically, let Q ~ IWT(I, m), then P = ΔQΔ follows a RIWT(m) distribution, where Δ is a diagonal matrix with ith diagonal element and Qii is the ith diagonal element of Q. In the RIW prior, the ρij have the same marginal distributions for all i ≠ j:
| (3) |
a beta distribution, on the interval [−1, 1], which is uniform if m = T + 1. Hence, the RIWT(T + 1) prior implies marginally uniform-distributed correlation coefficients. This prior is distinguished from the jointly uniform prior π(P) ∝ 1, which is a special case of the LKJ prior20. Actually the LKJ prior is equivalent, in a specific sense, to the restricted Wishart (RW) prior, which we now discuss. (Appendix A gives the proof.)
To define the RW prior, as for the RIW prior let Q* follow a Wishart distribution WT(I, m*), then P = ΔQ*Δ follows a restricted Wishart distribution RWT(m*) with degrees of freedom m*(> T − 1); again, Δ is a diagonal matrix with ith diagonal element . The RW prior has density π(P) ∝ |P|(m*−T−1)/2; if m* = T + 1, this is uniform on a compact subspace of the T(T − 1)/2 dimensional hypercube (−1, 1)T(T−1)/2. Also, the ρij for all i ≠ j follow the same beta distribution on [−1, 1], which is if m* = T + 1. Based on this, the marginal distributions of the correlation coefficients ρij in the jointly uniform prior tend to place density close to zero as the dimension T increases, which is the key difference from the marginally uniform prior. Furthermore, the conditional distribution of the correlation coefficient ρij given ρrs with (i, j) ≠ (r, s) in the RW prior also differs from that in the RIW prior.
Although the closed form for the conditional distribution of ρij given ρrs only exists for special cases, we can visualize these conditional distributions as in Figure 1, which compares the RIW and RW distributions when the dimension T is 3 or 10. As shown in Figure 1, the RIW distribution puts a marginally uniform prior on each pij while the marginal prior on the individual correlation p12 under the RW distribution (which is jointly uniform) concentrates around zero, more so as the dimension T increases. Figure 1 also shows that compared to the RW distribution, the RIW distribution puts more density close to 1 for ρ12 when both ρ23 and ρ13 are larger than 0.6. Interestingly, of 1,000,000 random draws from the RW distribution with T = 10, only two satisfied the condition that both ρ23 and ρ13 are larger than 0.8. By contrast, randomly drawing correlation matrices from the RIW distribution, 26,174 out of 1,000,000 satisfied this condition. The above findings may have implications for prior choice in applications. For example, in NMA, we might prefer the RIW prior to the RW prior because the RW prior places less density on all correlation elements ρij being large.
FIGURE 1.
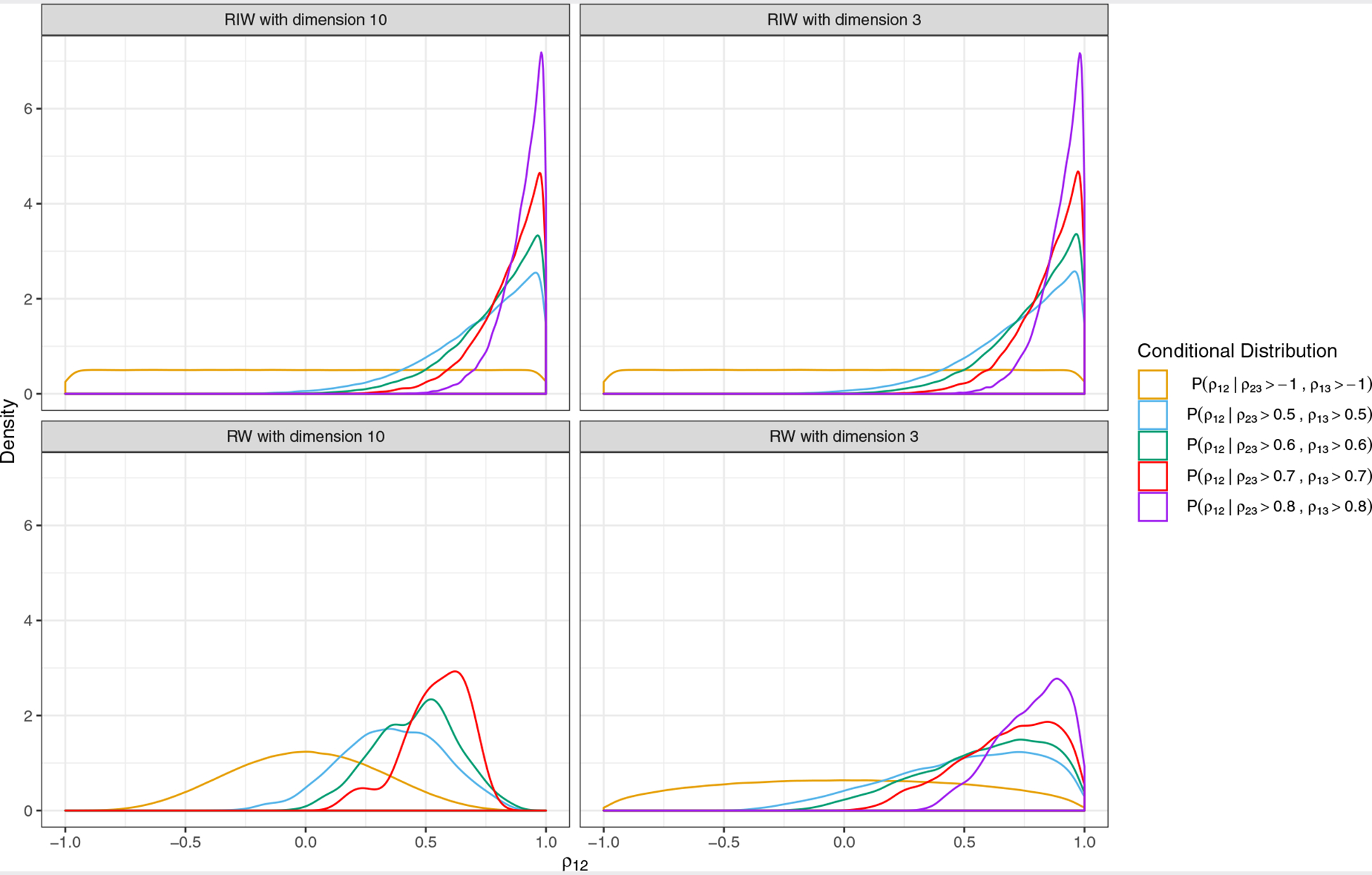
Conditional prior densities for ρ12 with dimension T = 3 (right panels) and dimension T = 10 (left panels). Each distribution is estimated from 1,000,000 random draws from the respective distribution (restricted inverse-Wishart in the first row and restricted Wishart in the second row), using the R function geom_density() with default settings to compute and draw kernel density estimates. Different colors denote different conditioning criteria; e.g., the purple line is the conditional distribution of ρ12 given ρ23 and ρ13 both larger than 0.8. We did not draw the purple density for the restricted Wishart with dimension 10 because only 2 out of 1,000,000 random draws had ρ23 and ρ13 both greater than 0.8.
3.2.2. Exchangeable correlation structure
To reduce model complexity, Lin et al.24 proposed an exchangeable structure (EQ prior) for P, where all off-diagonal elements ρij are assumed equal to a common value ρ. To keep P positive definite, ρ must be larger than , so we may specify a vague uniform prior for ρ on .
4. SIMULATION STUDIES
We conducted simulation studies to compare the performance of AB-NMA using different priors (IW, RW, RIW and EQ) in terms of bias and coverage probability, under different mechanisms for selecting treatment arms to be included in each study of the AB-NMA.
4.1. Simulation settings
We describe the simulation studies using three main steps: first, how we generated a complete data set; second, how we applied the missing data mechanisms to omit some arms from studies; and third, which estimands and priors we chose. In the simulation, we fit the AB-NMA with different priors to each simulated dataset, saved estimates of the estimands, and described the performance measures.
Each simulated NMA dataset {D1, … , DK} had binary outcome data Dk = {(rkt, nkt),t = 1, 2, 3} from 18 studies (K = 18) comparing three treatments (T = 3), denoted 1, 2, and 3. The number of patients nkt in each arm was fixed at 500. The number of simulated datasets in each setting was 1000. First, we generated complete datasets under the AB model specification in Equation (1) using the logit link and setting (μ1, μ2, μ3) = (μ2 + 0.5, μ2, μ2 − 0.5), (vk1, vk2, vk3)′ ~ MV N(0, Σ) where Σ = ΔPΔ with standard deviations (δ1, δ2, δ3) and correlation matrix P with entries (ρ12, ρ13, ρ23). In Scenario I, we chose (δ1, δ2, δ3) = (0.7, 0.4, 0.1), (μ1, μ2, μ3) = (−1.0, −1.5, −2.0) and . We also conducted additional simulations and analyses to confirm our findings under different correlation structures and μ’s. For these, we set (δ1, δ2, δ3) = (0.7, 0.4, 0.1) and considered three different values of μ2 ∈ {−0.5, −1.5, −2.5} with μ1 = μ2 + 0.5 and μ3 = μ2 − 0.5. We also considered three choices for P: high correlation scenario with a common between-treatment correlation ρ = 0.7, low correlation scenario with ρ = 0.2, and mixed between-treatment correlation scenario with (ρ12, ρ13, ρ23) = (0.7, 0.1, 0.4).
