Abstract
Mean residual life (MRL) is the remaining life expectancy of a subject who has survived to a certain time point and can be used as an alternative to hazard function for characterizing the distribution of a time-to-event variable. Inference and application of MRL models have primarily focused on full-cohort studies. In practice, case-cohort and nested case-control designs have been commonly used within large cohorts that have long follow-up and study rare diseases, particularly when studying costly molecular biomarkers. They enable prospective inference as the full-cohort design with significant cost-saving benefits. In this paper, we study the modeling and inference of a family of generalized MRL models under case-cohort and nested case-control designs. Built upon the idea of inverse selection probability, the weighted estimating equations are constructed to estimate regression parameters and baseline MRL function. Asymptotic properties of the proposed estimators are established and finite-sample performance is evaluated by extensive numerical simulations. An application to the New York University Women’s Health Study is presented to illustrate the proposed models and demonstrate a model diagnostic method to guide practical implementation.
Keywords: Counting process, Estimating equations, Inverse probability weighting, Model checking, Martingale residuals
1. Introduction
The mean residual life (MRL) is the remaining life expectancy given that a subject has survived to a certain time point. For a non-negative survival time T with finite expectation, the MRL at time t is m(t) = E(T − t|T > t). As alternatives to models based on the hazard function, those based on the MRL have a more intuitive explanation often leading to easier communication with patients. For instance, providing patients who have been on a treatment for a year with their remaining life expectancy is likely to be more informative than providing them with instantaneous hazards.
Various statistical models have been proposed to characterize the MRL function given covariates. Specifically, the proportional MRL model (Oakes and Dasu, 1990; Maguluri and Zhang, 1994) is,
| (1) |
where β is a vector of regression parameters characterizing the multiplicative effects of covariates on the MRL function, and m0(t) is an unknown baseline MRL function. The estimation and inference for model (1) have been developed to accommodate right censoring based on the counting process theory (Chen et al., 2005; Chen and Cheng, 2005). The additive MRL model (Chen and Cheng, 2006; Chen, 2007) is,
| (2) |
where the coefficients β characterize the additive change in remaining life expectancy per unit change in Z, and m0(t) is an unknown baseline MRL function. A quasi-partial score (QPS) estimation procedure has been proposed for the estimation of models (1) and (2) (Chen and Cheng, 2005, 2006; Chen, 2007).
More recently, Sun and Zhang (2009) proposed a class of generalized MRL models,
| (3) |
in which g(·) is a pre-specified link function to allow flexible model settings. Sun and Zhang (2009) proposed using the inverse probability of censoring weighting (IPCW) technique to develop estimating equations based on a zero-mean stochastic process. Model (3) can include the proportional MRL model and the additive MRL model as special cases. Moreover, model (3) has been extended to handle time-varying coefficients (Sun et al., 2012; Yang and Zhou, 2014). Note that the choice of g(·) link function can be flexible, but it needs to ensure that the MRL function is properly defined and satisfies the following conditions: (a) g(·) is twice continuously differentiable, (b) g(·) is strictly increasing, and (c) g{m0(t) + β′ 0Z} is a proper MRL function for all possible values of random covariates Z.
The MRL models described above have primarily been studied and applied in prospective cohort studies, where large sample size and long follow-up duration enable investigations on rare diseases and their complex mechanisms. Many large cohort studies store biological samples for future research. However, when studying costly molecular biomarkers, assembling detailed covariate information for the entire cohort often becomes time-consuming, expensive, and particularly cost-prohibitive for studying rare diseases.
Case-cohort (CC) design (Prentice, 1986) and nested case-control (NCC) design (Thomas, 1977) are widely considered as alternatives to the full-cohort design. In the CC design, a random sample of the full cohort is selected and named the subcohort. All incident cases (i.e. participants who developed the event of interest during follow-up regardless of being included in the subcohort or not) and the selected controls (i.e. participants who did not develop the event of interest) in the subcohort will be included in the CC analysis. The NCC design randomly samples a fixed number of controls for each case from the case’s risk set (i.e. the set of cohort participants free of event at the time of the case), and then assembles covariate information for all the cases and the selected controls. Compared with the NCC design, the CC design is deemed to be more efficient since the selected subcohort can be used for multiple different case groups (Kupper et al., 1975; Prentice, 1986). However the NCC design has the advantage of matching cases and controls on follow-up duration and can be extended to match on other confounders as well. Both designs are efficient and cost-effective in studying the relationship between exposures and diseases. However, for analysis of the CC and NCC studies, estimation and inference have been mostly dependent on the Cox proportional hazards model (Chen and Lo, 1999; Liu et al., 2010a,b; Scheike and Juul, 2004) and MRL modeling remains understudied. Recently, Ma et al. (2017) studied the proportional MRL model for CC studies using the QPS approach based on mean-zero process; however, the adoption of the proportional MRL models in NCC studies is yet to be developed due to its complex sampling mechanism.
Here we study modeling and inference for the generalized MRL model (3) under the CC and NCC designs. We propose a unified estimation procedure for both the CC and NCC designs and establish asymptotic properties for our proposed estimators. Statistical inference procedure based on bootstrap method (Efron, 1979) is adopted for the CC design, and perturbation resampling method (Cai and Zheng, 2013) is needed for the NCC design. A practical contribution also includes an R package to implement the proposed approaches.
In Section 2, we present the proposed estimating equations and inference procedures for model (3) under the CC and NCC designs. Section 3 reports the results from extensive simulation studies. In Section 4, we apply our approach to a real dataset from the New York University Women’s Health Study (NYUWHS), and present a model diagnostic method that can be readily used for model selection. Discussion and concluding remarks are given in Section 5. Additional simulations, regularity conditions, and technical proofs are provided in Appendix.
2. Estimation and Inference
Consider a full cohort with size n. Let Ti be the failure time and Ci be the censoring time for subject i = 1, 2, …, n. We assume Ti and Ci are conditionally independent given covariate Zi. The complete cohort data consist of n independently identically distributed random triplets , where and δi = I(Ti ≤ Ct). Let I(·) denote the indicator function throughout. In addition, denote and as the usual counting process and the at-risk process, respectively. We define filtration , where and .
Under the CC design, a subcohort with a fixed size is drawn randomly from the entire full cohort. Let γi = 1 indicate that subject i is included in the subcohort; 0 otherwise. All cases in the cohort and controls in the selected subcohort constitute the CC sample, which can be summarized as . Based upon the idea of inverse selection probabilities, the weight wi for each subject is defined as wi = δi + (1−δi)γi/p0, where is the proportion of subcohort to the full cohort.
Under the NCC design, for each case, m controls are randomly selected without replacement from the risk set excluding the case itself. The risk set at any time t is defined as . Following the notation of Samuelsen (1997), we define the “skeleton” filtration of the full-cohort as , where . Note that has all the information relevant to the NCC sampling and is contained in . Thus, the conditional probability that subject i is ever selected as a control given the skeleton filtration can be calculated as (Samuelsen, 1997),
| (4) |
Then, we define the weight for subject i under NCC studies as wi = δi + (1 − δi)γi/p0i, where γi is the indicator of whether subject i is ever selected as a control into the NCC study.
2.1. Estimation equations
Note that
| (5) |
is a martingale with respect to by the counting process and martingale theory (Fleming and Harrington 1991), where Λi(·) is the cumulative hazard function for subject i, β* and m*(·) are the true values of β and m0(·), respectively. Under the generalized MRL model (3), the relationship between the MRL function and the survival function can be easily derived as,
| (6) |
and the hazard function as
| (7) |
Motivated by the inverse probability weighting technique (Samuelsen, 1997) and the QPS estimation approach (Chen and Cheng, 2005, 2006), we propose the following unified estimation equations for model (3) under the CC and NCC designs. For 0 ≤ t ≤ τ,
| (8) |
| (9) |
where wi is the design-specific weight for subject i as defined above. Although equations (8) and (9) do not have martingale interpretation because wi’s are not predictable processes with respect to , they are mean-zero processes. Specifically, we have that because E[dMi(t; β*, m*)] = 0 and .
