Abstract
Background:
Alcohol use disorder (AUD) is one of the most common forms of substance use disorders with a strong contribution of genetic (50–60%) and environmental factors. Genome-wide association studies (GWAS) have identified a number of AUD-associated variants, including those in alcohol metabolism genes. These genetic variants may modulate gene expression, making individuals more susceptible to AUD. A long-term alcohol consumption can also change the transcriptome patterns of subjects via epigenetic modulations.
Methods:
To explore the interactive effect of genetic and epigenetic factors on AUD, we conducted a secondary analysis by integrating GWAS, copy number variation, brain transcriptome and DNA methylation data to unravel novel AUD-associated genes/variants. We applied the mega-analysis of odds ratio (MegaOR) method to prioritize AUD candidate genes (AUDgenes).
Results:
We identified a consensus set of 206 AUDgenes based on the multi-omics data. We demonstrated that these AUDgenes tend to interact with each other more frequent than chance expectation. Functional annotation analysis indicated that these AUDgenes were involved in substance dependence, synaptic transmission, glial cell proliferation, and enriched in neuronal and liver cells. We obtained a multidimensional evidence that AUD is a polygenic disorder influenced by both genetic and epigenetic factors as well as the interaction of them.
Conclusion:
We characterized multidimensional evidence of genetic, epigenetic, and transcriptomic data in AUD. We found that 206 AUD associated genes were highly expressed in liver, brain cerebellum, frontal cortex, hippocampus, and pituitary. Our studies provides important insights into the molecular mechanism of AUD and potential target genes for AUD treatment.
Keywords: GWAS, TWAS, eQTL, integrative study, alcohol use disorder, brain cerebellum, brain hippocampus, pituitary, glial cell
INTRODUCTION
Substance use disorders (SUDs) cost the United States over $200 billion a year [1]. Alcohol use disorder (AUD) is one of the most common forms of SUDs with a prevalence of 6.2% (National Survey on Drug Use and Health, or NIAAA, 2015), imposing a heavy burden to families and the society. According to the Diagnostic and Statistical Manual of Mental Disorders (DSM) criteria, AUD is diagnosed as a psychiatric disorder in which an individual is physically or psychologically dependent on alcohol. Twin studies have reported a genetic heritability of 50–60% for AUD [2]. In 2013, the American Psychiatric Association issued the fifth edition of the DSM criteria (DSM-V) and integrated alcohol dependence and alcohol abuse into one disorder, AUD, with mild, moderate, and severe sub-classifications. However, some studies published after 2013 still used alcohol abuse or dependence. Throughout this work, we use AUD but for each individual data set, we use alcohol abuse or dependence as stated in original studies.
While the etiology of AUD remains elusive, ethanol metabolism and interactions between a large number of genetic and environmental factors underlie AUD. Ethyl alcohol, or ethanol (C2H5OH),when consumed, is transported from blood to other organs including the brain [3]. Ethanol has acute molecular effects on excitatory and inhibitory synaptic transmission. A long-term consumption of alcohol could lead to AUD [4], which is also attributable to both genes and environmental factors including socio-cultural conditions [5]. Alcohol metabolism mainly occurs in liver and involves two enzymes, alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH), which break ethanol down to acetaldehyde and then to acetate [2].A recent genome-wide association study (GWAS) highlighted several alcohol metabolic-related genes that harbor AUD-associated variants [6]. However, the top variants in alcohol metabolic-related genes with genome-wide significance (p<5 × 10−8) can only account for a small portion of the heritability of AUD [6], implying that other AUD genetic risks factors are yet to be revealed [7 8]. The recent success in studying other complex diseases suggests that the integrative analysis of GWAS data and other types of regulatory annotations [e.g., expression quantitative trait loci (eQTL)] using methods such as the transcriptome-wide association study (TWAS) could reveal novel disease susceptibility loci [9–12]. In addition, copy number variations (CNVs) and rare variants have also been implied in AUD[13–15]. Several genes, such as the Serine Incorporator 2 (SERINC2) and alcohol dehydrogenase genes [15], were reported to carry rare variants that were associated with AUD in subjects of European descent. Alterations at the epigenetic level were also reported in AUD patients. For example, long-term alcohol consumption was reported to alter the transcriptome pattern and methylation levels of subjects, although the alternations in the brain were relatively small [16] and only nominally significant changes were observed [17 18]. With the accumulation of these insightful yet heterogeneous candidate variants and genes, integrative studies are needed for prioritizing candidate genes/variants underlying AUD.
Integrative analysis of high-throughput multi-omics data can prioritize disease-associated genes through a supervised or unsupervised fashion. The findings from previous studies have implied that the number of susceptibility genes for each complex disease is limited [19]. With this assumption, we recently developed an unsupervised machine learning approach named mega-analysis of odds ratio (MegaOR), aiming at identifying groups of genes that collectively lead to the maximum load of evidence across multi-omics data [20]. We have successfully implemented the method in studying the genetics of schizophrenia [20] and Crohn’s Disease [21]. In this study, we attempted to use MegaOR to identify AUD-associated genes. We collected six types of genome-wide omics data, each representing a type of molecular evidence associated with AUD. We also identified tissues that were most likely related to AUD and conducted TWAS for AUD in these tissues. Through the integrative analysis using MegaOR, we identified a set of candidate genes for AUD and characterized their roles through functional enrichment and drug-target crosstalk analyses.
METHODS
GWAS summary statistics
We downloaded the GWAS summary statistics from the latest and also the largest AUD study from the Million Veteran Program (MVP) [22 23]. The original GWA study collected genotype data from a total of 242,317 individuals (34,658 cases and 167,346 controls) with European ancestry. Alcohol dependence was diagnosed following the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) criteria, 4th edition. Before the meta-analysis, the consortium had imputed the genotype data using EAGLE2 [24] and Minimac3 [25] based on the 1000 Genomes Project Phase 3 European reference panel [26]. In total, the GWAS data included association results for 5,833,029 SNPs either genotyped or imputed (imputation INFO scores > 0.7 and minor allele frequency > 0.0005).
Gene expression data
We obtained the transcriptome data (by RNA-sequencing) of postmortem prefrontal cortex (PFC) tissues of 65 AUD patients and 73 healthy controls from a previous study [27]. All samples were European Australian males. Cases and controls were matched by age, sex, brain weight, brain pH, disease classification, and postmortem interval (PMI). Cases were diagnosed as AUD according to the DSM-IV criteria [27]. Quality control was conducted and differentially expressed genes were identified by fitting multiple linear regression models with covariates such as age, PMI, and RNA integrity number (RIN). Multiple testing correction was controlled at a false discovery rate (FDR) < 0.05.
Methylation data
DNA methylation data were generated with Illumina HumanMethylation450 using the postmortem prefrontal cortex (PFC) region in brains collected from 16 AUD patients and 16 healthy controls [18]. All samples were European Australian males. Cases and controls were matched by age, brain weight, brain pH, and postmortem interval (PMI). Cases were diagnosed as AUD according to the DSM-IV criteria. The methylation matrix was normalized by a beta-mixture quantile normalization method (BMIQ) using the R package limma. Probes with a detection p value > 0.05 in any subject were excluded. Batch effect that was likely caused by the use of multiple chips in the assays was detected and excluded using the ComBat function implemented by the R package sva. After quality control, 434,015 CpGs probes were retained for the following analysis. The residuals from the batch effect correction step was used to detect differentially methylated probes as well as differentially methylated genes (DMGs) based on the annotations of probes. To this end, the authors fitted empirical Bayesian linear models (implemented by the R package limma) for each probe by including age and PMI as the covariates. A FDR threshold of q < 0.05 was applied.