After generating the complete dataset, we omitted treatment arms to create partially missing data under MAR and MNAR with respect to absolute effects. For each missingness setting, we obtained two different sets of nine two-arm studies: one set compared treatments 1 and 2, and another compared treatments 2 and 3. To generate partially missing data under the MAR assumption, we first kept all treatment 2 data observed and ranked the studies in ascending order by rk2/nk2. Then, we made treatment 1 missing in the first nine studies in this ordering and treatment 3 missing in the last nine studies. This created datasets in which the results for the control arm (treatment 2) had improved over time (rk2/nk2 increased with k), while only studies with treatment 1 (more advanced regimen) versus 2 were available in the more recent period and studies with treatment 3 (less advanced regimen) versus 2 were available in the earlier period. Next, we used the following strategy to create missing data under MNAR with respect to absolute effects. We used mkt (k = 1, … , K and t = 1, … , T) to indicate missingness of the tth treatment in the kth study; mkt = 1 indicated missing and mkt = 0 indicated not missing. First, we assumed all treatment 2 data were observed, so mk2 = 0, k = 1, … , K. Then we determined the missingness of treatment 1 based on the data for all 3 treatments. After determining mk1, the missingness of treatment 3 followed automatically: mk3 = 1 − mk1. The model to generate the missingness indicators was:
| (4) |
where πk1 is the probability of treatment 1 being missing in study k. The parameters β0 and β1 were pre-defined to control the average number of studies without treatment 1 to be 9 in each scenario: β1 = 1 and β0 = −μ1 – μ2 – μ3. A continuity correction of 0.5 was applied to both r and n when rki was zero.
We focus on these estimands: the two kinds of log odds ratio comparing treatments i and j, and cLORij = μi – μj, the absolute risk of an event for treatment t (pt), the standard deviation of log-odds for treatment t (δt), and the correlation between treatment-specific (treatments i and j) log-odds (ρij). When applying the AB-IW model, we set the prior for the covariance matrix Σ to be IWT(I, T + 1). For models using the RW, RIW and EQ priors, we imposed independent uniform priors U(0, 5) on standard deviations δi, i = 1, … , T, then set the RWT(T + 1), RIWT(T + 1) prior for the correlation matrix P and the uniform prior for the correlation coefficient ρ, respectively. Clearly, the true standard deviations (δ1, δ2, δ3) = (0.7, 0.4, 0.1) were not close to the center of the prior U(0,5), nor were the true correlations close to the center of the RW, RIW, and EQ priors.
We implemented the models using Stan25 in conjunction with R26. We chose posterior mean and 95% equal tailed credible interval as point and interval estimates respectively. To measure the performance of different methods, we used bias and coverage probability of 95% credible interval.
4.2. Simulation results
Table 1 summarizes bias of the estimates and coverage probability (CP) of the 95% credible intervals given by four AB models (AB-IW, AB-RW, AB-RIW and AB-EQ) under different missingness settings (no missing, MAR, and MNAR). The table includes the log odds ratio comparing treatments i and j, mLORij and cLORij, the absolute risk of an event for treatment t (pt), the standard deviation of log-odds for treatment t (δt), and the correlation between treatment-specific (treatments i and j) log-odds (ρij).
TABLE 1.
Simulation results comparing data generated under Scenario I with (δ1, δ2, δ3) = (0.7, 0.4, 0.1), (μ1, μ2, μ3) = (−1.0, −1.5, −2.0) and Performance of AB models (IW, RW, RIW, and EQ) under different missingness mechanisms (No missing, MAR, MNAR) is shown: the bias of estimates (posterior mean) of cLORij, mLORij, pt, δt, and ρij and coverage probability of 95% credible intervals. MCMC errors are at the 0.01 level.
| Bias (Coverage Probability) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Truth | AB-IW | AB-RW | AB-RIW | AB-EQ | AB-IW | AB-RW | AB-RIW | AB-EQ | AB-IW | AB-RW | AB-RIW | AB-EQ |
| Scenario I | |||||||||||||
| Missing Setting | No missing | MAR | MNAR | ||||||||||
| cLOR12 | 0.50 | −0.001(0.976) | −0.003(0.970) | −0.003(0.962) | −0.003(0.964) | 0.243(0.900) | 0.136(0.954) | 0.087(0.964) | 0.032(0.962) | −0.226(0.884) | −0.186(0.921) | −0.172(0.918) | −0.151(0.916) |
| cLOR13 | 1.00 | 0.010(0.957) | 0.001(0.950) | 0.000(0.946) | 0.001(0.937) | 0.293(0.904) | 0.174(0.946) | 0.120(0.959) | 0.034(0.970) | −0.240(0.927) | −0.205(0.921) | −0.188(0.927) | −0.155(0.937) |
| cLOR23 | 0.50 | 0.011(0.990) | 0.004(0.965) | 0.004(0.961) | 0.004(0.960) | 0.050(1.000) | 0.039(0.977) | 0.033(0.980) | 0.003(0.978) | −0.014(0.999) | −0.019(0.970) | −0.017(0.976) | −0.004(0.965) |
| mLOR12 | 0.54 | −0.010(0.979) | 0.002(0.969) | 0.002(0.965) | 0.003(0.966) | 0.208(0.913) | 0.145(0.956) | 0.112(0.966) | 0.066(0.967) | −0.230(0.871) | −0.164(0.923) | −0.150(0.921) | −0.133(0.922) |
| mLOR13 | 1.07 | −0.012(0.962) | 0.012(0.957) | 0.014(0.958) | 0.015(0.945) | 0.221(0.950) | 0.187(0.944) | 0.148(0.961) | 0.072(0.971) | −0.280(0.898) | −0.181(0.927) | −0.162(0.933) | −0.133(0.936) |
| mLOR23 | 0.54 | −0.002(0.994) | 0.011(0.965) | 0.012(0.964) | 0.012(0.964) | 0.013(1.000) | 0.041(0.980) | 0.036(0.980) | 0.006(0.979) | −0.050(0.998) | −0.016(0.973) | −0.012(0.978) | −0.001(0.966) |
| p1 | 0.28 | 0.003(0.940) | 0.005(0.945) | 0.005(0.951) | 0.005(0.947) | 0.051(0.855) | 0.038(0.948) | 0.032(0.959) | 0.021(0.965) | −0.037(0.864) | −0.025(0.926) | −0.022(0.930) | −0.019(0.928) |
| p2 | 0.19 | 0.003(0.977) | 0.002(0.959) | 0.003(0.962) | 0.003(0.958) | 0.003(0.974) | 0.003(0.962) | 0.003(0.962) | 0.003(0.960) | 0.003(0.975) | 0.003(0.964) | 0.003(0.964) | 0.003(0.966) |
| p3 | 0.12 | 0.002(1.000) | 0.000(0.965) | 0.000(0.965) | 0.000(0.963) | 0.002(1.000) | −0.003(0.976) | −0.002(0.974) | 0.001(0.981) | 0.008(1.000) | 0.003(0.956) | 0.003(0.960) | 0.002(0.962) |
| δ1 | 0.70 | 0.023(0.964) | 0.052(0.948) | 0.064(0.936) | 0.067(0.934) | −0.044(0.980) | 0.086(0.969) | 0.131(0.957) | 0.132(0.950) | −0.038(0.975) | 0.063(0.957) | 0.077(0.956) | 0.079(0.958) |
| δ2 | 0.40 | 0.068(0.932) | 0.029(0.961) | 0.037(0.957) | 0.038(0.954) | 0.066(0.938) | 0.034(0.958) | 0.039(0.953) | 0.039(0.957) | 0.067(0.934) | 0.034(0.955) | 0.040(0.950) | 0.039(0.955) |
| δ3 | 0.10 | 0.202(0.000) | 0.004(0.980) | 0.005(0.981) | 0.005(0.979) | 0.296(0.000) | 0.035(0.977) | 0.041(0.975) | 0.041(0.977) | 0.295(0.000) | 0.031(0.988) | 0.032(0.986) | 0.032(0.982) |
| ρ12 | 0.67 | −0.170(0.927) | −0.100(0.961) | −0.067(0.960) | −0.060(0.955) | −0.454(0.754) | −0.330(0.955) | −0.270(0.971) | −0.180(0.982) | −0.359(0.786) | −0.247(0.939) | −0.200(0.948) | −0.146(0.959) |
| ρ13 | 0.44 | −0.316(0.914) | −0.196(0.986) | −0.091(0.989) | 0.162(0.820) | −0.430(1.000) | −0.406(1.000) | −0.351(0.999) | 0.042(0.970) | −0.411(1.000) | −0.363(1.000) | −0.280(1.000) | 0.076(0.956) |
| ρ23 | 0.67 | −0.511(0.098) | −0.293(0.974) | −0.228(0.983) | −0.060(0.955) | −0.623(0.293) | −0.546(0.983) | −0.508(0.989) | −0.180(0.982) | −0.593(0.211) | −0.471(0.973) | −0.423(0.984) | −0.146(0.959) |
All four AB models gave unbiased estimates and good coverage probabilities for the log odds ratios mLORij and cLORij and the absolute risks pt for complete datasets (“no missing”, i.e., no omitted arms). However, the AB-IW model produced biased estimates of the standard deviations δt and correlations ρij and intervals with low coverage probabilities. In particular, the AB-IW method had bias 0.202 for δ3, which was large relative to the true value δ3 = 0.1, while all AB-RW, AB-RIW and AB-EQ had almost no bias (0.005). Also, for the IW prior the bias of ρ23 (−0.511) was so large that this method estimated little posterior association between treatments 2 and 3, although the true correlation was 0.667. In summary, the IW prior gave upwardly biased estimates for small variances and correlation estimates biased towards zero, especially when the variance was small. In contrast, the AB-RW, AB-RIW and AB-EQ priors gave estimates of standard deviations and correlations with much smaller biases (albeit not exactly zero) with good coverage probability.
