Abstract
We present detailed comparative analyses to assess population-level differences in patterns of genetic deafness between European/American and Japanese cohorts with non-syndromic hearing loss. One thousand eighty-three audiometric test results (921 European/American and 162 Japanese) from members of 168 families (48 European/American and 120 Japanese) with non-syndromic hearing loss secondary to pathogenic variants in one of three genes (KCNQ4, TECTA, WFS1) were studied. Audioprofile characteristics, specific mutation types and protein domains were considered in the comparative analyses. Our findings support differences in audioprofiles driven by both mutation type (non-truncating vs. truncating) and ethnic background. The former finding confirms data that ascribe a phenotypic consequence to different mutation types in KCNQ4; the latter finding suggests that there are ethnic-specific effects (genetic and/or environmental) that impact gene-specific audioprofiles for TECTA and WFS1. Identifying the drivers of ethnic differences will refine our understanding of phenotype-genotype relationships and the biology of hearing and deafness.
Keywords: ADNSHL, audioprofiles, KCNQ4, TECTA, WFS1
1. Introduction
Hearing loss is the most common sensory defect, affecting roughly 466 million people worldwide (World Health Organization 2019). Its prevalence in newborns is 1.7 per 1000 births (Centers for Disease Control and Prevention 2018) but climbs dramatically to affect 50% of octogenarians (Fortnum et al. 2001; Morton and Nance 2006). Causality is broadly considered to be genetic and/or environmental, with the majority of congenital hearing loss in developed countries being genetic (Korver et al. 2017). Adult-onset hearing loss is considerably more complex and reflects the impact of genetic and environmental factors on auditory function (Vuckovic et al. 2018).
Over the past two decades, our understanding of monogenic hearing loss has increased considerably, with hundreds of studies reporting genetic causes for non-syndromic hearing loss and the phenotypic consequence of various genetic variants on auditory thresholds. These data have facilitated detailed phenotypic-genotypic studies that have provided insight into the biology of hearing and deafness. In addition, by constructing gene-specific audioprofiles (two-dimensional data showing threshold and frequency) it has been possible to gain insight into the natural history of different types of genetic hearing loss thereby allowing clinicians to prognosticate rate-of-decline of hearing thresholds for persons with given genetic causes for their hearing loss (Taylor et al. 2013).
By creating ethnicity-specific audioprofiles, it also is possible to compare auditory thresholds and rates of progression of hearing loss at given genetic loci across populations. We hypothesized that this type of targeted analysis would be valuable to identify the potential existence of population-specific genetic modifiers that impact a hearing loss phenotype. To test this hypothesis, we identified two geographically distinct cohorts, one European/American and the other Japanese, with autosomal dominant non-syndromic hearing loss (ADNSHL) secondary to pathogenic variants in one of three genes (KCNQ4, TECTA, WFS1). We selected KCNQ4 because other studies have identified a mutation-dependent effect on auditory thresholds (Hildebrand et al. 2008; Wasano et al. 2015; Watabe et al. 2013), and TECTA and WFS1 because while hearing loss associated with both of these genes can be progressive through the lifetime of a person, it does not progress to severe-to-profound deafness (Sloan-Heggen et al. 2016; Yasukawa et al. 2019).
2. Materials and Methods
Data Collection
Audiometric data and diagnosed causative variants were compiled for subjects with autosomal dominant non-syndromic hearing loss (ADNSHL) in one of three genes (KCNQ4, TECTA and WFS1) from two geographical populations (Europe/United States and Japan). The European/American data were obtained from AudioGene v4.0 (https://audiogene.eng.uiowa.edu). The Japanese data were obtained from the Clinical Next-Generation Sequencing Database, which contains the clinical and targeted genomic analysis data of over 8000 clinic deafness patients (Nishio and Usami 2017). Variants were classified according to ACMG criteria (Oza et al. 2018; Richards et al. 2015), and only individuals whose variants had classifications consistent with clinical diagnoses were included. Ethnicities of individuals were self-identified and presumed to correlate strongly with geographical population membership.
Audioprofiles
Audioprofiles were generated for six cohorts–a European/American cohort and a Japanese cohort for each of KCNQ4, TECTA and WFS1. Each audioprofile was created by first grouping audiometric test results into age ranges (0–19, 20–39, 40–59 and 60–99) by age at testing, then averaging hearing loss thresholds by frequency within the age ranges. Two-sample t-tests were performed for ages within each age range to ensure no significant population differences in age distribution which could account for observable audioprofile differences.
To mitigate bias (for example, toward overrepresented families or overrepresented individuals), account for intrafamilial variability, and make use of all data, a Monte Carlo approach was taken: A random audiometric test from a random individual was chosen from each family, and an audioprofile was constructed for the sample. This random sampling and audioprofile construction was repeated 1000 times to produce 1000 audioprofiles thereby sampling all audiometric results. Hearing thresholds at each frequency of the resulting audioprofiles were averaged to produce a single audioprofile reflective of all available data with all families equally weighted.