Gene-based association test
Gene-based p-values were calculated using Magma [28]. Specifically, for each gene, we considered all SNPs in the gene body and 50kb upstream and 35 kb downstream of the gene. Magma utilizes the mean of the χ2 statistic to obtain gene-based p-values and takes account of the gene length, SNP density, and local linkage disequilibrium (LD) structure. We used the 1000 Genome Project European panel as the reference panel for LD [26].
AUD related tissues
To identify the tissues that are most likely related with AUD, we applied two methods to determine the AUD-associated tissues, the decoding Tissue Specificity (deTS) package developed in our previous work [29] and the Functional Mapping and Annotation (FUMA) [30]. deTS is an R package that provides a panel with 47 tissues from the GTEx (v7) expression data and implements Fisher’s exact test to examine whether genes in a query list are enriched in a tissue. FUMA is a web service (https://fuma.ctglab.nl/), which contains two major functions: SNP2GENE and GENE2FUNC. We used the tissue specificity subfunction under the GENE2FUNC to discover whether our candidate gene lists were enriched in up- regulated differentially expressed genes (DEG) across 53 tissues from the GTEx (v7) expression data. We utilized the deTS and the FUMA tissue specificity function to identify disease-related tissues based on genes defined by the GWAS Magma results using different thresholds (i.e., pMagma< 0.05, pMagma< 0.01, pMagma < 5 × 10–3, pMagma < 0.001, pMagma < 5 × 10–4, pMagma < 10–4, pMagma < 5 × 10–5, pMagma < 10–5, pMagma < 5 × 10−6, and pMagma < genome-wide significance with Bonferroni corrections).
Fisher’s method
We used the Fisher’s method to combine p-values from AUD-associated tissues generated by Sherlock and MetaXcan [9 10], respectively into one Chi-squared distribution statistic (X 2), using the formula
where n is the number of AUD-associated tissues and pk is the p-value for the kth tissues. The sum of n independent chi-squared values, each with two degrees of freedom, follows a chi-squared distribution with 2n degrees of freedom.
Transcriptome wide association studies (TWAS)
TWAS detects gene-disease association based on imputed gene expression from genotype data or GWAS summary statistics. We applied the TWAS method, MetaXcan [9], which could take GWAS summary statistics as the input, to the AUD GWAS data using the tissues identified by deTS (see the Result section “Decode AUD related tissues”).
Combinatory results of eQTL and GWAS data (Sherlock)
Many GWAS signals were located in non-coding or genetic regulatory regions. We utilized the method Sherlock to combine tissue eQTL data and GWAS signals at the gene level [10]. Specifically, we selected the GTEx eQTL data from the AUD-related tissues identified by deTS (see the Result section “Decode AUD related tissues”).
Copy number variation (CNV)
We collected AD associated CNVs from a GWAS that was conducted using African American and European American populations [13]. Diagnosis of AD was followed DSM-IV (or DSM-III-R) criteria. The original study collected the genotypeing data of 6,045 subjects (3,243 cases and 2,802 controls) from three datasets: the Study of Addiction: Genetics and Environment (SAGE), the Collaborative Study on the Genetics of Alcoholism (COGA), the Center for Inherited Disease Research (CIDR), and the Genome- wide Association Study of Alcohol Use and Alcohol Use Disorder in Australian Twin-Families (OZALC). Overall, nine CNV regions were identified to be associated with AD. We mapped genes to these regions and denoted them as the CNV associated genes.
Mega-analysis of odds ratio (MegaOR)
We recently developed a method called MegaOR [20] with the aim of identifying groups of genes whose combinatory load of disease association was optimized across multiple dimension of evidence. Specifically, MegaOR takes as the input a multidimensional matrix of evidence, each column representing the association results with the investigated trait. To enable integration of different types of omics data, we requested the input matrix to be a binary matrix. Specifically, in each column of the evidence matrix, genes were labeled as 1 if they were determined as significantly associated with the trait or 0 otherwise. For instance, in the category of GWAS results, genes whose Magma p-values were < 0.05 were labeled 1 and other genes 0. In this way, categorical data such as CNVs and continuous data such as gene-based p-values could be integrated into one matrix for the following analyses. In addition, the dimension-specific thresholds were defined for each individual omics data to fully address the particular data features and were independent of other dimensions. For example, Odds Ratio (OR) in one dimension is represented by (the count of a given group of genes ∩ genes in one dimension labelled with 1) divided by (the count of a given group of genes ∩ genes in one dimension labelled with 0). MegaOR takes the binary matrix as input and defines a combined OR (cOR): , where OR represents the odds ratio in each dimension, d is the number of evidence, and μ represents the average OR across dimensions. The part is introduced as the penalty term to control deviation of individual dimensional ORs and balance them across multiple lines of evidence. An iterative optimization procedure was implemented to find the best set of genes (denoted by S) with the pre-defined size n. At the stable status, genes in S tended to have the optimized cOR. Further details can be found in our previous works [20 21].
Determine the threshold of the significant occurrence
For N iterative runs, we performed an exact test of a null hypothesis about the probability of occurrence in a Bernoulli experiment (binomial test). For each gene from the evidence matrix with d genes, the expected occurrence of one gene is S/d (S is the gene set size). If the observed occurrence of one gene is significantly greater than the expected occurrence (S/d), we could determine the threshold K for each N runs (the significant p value is defined as 0.05 adjusted by multiple testing corrections). For example, at the permutation of N = 100 times and the gene size of S=200, each gene in the dataset (d = 1000 genes) is expected to occur 20 times. If the null hypothesis is that the true probability of occurrence is no greater than 0.2 and observed number of one gene is 50 times, we could use R platform (3.5.2) command binom.test (50, 100, 0.2, alternative = “greater”) and obtain the p = 2.14 × 10−11. Thus, we would reject the null hypothesis at this observed occurrence. Finally, we iteratively search and find the smallest number (defined as threshold K) that would reject the null hypothesis.
RESULTS
Multi-dimensional evidence for AUD
We collected and processed six major categories of evidence: genes defined by GWAS results (Magma), genes defined by integrative analysis of GWAS and eQTL (Sherlock), TWAS (MetaXcan), differentially expressed genes (DEGs), differentially methylated genes (DMGs), and CNV genes. Among these data, Sherlock and MetaXcan results were calculated for different tissues and thus, each of them had multiple sets of results. Each of these multidimensional data sets were conducted using case-control design and each set of the nominated genes represented one kind of disease association with AUD.