For complete datasets, mis-estimating correlations between treatment-specific log-odds did not bias estimates of log odds ratios and absolute risks. However, in datasets with missing entries (arms omitted under MAR/MNAR), underestimation of correlations, especially for the star-shaped network structure used here, gave estimated relative effects with larger bias and worse coverage probability. Underestimation of correlations increased under MAR/MNAR compared to complete data for all priors considered; e.g., for AB-IW, the estimated bias for (ρ12, ρ13, ρ23) was (−0.17, −0.32, −0.51), (−0.45, −0.43, −0.62) and (−0.36, −0.41, −0.59) in the no missing, MAR, and MNAR scenarios, respectively. Also, while the AB-RIW and AB-EQ priors gave similar results under MNAR for mLORij and cLORij, the extra but reasonable assumption of equal correlations between treatment-specific log-odds reduced the bias of estimated correlations and log odds ratios under MAR. The performance of AB-RW prior is slightly worse than AB-RIW prior in terms of estimated relative effects and correlations because RW prior places less density on all correlation elements being large, as was mentioned regarding Figure 1.
We verified our findings using additional simulations, shown in Table 2, under various μ’s and correlation structures. The AB-RW prior gave smaller bias and higher coverage probability than the AB-IW prior for mLOR13 (cLOR13), the AB-RIW prior gave smaller bias and higher coverage probability than the AB-RW prior for mLOR13 (cLOR13), and the AB-EQ prior gave much smaller bias than the AB-RIW prior for mLOR13 (cLOR13) under MAR/MNAR. Also, AB-RIW and AB-EQ generally provided more reliable estimates of absolute risks (p1) than AB-IW. These results were in line with our findings in Table 1.
TABLE 2.
Additional simulations comparing performance (bias and coverage probability of 95% credible intervals) of AB-IW, AB-RIW and AB-EQ with respect to posterior mean of cLOR13, mLOR13 and p1 under MAR and MNAR using data generated with (μ1, μ2, μ3) = (μ2 + 0.5, μ2, μ2 − 0.5), (δ1, δ2, δ3) = (0.7, 0.4, 0.1), μ2 ∈ {−0.5, −1.5, −2.5} and correlation matrix Pchosen from low, high and mixed correlation scenarios. MCMC errors are at the 0.01 level.
| cLOR13: Bias (Coverage Probability) | mLOR13: Bias (Coverage Probability) | p1: Bias (Coverage Probability) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario | μ2 | Correlation structure | Truth | AB-IW | AB-RW | AB-RIW | AB-EQ | Truth | AB-IW | AB-RW | AB-RIW | AB-EQ | Truth | AB-IW | AB-RW | AB-RIW | AB-EQ |
| MAR | |||||||||||||||||
| 1 | −0.50 | High | 1.00 | 0.302(0.883) | 0.175(0.945) | 0.117(0.957) | 0.036(0.967) | 1.00 | 0.263(0.913) | 0.170(0.948) | 0.119(0.959) | 0.048(0.968) | 0.50 | 0.059(0.847) | 0.033(0.948) | 0.023(0.954) | 0.011(0.967) |
| 2 | −0.50 | Low | 1.00 | 0.090(0.979) | 0.058(0.981) | 0.045(0.978) | −0.063(0.978) | 1.00 | 0.061(0.983) | 0.053(0.980) | 0.042(0.977) | −0.045(0.977) | 0.50 | 0.017(0.961) | 0.011(0.976) | 0.009(0.971) | −0.009(0.976) |
| 3 | −0.50 | Mixed | 1.00 | 0.280(0.896) | 0.152(0.946) | 0.103(0.962) | 0.042(0.961) | 1.00 | 0.241(0.920) | 0.147(0.945) | 0.105(0.958) | 0.049(0.961) | 0.50 | 0.057(0.852) | 0.031(0.949) | 0.022(0.955) | 0.016(0.953) |
| 4 | −1.50 | High | 1.00 | 0.291(0.895) | 0.159(0.953) | 0.099(0.965) | 0.017(0.971) | 1.07 | 0.218(0.939) | 0.170(0.954) | 0.126(0.962) | 0.053(0.972) | 0.28 | 0.051(0.858) | 0.034(0.951) | 0.027(0.962) | 0.018(0.966) |
| 5 | −1.50 | Low | 1.00 | 0.093(0.982) | 0.051(0.979) | 0.035(0.977) | −0.069(0.977) | 1.07 | 0.043(0.990) | 0.087(0.980) | 0.081(0.977) | −0.011(0.976) | 0.28 | 0.020(0.965) | 0.024(0.977) | 0.024(0.977) | 0.007(0.979) |
| 6 | −1.50 | Mixed | 1.00 | 0.280(0.903) | 0.151(0.947) | 0.097(0.961) | 0.026(0.968) | 1.07 | 0.207(0.946) | 0.160(0.950) | 0.121(0.960) | 0.055(0.966) | 0.28 | 0.052(0.856) | 0.036(0.946) | 0.029(0.955) | 0.023(0.959) |
| 7 | −2.50 | High | 1.00 | 0.304(0.913) | 0.181(0.946) | 0.124(0.965) | 0.031(0.973) | 1.14 | 0.186(0.963) | 0.195(0.949) | 0.160(0.962) | 0.074(0.981) | 0.14 | 0.031(0.860) | 0.027(0.933) | 0.024(0.944) | 0.018(0.955) |
| 8 | −2.50 | Low | 1.00 | 0.108(0.985) | 0.059(0.983) | 0.043(0.980) | −0.075(0.978) | 1.14 | 0.026(0.995) | 0.124(0.981) | 0.123(0.975) | 0.017(0.980) | 0.14 | 0.015(0.969) | 0.024(0.971) | 0.025(0.968) | 0.012(0.980) |