Given a fixed β and pre-specified g(·), equation (8) can be rewritten as a first-order linear ordinary differential equation about m0(t), which leads to a closed form solution. Let denote the solution of equation (8) with respect to m0(t), and then it can be plugged back into equation (9). It is evident that equation (9) can be re-arranged and written as,
| (10) |
To solve equation (10) for β, the Newton-Raphson algorithm can be applied after calculating the Jacobian matrix for multidimensional case. We denote as the resulting estimator of β.
Remark 1: We conducted numerical simulations to compare the IPCW method (Sun and Zhang, 2009) and the QPS estimator (Chen and Cheng, 2005, 2006) under the full-cohort design with various censoring proportions (results in Appendix) and observed that the QPS estimator outperformed the IPCW estimator in terms of estimation stability and 95% confidence interval coverage probability, especially under the high censoring scenarios. Based on these observations, we developed our estimating procedure based on the QPS approach for the generalized MRL modeling in the CC and NCC designs.
2.2. Asymptotic properties
In this section, we summarize the asymptotic properties of our proposed estimators and defer the details of assumptions and proofs toAppendix. We first introduce some notations. Let
Theorem 1 Under the regularity conditions (C1) to (C4) stated in Appendix, we have:
The estimators and exist and are consistent.
- in distribution. Moreover, variance matrix components A and Σ can be consistently estimated by and Specifically, for the CC design,
where α⨂2 denotes ααT for a vector α. For the NCC design, is estimated differently as,
where , and .
Remark 2: The NCC sampling is a dynamic process and the probability of being selected as a control is neither a constant nor independent, even in the asymptotic sense. The asymptotic variance estimate derived under the NCC design explicitly contains the cross-product term between subject i and j as shown in the in Theorem 1.
2.3. Numerical variance estimation
The asymptotic variance formula and the plug-in estimators that are described above can be difficult to implement analytically. In practice, bootstrap method (Efron, 1979) is often adopted to compute the standard errors (SE) of estimators. Under the full-cohort design, estimating the SEs via the standard bootstrap method is straightforward. For the CC design, since the weight of each selected control is pre-specified, one can obtain the estimated SEs by implementing the bootstrapping within the selected subcohort.
Under the NCC design, however, the standard bootstrap approach cannot fully capture the complex correlation structure induced by repeated finite risk set sampling nor applicable for the proposed estimators. We thus employ the perturbation resampling method (Cai and Zheng, 2013) to estimate the SEs under the NCC design. Instead of resampling the selected subsample, the perturbation method approximates the variance of the IPW estimators by perturbing the weight of each subject. Specifically, the procedures are described below:
Generate n2 random realizations of Iil from a given distribution with E(Iil) = 1 and let I = {Iil, i = 1, …, n; l = 1, …, n}.
- Calculate the perturbed weights as , where
and V0ij = 1 denotes the jth subject is selected as a control for the ith subject. Replace the wi in (10) using the perturbed weights from step 2 and obtain the estimator .
Repeat steps 1–3 for M times and use the standard deviation of these to approximate the SE of .
3. Simulation Studies
We conducted simulations to evaluate the finite-sample performance of the proposed inference procedures and to compare their efficiency under the CC and NCC designs to the full-cohort analysis. We considered two models with g(t) = exp(t) and g(t) = t, which corresponded to the proportional MRL model and additive MRL model, respectively. To generate the data, the baseline function m0(t) was taken from the Hall-Wellner family, which was m0(t) = g−1 {(D1t + D2)+}, where D1 > −1, D2 > 0 and d+ denoted dI(d ≥ 0) for any quantity d. In our setting, we considered D1 = −0.5 and D2 = 0.5. Let Z1 be a Bernoulli random variable with success probability 0.5 and Z2 be a uniform random variable on (0, 1). The true parameters (β1, β2)′ were set to be (0, 0)′ for null effects and (0.2, 0.2)′ for moderate effects. Censoring time C was generated from the exponential distribution with parameter λ, which was chosen to yield different censoring proportions. We conducted 500 simulations under each setting.
The first set of simulations evaluated our proposed methods for the proportional MRL model under the full-cohort, CC, and NCC designs. To mimic a practical setting, we considered a cohort size of 1000 and 5000, and high censoring rates of 70%, 80% and 90%. In addition, a long follow-up duration was assumed as in many large epidemiology cohort studies and for stable estimation of the MRL models. We selected 1 control for each case in the NCC design and 30% random subcohort for the CC design. The SEs of estimators were computed based on the standard bootstrap method for the full-cohort and CC designs. The perturbation-based method was applied to estimate SEs under the NCC design, where the weights were generated from a Gamma distribution with shape and scale parameters of 1.
Table 1 and Table 2 summarize the results for the proportional MRL model over 1000 simulations for cohort size of 1000 and 5000. The biases of the estimators were small, SDs and SEs matched well, and coverage probability (CP) of 95% Wald-type confidence interval was close to the nominal level. The SDs and SEs decreased as sample size increased. The estimates from the CC and NCC designs lost some efficiency when compared to those obtained in the full-cohort analysis, but noted that these within-cohort sampling designs also would save cost by only using the partially sampled subcohort.
Table 1.
Simulation results under proportional MRL models
| N | β | censoring rate | Full† | CC (30%)‡ | NCC (1-to-1)§ | ||||
|---|---|---|---|---|---|---|---|---|---|
| β1 | β2 | β1 | β2 | β1 | β2 | ||||
| 1000 | (0,0) | 0.7 | Bias | −0.002 | −0.002 | 0.001 | 0.002 | −0.002 | −0.001 |
| SD | 0.060 | 0.105 | 0.079 | 0.129 | 0.072 | 0.122 | |||
| SE | 0.061 | 0.106 | 0.078 | 0.136 | 0.065 | 0.112 | |||
| CP | 95.0 | 94.7 | 93.9 | 96.2 | 92.0 | 93.2 | |||
| 1000 | (0,0) | 0.8 | Bias | −0.000 | −0.001 | −0.002 | −0.012 | −0.001 | 0.000 |
| SD | 0.071 | 0.129 | 0.097 | 0.156 | 0.092 | 0.155 | |||
| SE | 0.073 | 0.127 | 0.091 | 0.157 | 0.082 | 0.143 | |||
| CP | 95.5 | 94.2 | 93.5 | 94.6 | 92.7 | 92.5 | |||
| 1000 | (0,0) | 0.9 | Bias | 0.001 | −0.009 | 0.002 | 0.005 | −0.007 | −0.001 |
| SD | 0.086 | 0.153 | 0.092 | 0.164 | 0.114 | 0.202 | |||
| SE | 0.086 | 0.150 | 0.093 | 0.162 | 0.109 | 0.190 | |||
| CP | 95.0 | 94.5 | 95.8 | 95.3 | 93.2 | 94.4 | |||
| 1000 | (0.2,0.2) | 0.7 | Bias | 0.000 | −0.001 | −0.002 | 0.002 | −0.002 | −0.006 |
| SD | 0.048 | 0.087 | 0.063 | 0.104 | 0.057 | 0.093 | |||
| SE | 0.050 | 0.087 | 0.060 | 0.105 | 0.051 | 0.088 | |||
| CP | 95.1 | 95.3 | 94.2 | 95.9 | 91.7 | 93.4 | |||
| 1000 | (0.2,0.2) | 0.8 | Bias | 0.001 | −0.002 | 0.008 | 0.002 | −0.005 | −0.006 |
| SD | 0.057 | 0.105 | 0.073 | 0.125 | 0.069 | 0.123 | |||
| SE | 0.059 | 0.103 | 0.073 | 0.127 | 0.067 | 0.116 | |||
| CP | 95.3 | 95.4 | 95.3 | 95.4 | 94.2 | 94.1 | |||
| 1000 | (0.2,0.2) | 0.9 | Bias | −0.004 | −0.012 | −0.011 | −0.013 | −0.018 | −0.006 |
| SD | 0.079 | 0.136 | 0.092 | 0.154 | 0.103 | 0.182 | |||
| SE | 0.077 | 0.134 | 0.088 | 0.153 | 0.099 | 0.174 | |||
| CP | 94.6 | 94.8 | 94.3 | 94.5 | 94.2 | 95.2 | |||
SD: sample standard deviation; SE: mean of estimated standard error;
CP: empirical coverage probability of 95% confidence interval;
Full-cohort population
Case-Cohort design with 30% random sample from full-cohort population
Nested Case-Control design with 1 control for each case
Table 2.