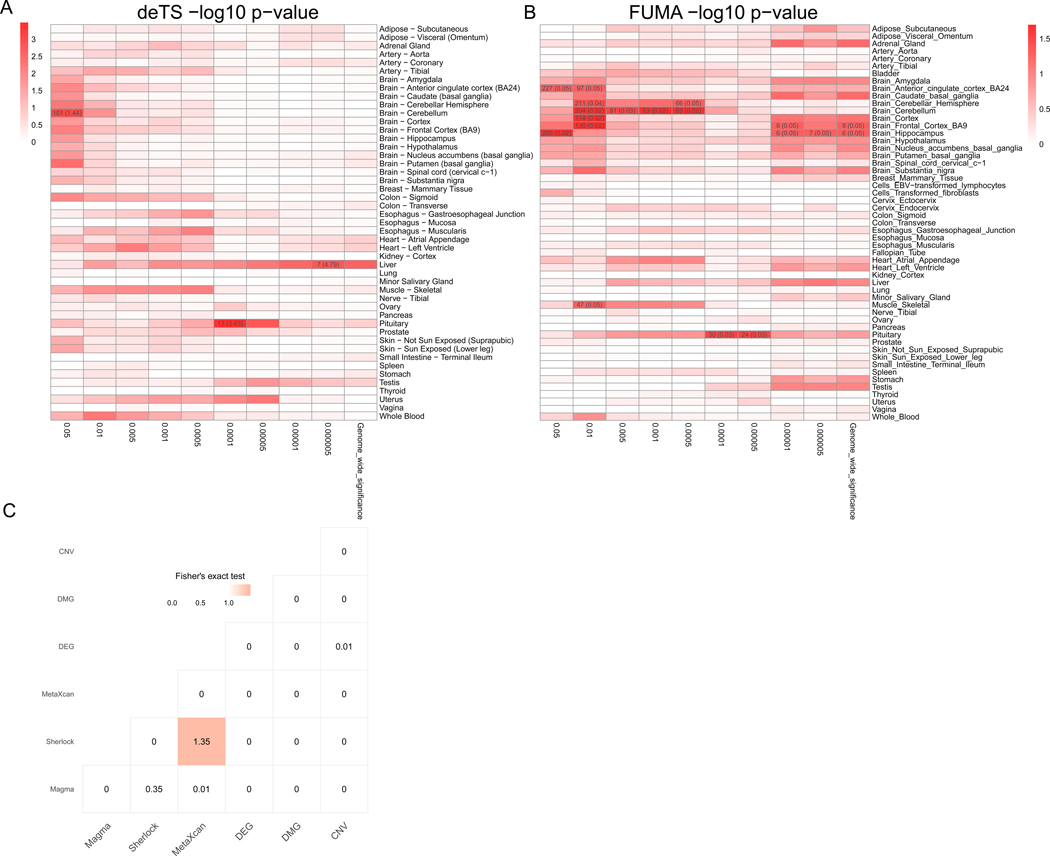
Previous studies have suggested that interpretation of disease-associated genetic variants are context specific, as genetic regulatory effects have a strong tissue specificity [9 31]. To identify the disease-related tissues of AUD, we first applied tissue-specific enrichment analysis (TSEA) of the AUD GWAS data and determined three tissues for AUD: pituitary (the most significant raw p = 3.2 × 10−4), brain cerebellum (raw p = 8.9 × 10−4), and liver (p = 1.5 × 10−3) (see below). We further applied a widely used online tool, Functional Mapping and annotation (FUMA), to validate the tissue-specific enrichment analysis by deTS [29 30]. As a result, FUMA identified four frequently occurred AUD-associated tissues: brain cerebellum (raw p = 0.02), brain frontal cortex (BA9) (raw p = 0.02), brain hippocampus (raw p = 0.02), and pituitary (raw p = 0.03). Although the AUD-associated tissues identified by FUMA only has nominal significance, the results from deTS and FUMA have relatively consistent signals (Figure 1A, 1B). Thus, we combined the tissues from two approaches and used five AUD-associated tissues in this study: brain cerebellum, frontal cortex (BA9), hippocampus, and pituitary as well as liver as the five AUD-associated tissues in this study. Therefore, we had candidate AUD-associated genes from a total of 14 types of evidence, including five result sets for Sherlock and MetaXcan, respectively, plus GWAS, DEGs, DMGs, and CNV genes. Considering the alternation in the brain are relatively small (20–50% changes from previous studies [16]), and only a few genomic-wide significant loci in GWAS. We thus applied the following threshold to balance the number of genes in each evidence and control the false discovery rate. Specifically, there were 270 Magma genes (FDR < 0.05), 243 Sherlock genes in brain cerebellum (raw p < 0.05), 104 Sherlock genes in brain frontal cortex (raw p < 0.05), 77 Sherlock gene in brain hippocampus (raw p < 0.05), 111 Sherlock genes in liver (raw p < 0.05), 159 Sherlock genes from pituitary (raw p < 0.05), 94 MetaXcan genes in brain cerebellum (raw p < 0.05), 57 MetaXcan genes in brain frontal cortex (BA9) (raw p < 0.05), 57 MetaXcan genes in brain hippocampus (raw p < 0.05), 82 MetaXcan gene in liver (raw p < 0.05), 88 MetaXcan gene in pituitary (raw p < 0.05), 116 DEGs (FDR < 0.05), 536 DMGs (FDR < 0.05), and 9 CNV regions (nominal p < 0.05) with 27 genes overlapped within these regions (Table S1). These data formed a matrix with 998 genes, each containing at least one source of association evidence. Among them, 898 (90.0%) genes contained only one type of evidence and no gene had more than 14 lines of evidence. To summarize the information from multi-tissues, we further used Fisher’s methods to merge the genes of Sherlock and MetaXcan from multiple disease-related tissues, resulting in 67 Sherlock genes and 30 MetaXcan genes with nominal pcombined< 0.05, respectively.
Figure 1. Overview of the multidimensional data for Alcohol use disorder (AUD) study.
(A) Tissue-Specific Enrichment Analysis (TSEA) of Magma genes. X-axis: groups of genes defined at different thresholds based on Magma p-value. Y-axis: 47 GTEx tissues used as the reference panel. Cells were highlighted in red proportional to the corresponding p-values. Cells with Bonferroni correction significance were further annotated with the numbers in each cell represented the number of shared genes between the Magma genes and the tissue-specific genes, followed by the odds ratio in braces. (B) FUMA tissue-specific enrichment analysis of Magma genes. X-axis: groups of genes defined at different thresholds based on Magma p-value. Y-axis: 53 GTEx tissues used as the reference panel. Cells were highlighted in red proportional to the corresponding p-values. Cells with nominal p value < 0.05 were further annotated with the numbers in each cell represented the number of shared genes between the Magma genes and the tissue-specific genes, followed by the raw p value in braces (C) Pair-wise comparison of the six types of omics data. The -log10 (p-value) from Fisher’s exact test was labeled for each cell. The shade of the cell was proportional to the p-value.
Decode AUD related tissues
To identify AUD related tissues, unbiased data analyses without a priori assumptions are needed. We used Magma genes to determine AUD related tissues, as these genes were defined without tissue information. We performed tissue-specific enrichment analysis using Magma genes defined at different thresholds (p < 0.05, p < 0.01, p < 5 × 10−3, p < 1 × 10−4, p < 5 × 10−4, p < 1 × 10−5, p < 5 × 10−5, p < 1 × 10−6, p < 5 × 10−6, and p < 2.7 × 10−6 genome-wide significance with Bonferroni corrections for 18,376 genes from Magma). As shown in Figure 1A and Figure1B, we identified five AUD-associated tissues from two computational approaches [30 31]. deTS identified three significant tissues: pituitary (raw p = 3.2 × 10−4 at p < 1 × 10−4 Magma threshold), brain cerebellum (raw p = 8.9 × 10−4 at p < 0.05 Magma threshold), and liver (p = 1.5 × 10−3 at p < 5 × 10−6 Magma threshold). FUMA identified four AUD-associated tissues with multiple occurrence: brain cerebellum (raw p = 0.02), brain frontal cortex (BA9) (raw p = 0.02), brain hippocampus (raw p = 0.02), and pituitary (raw p = 0.03). These findings were consistent with previous reports that liver is the tissue where alcohol is metabolized. Both molecular biology studies and GWAS reported the involvement of ADH and ALDH family genes in alcohol consumption. In addition, ethanol has the acute molecular effects on excitatory and inhibitory synaptic transmission and a long-term exposure to ethanol impacts the ability of adapting alcohol as neurotransmitter, whichcould lead to AUD liability [4]. Thus, liver and brain cerebellum, frontal cortex (BA9), hippocampus, and pituitary were most likely related to AUD, and thus we used these tissues for the following analyses (Sherlock and MetaXcan).
Pair-wise comparison of the multidimensional association data
As shown in Figure 1C, for the six types of data, we only observed a nominally significant correlation between Sherlock and MetaXcan genes (p = 4.45 × 10−2). This is not surprising because both methods measured the integrative signal of genetic variance and transcriptome changes. We failed to detect any correlation between Magma genes and Sherlock or between Magma and MetaXcan genes, even though all three used the GWAS summary statistics. These results implied that there was an independent information that could be obtained by integrating eQTL and genetically regulated expression (GReX) than using GWAS data only, proving the necessity to integrate these diverse evidence data. Surprisingly, although DEGs and DMGs were conducted on the same type of tissue in different studies, they showed limited associations with each other.