Quantitative Analysis
To determine the magnitude and significance of any differences between audioprofiles, an analysis was performed using multivariate analysis of covariance (MANCOVA), where dependent variables were hearing loss thresholds in dB at each frequency and the independent variable was population. Since the two population cohorts had different age distributions and hearing loss is strongly dependent on age, we treated age as a covariate. An ANCOVA was then performed for each frequency to investigate which frequencies contribute most to differences in overall hearing loss patterns.
A Monte Carlo approach was also used for the quantitative analyses. For each family, data from one audiometric test of one individual were randomly sampled and the quantitative analysis performed. The quantitative analysis was repeated 1000 times and the median MANCOVA and ANCOVA p-values from all repetitions were reported as the p-values.
Subset Analysis
To determine possible drivers of any observed differences between the two populations, subjects within the cohorts having shared characteristics were grouped into smaller cohorts. For example, the prevalence of certain mutations (truncating, i.e. frameshift, nonsense vs. non-truncating, i.e. missense, in-frame indel) may vary between populations and account for differences in audioprofiles. The aforementioned analyses were repeated on these smaller cohorts to study particular hypothesized drivers of differences and their effects.
3. Results
Dataset Composition
The study dataset comprised 1083 audiograms (921 European/American, 162 Japanese) from 519 individuals (357, 162) belonging to 168 families (48, 120). Each audiogram included hearing loss thresholds at up to seven frequencies (125 Hz, 250 Hz, 500 Hz, 1000 Hz, 2000 Hz, 4000 Hz, 8000 Hz) for each ear and age at testing. By gene, 70 families (15 European/American, 55 Japanese) had a diagnosed cause of hearing loss in KCNQ4, 41 (13, 28) in TECTA and 57 (20, 37) in WFS1 (Table 1). All causative variants and their ACMG classifications are listed in Table S1.
Table 1.
Counts of audiometric test results by gene, mutation effect and population. Distinct family counts in parentheses.
| Gene | Mutation Effect | Total | Audiograms European/American | Audiograms Shinshu |
|---|---|---|---|---|
| KCNQ4 | Deletion | 67 (30) | 26 (1) | 41 (29) |
| Frameshift | 66 (29) | 26 (1) | 40 (28) | |
| Insertion | 1 (1) | 0 | 1 (1) | |
| Missense | 523 (37) | 495 (13) | 28 (24) | |
| Nonsense | 2 (1) | 2 (1) | 0 | |
| TECTA | Deletion | 62 (2) | 62 (2) | 0 |
| Frameshift | 0 | 0 | 0 | |
| Insertion | 0 | 0 | 0 | |
| Missense | 152 (41) | 115 (13) | 37 (28) | |
| Nonsense | 0 | 0 | 0 | |
| WFS1 | Deletion | 0 | 0 | 0 |
| Frameshift | 0 | 0 | 0 | |
| Insertion | 0 | 0 | 0 | |
| Missense | 337 (57) | 283 (20) | 54 (37) | |
| Nonsense | 0 | 0 | 0 |
Note that an audiometric test result may be counted for multiple rows, since the associated mutation may have multiple effects (e.g, both deletion and frameshift).
Analysis by Gene
Population-based audioprofiles are presented in Fig. 1. KCNQ4 audioprofiles showed a steady, approximately even progression of hearing loss across all frequencies by age in both population cohorts with no notable population-specific differences. By comparison, TECTA audioprofiles showed progressive hearing loss at low and high frequencies in the European/American cohort with hearing preserved in the middle frequencies, while in the Japanese cohort hearing tended to be preserved at both the low and middle frequencies. In both populations, WFS1 audioprofiles showed progressive hearing loss at all frequencies, with a sharp age-related increase in hearing loss at high frequencies that occurred earlier in the European/American cohort as compared to the Japanese cohort. No significant differences (p<0.05) in population age distributions were identified that could account for these observed differences (Table S2). Some anomalies are observed where hearing loss appeared less severe in an older age group versus a younger one; these differences presumably reflect limited data for those age groups as the error bars of their audioprofile lines largely overlap.
Fig. 1.