Simulation results under proportional MRL models
| N | β | censoring rate | Full† | CC (30%)‡ | NCC (1-to-l)§ | ||||
|---|---|---|---|---|---|---|---|---|---|
| β1 | β2 | β1 | β1 | β2 | β1 | ||||
| 5000 | (0,0) | 0.7 | Bias | 0.000 | 0.001 | 0.000 | −0.003 | 0.000 | −0.001 |
| SD | 0.027 | 0.048 | 0.034 | 0.060 | 0.033 | 0.054 | |||
| SE | 0.027 | 0.047 | 0.035 | 0.060 | 0.029 | 0.051 | |||
| CP | 94.6 | 94.0 | 95.3 | 94.4 | 92.0 | 93.6 | |||
| 5000 | (0,0) | 0.8 | Bias | 0.000 | 0.002 | −0.002 | −0.001 | −0.001 | 0.002 |
| SD | 0.033 | 0.058 | 0.040 | 0.072 | 0.040 | 0.072 | |||
| SE | 0.032 | 0.056 | 0.041 | 0.070 | 0.037 | 0.064 | |||
| CP | 94.9 | 94.4 | 95.4 | 95.0 | 92.4 | 92.1 | |||
| 5000 | (0,0) | 0.9 | Bias | −0.001 | 0.003 | 0.002 | 0.000 | 0.001 | 0.003 |
| SD | 0.043 | 0.076 | 0.050 | 0.083 | 0.056 | 0.098 | |||
| SE | 0.042 | 0.074 | 0.048 | 0.083 | 0.052 | 0.091 | |||
| CP | 94.8 | 94.7 | 94.5 | 95.1 | 93.6 | 92.6 | |||
| 5000 | (0.2,0.2) | 0.7 | Bias | 0.000 | 0.001 | −0.001 | −0.001 | −0.003 | −0.004 |
| SD | 0.023 | 0.040 | 0.027 | 0.047 | 0.023 | 0.042 | |||
| SE | 0.022 | 0.038 | 0.027 | 0.047 | 0.023 | 0.039 | |||
| CP | 94.6 | 94.2 | 95.0 | 94.9 | 93.9 | 93.6 | |||
| 5000 | (0.2,0.2) | 0.8 | Bias | 0.000 | 0.002 | −0.001 | −0.001 | −0.002 | 0.002 |
| SD | 0.026 | 0.046 | 0.031 | 0.054 | 0.032 | 0.056 | |||
| SE | 0.026 | 0.045 | 0.032 | 0.056 | 0.030 | 0.052 | |||
| CP | 95.1 | 94.8 | 95.4 | 96.1 | 93.8 | 93.4 | |||
| 5000 | (0.2,0.2) | 0.9 | Bias | 0.001 | 0.004 | −0.003 | 0.000 | −0.006 | −0.009 |
| SD | 0.036 | 0.063 | 0.043 | 0.071 | 0.050 | 0.088 | |||
| SE | 0.036 | 0.062 | 0.042 | 0.072 | 0.047 | 0.082 | |||
| CP | 95.6 | 95.2 | 94.0 | 95.3 | 93.4 | 93.2 | |||
SD: sample standard deviation; SE: mean of estimated standard error;
CP: empirical coverage probability of 95% confidence interval;
Full-cohort population
Case-Cohort design with 30% random sample from full-cohort population
Nested Case-Control design with 1 control for each case
In Table 3 and Table 4, we summarized the simulation results for the additive MRL models under full cohort size of 1000 and 5000 with high censoring rates of 70%, 80%, and 90%. Note that the additive MRL model does not ensure positive baseline MRL functions at any time, which could make the estimating procedure unstable when the sample size is small. In order to have meaningful estimates from the estimating equations, by the convention of Reid (1981) and James (1986), the longest observation was always assumed to be a true event. We found that this single data-point change at the tail had minimal impact on the estimation. In general, the biases were small and the SDs of the estimates were close to the average of SEs. The 95% confidence interval coverage probabilities were around the nominal level, indicating that the bootstrapping and perturbation resampling methods worked well for the CC and NCC designs, respectively. The overall performance improved with increasing sample size. Figures 1 and 2 visually demonstrate the performance of the full-cohort, CC, and NCC designs, specifically on efficiency.
Table 3.
Simulation results under additive MRL models
| N | β | censoring rate | Full† | CC (30%)‡ | NCC (1-to-l)§ | ||||
|---|---|---|---|---|---|---|---|---|---|
| β1 | β2 | β1 | β1 | β2 | β1 | ||||
| 1000 | (0,0) | 0.7 | Bias | −0.001 | −0.001 | 0.001 | 0.003 | −0.002 | 0.000 |
| SD | 0.045 | 0.070 | 0.057 | 0.096 | 0.054 | 0.095 | |||
| SE | 0.045 | 0.079 | 0.057 | 0.099 | 0.051 | 0.089 | |||
| CP | 94.9 | 94.4 | 94.2 | 95.9 | 93.5 | 92.8 | |||
| 1000 | (0,0) | 0.8 | Bias | 0.000 | −0.001 | 0.002 | −0.004 | −0.002 | −0.002 |
| SD | 0.059 | 0.106 | 0.078 | 0.126 | 0.053 | 0.096 | |||
| SE | 0.060 | 0.105 | 0.073 | 0.127 | 0.054 | 0.094 | |||
| CP | 95.4 | 94.8 | 94.1 | 95.3 | 92.7 | 94.0 | |||
| 1000 | (0,0) | 0.9 | Bias | 0.004 | 0.000 | 0.009 | 0.017 | 0.000 | 0.012 |
| SD | 0.080 | 0.143 | 0.088 | 0.168 | 0.103 | 0.187 | |||
| SE | 0.079 | 0.139 | 0.088 | 0.154 | 0.106 | 0.193 | |||
| CP | 95.8 | 94.8 | 95.6 | 93.9 | 94.0 | 93.0 | |||
| 1000 | (0.2,0.2) | 0.7 | Bias | 0.010 | 0.008 | 0.007 | 0.017 | 0.010 | 0.014 |
| SD | 0.066 | 0.114 | 0.087 | 0.147 | 0.080 | 0.135 | |||
| SE | 0.067 | 0.114 | 0.085 | 0.145 | 0.078 | 0.135 | |||
| CP | 96.0 | 94.8 | 95.5 | 95.3 | 94.6 | 95.0 | |||
| 1000 | (0.2,0.2) | 0.8 | Bias | 0.010 | 0.007 | 0.014 | 0.013 | 0.007 | 0.012 |
| SD | 0.088 | 0.158 | 0.117 | 0.204 | 0.114 | 0.192 | |||
| SE | 0.091 | 0.155 | 0.115 | 0.194 | 0.112 | 0.194 | |||
| CP | 95.7 | 95.1 | 95.5 | 95.8 | 94.7 | 95.3 | |||
| 1000 | (0.2,0.2) | 0.9 | Bias | −0.006 | −0.010 | −0.007 | 0.010 | −0.012 | 0.005 |
| SD | 0.138 | 0.229 | 0.161 | 0.282 | 0.167 | 0.305 | |||
| SE | 0.123 | 0.212 | 0.138 | 0.240 | 0.196 | 0.359 | |||
| CP | 92.9 | 94.8 | 91.5 | 92.2 | 95.6 | 93.6 | |||
SD: sample standard deviation; SE: mean of estimated standard error;
CP: empirical coverage probability of 95% confidence interval;
Full-cohort population
Case-Cohort design with 30% random sample from full-cohort population
Nested Case-Control design with 1 control for each case
Table 4.