AUD associated genes (AUDgenes) identified by MegaOR
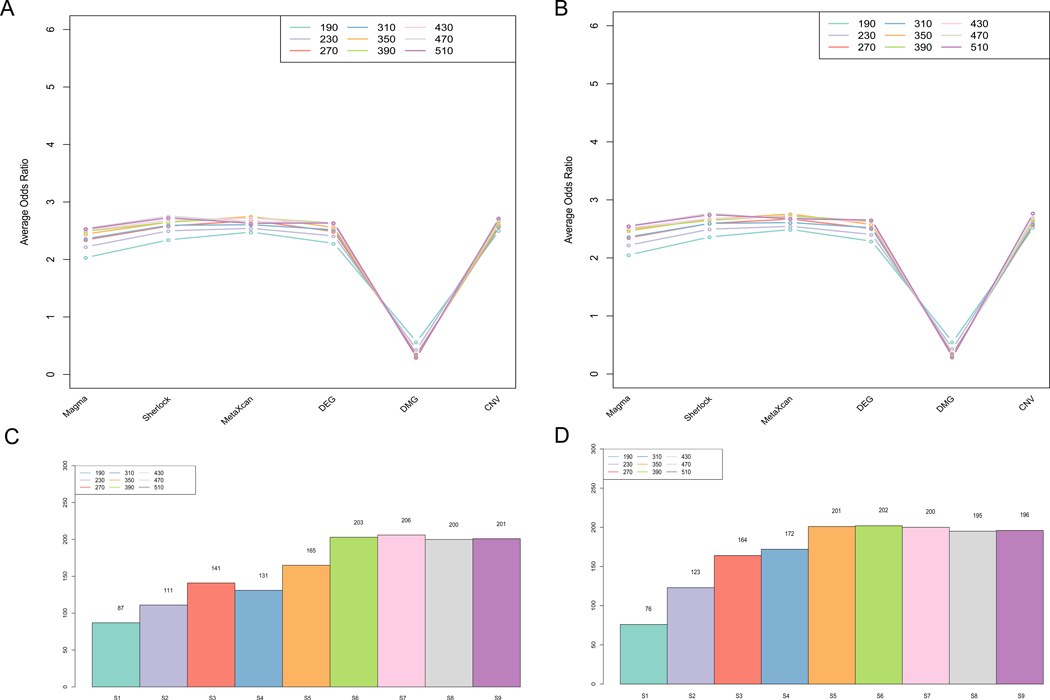
To prioritize candidate genesthat contain optimal evidence, we conducted the analyses using MegaOR for the 998 genes (evidence-only matrix with 6 dimension). Due to the intrinsic configuration, MegaOR requires to predefine the set size as one of the inputs. Thus, we tested multiple set sizes assuming that the number of authentic AUD-associated genes were at the scale of hundreds, as with most complex diseases [19]. Specifically, we tested nine setsfor each matrix, which contains 20% - 50% of the whole size of the evidence-only matrix (S = 190, 230, 270, 310, 350, 390, 430, 470, and 510 for the evidence-only matrix) (Figure 2). To test the robustness of our MegaOR algorithm, we initiated different numbers (100 and 1000) of runs and obtained sets of candidate genes with the same size because the sets of genes with the maximum load of evidence were not unique at a fixed set size. To obtain the significantly occurred genes from the iterative runs, we used a binomial test to dynamically select genes that occurred in more than N runs in 9 set sizes and defined these genes as the consensus genes (i.e., AUDgenes). For example in 100 runs, we named the genes at each set size as S1 (n = 190, AUDgenes: 87), S2 (n = 230, AUDgenes: 111), S3 (n = 270, AUDgenes: 141), S4 (n = 310, AUDgenes: 131), S5 (n = 350, AUDgenes: 165), S6 (n = 390, AUDgenes: 203), S7 (n = 430, AUDgenes: 206), S8 (n = 470, AUDgenes: 200), and S9 (n = 510, AUDgenes: 201). For a 1000 runs set, the number of AUDgenes in the consensus sets ranged from 76 (S1, n = 190) to 202 (S6, n = 390) (Figure 2B). In both matrices, a converged stable status of AUDgenes were found from S6 to S10 in both 100 and 1000 run sets (Figure 2C and 2D). Interestingly, we observed a peak at S7 in 100 run set. These 206 genes of AUDgenes reached the local maximum load of evidence among the tested geneset size under controlled false discovery rate (see Methods). These 206 AUDgenes came from six types of evidence, including 73 GWAS genes, 31 Sherlock genes, 22 MetaXcan genes, 84 DEGs, 19 DMGs, and 25 CNV (Table S2).
Figure 2. Summary of AUD candidate genes identified by MegaOR.
(A) Odds Ratio (OR) distribution for each type of evidence in each set size for the evidence matrix with 998 genes in 100 runs. Each dot indicates the average OR in the corresponding evidence type from 100 sets resulted from MegaOR (see main text). (B) OR distribution in 1000 runs. (C) Distribution of AUDgenes at the stable stage for each set size in 100 runs. (D) Distribution of AUDgenes at each set size in 1000 runs.
AUDgenes tended to interact with each other more often than expected
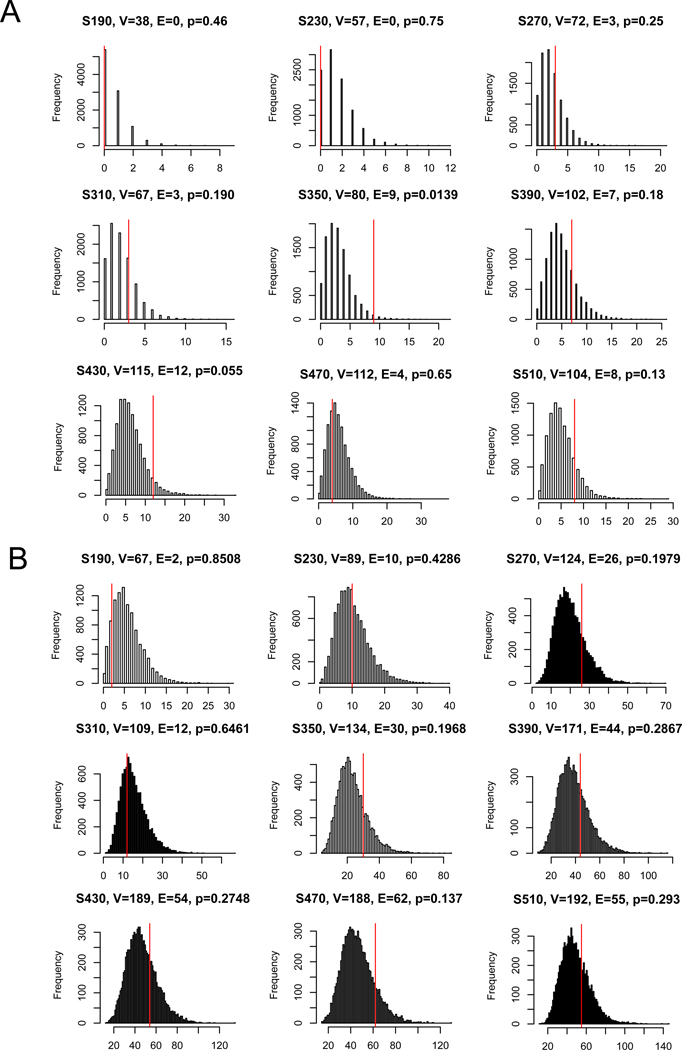
Following the theories and observations in network medicine [32], disease genes tended to have more co-expression than observed among random genes in the human interactome, mainly because genes underlying the same diseases are often involved in related biological pathways in disease-relevant tissues. We tested whether AUDgenes had significantly more interactions than the random expectation using two curated human PPI networks: influence graph [33], and a combined dataset of HPRD and STRING (MAGI) [34]. These reference networks were described in previous study [21]. As shown in Figure 3, for each set of AUDgenes from evidence-only matrix, we resampled 10,000 random gene sets in the same set sizes. The proportion of the random gene sets that had interactions exceeding the interactions among AUDgenes was used to calculate an empirical p value. We performed this analysis in each of the two curated PPI network respectively. Due to the consensus AUDgenes obtained from multiple discrete sources and tissues, the AUDgenes only showed slight significantly more PPIs than those from random gene sets in most cases.