| 9 | −2.50 | Mixed | 1.00 | 0.289(0.919) | 0.168(0.956) | 0.115(0.966) | 0.027(0.971) | 1.14 | 0.172(0.977) | 0.183(0.954) | 0.151(0.966) | 0.066(0.981) | 0.14 | 0.031(0.870) | 0.027(0.935) | 0.024(0.948) | 0.020(0.951) |
| MNAR | |||||||||||||||||
| 1 | −0.50 | High | 1.00 | −0.252(0.914) | −0.208(0.905) | −0.189(0.913) | −0.153(0.926) | 1.00 | −0.261(0.897) | −0.197(0.903) | −0.179(0.909) | −0.148(0.922) | 0.50 | −0.052(0.853) | −0.041(0.907) | −0.038(0.914) | −0.033(0.920) |
| 2 | −0.50 | Low | 1.00 | −0.250(0.897) | −0.250(0.884) | −0.248(0.889) | −0.227(0.895) | 1.00 | −0.256(0.880) | −0.228(0.880) | −0.226(0.885) | −0.209(0.891) | 0.50 | −0.054(0.836) | −0.051(0.876) | −0.051(0.879) | −0.047(0.890) |
| 3 | −0.50 | Mixed | 1.00 | −0.222(0.942) | −0.178(0.923) | −0.164(0.930) | −0.141(0.933) | 1.00 | −0.231(0.927) | −0.168(0.922) | −0.155(0.928) | −0.136(0.932) | 0.50 | −0.049(0.862) | −0.038(0.915) | −0.036(0.917) | −0.034(0.920) |
| 4 | −1.50 | High | 1.00 | −0.244(0.917) | −0.207(0.905) | −0.188(0.906) | −0.155(0.925) | 1.07 | −0.285(0.870) | −0.185(0.910) | −0.165(0.913) | −0.137(0.925) | 0.28 | −0.037(0.860) | −0.025(0.916) | −0.022(0.916) | −0.019(0.921) |
| 5 | −1.50 | Low | 1.00 | −0.239(0.904) | −0.243(0.896) | −0.241(0.894) | −0.223(0.902) | 1.07 | −0.274(0.871) | −0.198(0.906) | −0.195(0.897) | −0.182(0.912) | 0.28 | −0.037(0.846) | −0.029(0.912) | −0.028(0.914) | −0.027(0.923) |
| 6 | −1.50 | Mixed | 1.00 | −0.204(0.937) | −0.170(0.923) | −0.154(0.932) | −0.130(0.937) | 1.07 | −0.247(0.903) | −0.148(0.921) | −0.132(0.934) | −0.111(0.936) | 0.28 | −0.033(0.885) | −0.021(0.928) | −0.018(0.934) | −0.017(0.930) |
| 7 | −2.50 | High | 1.00 | −0.253(0.926) | −0.234(0.916) | −0.214(0.923) | −0.177(0.929) | 1.14 | −0.336(0.860) | −0.205(0.925) | −0.183(0.924) | −0.152(0.941) | 0.14 | −0.021(0.875) | −0.012(0.932) | −0.009(0.938) | −0.008(0.940) |
| 8 | −2.50 | Low | 1.00 | −0.236(0.910) | −0.252(0.907) | −0.252(0.901) | −0.233(0.908) | 1.14 | −0.311(0.855) | −0.193(0.932) | −0.189(0.933) | −0.178(0.938) | 0.14 | −0.020(0.855) | −0.011(0.936) | −0.011(0.929) | −0.010(0.939) |
| 9 | −2.50 | Mixed | 1.00 | −0.229(0.936) | −0.210(0.930) | −0.193(0.936) | −0.162(0.943) | 1.14 | −0.310(0.882) | −0.179(0.933) | −0.160(0.943) | −0.135(0.950) | 0.14 | −0.020(0.873) | −0.010(0.937) | −0.008(0.935) | −0.008(0.941) |
5. CASE STUDIES
One pitfall of AB-NMA is that the data may not provide enough information about the variances of log-odds for some treatments or about correlations between treatment-specific log-odds. In a Bayesian analysis, such a lack of information may cause the posterior to be dominated by prior information. This section uses three examples with different levels of information to examine the performance of different AB methods. Here, we focus more on posterior distributions of standard deviations than on correlations for two reasons. First, Figure 1 shows, for the RIW prior, that the positive definiteness property already imposes strong prior information on relationships among individual correlations and the IW and EQ priors make more strict assumptions than the RIW prior, as shown in Sections 3.1 and 3.2.2, respectively. Second, NMAs commonly provide little information about correlations and no practical remedy is available to avoid the influence of the prior on the posterior. We considered seven models in these case studies: AB-IW, AB-RW, AB-RIW, AB-EQ, AB-RW-EV, AB-RIW-EV and AB-EQ-EV. The latter three new models were based on AB-RW, AB-RIW and AB-EQ with the further assumption that all δi(i = 1, … , T) were equal to δ, with a uniform prior U(0, 5).
5.1. Example 1: smoking abstinence data
Mills et al.27 summarized results of 101 trials with 31,321 individuals comparing 4 interventions for the primary outcome of abstinence from smoking at least 4 weeks post-target quit date. Figure 2a shows the network plot of the data with treatments 1–4 being control, nicotine replacement therapy (NRT), bupropion, and varenicline, respectively. We used the AB-IW, AB-RW, AB-RIW, and AB-EQ models to analyze this dataset.
FIGURE 2.
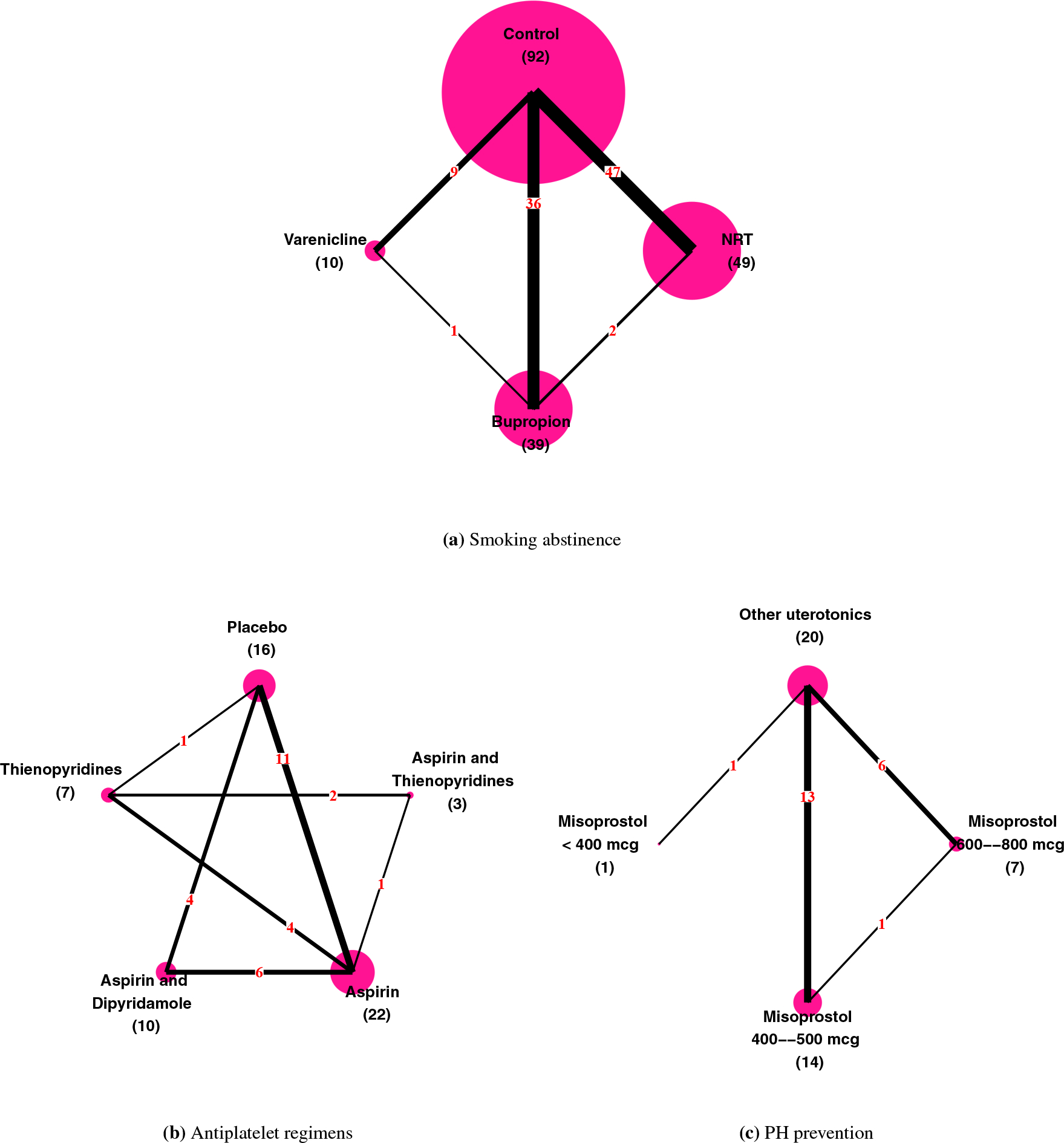
Network plots of the three example datasets. Each node in a plot stands for a treatment and each edge represents a direct comparison between two treatments. Vertex size is proportional to the number (in parenthesis) of direct comparisons containing that treatment; edge thickness is proportional to the number (in red) of direct comparisons.