Simulation results under additive MRL models
| N | β | censoring rate | Full† | CC (30%)‡ | NCC (1-to-l)§ | ||||
|---|---|---|---|---|---|---|---|---|---|
| β1 | β2 | β1 | β1 | β2 | β1 | ||||
| 5000 | (0,0) | 0.7 | Bias | 0.000 | 0.000 | 0.000 | −0.002 | 0.001 | −0.001 |
| SD | 0.020 | 0.035 | 0.025 | 0.044 | 0.025 | 0.042 | |||
| SE | 0.020 | 0.034 | 0.025 | 0.044 | 0.024 | 0.041 | |||
| CP | 94.6 | 94.2 | 95.9 | 94.9 | 93.4 | 94.2 | |||
| 5000 | (0,0) | 0.8 | Bias | 0.000 | 0.001 | −0.001 | 0.000 | −0.001 | 0.002 |
| SD | 0.026 | 0.047 | 0.032 | 0.059 | 0.032 | 0.060 | |||
| SE | 0.026 | 0.045 | 0.033 | 0.057 | 0.033 | 0.057 | |||
| CP | 94.8 | 94.5 | 95.3 | 94.5 | 95.2 | 93.5 | |||
| 5000 | (0,0) | 0.9 | Bias | 0.000 | 0.004 | 0.002 | 0.001 | 0.001 | 0.005 |
| SD | 0.039 | 0.068 | 0.045 | 0.074 | 0.052 | 0.091 | |||
| SE | 0.038 | 0.066 | 0.044 | 0.076 | 0.050 | 0.088 | |||
| CP | 94.8 | 94.7 | 94.4 | 95.7 | 94.6 | 94.2 | |||
| 5000 | (0.2,0.2) | 0.7 | Bias | 0.004 | 0.005 | 0.005 | 0.004 | 0.005 | 0.006 |
| SD | 0.029 | 0.050 | 0.038 | 0.066 | 0.035 | 0.059 | |||
| SE | 0.029 | 0.049 | 0.037 | 0.063 | 0.033 | 0.058 | |||
| CP | 95.0 | 93.7 | 95.0 | 94.4 | 93.9 | 94.7 | |||
| 5000 | (0.2,0.2) | 0.8 | Bias | 0.004 | 0.006 | 0.009 | 0.001 | 0.005 | 0.008 |
| SD | 0.039 | 0.067 | 0.051 | 0.083 | 0.048 | 0.084 | |||
| SE | 0.039 | 0.065 | 0.049 | 0.082 | 0.048 | 0.082 | |||
| CP | 94.7 | 94.1 | 95.0 | 94.8 | 94.8 | 94.2 | |||
| 5000 | (0.2,0.2) | 0.9 | Bias | 0.000 | 0.006 | −0.001 | 0.000 | −0.003 | 0.002 |
| SD | 0.063 | 0.105 | 0.074 | 0.121 | 0.082 | 0.135 | |||
| SE | 0.058 | 0.098 | 0.067 | 0.114 | 0.079 | 0.136 | |||
| CP | 94.1 | 94.0 | 93.0 | 93.7 | 94.1 | 95.4 | |||
SD: sample standard deviation; SE: mean of estimated standard error;
CP: empirical coverage probability of 95% confidence interval;
Full-cohort population
Case-Cohort design with 30% random sample from full-cohort population
Nested Case-Control design with 1 control for each case
Fig. 1.
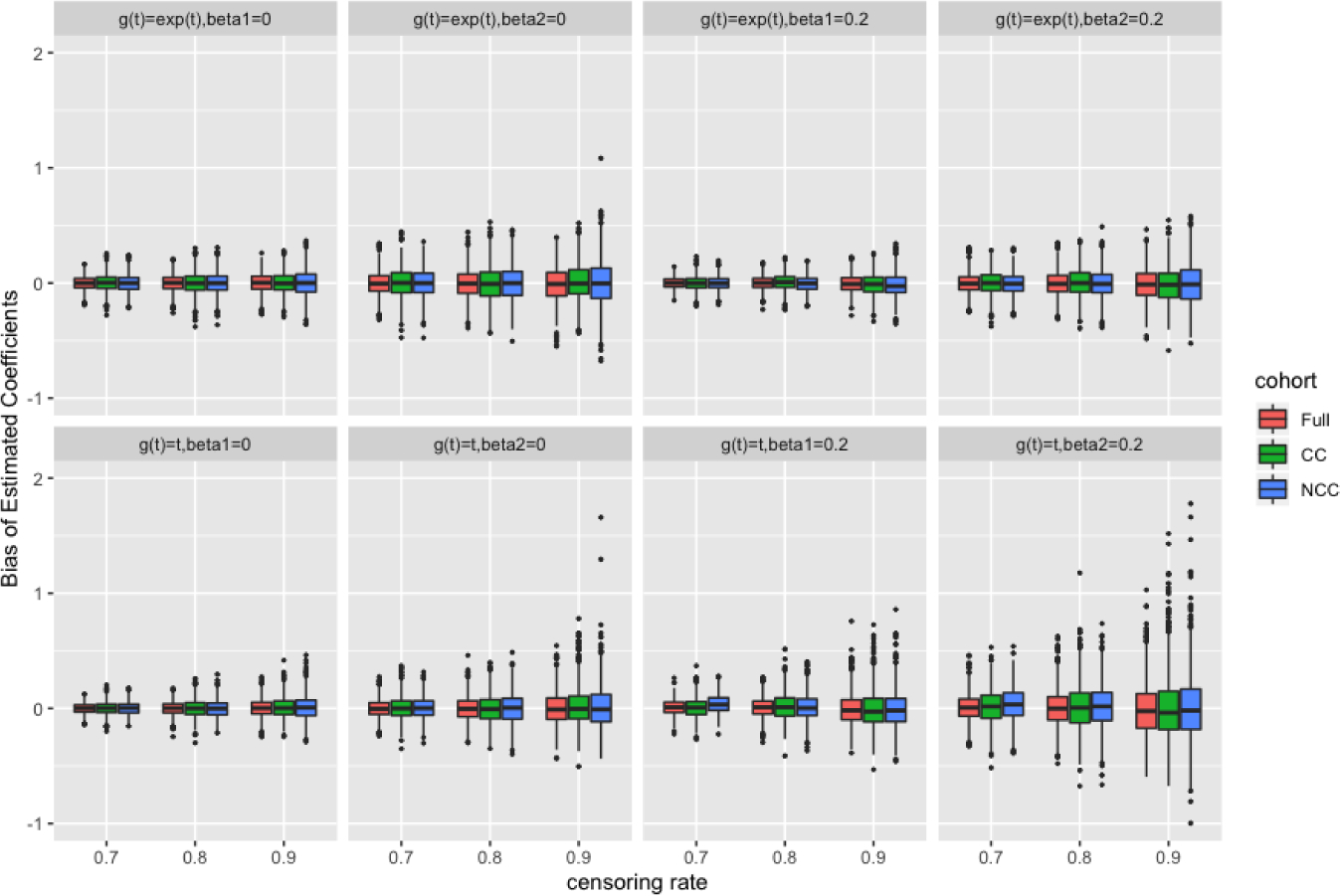
Bias of estimated coefficients with sample size of 1000
Fig. 2.
