Abstract
The snow sheep, Ovis nivicola, which is endemic to the mountain ranges of northeastern Siberia, are well adapted to the harsh cold climatic conditions of their habitat. In this study, using long reads of Nanopore sequencing technology, whole-genome sequencing, assembly, and gene annotation of a snow sheep were carried out. Additionally, RNA-seq reads from several tissues were also generated to supplement the gene prediction in snow sheep genome. The assembled genome was ∼2.62 Gb in length and was represented by 7,157 scaffolds with N50 of about 2 Mb. The repetitive sequences comprised of 41% of the total genome. BUSCO analysis revealed that the snow sheep assembly contained full-length or partial fragments of 97% of mammalian universal single-copy orthologs (n = 4,104), illustrating the completeness of the assembly. In addition, a total of 20,045 protein-coding sequences were identified using comprehensive gene prediction pipeline. Of which 19,240 (∼96%) sequences were annotated using protein databases. Moreover, homology-based searches and de novo identification detected 1,484 tRNAs; 243 rRNAs; 1,931 snRNAs; and 782 miRNAs in the snow sheep genome. To conclude, we generated the first de novo genome of the snow sheep using long reads; these data are expected to contribute significantly to our understanding related to evolution and adaptation within the Ovis genus.
Keywords: snow sheep, PromethION sequencing, de novo assembly, annotation
Introduction
The Ovis genus is characterized by its abundance of species and subspecies that inhabit various habitats around the world. The most recognized between scientists is the classification, suggested by (Nadler et al. 1973), which distributes the wild Ovis in seven species: Ovis ammon, O. musimon, O. orientalis, O. vignei, O. dalli, O. canadensis, and O. nivicola. This classification was used by Rezaei et al. (2010) who studied the taxonomy of wild Ovis using CytB sequence. However, the International Council for Game and Wildlife Conservation uses the classification as suggested by Raul Valdez (1982), who classified wild Ovis in six species. Ovis nivicola, also known as snow sheep (fig. 1A), is endemic to the mountain ranges of northeastern Siberia between the Lena River in the west and the Chukotka and Kamchatka peninsula in the east. A secluded subspecies of snow sheep is also found more west on the Putorana Plateau (Zheleznov-Chukotskii 1994). Due to intensified anthropogenic activities, snow sheep populations have retreated to even higher mountain areas in recent times. The remoteness and harsh climatic conditions of their habitat make these wild sheep the ideal candidates to study mammalian adaptation to extreme environments with regard to elevation and temperature. At the same time, these factors might also be among the reasons why only few genetic studies on snow sheep have been conducted so far. Whereas reference genomes for domestic sheep (Archibald et al. 2010) as well as the wild sheep species O. canadensis (Miller et al. 2015) and O. ammon (Yang et al. 2017) have been available for several years now, only a mitochondrial reference genome has been published for snow sheep (Dotsev et al. 2019). Investigations of the genetics of different snow sheep populations have mainly been carried out on microsatellite (Deniskova et al. 2018) and single nucleotide polymorphism genotype level (Deniskova et al. 2016; Medvedev et al. 2017; Dotsev et al. 2018) so far. In the present study, short-read Illumina and long-read Nanopore sequence data were generated from snow sheep samples and used for de novo assembly of the snow sheep genome. This assembly will build the basis for whole-genome sequence-based studies on the genetic characteristics of snow sheep that could also shed light on their demographic history and adaptation to the extreme cold environments.
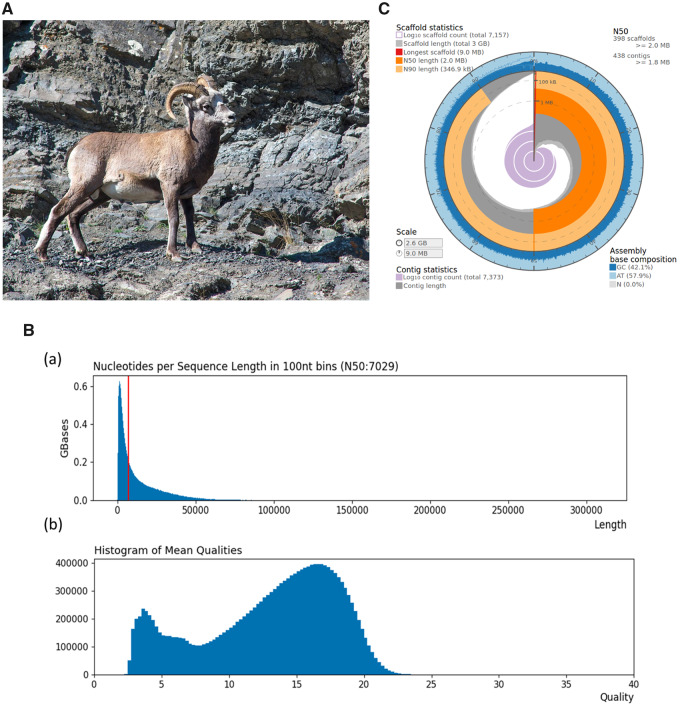
Fig. 1.