Table 3 summarizes results for marginal log odds ratios, mLORij, comparing treatments i and j, the absolute risk of treatment t (pt), the standard deviation of log-odds for treatment t (δt), the correlation between treatment-specific (treatments i and j) log-odds (ρij), the probability that treatment t ranks ith (Rankti; a higher rank means a larger proportion with events), and the deviance information criterion (DIC)28 using the different models. The four AB methods (AB-IW, AB-RW, AB-RIW, and AB-EQ) gave similar results except for the log odds ratio of bupropion versus varenicline (mLOR34) and NRT versus varenicline (mLOR24). Although the AB-RIW and AB-EQ models gave slightly different posterior means of the log odds ratios, they led to the same conclusions: all active therapies (NRT, bupropion, and varenicline) increased smoking abstinence compared to control in the short term, and varenicline was more effective than NRT or bupropion. The rank probabilities also indicated that varenicline had the best results (Rank41 > 0.9) and control had the worst (Rank14 = 1). Because this dataset was large and provided enough information — 101 trials comparing four treatments, each included in at least 9 trials (varenicline was in 9 trials) — it is not surprising that these AB methods gave similar conclusions.
TABLE 3.
Smoking abstinence data: comparison of posterior means and 95% credible intervals under 4 models, specifically mLORij comparing the ith and jth treatment, absolute risk of events for the tth treatment (pt), standard deviation of log-odds for the tth treatment (δt), correlation between treatment-specific (treatments i and j) log-odds (ρij), and the ith rank probability of the tth treatment (Rankti). Regimen labels: (1) control, (2) nicotine replacement therapy, (3) bupropion, (4) varenicline.
| Point Estimate (95% Credible Interval) | ||||
|---|---|---|---|---|
| Parameter | AB-IW | AB-RW | AB-RIW | AB-EQ |
| mLOR12 | −0.58 (−0.74, −0.43) | −0.59 (−0.75, −0.42) | −0.58 (−0.73, −0.43) | −0.57 (−0.71,−0.43) |
| mLOR13 | −0.67 (−0.83,−0.51) | −0.68 (−0.83, −0.53) | −0.67 (−0.80, −0.53) | −0.66 (−0.81,−0.51) |
| mLOR14 | −0.90 (−1.22,−0.57) | −0.89 (−1.15,−0.59) | −0.94 (−1.16,−0.71) | −0.93 (−1.16,−0.73) |
| mLOR23 | −0.09 (−0.30, 0.12) | −0.09 (−0.30,0.11) | −0.09 (−0.28, 0.11) | −0.09 (−0.28, 0.10) |
| mLOR24 | −0.32 (−0.66, 0.02) | −0.30 (−0.59, 0.01) | −0.36 (−0.61,−0.11) | −0.36 (−0.61,−0.13) |
| mLOR34 | −0.23 (−0.57,0.11) | −0.20 (−0.49, 0.10) | −0.27 (−0.51,−0.03) | −0.27 (−0.52, −0.04) |
| Rank11 | 0.00 | 0.00 | 0.00 | 0.00 |
| Rank21 | 0.01 | 0.01 | 0.00 | 0.00 |
| Rank31 | 0.08 | 0.08 | 0.01 | 0.01 |
| Rank41 | 0.90 | 0.91 | 0.98 | 0.99 |
| p1 | 0.28 (0.24, 0.32) | 0.28(0.24,0.31) | 0.28(0.24,0.31) | 0.28 (0.24, 0.31) |
| p2 | 0.41 (0.36, 0.45) | 0.41 (0.36,0.45) | 0.41 (0.36,0.45) | 0.41 (0.36,0.45) |
| p3 | 0.43 (0.38, 0.48) | 0.43 (0.39,0.48) | 0.43 (0.38, 0.47) | 0.43 (0.38, 0.47) |
| p4 | 0.49(0.41,0.56) | 0.48 (0.42, 0.55) | 0.50 (0.44, 0.55) | 0.50 (0.45, 0.55) |
| δ | · | · | · | · |
| δ1 | 0.89(0.76, 1.06) | 0.88(0.74, 1.04) | 0.91 (0.77, 1.08) | 0.91 (0.77, 1.08) |
| δ2 | 0.83(0.66, 1.03) | 0.82(0.66, 1.02) | 0.85(0.68, 1.06) | 0.87 (0.69, 1.08) |
| δ3 | 0.79 (0.64, 0.97) | 0.76 (0.62, 0.94) | 0.78 (0.63,0.96) | 0.78 (0.63, 0.97) |
| δ4 | 0.54 (0.32, 0.90) | 0.42 (0.23, 0.75) | 0.49 (0.27, 0.82) | 0.48 (0.28, 0.81) |
| ρ | · | · | · | 0.87 (0.78, 0.93) |
| ρ12 | 0.80 (0.65, 0.89) | 0.78(0.62,0.89) | 0.83(0.69,0.92) | · |
| ρ13 | 0.85 (0.73, 0.93) | 0.89 (0.74, 0.97) | 0.92(0.81,0.98) | · |
| ρ14 | 0.57 (−0.15, 0.89) | 0.60 (−0.14,0.94) | 0.86 (0.48, 0.98) | · |
| ρ23 | 0.76(0.51,0.90) | 0.68 (0.32, 0.94) | 0.83 (0.57, 0.98) | · |
| ρ24 | 0.51 (−0.21,0.86) | 0.47 (−0.31, 0.92) | 0.79 (0.33, 0.98) | · |
| ρ34 | 0.53 (−0.18,0.87) | 0.53 (−0.25, 0.95) | 0.86 (0.42, 0.99) | · |
| DIC | 331.38 | 332.70 | 332.40 | 330.30 |
| 185.18 | 188.61 | 191.57 | 188.71 | |
| pD | 146.20 | 146.09 | 146.83 | 141.59 |
5.2. Example 2: serious vascular events prevention data
This dataset, reported by Thijs et al.29, consisted of 24 antiplatelet trials involving 42,688 patients after transient ischaemic attack (TIA) or stroke and compared 5 regimens: 1) placebo, 2) aspirin (ASA) plus thienopyridines (THIENO), 3) aspirin, 4) aspirin plus dipyridamole (DP), and 5) thienopyridines. The outcome was occurrence of a serious vascular event, including myocardial infarction and vascular death after TIA or stroke. Figure 2b shows the network plot; ASA plus thienopyridines was included in only 3 trials. As we had limited information about the variance of the effect of ASA plus THIENO, one may wonder how its posterior distribution was influenced by its prior. Hence, we added three models: 1) the AB-RW model assuming equal variances (AB-RW-EV), 2) the AB-RIW model assuming equal variances (AB-RIW-EV), and 3) the AB-EQ model assuming equal variances (AB-EQ-EV).
Table 4 summarizes the results. For the absolute proportion of serious vascular events for ASA plus THIENO (p2), which had limited information, both the AB-RIW and AB-EQ methods gave an unrealistically high upper bound for the 95% credible interval of δ2 (RIW: 0.19 to 3.67; EQ: 0.23 to 2.73). This resulted in wide credible intervals for the absolute risk p2 (RIW: 0.06 to 0.53; EQ: 0.11 to 0.48) and potentially biased estimates. Such wide intervals meant that we could not find any significant relative effects involving ASA plus THIENO; for example, using AB-RIW, mLOR12 = 0.24 with 95% CI (−1.52, 1.35). However, assuming equal variances (the AB-RIW-EV and AB-EQ-EV methods) narrowed the credible intervals of p2 and δ2. Using the AB-IW model, standard deviations δ1 and δ4 were potentially overestimated when comparing with the other two separation strategy priors (RIW and EQ), which caused disruptions in estimating absolute risks and relative risks related to the placebo and aspirin plus DP arms. Moreover, correlations ρ12, ρ23, ρ24, and ρ25 were potentially underestimated by both AB-IW and AB-RW-EV models because of the lack of comparisons between these treatments. Although the AB-IW and AB-RW-EV models could also control the credible intervals of δt and pt, DIC indicated that we may prefer AB-RIW-EV/AB-EQ-EV to AB-IW/AB-RW-EV (DIC: 74.91/74.55 vs. 89.97/87.08) and the DIC differences here were large enough to be of practical importance (larger than 5 units).
TABLE 4.