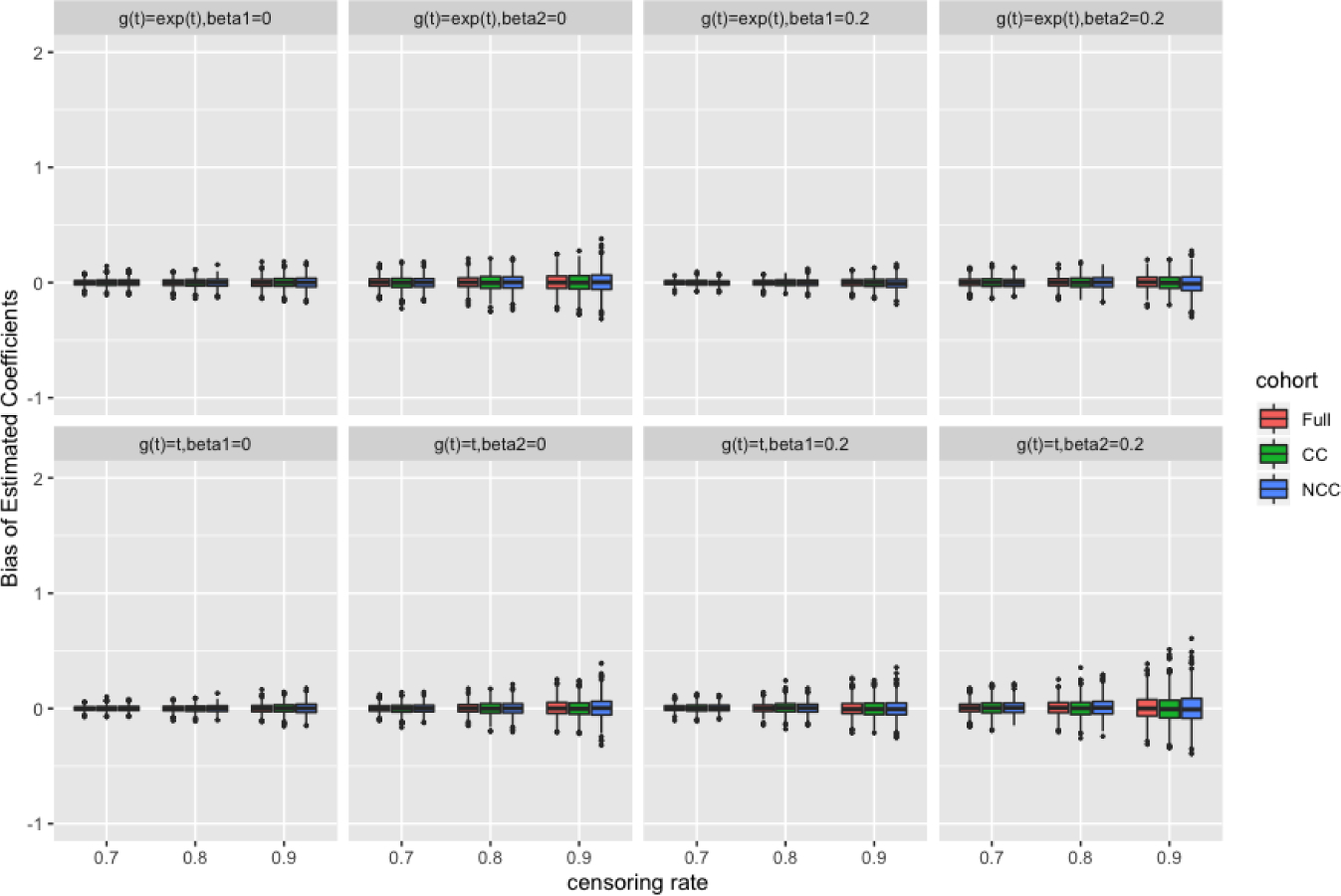
Bias of estimated coefficients with sample size of 5000
4. Application
4.1. Data analysis
The proposed models and estimation methods were demonstrated through a study conducted in the NYUWHS, a prospective cohort study that enrolled 14,274 healthy women aged 35–65 between 1985 and 1991 at a breast cancer screening center in New York City. One primary interest of the study was to investigate the association between endogenous sex hormones and breast cancer risk. At enrollment, participants completed a questionnaire on lifestyle and reproductive history. Blood samples were collected and stored for all participants. Every two to four years, study participants were asked to update information on their health conditions by completing a questionnaire. Within the NYUWHS, multiple nested case-control studies on breast cancer have been conducted (Scarmo et al., 2013; Clendenen et al., 2015).
The cohort dataset we considered here was from the NYUWHS including 6,610 women who were less than 50 years of age at enrollment and used in a recent study for breast cancer risk prediction in younger women (Ge et al., 2018). In this cohort, approximately 12% of the study participants developed breast cancer during the follow-up period (Mean = 21.3 years, SD = 4.8 years). We considered risk factors that are included in the Gail model (Breast Cancer Risk Assessment Tool) (Gail et al., 1989): age, race, age at menarche, number of breast biopsies, age at first live birth, and number of first-degree relatives with breast cancer. The proportional MRL model and the additive MRL model were applied to evaluate the multiplicative and the additive effects of these risk factors on the MRL function of time to breast cancer occurrence. Within this cohort, we conducted NCC sampling with one matched control, and CC sampling with 20% of the full-cohort size. In order to obtain empirical results, we ran 200 times for both the CC and NCC sampling, and reported the mean of point estimates, as well as the mean of estimated standard error of . The SEs were computed using the same approach as in the simulation studies.
The results are summarized in Table 5. Under both the proportional and additive MRL models, the estimates from the CC and NCC designs were similar to the ones from full cohort. Specifically, age at first live birth, number of breast biopsies, and number of first-degree relatives with breast cancer had significant effects on shortening the expected residual disease-free time, which were consistent with the effects observed in the Gail model. As expected, we observed that SEs of the estimators in the CC and NCC designs were larger than that in the full-cohort design.
Table 5.
Analysis of NYUWHS data
| Full‡ | CC (20%)§ | NCC (1-to-1)¶ | ||||
|---|---|---|---|---|---|---|
| Covariate | Estimate | SE | Estimate | SE | Estimate | SE |
| g(t) = exp(t) | ||||||
| race | −0.0084* | 0.0025 | −0.0083* | 0.0030 | −0.0085* | 0.0032 |
| age at menarche | −0.0027 | 0.0018 | −0.0027 | 0.0022 | −0.0027 | 0.0024 |
| age at 1st live birth | −0.0070* | 0.0015 | −0.0071* | 0.0018 | −0.0068* | 0.0020 |
| # of breast biopsies | −0.0116* | 0.0032 | −0.0116* | 0.0038 | −0.0116* | 0.0040 |
| # of relatives† | −0.0188* | 0.0041 | −0.0191* | 0.0051 | −0.0182* | 0.0053 |
| g(t) = t | ||||||
| race | −2.8210* | 1.0297 | −2.7862* | 1.1421 | −3.5018* | 1.4782 |
| age at menarche | −0.8995 | 0.6284 | −0.8603 | 0.6994 | −1.0138 | 0.9348 |
| age at 1st live birth | −2.7701* | 0.6331 | −2.8038* | 0.6993 | −3.1273* | 0.8129 |
| # of breast biopsies | −2.3625* | 0.5487 | −2.3372* | 0.6191 | −2.8184* | 0.9047 |
| # of relatives† | −3.8616* | 0.6153 | −3.8675* | 0.6984 | −4.4870* | 1.1350 |
SE: Average of estimated standard error
The coefficient is statistically significant at 0.05 significant level
Number of first-degree relatives diagnosed with breast cancer
Full-cohort population
Case-Cohort design with 20% random sample from full-cohort population
Nested Case-Control design with 1 control for each case
The interpretation of the additive MRL model is straightforward. For example, in the full-cohort, keeping all other covariates fixed, each additional breast biopsy decreased the expected remaining disease-free time by approximately 2.363 years. The average reduction of MRL estimated from the CC and NCC designs were 2.337 years and 2.818 years, respectively. For the proportional MRL model under the full-cohort design, keeping all other covariates fixed, each additional breast biopsy shortened the expected residual disease-free time by around 1.15%. The estimated percentage under the CC and NCC designs were both 1.15%.
4.2. Model diagnosis
The generalized MRL framework provides flexible model options to study the association between covariates and MRL time under the full-cohort, CC, and NCC designs. For its practical use, some model diagnostic methods would be useful for model selection. Several graphical and numerical model diagnostic approaches have been proposed based on martingale residuals (Lin et al., 1993), but mainly focused on full-cohort data. We adopt the approaches from Lin et al. (1993) and propose weighted test statistics and processes for model selection in the generalize MRL models for CC and NCC studies.