Figure 3. Distribution of protein-protein interactions (PPIs) among AUDgenes.
The analysis was conducted using the MAGI (A) and influence graph reference panel (B). In each panel, the distribution of the intersections was displayed for 10,000 randomly selected gene sets, each with the same set sizes as the query set. X-axis is the number of interactions. Y-axis is the frequency of the interactions. In the title of each panel, V denoted the number of AUDgenes that were annotated in the corresponding PPI network; E denoted the number of interactions among these AUDgenes; and the p-value was the empirical rank p- value. The vertical red line indicates the number of interactions observed for the actual AUDgenes.
Functional enrichment analysis of AUDgenes
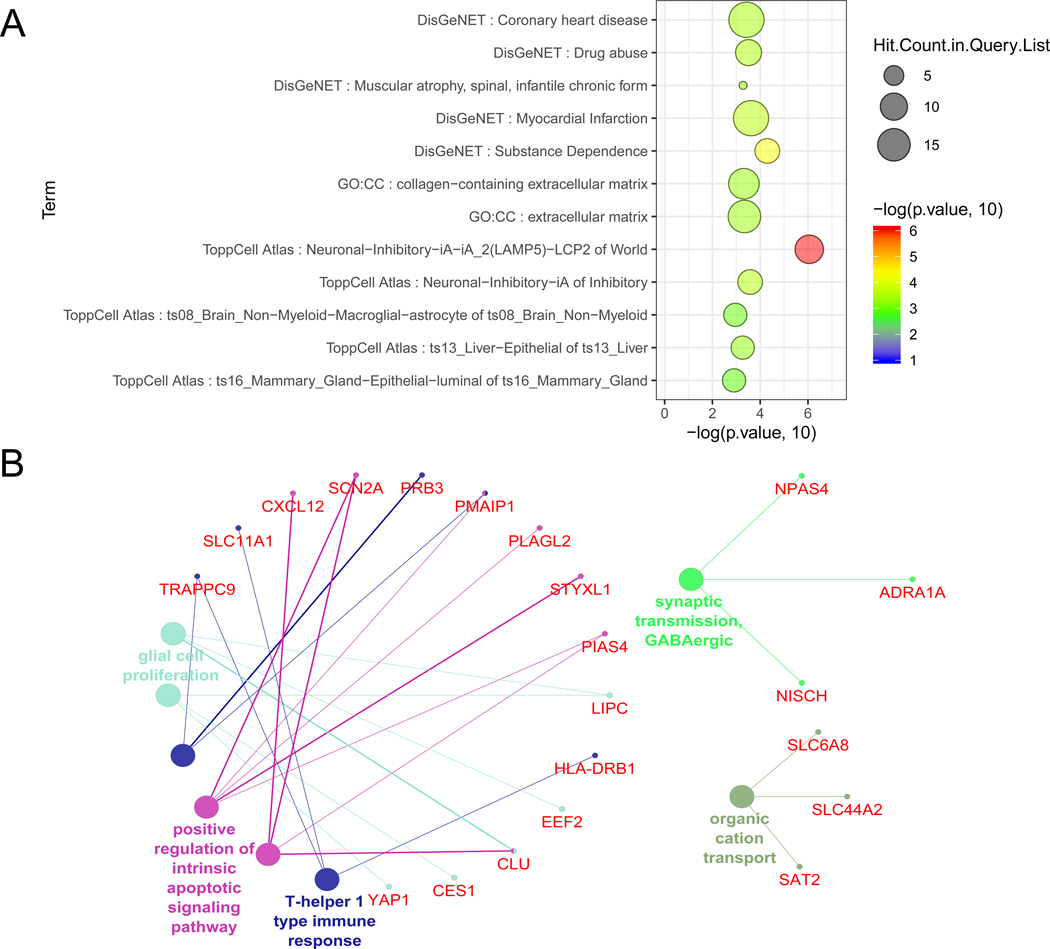
To identify the biological roles of the AUDgenes, we conducted gene set enrichment analysis using ToppFun (https://toppgene.cchmc.org/, accessed 11/23/2019). We focused on Gene Ontology (GO) terms and gene sets from the DisGeNET [35]. As shown in Figure 4A, 206 AUDgenes were significantly enriched in cellular components for extracellular matrix (GO: 0031012, FDR < 0.095) and collagen- containing extracellular matrix (GO: 0062023, FDR < 0.095). DisGeNET is a database integrating data from expert curated repositories, GWAS catalogue, animal models, and scientific literature [35]. AUDgenes were found to be significantly enriched in Substance Dependence (Unified Medical Language System (UMLS) ID: C0038580, FDR <0.092) and Drug abuse (Unified Medical Language System (UMLS) ID: C0013146, FDR <0.092). We further used the Cytoscape plug-in ClueGO to illustrate the relationship between genes and GO terms [36 37]. GO terms with at least three AUDgenes were demonstrated for GO Biological Process (BP, Figure 4B), and biological process for synaptic transmission GABAergic (GO: 0051932, Bonferroni corrected p = 1.90 × 10−2), glia cell proliferation (GO: 0014009, Bonferroni corrected p = 3.6 × 10−2), and positive regulation of intrinsic apoptotic signaling pathway (GO: 2001244, Bonferroni corrected p = 2.7 × 10−2) (Figure 4B). Synapse is a critical location involving activities of several neuro-transmitters such as glutamatergic, GABA, and dopamine [38], chronic drinking could alter the neural communication and synaptic plasticity [39], and the dysregulation of synapse transmission is the key mechanism of substance dependence. Previous studies have shown that the chronic exposure of ethanol would trigger the cellular apoptosis in immune system and central nervous system [40]. It is interesting to find glia cell proliferation might be involved in AUD. Glad cells (astrocytes, oligodendrocytes, and microglia) are non-neuronal cells, representing the most abundant cell groups in human central nervous system. They maintain homeostasis and provide support and protection for neurons [41]. Previous studies have shown that ethanol could influence the astrocytes density, induce ethanol-seeking behavior, and induce inflammation in glia cells [42–44]. Collectively, these results indicated that our AUDgenes were likely to play multiple diverse roles in AUD.
Figure 4. Functional enrichment analysis of the AUDgenes.
(A) Bubble plot of the functional enrichment results using 206 AUDgenes obtained. X-axis was the –log10 (raw p-value). Y-axis indicated the enriched gene sets with FDR < 0.10. Circle size was proportional to the shared AUDgenes with genes from the corresponding gene set. Circle color was proportional to the raw p-value. (B) Enrichment analysis results using the ClueGO method in Cytoscape for GO Biological Process [37]. Each dot represented a gene (name in red) or a GO term (name in other colors). Dots in the same color were considered from the same functional group by ClueGO annotation. Each edge indicated the gene was a component gene of the linked GO term.