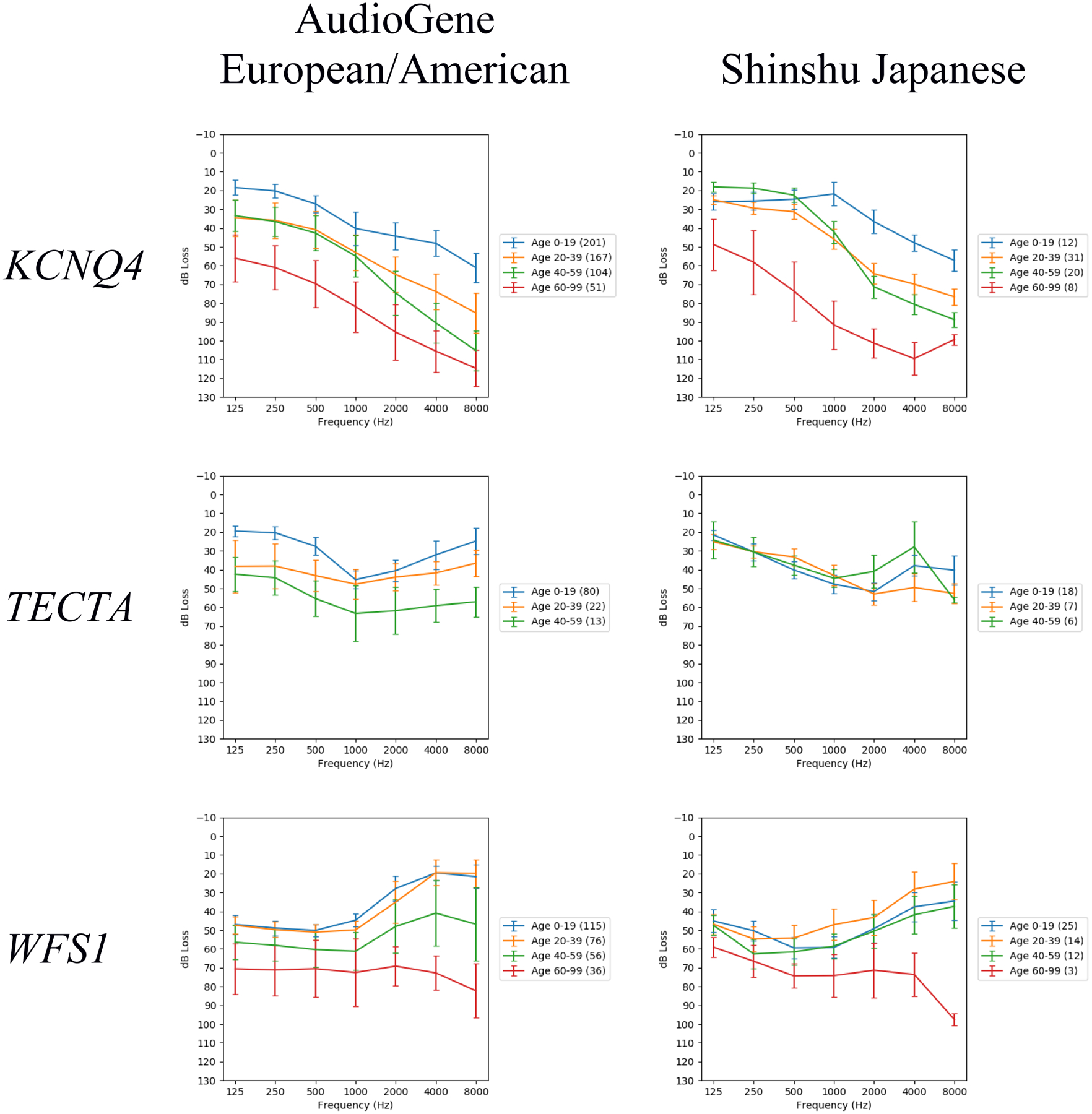
Audioprofiles for each pairing of population (European/American, Japanese) and gene (KCNQ4, TECTA, WFS1). The vertical bars indicate standard error of the mean. The number of audiometric test results within each age range is indicated in the legend by the numbers in parentheses. These audioprofiles are each the average of 1000 audioprofiles generated by random family member sampling.
MANCOVA was performed to determine the significance of the differences observed between populations for all gene-specific cohorts, with median population p-values for KCNQ4, TECTA and WFS1 of 0.27, 0.012 and 0.0035, respectively (Table 2). Post hoc ANOVAs of individual frequencies identified the greatest difference for TECTA at 250 Hz and 8000 Hz (Table 3). For WFS1, the greatest difference was seen at 2000 Hz. Little differences were seen for KCNQ4, consistent with the negative results from MANCOVA.
Table 2.
Distribution of population p-values from MANCOVA of 1000 random family samples for each gene.
| KCNQ4 | TECTA | WFS1 | |
|---|---|---|---|
| Population p-value distribution* | Min. :0.0000256 1st Qu.:0.1296056 |
Min. :0.0001179 1st Qu.:0.0044867 |
Min. :5.130e-06 1st Qu.:7.934e-04 |
| Median :0.2725848 | Median :0.0119179 | Median :3.452e-03 | |
| Mean :0.3151173 3rd Qu.:0.4634762 |
Mean :0.0239117 3rd Qu.:0.0290326 |
Mean :1.044e-02 3rd Qu.:1.092e-02 |
|
| Max. :0.9375881 | Max. :0.2961256 | Max. :2.388e-01 |
The median p-value was taken to be the most informative. Significant p-values (p<0.05/3) bolded.