In the NYUWHS application, we used the standardized empirical score process to conduct model selection graphically. Figure 3 and Figure 4 show the results for the proportional and additive MRL models in full cohort, respectively. The standardized score process in the proportional MRL was smaller than that in the additive MRL model. Another method is to use the value as a numerical measure to assess the overall fit of the model. The larger value of means the higher chance of violation of the specified model assumption. For the full-cohort data from the NYUWHS application, the measures were 0.0656 vs. 0.4402 for the proportional and additive MRL models, respectively, and thus we concluded the proportional MRL model fitted the data better in this application. For the CC and NCC designs with 200 times sampling, we observed the same conclusion, as the measure was 0.033 vs. 0.401 (CC) and 0.032 vs. 0.445 (NCC) for the proportional and additive MRL models.
Fig. 3.
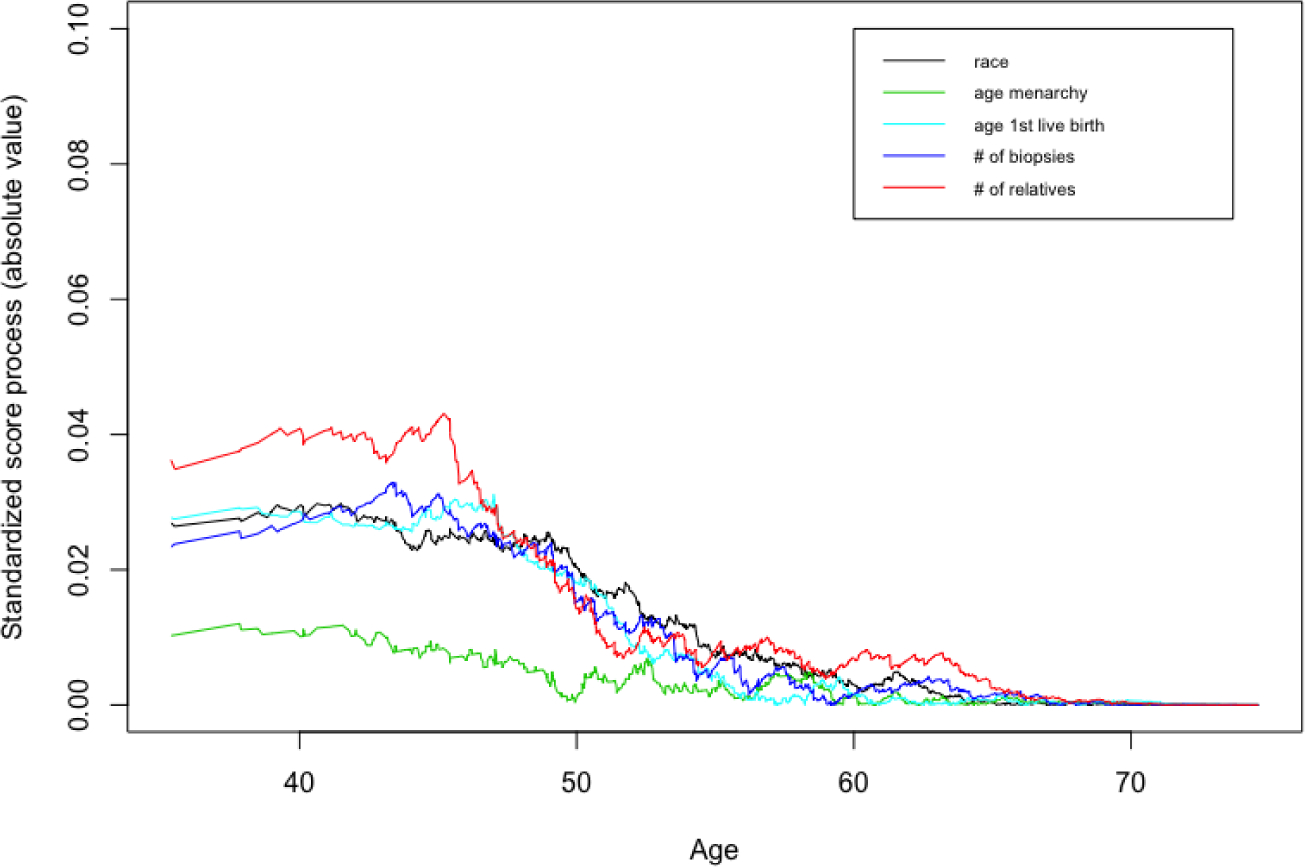
Model checking: proportional MRL model
Fig. 4.
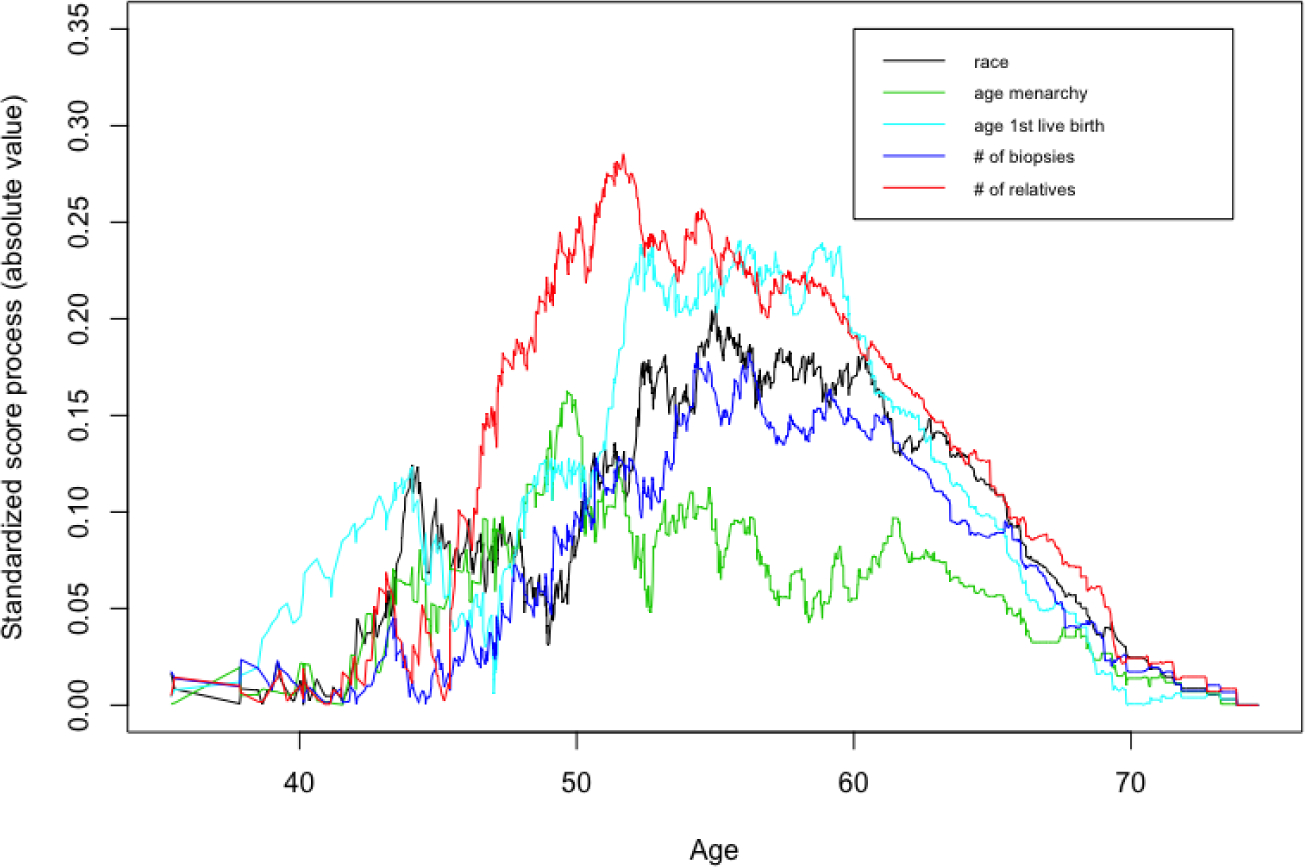
Model checking: additive MRL model
Although we do not study asymptotic properties of these model diagnostic measures for the generalized MRL models in this paper, we evaluated their empirical performance using simulations. Specifically, when the true model was proportional MRL model with sample size of 5000, β value of (0.2,0.2), and 80% censoring proportion, the proportions of numerical measure indicating the correct model were 97.4%, 93.0%, and 97.8% under the full-cohort, CC, and NCC designs, respectively. When the true model was additive MRL model with the same sample size, effect size, and censoring proportion, the proportions of numerical measure indicating the additive MRL model were 52.6%, 46.0%, and 63.6% under the full-cohort, CC, and NCC designs, respectively. We observed that the performance of the numerical measure was dependent on sample size, censoring rate, and effect size.