DISCUSSION
In this work, we characterized multidimensional evidence of genetic, epigenetic, and transcriptomic data in AUD. We found that AUD associated genes mainly expressed in liver and several brain regions including the hippocampus, the cerebellum, the brain frontal cortex (BA9), and the pituitary. Our study identified 206 candidate genes that were associated with AUD from multidimensional data analysis. These AUDgenes were enriched in Substance Dependence, synapse transmission, and glial cell proliferation, providing insights for the understanding the function of AUD related tissues and genes.
We found that AUDgenes were mostly expressed in liver and brain regions such as the cerebellum, the brain frontal cortex, the brain hippocampus, and the pituitary. Liver is known to be the alcohol-metabolizing factory. The ability of metabolizing alcohol is highly related with the amount of alcohol consumption and thus, it was expected that liver was found where GWAS signals were enriched. Ethanol has an ubiquity of distribution of molecular targets in neurons (such as GABergic, glutamatergic, and dopaminergic neurons) and synapses (including GABA, NMDA, AMPA, and D1 receptors ) throughout the brain [4]. We found four brain regions that were particularly enriched with GWAS signals utilizing two tissue-specificity enrichment methods. Brain cerebellum plays an important role in motor control by contributing to coordination, precision, and accurate timing. Recently, brain cerebellum was highlighted as an important neuroanatomical region in alcohol consumption model mice [45]. Some drugs targeting this region have been successfully reduced the alcohol consumption in model mice [46]. The function of brain frontal cortex is featured by complex cognitive behavior, personality expression, decision making, and moderating social behavior. This brain region is in the brain reward pathway and has been widely used for understanding the neurobiological mechanisms of AUD [18 27]. The hippocampus is an important part of the brain reward system and is critical for declarative memory, i.e., memory of facts (persons, places, or things) [47]. Along with the amygdala, the hippocampusestablishes memories of drug experiences and such kinds of memories help to create a conditioned response (intense craving) and subsequently increase the risk of relapse [48]. Previous study did not observe a significant change of the hippocampal volume between groups with high-genetic risk for addiction versus group with low-risk [49], although hippocampal volume alterations were proved to be correlated with alcohol consumption [50]. Thus, the mediated effects of alcohol × gene interaction might affect hippocampus volume. The pituitary gland is an endocrine gland at the bottom part of the hypothalamus brain region. A previous study revealed that alcohol affected hormonal pathways that can influence alcohol-seeking behavior by dysregulating the Hypothalamic Pituitary Adrenal (HPA) axis using an animal model [51–53]. Collectively, these tissues were all biologically related to AUD and might provide as candidate tissues for treatment in future studies.
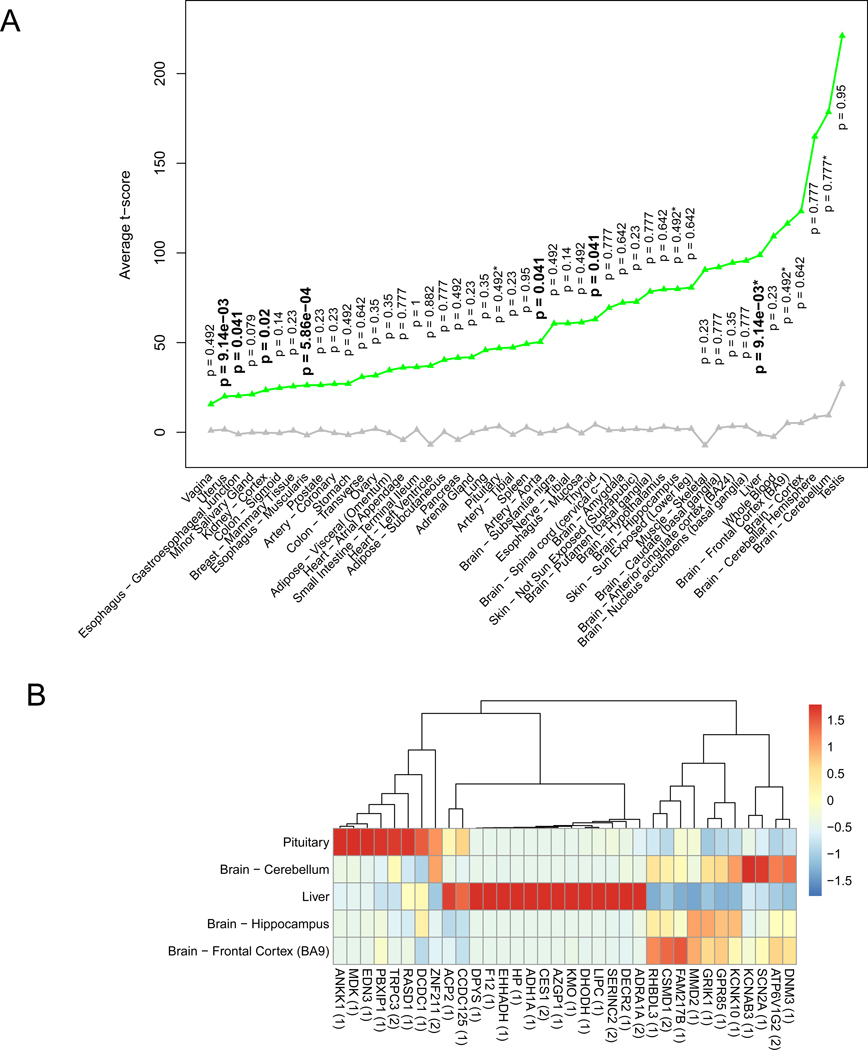
Considering that the five tissues were identified using GWAS genes, we looked back the tissue- specific expression of these 206 AUDgenes. We found that AUDgenes tended to be highly expressed in liver and all brain regions (Figure 5A). Because AUDgenes were prioritized based on multidimensional data, we particularly extracted these 206 AUDgenes and only 34 genes were from Magma. Exploration of the expression preference of these genes in GTEx V7 showed two clearly distinctive clusters: a 15-gene cluster (ADRA1A, DECR2, SERINC2, LIPC, DHODH, KMO, AZGP1, CES1, ADH1A, HP, EHHADH, F12, DPYS, CCDC125, and ACP2) highly expressed in liver and a 19-gene cluster highly expressed in brain regions (Figure 5B).
Figure 5. AUDgenes tissue enrichment analysis.
(A) Line chart for the average t-scores of tissue- specificity for 206 AUDgenes. The t-score was pre-calculated by the deTS package to measure tissue specificity for each gene. X-axis was the 47 tissues; y-axis was the average t-scores for the AUDgenes in each tissue. Green triangles represented AUDgenes and grey triangle for the other genes in the corresponding tissue. Tissues were ordered by the mean t-scores of the AUDgenes. P-values along the green triangle were from the tissue-specific enrichment analysis for each tissue, with significant ones in bold (Benjamini–Hochberg adjusted p < 0.05). *: the five AUD-related tissues. (B) Heatmap of the shared genes between 206 AUDgenes and tissue-specific genes from five tissues (liver and brain cerebellum, frontal cortex BA9, hippocampus, and pituitary). RPKM values of genes were normalized across the five tissues. The number in the bracket after each gene symbol indicated the number of lines of evidence that the gene had in the evidence matrix.
We explored whether our AUDgenes were also candidate drug targets. To this end, we queried the Therapeutic Target Database (TTD) (http://bidd.nus.edu.sg/group/cjttd/, access date: 2/5/2019) and DrugBank (https://www.drugbank.ca/, access date: 2/5/2019) to identify Food and Drug Administration (FDA) approved drugs that were used for AUD [54 55]. For TTD, we used “alcohol use disorder”, “alcohol dependence”, or “alcohol abuse” as the keywords; for DrugBank, we used “drugs used in alcohol dependence” as the keyword to search for medication target genes for AUD from TTD and DrugBank, respectively. We found eight drugs from TTD and four drugs from DrugBank, with four FDA approved medication for AUD: Acamprosate, Calcium carbimide, Disulfiram, and Naltrexone. These drugs had 31 target genes (Table S3). Although we failed to find an overlapping between our AUDgenes and current known AUD drug targets, we found 45 out of 206 AUDgenes were labelled as “DRUGGABLE GENOME” from the drug gene interaction database (http://www.dgidb.org/, access date: 11/25/2019) (Table S4) [56].