Table 3.
Median p-values from per-frequency ANOVAs of 1000 random family samples for each gene.
| KCNQ4 | TECTA | WFS1 | |
|---|---|---|---|
| 125 Hz | 0.37 | 0.21 | 0.51 |
| 250 Hz | 0.49 | 0.058 | 0.39 |
| 500 Hz | 0.31 | 0.11 | 0.45 |
| 1000 Hz | 0.48 | 0.66 | 0.51 |
| 2000 Hz | 0.32 | 0.12 | 0.097 |
| 4000 Hz | 0.34 | 0.28 | 0.17 |
| 8000 Hz | 0.39 | 0.0092 | 0.54 |
Analysis by Gene and Mutation Effect
Audiometric data were sub-grouped by mutation type, restricting the analysis to KCNQ4 missense and TECTA missense cohorts based on data availability (Table 1). WFS1 was not included as all WFS1 mutations were missense, so the analysis would be redundant to the analysis by gene. Again, the TECTA European/American cohort showed progressive hearing loss at the low and high frequencies with stable hearing loss in the middle frequencies, while the Japanese cohort showed stable hearing loss at both low and middle frequencies (Fig. 2). MANCOVA showed a significant difference in hearing loss between populations for TECTA missense (median p=0.012) (Table S3). Post hoc ANOVAs of TECTA missense data showed the greatest difference between populations at both 250 Hz and 8000 Hz (Table S4).
Fig. 2.
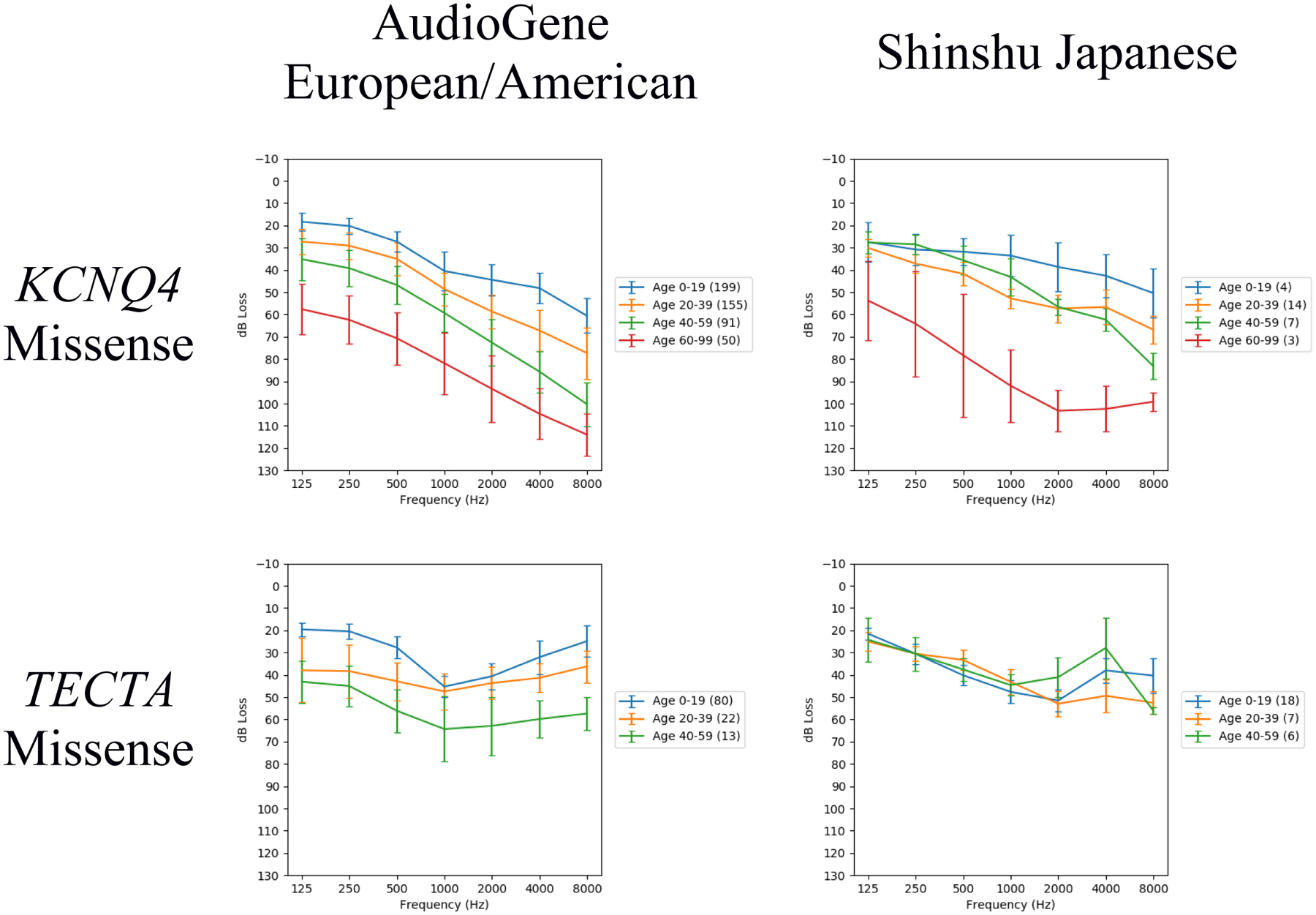
Audioprofiles for each pairing of population (AudioGene European/American and Shinshu Japanese) and gene mutation effect group. Only gene mutation effect groups with a non-negligible number of audiometric test results and distinct families for both populations are shown. The number of audiometric test results within each age range is indicated in the legend by the numbers in parentheses. These audioprofiles are each the average of 1000 audioprofiles generated by random family member sampling.