5. Discussion
In this manuscript, we developed unified inference procedures for the generalized MRL models under CC and NCC designs. The proposed models and inference procedures expand the analytical toolbox for these commonly used within-cohort sampling designs. In addition, we presented some model diagnostic and selection tools for the MRL models in full-cohort, CC, and NCC studies. Our numerical results support the use of the procedures, but the theoretical properties of which warrant further investigation.
The SEs of the proposed estimators were estimated using the bootstrapping method for CC data and the perturbation resampling approach for NCC data. Note that the perturbation approach requires the generation of n2 random realization matrix and subsequent matrix calculations, and it does tend to be computationally heavier comparing to the bootstrap method.
Both CC and NCC designs have cost-saving benefits as compared to the full-cohort design. In simulation studies and the real data example, we found estimators performed consistently across the three study designs when the data structure supported the application of MRL modeling. Based on the inverse probability weighting approach, other cohort sampling designs including counter-matching design and quota-matching design can be considered as well. Moreover, the generalized MRL models with time-varying coefficients under full cohort can also be extended to CC and NCC designs in future research.
In practice, left truncation may occur in epidemiological cohort studies. To incorporate the left truncation, controls need to be drawn from an adjusted risk set defined as , where Li denotes the left-truncation time for subject i. Besides, it is not rare to see that some patients may have relative longer survival time comparing to others in a cohort. If only few subjects have long survival time, the results may be biased and the restricted MRL should be considered.
Remark: An R package gmrl is available at https://github.com/pengjin0105/gmrl
Acknowledgements
The authors would like to thank the Associate Editor and two referees for their valuable suggestions. The research was partially supported by NIH grants RO1 CA178949.
Appendix
Simulation study
We conducted numerical simulations to compare the IPCW method and the QPS estimator under the full cohort of 1000 subjects when censoring rates were approximately 10%, 30% and 80%. A total of 500 simulations were conducted for both the proportional and additive MRL models. We reported the bias, the standard deviation (SD) of the estimates, the average of estimated standard error (SE) and the coverage rate (CP) of 95% Wald-type confidence intervals (see results in Table 6). The SEs of the estimates were calculated through standard bootstrap method. Based on the simulation results, the two estimators had similar performance when censoring probability was low. The biases were all small and the means of estimated SEs were close to the empirical SDs of the parameter estimators. The 95% Wald-type confidence intervals had proper coverage rate. However, when the censoring rate was 80%, the IPCW performance dropped and underestimated SEs, which led to low coverage probabilities.
Table 6.
Comparison between IPCW estimator and QPS estimator
| censoring rate | IPCW estimator† | QPS estimator‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|
| β = (0, 0) | β = (0.2, 0.2) | β = (0, 0) | β = (0.2, 0.2) | ||||||
| g(t) =exp(t) | |||||||||
| 0.1 | Bias | 0.000 | −0.006 | 0.000 | −0.004 | 0.000 | 0.003 | 0.000 | 0.002 |
| SI) | 0.038 | 0.065 | 0.032 | 0.049 | 0.039 | 0.065 | 0.033 | 0.056 | |
| SE | 0.037 | 0.065 | 0.030 | 0.051 | 0.038 | 0.066 | 0.032 | 0.056 | |
| CP | 94.0 | 94.6 | 92.7 | 94.8 | 94.2 | 96.2 | 94.2 | 95.5 | |
| 0.3 | Bias | −0.003 | −0.004 | −0.001 | −0.002 | 0.001 | −0.004 | 0.001 | −0.003 |
| SD | 0.047 | 0.078 | 0.037 | 0.059 | 0.043 | 0.075 | 0.036 | 0.063 | |
| SE | 0.046 | 0.080 | 0.035 | 0.060 | 0.042 | 0.073 | 0.036 | 0.062 | |
| CP | 94.4 | 94.6 | 93.7 | 93.9 | 94.2 | 94.1 | 95.1 | 94.6 | |
| 0.8 | Bias | 0.001 | 0.004 | 0.006 | 0.005 | −0.001 | −0.001 | 0.001 | −0.002 |
| SD | 0.273 | 0.459 | 0.139 | 0.237 | 0.071 | 0.129 | 0.057 | 0.105 | |
| SE | 0.245 | 0.373 | 0.138 | 0.225 | 0.073 | 0.127 | 0.059 | 0.103 | |
| CP | 86.2 | 86.0 | 92.6 | 93.4 | 95.5 | 94.2 | 95.3 | 95.4 | |
| g(t) = t | |||||||||
| 0.1 | Bias | 0.000 | −0.002 | −0.001 | −0.004 | 0.000 | 0.002 | 0.004 | 0.007 |
| SD | 0.013 | 0.022 | 0.024 | 0.044 | 0.020 | 0.034 | 0.030 | 0.050 | |
| SE | 0.013 | 0.022 | 0.023 | 0.043 | 0.020 | 0.034 | 0.029 | 0.051 | |
| CP | 94.0 | 94.8 | 94.6 | 93.0 | 93.9 | 96.1 | 94.0 | 95.6 | |
| 0.3 | Bias | −0.001 | −0.001 | −0.004 | −0.006 | 0.001 | −0.002 | 0.001 | −0.002 |
| SI) | 0.016 | 0.028 | 0.030 | 0.057 | 0.024 | 0.042 | 0.034 | 0.060 | |
| SE | 0.016 | 0.028 | 0.029 | 0.057 | 0.024 | 0.042 | 0.034 | 0.058 | |
| CP | 94.6 | 94.6 | 93.6 | 93.8 | 94.4 | 94.3 | 94.3 | 94.1 | |
| 0.8 | Bias | 0.000 | 0.001 | −0.098 | −0.091 | 0.001 | −0.002 | 0.010 | 0.006 |
| SD | 0.106 | 0.184 | 0.139 | 0.222 | 0.059 | 0.102 | 0.094 | 0.158 | |
| SE | 0.091 | 0.142 | 0.116 | 0.179 | 0.058 | 0.101 | 0.084 | 0.146 | |
| CP | 84.8 | 84.8 | 78.8 | 85.4 | 94.3 | 94.9 | 92.8 | 93.8 | |
SD: sample standard deviation; SE: mean of estimated standard error;
CP: empirical coverage probability of 95% confidence interval;
Inverse probability censoring weighted estimator
Quasi-partial score estimator
Regularity conditions
Let , and be the limits of , and , respectively. We assume the following regularity conditions:
(C1) sup supp(F) ≤ sup supp(G), where F(·) and G(·) are the distribution functions of T and C, respectively;
(C2) Z is bounded;
(C3) m*(t) is continuously differentiable on [0,τ];
(C4) is nonsingular;
Proof of Theorem 1(i)
First, we want to establish the consistency of the estimators and . Condition (C3) implies that m0(t) is of bounded variation on [0, τ]. Define for any ϵ > 0 and we have . By the strong law of large numbers and the fact that E{widMi(t)} = 0, for large n, t ∈ [0, τ], , and sufficiently large θ,
| (11) |
| (12) |
By (11), (12), and the monotonicity and continuity of g function, for any t ∈ [0, τ] and , there exists a unique that satisfies
| (13) |
Note that (11) and (12) hold for any θ > 0 when and only when β = β*. Then we have that converges to m0(t; β) uniformly in t ∈ [0, τ] and β in a compact set which contains the true parameter β*, and m0(t; β*) = m*(t). Thus, to prove the existence and uniqueness of and , it suffices to show that there exists a unique solution to U(β) = 0. Take derivative of (13) with respect to β, we have
which is a first-order linear ordinary differential equation about . The solution is
where
Let . We have
Thus, converges in probability to a nonrandom A. It is easy to check that U(β*) → 0 almost surely, and A is nonsingular by (C4). The convergence of and the continuity of A(β) imply that we can find a small neighborhood of β* in which is nonsingular when n is large enough. Therefore, it follows from the inverse function theorem that within a small neighborhood of β*, there exists a unique solution to U(β) = 0 for sufficiently large n. Thus, there exists unique estimators and . Since is strongly consistent to β*, then it follows the uniform convergence of to m0(t; β) that almost surely in [0, τ].