We applied a relatively stringent threshold to define significant genes in each omics data, and thus obtained rigorous but a limited number of genes from each analysis. First, for genetics related evidence such as Magma, Sherlock, and TWAS, we found that only a few genes reached genome-wide significance. For example, with the original GWAS summary statistics, there were less than 10 significant loci with genome-wide significance and 40 genes with nominal pMagma < 5 × 10-6. Notably, this data set was by far the largest GWAS for AUD, with 34,658 cases and 167,346 controls. The secondary analyses of the GWAS summary statistics also had a limited number of significant genes (adjusted pSherlock or pMetaXcan< 5 × 10−2). We only identified less than 100 genes from Sherlock and MetaXcan analysis. These results were not surprising, considering the long reported issues of missing heritability between observed heritability and heritability explained by the genome-wide significant variants [7 57 58]. It was likely that a large number of small-effect common variants were underling the disease susceptibility [7 8 59] and hence, we chose less stringent thresholds. Second, the changes of gene expression and DNA methylation level were relatively small (20–50% changes) as reported in previous studies [16–18]. This was likely due to the small size of brain tissues samples. Considering these facts, we employed less stringent thresholds when defining omics-specific genes to control the potential false negatives at the expense of increased false positives. Future studies with large sample sizes and cell-type level data are warranted.
CONCLUSIONS
In summary, we characterized multidimensional evidence of genetic, epigenetic, and transcriptomic data in AUD. We found that AUD associated genes tend to be specifically and highly expressed in liver and brain cerebellum, frontal cortex, hippocampus, and pituitary. Our research revealed and prioritized 206 AUDgenes or AUD susceptibility genes that interact with environments factors. The functions of these AUDgenes were enriched in substance dependence, synaptic transmission, cellular apoptosis, and glial cell proliferation. Our research characterized the AUD related tissues and major affected genes.
Supplementary Material
Table S1. Summary of data sources.
Table S2. Evidence matrix for the 206 AUDgenes.
Table S3. FDA approved AUD drugs and their target genes obtained from Therapeutic Target Database and DrugBank.
Table S4. Druggable genes among AUDgenes from the drug gene interaction database.
ACKNOWLEDGMENTS
We would like to thank Drs. Hang Zhou and Joel Gelernter for providing us the alcohol use disorder summary statistics for European descents and valuable suggestions. We would like to thank the members of Bioinformatics and Systems Medicine Laboratory (BSML) for valuable discussion. We would like to thank two anonymous reviewers gave us critical and helpful suggestions.
FUNDING
This work was supported by National Institutes of Health grants [R01LM012806].
Footnotes
DATA AVAILABILITY STATEMENT
Publicly available datasets were analyzed in this study. The gene expression and methylation dataset were generated in our previous work. Other data could be accessed from public resources described in Methods.
STATEMENT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.NIDA. https://www.drugabuse.gov/related-topics/trends-statistics. Secondary https://www.drugabuse.gov/related-topics/trends-statistics April 24 2017.
- 2.Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet 2009;126(1):91–9 doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuckit MA. Alcohol-use disorders. Lancet 2009;373(9662):492–501 doi: 10.1016/S0140-6736(09)60009-X. [DOI] [PubMed] [Google Scholar]
- 4.Abrahao KP, Salinas AG, Lovinger DM. Alcohol and the Brain: Neuronal Molecular Targets, Synapses, and Circuits. Neuron 2017;96(6):1223–38 doi: 10.1016/j.neuron.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young-Wolff KC, Enoch MA, Prescott CA. The influence of gene-environment interactions on alcohol consumption and alcohol use disorders: a comprehensive review. Clin Psychol Rev 2011;31(5):800–16 doi: 10.1016/j.cpr.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu SA, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen LS, Clarke TK, Chou YL, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, Hartmann AM, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Jan Hottenga J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Lai D, Ligthart L, Loukola A, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Meyers JL, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang JC, Webb BT, Wedow R, Wetherill L, Wills AG, andMe Research T, Boardman JD, Chen D, Choi DS, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye MA, Gabel W, Hayward C, Ising M, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Mannisto S, Muller-Myhsok B, Murray AD, Nurnberger JI, Palotie A, Preuss U, Raikkonen K, Reynolds MD, Ridinger M, Scherbaum N, Schuckit MA, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma DI, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AM, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer CJ, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak VM, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes HH, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nothen MM, Palmer AA, Pedersen NL, Penninx B, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen PH, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 2018;21(12):1656–69 doi: 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edenberg HJ. Common and rare variants in alcohol dependence. Biol Psychiatry 2011;70(6):498–9 doi: 10.1016/j.biopsych.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Bierut LJ. Identifying genetic variation for alcohol dependence. Alcohol Res 2012;34(3):274–81 [PMC free article] [PubMed] [Google Scholar]
- 9.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, Stahl EA, Huckins LM, Consortium GT, Nicolae DL, Cox NJ, Im HK. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018;9(1):1825 doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He X, Fuller CK, Song Y, Meng Q, Zhang B, Yang X, Li H. Sherlock: detecting gene- disease associations by matching patterns of expression QTL and GWAS. Am J Hum Genet 2013;92(5):667–80 doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marigorta UM, Denson LA, Hyams JS, Mondal K, Prince J, Walters TD, Griffiths A, Noe JD, Crandall WV, Rosh JR, Mack DR, Kellermayer R, Heyman MB, Baker SS, Stephens MC, Baldassano RN, Markowitz JF, Kim MO, Dubinsky MC, Cho J, Aronow BJ, Kugathasan S, Gibson G. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat Genet 2017;49(10):1517–21 doi: 10.1038/ng.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, Schizophrenia Working Group of the Psychiatric Genomics C, McCarroll S, Neale BM, Ophoff RA, O’Donovan MC, Crawford GE, Geschwind DH, Katsanis N, Sullivan PF, Pasaniuc B, Price AL. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat Genet 2018;50(4):538–48 doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulovari A, Liu Z, Zhu Z, Li D. Genome-wide meta-analysis of copy number variations with alcohol dependence. Pharmacogenomics J 2018;18(3):398–405 doi: 10.1038/tpj.2017.35. [DOI] [PubMed] [Google Scholar]