Serious vascular events prevention data: comparison of posterior mean and 95% credible interval under 7 different models, specifically marginal log odds ratio mLORij comparing the ith and jth treatments, absolute risk of events for the tth treatment (pt), standard deviation of log-odds for tth treatment (δt), and correlation between treatment-specific (treatments i and j) log-odds (ρij). Regimen labels: (1) placebo, (2) aspirin plus thienopyridines, (3) aspirin, (4) aspirin plus dipyridamole, (5) thienopyridines.
| Point Estimate (95% Credible Interval) | |||||||
|---|---|---|---|---|---|---|---|
| Parameter | AB-IW | AB-RW | AB-RIW | AB-EQ | AB-RW-EV | AB-RIW-EV | AB-EQ-EV |
| mLOR12 | 0.82(0.03, 1.51) | 0.66 (−0.90, 1.49) | 0.24 (−1.52, 1.35) | 0.14 (−1.32, 0.75) | 0.80(0.28, 1.34) | 0.39(0.16,0.89) | 0.35 (0.16, 0.56) |
| mLOR13 | 0.28 (0.00, 0.57) | 0.27 (0.06, 0.50) | 0.22 (0.08, 0.38) | 0.22 (0.08, 0.38) | 0.23 (0.04, 0.46) | 0.17(0.07,0.28) | 0.17(0.07,0.28) |
| mLOR14 | 0.48 (0.06, 0.90) | 0.48(0.16,0.77) | 0.44 (0.26, 0.61) | 0.44 (0.27, 0.60) | 0.45(0.15,0.76) | 0.41 (0.27, 0.55) | 0.41 (0.28,0.54) |
| mLOR15 | 0.40 (−0.06, 0.85) | 0.38 (−0.13, 0.81) | 0.30 (0.06, 0.57) | 0.30 (0.09, 0.53) | 0.36 (0.06, 0.69) | 0.25(0.10,0.41) | 0.25(0.11,0.40) |
| mLOR23 | −0.54 (−1.23, 0.23) | −0.39 (−1.24, 1.18) | −0.02 (−1.13, 1.74) | 0.08 (−0.50, 1.53) | −0.57 (−1.10,−0.07) | −0.21 (−0.70,−0.01) | −0.18 (−0.36,−0.01) |
| mLOR24 | −0.34 (−1.09,0.49) | −0.18 (−1.05, 1.39) | 0.20 (−0.91, 1.96) | 0.30 (−0.31, 1.75) | −0.35 (−0.92, 0.20) | 0.02 (−0.48, 0.27) | 0.06 (−0.17, 0.26) |
| mLOR25 | −0.42 (−1.20,0.40) | −0.28 (−1.26, 1.30) | 0.06 (−1.09, 1.80) | 0.15 (−0.42, 1.57) | −0.43 (−1.03,0.09) | −0.13 (−0.63,0.07) | −0.10 (−0.28, 0.06) |
| mLOR34 | 0.21 (−0.20,0.60) | 0.21 (−0.11, 0.51) | 0.22 (0.03, 0.38) | 0.22 (0.05, 0.38) | 0.22 (−0.05, 0.50) | 0.24(0.10,0.37) | 0.24 (0.10, 0.36) |
| mLOR35 | 0.13 (−0.32, 0.56) | 0.11 (−0.40,0.55) | 0.08 (−0.13, 0.31) | 0.07 (−0.09, 0.25) | 0.13 (−0.15,0.43) | 0.08 (−0.04, 0.21) | 0.08 (−0.03, 0.19) |
| mLOR45 | −0.08 (−0.62, 0.46) | −0.10 (−0.64, 0.40) | −0.14 (−0.40, 0.15) | −0.15 (−0.37,0.10) | −0.09 (−0.45, 0.28) | −0.16 (−0.33, 0.03) | −0.16 (−0.32, 0.02) |
| p1 | 0.21 (0.17,0.25) | 0.21 (0.18,0.24) | 0.20(0.17,0.23) | 0.20(0.17,0.23) | 0.20(0.17,0.24) | 0.19(0.16,0.22) | 0.19(0.16,0.22) |
| p2 | 0.11 (0.06,0.20) | 0.13(0.06,0.39) | 0.18 (0.06, 0.53) | 0.19(0.11,0.48) | 0.10(0.06,0.15) | 0.14(0.09,0.17) | 0.14(0.12,0.18) |
| p3 | 0.17(0.14,0.20) | 0.17(0.14,0.20) | 0.17(0.14,0.20) | 0.17(0.14,0.20) | 0.17(0.14,0.19) | 0.17(0.14,0.19) | 0.17(0.14,0.19) |
| p4 | 0.14(0.10,0.19) | 0.14(0.11,0.18) | 0.14(0.11,0.17) | 0.14(0.11,0.17) | 0.14(0.11,0.17) | 0.14(0.11,0.16) | 0.14(0.11,0.16) |
| p5 | 0.15(0.11,0.21) | 0.15(0.11,0.23) | 0.16(0.12,0.20) | 0.16(0.12,0.19) | 0.15(0.12,0.18) | 0.15(0.13,0.18) | 0.16(0.13,0.18) |
| δ | · | · | · | · | 0.37 (0.27, 0.49) | 0.42 (0.30, 0.59) | 0.42 (0.30, 0.59) |
| δ1 | 0.42 (0.28, 0.65) | 0.28(0.13,0.49) | 0.36(0.19,0.61) | 0.36(0.19,0.62) | · | · | · |
| δ2 | 0.55 (0.30, 1.05) | 0.86(0.14,3.63) | 0.93(0.19,3.67) | 0.82 (0.23, 2.73) | · | · | · |
| δ3 | 0.52 (0.36, 0.75) | 0.46 (0.30, 0.68) | 0.51 (0.34,0.77) | 0.52 (0.35, 0.77) | · | · | · |
| δ4 | 0.48 (0.29, 0.79) | 0.34(0.12,0.74) | 0.40(0.18,0.73) | 0.41(0.19,0.73) | · | · | · |
| δ5 | 0.57 (0.35, 0.94) | 0.60(0.29, 1.27) | 0.55 (0.31, 1.00) | 0.54 (0.32, 0.92) | · | · | · |
| ρ | · | · | · | 0.97(0.85, 1.00) | · | · | 0.98 (0.90, 1.00) |
| ρ12 | 0.05 (−0.62, 0.66) | 0.07 (−0.71, 0.79) | 0.73 (−0.95, 1.00) | · | 0.10 (−0.68, 0.80) | 0.90 (−0.24, 1.00) | · |
| ρ13 | 0.43 (−0.12,0.79) | 0.68(0.11,0.96) | 0.96(0.81, 1.00) | · | 0.70(0.17,0.96) | 0.98 (0.88, 1.00) | · |
| ρ14 | 0.25 (−0.37, 0.72) | 0.42 (−0.33, 0.91) | 0.95 (0.68, 1.00) | · | 0.48 (−0.28, 0.93) | 0.97 (0.83, 1.00) | · |
| ρ15 | 0.23 (−0.43, 0.73) | 0.33 (−0.43, 0.87) | 0.92 (0.54, 1.00) | · | 0.40 (−0.31, 0.89) | 0.96(0.78, 1.00) | · |
| ρ23 | 0.07 (−0.67, 0.73) | 0.10 (−0.68, 0.80) | 0.73 (−0.94, 1.00) | · | 0.14 (−0.63, 0.81) | 0.90 (−0.23, 1.00) | · |
| ρ24 | 0.04 (−0.65, 0.68) | 0.04 (−0.73, 0.77) | 0.72 (−0.94, 1.00) | · | 0.08 (−0.70, 0.79) | 0.90 (−0.23, 1.00) | · |
| ρ25 | 0.07 (−0.67, 0.76) | 0.07 (−0.70, 0.80) | 0.71 (−0.90, 1.00) | · | 0.09 (−0.69, 0.81) | 0.90 (−0.22, 1.00) | · |
| ρ34 | 0.35 (−0.27, 0.78) | 0.50 (−0.18, 0.92) | 0.95 (0.69, 1.00) | · | 0.56 (−0.10, 0.94) | 0.97 (0.83, 1.00) | · |
| ρ35 | 0.32 (−0.38, 0.79) | 0.39 (−0.34, 0.88) | 0.92 (0.56, 1.00) | · | 0.48 (−0.15, 0.90) | 0.97 (0.79, 1.00) | · |
| ρ45 | 0.18 (−0.54, 0.74) | 0.21 (−0.59, 0.83) | 0.91 (0.44, 1.00) | · | 0.28 (−0.49, 0.86) | 0.96(0.73, 1.00) | · |
| DIC | 89.97 | 88.56 | 79.99 | 79.11 | 87.08 | 74.91 | 74.55 |
| 48.23 | 49.77 | 46.60 | 46.48 | 49.71 | 46.27 | 46.19 | |
| pD | 41.74 | 38.78 | 33.40 | 32.62 | 37.38 | 28.64 | 28.37 |
5.3. Example 3: postpartum haemorrhage prevention data
Hofmeyr et al.30 reviewed 25 studies on prevention of postpartum haemorrhage (blood loss ≥ 1000 ml). This NMA compared four regimens: 1) other uterotonics, 2) misoprostol 600–800 mcg, 3) misoprostol 400–500 mcg, and 4) misoprostol <400 mcg as in Figure 2c. Table 5 summarizes the results; we focus on δ4. As Figure 2c shows, only one trial directly compared treatment 4 with other treatments; therefore, for the AB-EQ method, δ4’s posterior (mean 2.48; 95% CI, 0.19, 4.85) was dominated by the U(0,5) prior. However, this did not imply that AB-EQ was inferior to AB-EQ-EV. When confronted with a low-information situation, both the AB-EQ and AB-EQ-EV (or the AB-RIW and AB-RIW-EV) methods were useful: AB-EQ alerted us to a lack of information, while AB-EQ-EV showed the consequences of an extra assumption.