Because no population-specific differences in KCNQ4 audioprofiles were observed, we also completed an analysis by mutation type by combining cohorts into either one of two inter-population cohorts: one comprising subjects with KCNQ4 truncating mutations and the other comprising subjects with KCNQ4 missense mutations. Subjects with KCNQ4 truncating mutations showed more rapid progression of high-frequency hearing loss with age than did subjects with KCNQ4 missense mutations, with significant differences at nearly all frequencies (Fig. 3; Table S5; Table S6).
Fig. 3.
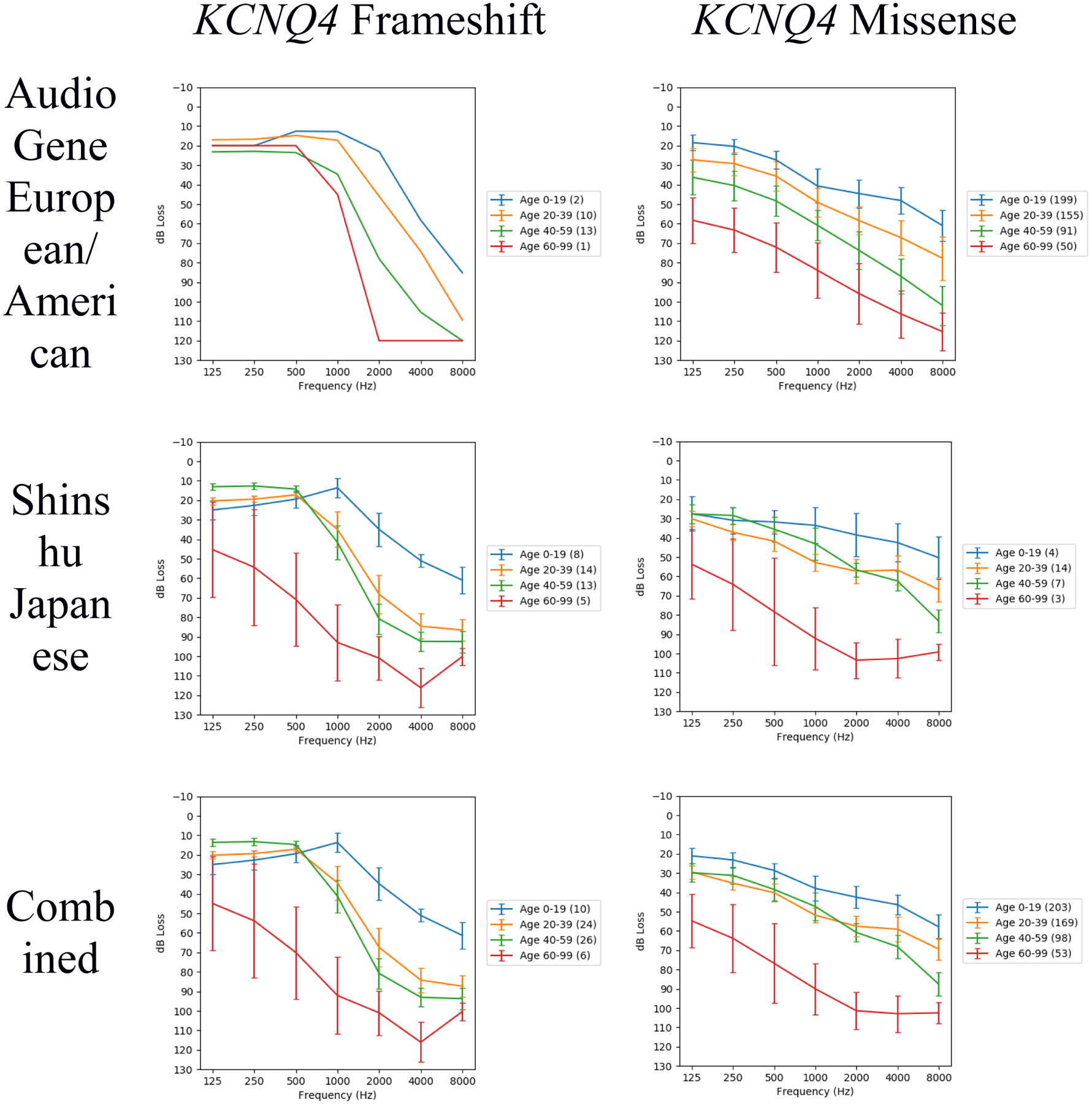
Audioprofiles for subjects with hearing loss attributed to either KCNQ4 frameshift or KCNQ4 missense mutations, with subjects separated by population or combined from both populations. The number of audiometric test results within each age range is indicated in the legend by the numbers in parentheses. These audioprofiles are each the average of 1000 audioprofiles generated by random family member sampling. Note there is only one European/American KCNQ4 frameshift family; this accounts for the similarity between the Japanese and combined populations for KCNQ4 frameshift.