- 14.Zuo L, Wang KS, Zhang XY, Li CS, Zhang F, Wang X, Chen W, Gao G, Zhang H, Krystal JH, Luo X. Rare SERINC2 variants are specific for alcohol dependence in individuals of European descent. Pharmacogenet Genomics 2013;23(8):395–402 doi: 10.1097/FPC.0b013e328362f9f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo L, Zhang H, Malison RT, Li CS, Zhang XY, Wang F, Lu L, Lu L, Wang X, Krystal JH, Zhang F, Deng HW, Luo X. Rare ADH variant constellations are specific for alcohol dependence. Alcohol Alcohol 2013;48(1):9–14 doi: 10.1093/alcalc/ags104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology 2006;31(7):1574–82 doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Wang F, Xu H, Liu Y, Liu J, Zhao H, Gelernter J. Differentially co-expressed genes in postmortem prefrontal cortex of individuals with alcohol use disorders: influence on alcohol metabolism-related pathways. Hum Genet 2014;133(11):1383–94 doi: 10.1007/s00439-014-1473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Xu H, Zhao H, Gelernter J, Zhang H. DNA co-methylation modules in postmortem prefrontal cortex tissues of European Australians with alcohol use disorders. Sci Rep 2016;6:19430 doi: 10.1038/srep19430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol 2005;34(5):1129–37 doi: 10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- 20.Jia P, Chen X, Xie W, Kendler KS, Zhao Z. Mega-analysis of Odds Ratio: A Convergent Method for a Deep Understanding of the Genetic Evidence in Schizophrenia. Schizophr Bull 2018. doi: 10.1093/schbul/sby085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai Y, Pei G, Zhao Z, Jia P. A Convergent Study of Genetic Variants Associated With Crohn’s Disease: Evidence From GWAS, Gene Expression, Methylation, eQTL and TWAS. Frontiers in Genetics 2019;10(318) doi: 10.3389/fgene.2019.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 2019;10(1):1499 doi: 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia P, Dai Y, Hu R, Pei G, Manuel AM, Zhao Z. TSEA-DB: a trait-tissue association map for human complex traits and diseases. Nucleic Acids Res 2019. doi: 10.1093/nar/gkz957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, A LP. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016;48(11):1443–48 doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet 2016;98(1):116–26 doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature 2015;526(7571):68–74 doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor M, Wang JC, Farris SP, Liu Y, McClintick J, Gupta I, Meyers JL, Bertelsen S, Chao M, Nurnberger J, Tischfield J, Harari O, Zeran L, Hesselbrock V, Bauer L, Raj T, Porjesz B, Agrawal A, Foroud T, Edenberg HJ, Mayfield RD, Goate A. Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry 2019;9(1):89 doi: 10.1038/s41398-019-0384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized geneset analysis of GWAS data. PLoS Comput Biol 2015;11(4):e1004219 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pei G, Dai Y, Zhao Z, Jia P. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics 2019. doi: 10.1093/bioinformatics/btz138[published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8(1):1826 doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, Nih/Nci, Nih/Nhgri, Nih/Nimh, Nih/Nida, Biospecimen Collection Source Site N, Biospecimen Collection Source Site R, Biospecimen Core Resource V, Brain Bank Repository-University of Miami Brain Endowment B, Leidos Biomedical-Project M, Study E, Genome Browser Data I, Visualization EBI, Genome Browser Data I, Visualization-Ucsc Genomics Institute UoCSC, Lead a, Laboratory DA, Coordinating C, management NIHp, Biospecimen c, Pathology, e QTLmwg, Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature 2017;550(7675):204–13 doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12(1):56–68 doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding J, McConechy MK, Horlings HM, Ha G, Chun Chan F, Funnell T, Mullaly SC, Reimand J, Bashashati A, Bader GD, Huntsman D, Aparicio S, Condon A, Shah SP. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat Commun 2015;6:8554 doi: 10.1038/ncomms9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hormozdiari F, Penn O, Borenstein E, Eichler EE. The discovery of integrated gene networks for autism and related disorders. Genome Res 2015;25(1):142–54 doi: 10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinero J, Bravo A, Queralt-Rosinach N, Gutierrez-Sacristan A, Deu-Pons J, Centeno E, Garcia-Garcia J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45(D1):D833–D39 doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498–504 doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25(8):1091–3 doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee N Neurotransmitters in alcoholism: A review of neurobiological and genetic studies. Indian J Hum Genet 2014;20(1):20–31 doi: 10.4103/0971-6866.132750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroener S, Mulholland PJ, New NN, Gass JT, Becker HC, Chandler LJ. Chronic alcohol exposure alters behavioral and synaptic plasticity of the rodent prefrontal cortex. PLoS One 2012;7(5):e37541 doi: 10.1371/journal.pone.0037541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasala S, Barr T, Messaoudi I. Impact of Alcohol Abuse on the Adaptive Immune System. Alcohol Res 2015;37(2):185–97 [PMC free article] [PubMed] [Google Scholar]
- 41.Sherwood CC, Stimpson CD, Raghanti MA, Wildman DE, Uddin M, Grossman LI, Goodman M, Redmond JC, Bonar CJ, Erwin JM, Hof PR. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci U S A 2006;103(37):13606–11 doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adermark L, Bowers MS. Disentangling the Role of Astrocytes in Alcohol Use Disorder. Alcohol Clin Exp Res 2016;40(9):1802–16 doi: 10.1111/acer.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He J, Crews FT. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp Neurol 2008;210(2):349–58 doi: 10.1016/j.expneurol.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bull C, Freitas KC, Zou S, Poland RS, Syed WA, Urban DJ, Minter SC, Shelton KL, Hauser KF, Negus SS, Knapp PE, Bowers MS. Rat nucleus accumbens core astrocytes modulate reward and the motivation to self-administer ethanol after abstinence. Neuropsychopharmacology 2014;39(12):2835–45 doi: 10.1038/npp.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan JS, Mohr C, Rossi DJ. Opposite actions of alcohol on tonic GABA(A) receptor currents mediated by nNOS and PKC activity. Nat Neurosci 2013;16(12):1783–93 doi: 10.1038/nn.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan JS, Nipper MA, Richardson BD, Jensen J, Helms M, Finn DA, Rossi DJ. Pharmacologically Counteracting a Phenotypic Difference in Cerebellar GABAA Receptor Response to Alcohol Prevents Excessive Alcohol Consumption in a High Alcohol- Consuming Rodent Genotype. J Neurosci 2016;36(35):9019–25 doi: 10.1523/JNEUROSCI.0042-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubin RD, Watson PD, Duff MC, Cohen NJ. The role of the hippocampus in flexible cognition and social behavior. Front Hum Neurosci 2014;8:742 doi: 10.3389/fnhum.2014.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belujon P, Grace AA. Hippocampus, amygdala, and stress: interacting systems that affect susceptibility to addiction. Ann N Y Acad Sci 2011;1216:114–21 doi: 10.1111/j.1749-6632.2010.05896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hill SY, De Bellis MD, Keshavan MS, Lowers L, Shen S, Hall J, Pitts T. Right amygdala volume in adolescent and young adult offspring from families at high risk for developing alcoholism. Biol Psychiatry 2001;49(11):894–905 [DOI] [PubMed] [Google Scholar]
- 50.Makris N, Oscar-Berman M, Jaffin SK, Hodge SM, Kennedy DN, Caviness VS, Marinkovic K, Breiter HC, Gasic GP, Harris GJ. Decreased volume of the brain reward system in alcoholism. Biol Psychiatry 2008;64(3):192–202 doi: 10.1016/j.biopsych.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rachdaoui N, Sarkar DK. Effects of alcohol on the endocrine system. Endocrinol Metab Clin North Am 2013;42(3):593–615 doi: 10.1016/j.ecl.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koob GF, Le Moal M. Drug abuse: hedonic homeostatic dysregulation. Science 1997;278(5335):52–8 doi: 10.1126/science.278.5335.52. [DOI] [PubMed] [Google Scholar]
- 53.Roberts AJ, Heyser CJ, Cole M, Griffin P, Koob GF. Excessive ethanol drinking following a history of dependence: animal model of allostasis. Neuropsychopharmacology 2000;22(6):581–94 doi: 10.1016/S0893-133X(99)00167-0. [DOI] [PubMed] [Google Scholar]
- 54.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46(D1):D1074–D82 doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang Q, Fu T, Zhang X, Cui X, Tu G, Zhang Y, Li S, Yang F, Sun Q, Qin C, Zeng X, Chen Z, Chen YZ, Zhu F. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res 2018;46(D1):D1121–D27 doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res 2018;46(D1):D1068–D73 doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature 2009;461(7265):747–53 doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010;11(6):446–50 doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci U S A 2014;111(49):E5272–81 doi: 10.1073/pnas.1419064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of data sources.
Table S2. Evidence matrix for the 206 AUDgenes.
Table S3. FDA approved AUD drugs and their target genes obtained from Therapeutic Target Database and DrugBank.
Table S4. Druggable genes among AUDgenes from the drug gene interaction database.