TABLE 5.
Postpartum haemorrhage prevention data: comparison of posterior mean and 95% credible interval under 7 different models, specifically marginal log odds ratio mLORij comparing the ith and jth treatments, absolute risk of events for the tth treatment (pt), standard deviation of log-odds for tth treatment (δt). Regimen labels: (1) other uterotonics, (2) misoprostol 600–800 mcg, (3) misoprostol 400–500 mcg, (4) misoprostol < 400 mcg.
| Point Estimate (95% Credible Interval) | |||||||
|---|---|---|---|---|---|---|---|
| AB-IW | AB-RW | AB-RIW | AB-EQ | AB-RW-EV | AB-RIW-EV | AB-EQ-EV | |
| mLOR12 | −0.10 (−0.78, 0.57) | −0.21 (−1.38,0.69) | −0.16 (−0.83, 0.42) | −0.16 (−0.80, 0.38) | −0.13 (−0.81, 0.56) | −0.17 (−0.53, 0.25) | −0.17 (−0.49, 0.21) |
| mLOR13 | −0.21 (−0.73, 0.29) | −0.21 (−0.83, 0.35) | −0.25 (−0.73, 0.20) | −0.26 (−0.76, 0.20) | −0.14 (−0.51, 0.25) | −0.18 (−0.45, 0.11) | −0.18 (−0.45, 0.10) |
| mLORl4 | 0.14 (−3.08,3.39) | −0.70 (−4.44, 2.76) | −0.59 (−4.40, 2.75) | 0.73 (−1.27, 3.19) | 0.08 (−2.61, 3.26) | 0.05 (−2.76, 3.31) | 1.13 (−0.74,3.78) |
| mLOR23 | −0.11 (−0.87,0.68) | 0.00 (−1.01, 1.24) | −0.09 (−0.78, 0.67) | −0.10 (−0.77, 0.64) | −0.01 (−0.75, 0.73) | −0.01 (−0.47, 0.43) | −0.02 (−0.44, 0.39) |
| mLOR24 | 0.23 (−3.00, 3.52) | −0.48 (−4.25,3.15) | −0.44 (−4.24, 2.96) | 0.89 (−1.17, 3.42) | 0.22 (−2.53, 3.46) | 0.21 (−2.60, 3.50) | 1.30 (−0.62, 3.98) |
| 1T1LOR34 | 0.35 (−2.87, 3.62) | −0.49 (−4.20, 3.02) | −0.34 (−4.18, 3.05) | 0.99 (−1.07, 3.47) | 0.22 (−2.48, 3.41) | 0.23 (−2.58, 3.49) | 1.32 (−0.57, 3.96) |
| p1 | 0.05 (0.03, 0.09) | 0.05 (0.03, 0.09) | 0.06(0.03,0.10) | 0.06(0.03,0.10) | 0.05 (0.03, 0.08) | 0.05 (0.03, 0.09) | 0.05 (0.03, 0.09) |
| p2 | 0.06(0.03,0.12) | 0.07(0.03,0.17) | 0.07(0.03,0.14) | 0.07(0.03,0.14) | 0.06(0.03,0.11) | 0.06(0.03,0.11) | 0.06(0.03,0.11) |
| p3 | 0.06(0.03,0.12) | 0.06(0.03,0.12) | 0.07(0.04,0.14) | 0.07(0.04,0.14) | 0.06 (0.03, 0.09) | 0.06(0.04,0.11) | 0.06(0.04,0.11) |
| p4 | 0.09 (0.00, 0.52) | 0.18(0.00,0.80) | 0.19(0.00,0.79) | 0.04(0.00,0.18) | 0.09 (0.00, 0.39) | 0.10(0.00,0.43) | 0.03(0.00,0.10) |
| δ | · | · | · | · | 1.15(0.82, 1.61) | 1.25(0.86,1.81) | 1.25(0.86,1.82) |
| δ1 | 1.23(0.82,1.81) | 1.20(0.80,1.79) | 1.32 (0.86, 2.01) | 1.32 (0.86, 2.00) | · | · | · |
| δ2 | 1.17(0.64,2.00) | 1.33(0.64,2.84) | 1.32(0.71,2.38) | 1.33(0.71,2.40) | · | · | · |
| δ3 | 1.32(0.82,2.08) | 1.34(0.81,2.18) | 1.46 (0.88, 2.34) | 1.48 (0.89, 2.37) | · | · | · |
| δ4 | 1.14(0.38,3.19) | 2.48(0.16,4.86) | 2.50(0.13,4.88) | 2.48(0.19,4.85) | · | · | · |
| D1C | 68.83 | 70.62 | 68.03 | 67.86 | 69.25 | 65.09 | 64.68 |
| 39.42 | 40.10 | 40.66 | 40.30 | 39.97 | 39.82 | 39.70 | |
| pD | 29.41 | 30.52 | 27.37 | 27.55 | 29.28 | 25.28 | 24.98 |
6. SUMMARY AND DISCUSSION
This paper evaluated different prior distributions for the covariance matrix in an AB-NMA, including the IW prior and a separation strategy with uniform priors on standard deviations and the RW, RIW or EQ prior on correlations. We compared their performance using extensive simulation studies with data generated under different mechanisms for selecting each study’s treatments. Separation strategies with a RIW or EQ prior on correlations performed much better than the IW prior in all situations in terms of bias and coverage probability of log odds ratios, absolute risks, variances and correlations. The commonly used IW prior often overestimated variances and underestimated correlations, which can lead to substantial bias for log odds ratios and absolute effects, especially under MNAR. The separation strategy with the EQ prior gave relatively small biases for log odds ratios under all conditions considered. These findings suggest that the separation strategy with the equal correlation prior is a much better choice of default vague prior in AB-NMA than the widely-used IW prior.
We conducted three case studies and compared separation strategies to the IW prior in terms of DIC. In the meta-analysis of serious vascular event prevention, the RIW and EQ priors had noticeably improved DIC compared to the IW and RW priors. However, estimating treatment-specific variances of log-odds in AB-NMA can be encumbered due to lack of information (i.e., most treatments are included in only a few trials). Here we proposed a straightforward but perhaps overly simple solution of assuming the treatment-specific variances are equal. This equal-variances assumption gave narrower credible intervals for treatment effects in the NMAs of serious vascular event prevention and postpartum haemorrhage prevention, and improved model fits. Thus, we suggest that NMA users consider sensitivity analyses with different assumptions on variance (homogeneous and heterogeneous variance assumptions) when using the separation strategy with the equal correlation prior.
Due to space limitation, we compared only four covariance priors. Other potential choices include the Cholesky decomposition19 and the spherical decomposition31. These methods further decompose the correlation matrix P as L′L, where L is a T × T upper-triangular matrix. Then we can place a weakly informative prior on L through a spherical parameterization. Technical details and applications to multivariate meta-analyses are in Lu and Ades4, Wei and Higgins32, and Lin et al.33. However, using this approach, the marginal distributions of the ρij depend strongly on the indexes i and j; see, e.g., Figure 3 in Wei and Higgins32. The different marginal distributions lack statistical or clinical interpretations and meta-analysts may reasonably be concerned about the potential impact of using different marginal priors for different correlations.
We evaluated the performance of a Bayesian analysis with different prior specifications with simulations from a frequentist perspective (e.g., bias and coverage probability); one may argue that this may be philosophically inappropriate34. Nevertheless, this does not affect the goal of this article, i.e., providing some practical recommendations of prior specifications for the Bayesian AB-NMA with less bias and better coverage probability.
In the continuing debate11,12 between proponents of the AB and CB approaches, White et al.13 recently compared both approaches and concluded that ‘both AB and CB models are suitable for the analysis of NMA data, but using random study intercepts requires a strong rationale such as relating treatment effects to study intercepts’. Perhaps such a rationale exists: Houwelingen et al.36,35 pointed out that the assumption of exchangeable absolute treatment risks is reasonable in most meta-analyses, while Béliveau et al.37 also mentioned that disconnected networks could benefit from being analyzed using random study intercepts. But perhaps this assumption appeared to be important only by accident, because of the choice of prior distributions: in our simulation study (Table 1), the separation strategy approach (AB-EV), compared with AB-IW, substantially reduced bias and potentially reduced the risk of using random study intercepts by improving estimation of the correlations between different treatments’ random effects of log-odds.
While the most important variance parameter in a CB-NMA is the heterogeneity of the treatment contrasts13, in the AB approach the variances of log-odds and correlations between treatment-specific log-odds are crucial, because: 1) variances of log-odds are needed to derive absolute risks and a much wider range of estimands, which is a key advantage of the AB approach; 2) correlations are critical to keep contrasts (relationships) between treatments stable, which can reduce the risk of assuming random study intercepts. Also, the AB-EQ-EV prior discussed in Section 5, with homogeneous variance of treatment specific log-odds and homogeneous correlations, is equivalent to assuming homogeneous variance of treatment contrasts in the CB approach9, in terms of the covariance structure.