Analysis by Structural Features
Differences in the prevalences of mutations affecting particular gene structural features between populations could account for the significant hearing loss differences seen between populations. To investigate the possible impact of domain-specific mutation effects an analysis by UniProt protein domain was completed for KCNQ4 and TECTA (Table S7; Table S8). KCNQ4-affected protein domains showed significantly different distributions between the populations (p=2.85e-03), while TECTA-affected protein domains did not (p=0.454). WFS1 has no reported protein domains on UniProt, so this analysis was not performed for WFS1.
Analysis by Variant-Specific Hearing Loss Progressivity
Many TECTA variants implicated in hearing loss are known to cause either progressive or stable hearing loss specifically (Yasukawa et al. 2019). A difference in the distribution of progressive versus stable variants between the two populations could explain the observed hearing loss differences. Each TECTA variant included in this study was therefore categorized as progressive, stable or unknown (Table S9), and a population-based analysis was completed. No significant difference in variant progressivity was seen between the two populations (Table S10).
4. Discussion
In this study, we used European/American and Japanese cohorts to show that ethnic-based characteristic differences impact some gene-specific audioprofiles. We studied KCNQ4, TECTA and WFS1. With KCNQ4, we validated differences in the degree of high-frequency hearing loss between cohorts with loss-of-function mutations as compared to missense mutations, which replicate with independent Japanese data earlier findings that mutation type impacts the KCNQ4-hearing loss phenotype (Hildebrand et al. 2008). We did not, however, observe ethnic-specific phenotypic differences. This finding suggests that KCNQ4 protein domain membership, which does differ between populations (Table S7), has little bearing on phenotype. For both TECTA and WFS1, in contrast, we did observe ethnic- specific differences in hearing loss thresholds demonstrating a population-based effect that reflects the impact of genetic modifiers (genetic background) and/or environmental factors on auditory phenotype.
Some sample characteristics differed significantly between the European/American and Japanese cohorts— particularly the distributions of family size and the number of audiograms per individual since we used preexisting data that were collected in various ways. These potentially biasing differences posed a challenge for comparative analysis, which we addressed by a repeated random-sampling procedure to ensure families and individuals were weighted equally within the populations under comparison to eliminate bias while making use of all available data. Randomly sampling family members also accounts for the possibility of intrafamilial variability, as has been observed with WFS1 sensorineural hearing loss (Tranebjaerg et al. 1993), by ensuring all family members are represented in the analysis. The procedure identified a difference in high-frequency hearing loss between individuals with KCNQ4 non-truncating vs. truncating mutations, replicating earlier findings that KCNQ4 truncating mutations cause more severe hearing loss and supporting the validity of the procedure. Still, differences in the number of families and the number of audiometric test results between populations, especially for KCNQ4 and TECTA, mean that intrapopulation variability and intrafamilial variability are captured to differing degrees between the populations, which is a limitation of this study. Differences in distributions of age at audiometry potentially introduce age-related hearing loss as a factor in the comparative analysis. While no significant differences were seen in age distributions (Table S2), it is possible that age differences could nonetheless explain some portion of the observed hearing loss differences, especially for the large 60–99 age range.
Neither mutation- nor domain-specific differences were observed with TECTA and WFS1, supporting the presence of underlying consequential ethnic-specific differences. TECTA encodes alpha-tectorin, one of approximately 50 proteins in the tectorial membrane (unpublished data). To explore the possibility that variants in other tectorial membrane-associated genes modulate the effect of the primary pathogenic TECTA variant we searched the 49 genes for variants common in one population (MAF>=5%) but not in the other (MAF<=1%). Combined Annotation-Dependent Depletion (CADD) scores (Rentzsch et al. 2019) were generated and used to rank-order the variants meeting these criteria (Table 4). A highly ranked variant with an especially high common MAF was identified in COL6A5: the p.I1114M common polymorphism (CADD=21.1), which could subtly affect the material properties of the tectorial membrane, potentially affecting mechanical excitation over a broad range of frequencies (Sellon et al. 2015).