Proof of Theorem 1(ii)
In this section, we first prove the theorem 1(ii) under the CC design, then prove the theorem under the NCC design following the proof from Lu and Liu (2012). We know from equation (8) that
Subtract the above two equations and use Taylor expansion, we have,
Hence, following the first-order ordinary differential equation,
Let and we have
By using Taylor expansion again in , we have
Thus,
As we defined in our manuscript, let be the σ-field generated by and be the σ-field generated by . We denote
It is evident that and . Following the proof in Lu and Tsiatis (2006), . Condition on , {ηi(1 – γi/p0i), i = 1, …, n} and the first term of are uncorrelated. Therefore, is asymptotically normal with mean zero and variance-covariance Σ = Σ1 + Σ2. By Taylor expansion and consistency of , it follows
thus .
Under the nested case-control design, the asymptotic distribution of is more difficult to derive because the NCC sampling scheme is a dynamic process. The probability of being selected as a control is neither a constant not independent. Thus, we consider the idea of central limit theory for asymptotically negatively dependent random variables (Zhang, 2000), which has been used in the proof of Lu and Liu (2012). Based on the following asymptotical representation, we have
By martingale central limit theorem, as n → ∞. Since , it is evident that U1(β*) and U2(β*) are uncorrelated. However because of the NCC sampling scheme, wi and wj (i ≠ j) are correlated even after conditioning on . Since , then . Thus, the conditional variance of can be written as
According to Samuelsen (1997), for , where . Thus, with some algebra, the can be written as
where , . According to strong law of large numbers, we have where
where
Thus, by the central limit theory for asymptotically negatively dependent random variables (Zhang, 2000), we have as n → ∞, and
in distribution as n → ∞. It is easy to see that Σ1 + Σ2 = Σ. Follow by Taylor expansion and consistency of , we have .
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Cai T, Zheng Y (2013) Resampling Procedures for Making Inference under Nested Case-control Studies. Journal of the American Statistical Association 108(504):1532–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Lo SH (1999) Case-Cohort and Case-Control Analysis with Cox’s Model. Biometrika 86(4):755–764 [Google Scholar]
- Chen YQ (2007) Additive Expectancy Regression. Journal of the American Statistical Association 102(477):153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, Cheng S (2005) Semiparametric regression analysis of mean residual life with censored survival data. Biometrika 92(1):19–29 [Google Scholar]
- Chen YQ, Cheng S (2006) Linear Life Expectancy Regression with Censored Data. Biometrika 93(2):303–313 [Google Scholar]
- Chen YQ, Jewell NP, Lei X, Cheng SC (2005) Semiparametric Estimation of Proportional Mean Residual Life Model in Presence of Censoring. Biometrics 61(1):170–178 [DOI] [PubMed] [Google Scholar]
- Clendenen TV, Ge W, Koenig KL, Axelsson T, Liu M, Afanasyeva Y, Andersson A, Arslan AA, Chen Y, Hallmans G, Lenner P, Kirchhoff T, Lundin E, Shore RE, Sund M, Zeleniuch-Jacquotte A (2015) Genetic Polymorphisms in Vitamin D Metabolism and Signaling Genes and Risk of Breast Cancer: A Nested Case-Control Study. PLOS ONE 10(10):e0140478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B (1979) Bootstrap Methods: Another Look at the Jackknife. The Annals of Statistics 7(1):1–26 [Google Scholar]
- Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, Mulvihill JJ (1989) Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. Journal of the National Cancer Institute 81(24):1879–1886 [DOI] [PubMed] [Google Scholar]
- Ge W, Clendenen TV, Afanasyeva Y, Koenig KL, Agnoli C, Brinton LA, Dorgan JF, Eliassen AH, Falk RT, Hallmans G, Hankinson SE, Hoffman-Bolton J, Key TJ, Krogh V, Nichols HB, Sandler DP, Schoemaker MJ, Sluss PM, Sund M, Swerdlow AJ, Visvanathan K, Liu M, Zeleniuch-Jacquotte A (2018) Circulating anti-Müllerian hormone and breast cancer risk: A study in ten prospective cohorts. International Journal of Cancer 142(11):2215–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James IR (1986) On Estimating Equations with Censored Data. Biometrika 73:35–42 [Google Scholar]
- Kupper LL, McMichael AJ, Spirtas R (1975) A Hybrid Epidemiologic Study Design Useful in Estimating Relative Risk. Journal of the American Statistical Association 70(351a):524–528, URL 10.1080/01621459.1975.10482466 [DOI] [Google Scholar]
- Lin DY, Wei LJ, Ying Z (1993) Checking the Cox Model with Cumulative Sums of Martingale-Based Residuals. Biometrika 80(3):557–572 [Google Scholar]
- Liu M, Lu W, Shore RE, Zeleniuch-Jacquotte A (2010a) Cox regression model with time-varying coefficients in nested case-control studies. Biostatistics 11(4):693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lu W, Tseng Ch (2010b) Cox Regression in Nested Case-Control Studies with Auxiliary Covariates. Biometrics 66(2):374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Liu M (2012) On estimation of linear transformation models with nested case-control sampling. Lifetime Data Anal 18(1):80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Tsiatis AA (2006) Semiparametric transformation models for the case-cohort study. Biometrika 93(1):207–214 [Google Scholar]
- Ma H, Shi J, Zhou Y (2017) Proportional Mean Residual Life Model with Censored Survival Data under Case-cohort Design. arXiv:170801634 [math, stat] [Google Scholar]
- Maguluri G, Zhang CH (1994) Estimation in the Mean Residual Life Regression Model. Journal of the Royal Statistical Society: Series B (Methodological) 56(3):477–489 [Google Scholar]
- Oakes D, Dasu T (1990) A note on residual life. Biometrika 77(2):409–410 [Google Scholar]
- Prentice RL (1986) A Case-Cohort Design for Epidemiologic Cohort Studies and Disease Prevention Trials. Biometrika 73(1):1–11 [Google Scholar]
- Reid N (1981) Influence Functions for Censored Data Ann Statist. 9(1):78–92 [Google Scholar]
- Samuelsen S (1997) A pseudolikelihood approach to analysis of nested case-control studies. Biometrika 84(2):379–394 [Google Scholar]
- Scarmo S, Afanasyeva Y, Lenner P, Koenig KL, Horst RL, Clendenen TV, Arslan AA, Chen Y, Hallmans G, Lundin E, Rinaldi S, Toniolo P, Shore RE, Zeleniuch-Jacquotte A (2013) Circulating levels of 25-hydroxyvitamin D and risk of breast cancer: a nested case-control study. Breast Cancer Research 15(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheike TH, Juul A (2004) Maximum likelihood estimation for Cox’s regression model under nested case-control sampling. Biostatistics 5(2):193–206 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang Z (2009) A Class of Transformed Mean Residual Life Models With Censored Survival Data. Journal of the American Statistical Association 104(486):803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Song X, Zhang Z (2012) Mean residual life models with time-dependent coefficients under right censoring. Biometrika 99(1):185–197 [Google Scholar]
- Thomas DC (1977) Addendum to “methods of cohort analysis: appraisal by application to asbestos mining” by Liddell FDK, McDonald JC, and Thomas DC Journal of the Royal Statistical Society: Series A (General) 140(4):483–485 [Google Scholar]
- Yang G, Zhou Y (2014) Semiparametric varying-coefficient study of mean residual life models. Journal of Multivariate Analysis 128:226–238 [Google Scholar]
- Zhang LX (2000) A Functional Central Limit Theorem for Asymptotically Negatively Dependent Random Fields. Acta Mathematica Hungarica 86(3):237–259 [Google Scholar]