Several other issues deserve further exploration and discussion. First, the uniform prior on standard deviations δi, i = 1, … , T in the AB model may cause upward bias when the true standard deviation is low. Second, if certain treatments are included in only a few trials in a NMA, the posteriors of their standard deviations may be dominated by their priors, which could lead to wide credible intervals for both the standard deviations and absolute risks, and thus bias the estimated absolute risks. In some NMAs, the assumption of equal variances might be too strong. Alternatively, one may model the standard deviations of study-specific log-odds of different treatments as random draws from a common distribution and thus allow borrowing strength in the estimation to shrink them in a data-dependent manner. The idea of extrapolation38,39 could also be applied to incorporate external evidence about standard deviations. Third, as mentioned in Sections 1, the T×T covariance matrix in the AB model is closely related to the CB model’s (T − 1)×(T − 1) covariance matrix, so we can probably apply corresponding RIW and EQ priors in the CB approach. Fourth, since the posterior distributions of some parameters (e.g., correlation coefficients ρij and marginal absolute risks pi) could be skewed, posterior medians might be better summaries than posterior means. Finally, it seems that all priors considered here may systematically underestimate the correlations. Further research on alternative priors may be fruitful.
Supplementary Material
Acknowledgement
This research was supported in part by NIH NLM R01LM012982.
Footnotes
Conflict of interest
The authors declare no potential conflict of interests.
Supplemental material
Supplemental material, including code to support our findings for this article have been submitted to the journal and will be available online.
References
- 1.Mills EJ, Ioannidis JPA, Thorlund K, Schünemann HJ, Puhan MA, Guyatt GH. How to use an article reporting a multiple treatment comparison meta-analysis. JAMA 2012; 308(12): 1246–1253. [DOI] [PubMed] [Google Scholar]
- 2.Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Annals of Internal Medicine 2015; 162(11): 777–784. [DOI] [PubMed] [Google Scholar]
- 3.Lu G, Ades AE. Combination of direct and indirect evidence in mixed treatment comparisons. Statistics in Medicine 2004; 23(20): 3105–3124. [DOI] [PubMed] [Google Scholar]
- 4.Lu G, Ades AE. Modeling between-trial variance structure in mixed treatment comparisons. Biostatistics 2009; 10(4): 792–805. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Carlin BP, Neaton JD, et al. Network meta-analysis of randomized clinical trials: reporting the proper summaries. Clinical Trials 2014; 11(2): 246–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucher HC, Guyatt GH, Griffith LE, Walter SD. The results of direct and indirect treatment comparisons in meta-analysis of randomized controlled trials. Journal of Clinical Epidemiology 1997; 50(6): 683–691. [DOI] [PubMed] [Google Scholar]
- 7.Rücker G, Schwarzer G. Ranking treatments in frequentist network meta-analysis works without resampling methods. BMC Medical Research Methodology 2015; 15(1): 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong H, Chu H, Zhang J, Carlin BP. A Bayesian missing data framework for generalized multiple outcome mixed treatment comparisons. Research Synthesis Methods 2016; 7(1): 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G, Ades AE. Assessing evidence inconsistency in mixed treatment comparisons. Journal of the American Statistical Association 2006; 101(474): 447–459. [Google Scholar]
- 10.Lin L, Chu H, Hodges JS. Sensitivity to excluding treatments in network meta-analysis. Epidemiology 2016; 27(4): 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias S, Ades AE. Absolute or relative effects? Arm-based synthesis of trial data. Research Synthesis Methods 2016; 7(1): 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong H, Chu H, Zhang J, Carlin BP. Rejoinder to the discussion of “a Bayesian missing data framework for generalized multiple outcome mixed treatment comparisons”, by S. Dias and A.E. Ades. Research Synthesis Methods 2016; 7(1): 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White IR, Turner RM, Karahalios A, Salanti G. A comparison of arm-based and contrast-based models for network meta-analysis. Statistics in Medicine 2019; 38(27): 5197–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnard J, McCulloch R, Meng XL. Modeling covariance matrices in terms of standard deviations and correlations, with application to shrinkage. Statistica Sinica 2000; 10(4): 1281–1311. [Google Scholar]
- 15.Caldwell DM, Ades AE, Higgins JPT. Simultaneous comparison of multiple treatments: combining direct and indirect evidence. BMJ 2005; 331(7521): 897–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelman A Prior distributions for variance parameters in hierarchical models. Bayesian Analysis 2006; 1(3): 515–534. [Google Scholar]
- 17.Alvarez I, Niemi J, Simpson M. Bayesian inference for a covariance matrix. Conference on Applied Statistics in Agriculture 2014; 26(1): 71–82. [Google Scholar]
- 18.O’Malley AJ, Zaslavsky AM. Domain-level covariance analysis for multilevel survey data with structured nonresponse. Journal of the American Statistical Association 2008; 103(484): 1405–1418. [Google Scholar]
- 19.Chen Z, Dunson DB. Random effects selection in linear mixed models. Biometrics 2003; 59(4): 762–769. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowski D, Kurowicka D, Joe H. Generating random correlation matrices based on vines and extended onion method. Journal of Multivariate Analysis 2009; 100(9): 1989–2001. [Google Scholar]
- 21.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics 1988; 44(4): 1049–1060. [PubMed] [Google Scholar]
- 22.Agresti A Categorical Data Analysis. Third edition John Wiley & Sons; Hoboken, NJ: 2013. [Google Scholar]
- 23.Huang A, Wand MP. Simple marginally noninformative prior distributions for covariance matrices. Bayesian Analysis 2013; 8(2): 439–452. [Google Scholar]
- 24.Lin L, Zhang J, Hodges JS, Chu H. Performing arm-based network meta-analysis in R with the pcnetmeta package. Journal of Statistical Software 2017; 80(5): 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter B, Gelman A, Hoffman MD, et al. Stan: a probabilistic programming language. Journal of Statistical Software 2017; 76(1): 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 27.Mills EJ, Wu P, Spurden D, Ebbert JO, Wilson K. Efficacy of pharmacotherapies for short-term smoking abstinance: a systematic review and meta-analysis. Harm Reduction Journal 2009; 6(1): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002; 64(4): 583–639. [Google Scholar]
- 29.Thijs V, Lemmens R, Fieuws S. Network meta-analysis: simultaneous meta-analysis of common antiplatelet regimens after transient ischaemic attack or stroke. European Heart Journal 2008; 29(9): 1086–1092. [DOI] [PubMed] [Google Scholar]
- 30.Hofmeyr GJ, Gülmezoglu AM, Novikova N, Linder V, Ferreira S, Piaggio G. Misoprostol to prevent and treat postpartum haemorrhage: a systematic review and meta-analysis of maternal deaths and dose-related effects. Bulletin of the World Health Organization 2009; 87(9): 666–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinheiro JC, Bates DM. Unconstrained parametrizations for variance-covariance matrices. Statistics and Computing 1996; 6(3): 289–296. [Google Scholar]
- 32.Wei Y, Higgins JPT. Bayesian multivariate meta-analysis with multiple outcomes. Statistics in Medicine 2013; 32(17): 2911–2934. [DOI] [PubMed] [Google Scholar]
- 33.Lin L, Chu H. Bayesian multivariate meta-analysis of multiple factors. Research Synthesis Methods 2018; 9(2): 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senn S Trying to be precise about vagueness. Statistics in Medicine 2007; 26(7): 1417–1430. [DOI] [PubMed] [Google Scholar]
- 35.Senn S Hans van Houwelingen and the art of summing up. Biometrical Journal 2010; 52(1): 85–94. [DOI] [PubMed] [Google Scholar]
- 36.Van Houwelingen HC, Zwinderman KH, Stijnen T. A bivariate approach to meta-analysis. Statistics in Medicine 1993; 12(24): 2273–2284. [DOI] [PubMed] [Google Scholar]
- 37.Béliveau A, Goring S, Platt RW, Gustafson P. Network meta-analysis of disconnected networks: how dangerous are random baseline treatment effects? Research Synthesis Methods 2017; 8(4): 465–474. [DOI] [PubMed] [Google Scholar]
- 38.Turner RM, Davey J, Clarke MJ, Thompson SG, Higgins JPT. Predicting the extent of heterogeneity in meta-analysis, using empirical data from the Cochrane Database of Systematic Reviews. International Journal of Epidemiology 2012; 41(3): 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner RM, Domínguez-Islas CP, Jackson D, Rhodes KM, White IR. Incorporating external evidence on between-trial heterogeneity in network meta-analysis. Statistics in Medicine 2019; 38: 1321–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