Table 4.
Variants common (MAF>=5%) in either the Japanese or European/American population but rare (MAF<=1%) in the other. The variant with the highest common MAF, bolded, was identified as a variant of interest.
| Gene | rsID | Chr. | Position | HGVS. p | Japanese MAF | European/American MAF | Consequence | CADD Score |
|---|---|---|---|---|---|---|---|---|
| COL9A2 | rs12077871 | 1 | 40773150 | p.Gln326* | 0.1138 | 0.0026 | stop gained | 38 |
| OTOA | rs200988634 | 16 | 21747639 | p.Glu801* | 0.2787 | 0.0025 | stop gained | 36 |
| COL5A1 | rs2229817 | 9 | 137726950 | p.Thr1757Met | 0.069 | 0.0018 | missense variant | 26.6 |
| OTOGL | rs79711087 | 12 | 80655832 | p.His658Leu | 0.056 | 0.0001 | missense variant | 23.5 |
| OTOG | rs7130190 | 11 | 17580175 | p.Thr375Ser | 0.0051 | 0.142 | missense variant | 23.4 |
| ANXA2 | rs17845226 | 15 | 60653205 | p.Val116Leu | 0.0001 | 0.1289 | missense variant | 22.2 |
| COL6A5 | rs1353613 | 3 | 130114082 | p.Ile1114Met | 0.4133 | 0.01 | missense variant | 21.1 |
| DSP | rs28763961 | 6 | 7569480 | p.Tyr494Phe | 0.0957 | 0.0025 | missense variant | 19.46 |
| OTOG | rs116947228 | 11 | 17618546 | p.Arg1237His | 0.1004 | 0.0023 | missense variant | 18.45 |
| EDFHD1 | rs112941683 | 2 | 233498506 | p.Ala31Val | 0.005 | 0.0505 | missense variant | 15.7 |
| IQGAL1 | rs2301831 | 15 | 91017718 | p.Ile859Ile | 0.1407 | 0.0032 | splice region variant; synonymous variant | 14.91 |
| ACAN | rs74505897 | 15 | 89401379 | p.Leu1855Phe | 0.0645 | 0.0016 | missense variant | 14.58 |
| TMPRSS9 | rs117767265 | 19 | 2405456 | p.Ala252Val | 0.057 | 0.0023 | missense variant | 13.42 |
Variants obtained from Japanese Multi Omics Reference Panel (jMorp) (Tadaka et al. 2017) release 202001 and ranked by CADD v1.4 score; only those with CADD score >=12.37 shown (Kircher et al. 2014). Genomic coordinates are per the GRCh37/hg19 genome assembly. Japanese MAFs are ToMMo 4.7KJPN Allele Frequency Panel v20190826 MAFs; European/American MAFs are gnomAD 2.1 non-Finnish European MAFs.
WFS1 encodes wolframin; however, identifying ethnic-specific variation in other genes that could potentially drive the observed differences in WFS1 audioprofiles is precluded by our limited knowledge of the biophysical role of wolframin in hearing loss. We observed the greatest differences at 2000 Hz and 4000 Hz, which could reflect the impact of a genetic modifier that alters the upper bound of the range of frequencies affected by WFS1 NSHL.
In summary, we have identified ethnic-specific differences for two genetic types of ADNSHL. These results should be expanded to other populations and to other genes. Validating ethnic-specific differences will provide novel insights into genetic hearing loss, refine our understanding of the biology of hearing and deafness, and may offer new ways to moderate specific genetic types of hearing loss.
Supplementary Material
Acknowledgments
This research was funded in part by National Institute on Deafness and Other Communication Disorders R01s DC002842 (RJS) and DC012049 (TLC, RJS).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest: The authors declare that they have no conflict of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the University of Iowa Institutional Review Board and the Shinshu University Ethical Committee, and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- Centers for Disease Control and Prevention (2018) Hearing Loss in Children. https://www.cdc.gov/ncbddd/hearingloss/ehdi-data2015.html. Accessed May 18, 2018
- Choi BY et al. (2014) Whole-exome sequencing identifies a novel genotype-phenotype correlation in the entactin domain of the known deafness gene TECTA PLoS One 9:e97040–e97040 doi: 10.1371/journal.pone.0097040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin RWJ et al. (2008) Mid-frequency DFNA8/12 hearing loss caused by a synonymous TECTA mutation that affects an exonic splice enhancer European Journal of Human Genetics 16:1430–1436 doi: 10.1038/ejhg.2008.110 [DOI] [PubMed] [Google Scholar]
- Fortnum HM, Summerfield AQ, Marshall DH, Davis AC, Bamford JM (2001) Prevalence of permanent childhood hearing impairment in the United Kingdom and implications for universal neonatal hearing screening: questionnaire based ascertainment study BMJ 323:536–540 doi: 10.1136/bmj.323.7312.536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand MS et al. (2011) DFNA8/12 caused by TECTA mutations is the most identified subtype of nonsyndromic autosomal dominant hearing loss Hum Mutat 32:825–834 doi: 10.1002/humu.21512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand MS et al. (2008) Audioprofile-directed screening identifies novel mutations in KCNQ4 causing hearing loss at the DFNA2 locus Genet Med 10:797–804 doi: 10.1097/GIM.0b013e318187e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants Nature Genetics 46:310 doi:10.1038/ng.289210.1038/ng.2892https://www.nature.com/articles/ng.2892#supplementary-informationhttps://www.nature.com/articles/ng.2892#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver AMH et al. (2017) Congenital hearing loss Nat Rev Dis Primers 3:16094–16094 doi: 10.1038/nrdp.2016.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer NC, Nishimura CJ, McMordie S, Smith RJH (2007) Audioprofiling identifies TECTA and GJB2- related deafness segregating in a single extended pedigree Clinical Genetics 72:130–137 doi: 10.1111/j.1399-0004.2007.00828.x [DOI] [PubMed] [Google Scholar]
- Moreno-Pelayo MA et al. (2001) A cysteine substitution in the zona pellucida domain of α-tectorin results in autosomal dominant, postlingual, progressive, mid frequency hearing loss in a Spanish family Journal of Medical Genetics 38:e13 doi: 10.1136/jmg.38.5.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE (2006) Newborn Hearing Screening — A Silent Revolution New England Journal of Medicine 354:2151–2164 doi: 10.1056/NEJMra050700 [DOI] [PubMed] [Google Scholar]
- Nishio S-y Usami S-i (2017) The Clinical Next- Generation Sequencing Database: A Tool for the Unified Management of Clinical Information and Genetic Variants to Accelerate Variant Pathogenicity Classification Hum Mutat 38:252–259 doi: 10.1002/humu.23160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza AM et al. (2018) Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss Hum Mutat 39:1593–1613 doi: 10.1002/humu.23630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga RF, de Brouwer APM, Huygen PLM, Kunst HPM, Kremer H, Cremers CWRJ (2006) A novel TECTA mutation in a Dutch DFNA8/12 family confirms genotype-phenotype correlation J Assoc Res Otolaryngol 7:173–181 doi: 10.1007/s10162-006-0033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019) CADD: predicting the deleteriousness of variants throughout the human genome Nucleic Acids Res 47:D886–D894 doi: 10.1093/nar/gky1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology Genetics in Medicine 17:405–423 doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagong B. et al. Two novel missense mutations in the TECTA gene in Korean families with autosomal dominant nonsyndromic hearing loss. [PubMed] [Google Scholar]
- Sellon JB, Farrahi S, Ghaffari R, Freeman DM (2015) Longitudinal spread of mechanical excitation through tectorial membrane traveling waves Proc Natl Acad Sci U S A 112:12968–12973 doi: 10.1073/pnas.1511620112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM et al. (2016) Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss Hum Genet 135:441–450 doi: 10.1007/s00439-016-1648-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadaka S et al. (2017) jMorp: Japanese Multi Omics Reference Panel Nucleic Acids Res 46:D551–D557 doi: 10.1093/nar/gkx978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KR et al. (2013) AudioGene: predicting hearing loss genotypes from phenotypes to guide genetic screening Hum Mutat 34:539–545 doi: 10.1002/humu.22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L, Barrett T, Rendtorff ND (1993) WFS1- Related Disorders In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (eds) GeneReviews((R)). Seattle (WA), [Google Scholar]
- Vuckovic D et al. (2018) Whole-genome sequencing reveals new insights into age-related hearing loss: cumulative effects, pleiotropy and the role of selection Eur J Hum Genet 26:1167–1179 doi: 10.1038/s41431-018-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasano K, Mutai H, Obuchi C, Masuda S, Matsunaga T (2015) A novel frameshift mutation in KCNQ4 in a family with autosomal recessive non-syndromic hearing loss Biochemical and Biophysical Research Communications 463:582–586 doi: 10.1016/j.bbrc.2015.05.099 [DOI] [PubMed] [Google Scholar]
- Watabe T, Matsunaga T, Namba K, Mutai H, Inoue Y, Ogawa K (2013) Moderate hearing loss associated with a novel KCNQ4 non-truncating mutation located near the N-terminus of the pore helix Biochemical and Biophysical Research Communications 432:475–479 doi: 10.1016/j.bbrc.2013.01.118 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2019) Deafness and hearing loss. https://www.who.int/news-room/fact-sheets/detail/deafness-and-hearing-loss. Accessed July 22, 2019 [Google Scholar]
- Yasukawa R et al. (2019) The Prevalence and Clinical Characteristics of TECTA-Associated Autosomal Dominant Hearing Loss Genes (Basel) 10:744 doi: 10.3390/genes10100744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