(A) Photograph of an adult Ovis nivicola from Suntar Hayata Ridge (Eastern Siberia). (B) Length distribution of Nanopore sequencing reads (a) and distribution of average read quality of Nanopore sequencing reads (b). (C) Visualization of the statistics of the snow sheep assembly (https://github.com/rjchallis/assembly-stats): the inner radius (highligthed in red color) represents the length of the longest scaffold, the radial axis from circumference indicates the scaffold length, the dark and light orange arcs represent the N50 and N90 scaffold lengths, respectively. The outermost circular layer (in shades of blue) shows the base composition at the given coverage (represented in terms of percentage) of the genome.
Materials and Methods
Sample Collection and Processing
For whole-genome sequencing using PromethION and Illumina sequencing data, the muscle sample was collected from a healthy male. For RNA sequencing, samples of five different tissues (skin, cerebrum, heart, liver, large intestine) were taken from three different snow sheep from the Suntar-Hayat and Central Verkhoyansk Ridges in Russia. The sampling locations were selected after observations from the air in 2008–2010 and covered an area between 63° and 71° north latitude, and between 128° and 139° east longitude. The samples were placed in liquid nitrogen immediately after the collection. They were later stored at −80 °C for later analysis.
Construction of Sequencing Libraries
Libraries for long-read sequencing were prepared from ∼3 µg of unsheared genomic DNA, following the protocol of Oxford Nanopore’s LSK109 kit (ONT, Oxford, UK). First DNA was end-repaired and A-tailed using the Ultra II end-repair module (New England Biolabs, Ipswich MA) for 10 min at 20 °C. Enzymes were inactivated at 65 °C for 5 min and DNA was purified with 1× volumes of Ampure XP beads (Beckman Coulter, Brea CA). End-repaired DNA was eluted from the beads at 55 °C for 20 min. Then sequencing adapter with attached motor protein was ligated to the DNA fragments using T4 Quick ligation module (New England Biolabs, Ipswich MA) and components from LSK109 sequencing kit. Adapted library was purified with 0.45 volumes of Ampure beads and eluted in LSK109 elution buffer for 20 min at 37 °C. Approximately 400 ng of library was loaded on a PromethION flowcell and run for 72 h on a PromethION beta sequencer (ONT, Oxford, UK).
The library construction for the Illumina sequencing was carried out with 200 ng of genomic DNA following the Illumina library preparation protocols (NexteraFlex DNA library kit, Illumina, San Diego, CA). The resulting amplified library was quantified and controlled on an Agilent Bioanalyser 2100 (Agilent, Santa Clara, CA) and sequenced in 2 × 100 bp paired-end mode on an Illumina HiSeq1500, yielding ∼200 Mio read pairs.
For RNA-seq library construction, total RNA was prepared from tissue of hunted animals. Small tissue pieces were collected into tubes containing solution D (Chomczynski and Sacchi 1987). Preserved tissue was stored and transported at −80 °C, homogenized in trizol (Silent Crusher, Heidolph, Germany) and extracted on a Maxwell RSC48 device using miRNA tissue kit (both Promega, Madison, WI). Prior to constructing RNA-seq libraries, total RNA was analyzed on Bioanalyser (Agilent), and samples with least degraded RNA (RNA Integrity Number > 6) were used for RNA-seq library generation with Lexogen sense mRNA (Lexogen Vienna, Austria) and Nugen Complete (Redwood City, San Francisco, CA) RNA-seq kits. Libraries were sequenced on Illumina HiSeq1500.
Genome Assembly
Porechop, version 0.2.4, was used for adapter trimming and Guppy, version 3.5.1, was used for base calling. After evaluating a number of established and upcoming long-read assemblers, wtdbg2 (redbean), version 2.5 (Ruan and Li 2019), with default settings for Oxford Nanopore reads (option -x ont) but with shorter acceptable reads (-L 2000) was chosen based on giving by far the most contiguous assemblies. The assemblies generated from the base-called reads using different Guppy versions were polished with up to three rounds of Racon, version 1.3.3. (Vaser et al. 2017), from which consistently round 2 was chosen based on having the highest count of Illumina reads mapping to it, which implied that round 3 is overcorrecting. The Illumina reads were further used for one round of polishing with Pilon, version 1.23 (Walker et al. 2014).
Annotation of Repeat Sequences
RepeatModeler, version 1.0.11 (Smit and Hubley 2008–2015), was used to identify a de novo repeat genomic sequences in the snow sheep genome. Subsequently, this custom repeat library was used in RepeatMasker, version 4.0.9 (Smit et al. 2013–2015; Tarailo-Graovac and Chen 2004), to soft-mask the identified repeat families. Additionally, the trf tool, version 4.07b (Benson 1999), was employed to predict the tandem repeats.
Scaffolding Using RNA-Seq Data
To further improve the assembly, RNA-seq-based approach as implemented in AGOUTI, version 0.3.3, was used (Zhang et al. 2016). For this purpose, the paired-end RNA-seq data sets from five tissues were aligned to the assembled snow sheep genome using Hisat2, version 2.1.0 (Kim et al. 2015), aligner with default parameters. Augustus, version 3.3.3 (Stanke and Waack 2003), was used to predict gene models from the snow sheep assembly using configuration parameters trained on Homo sapiens (human). The name-sorted bam file and gene models were used as inputs in AGOUTI (Zhang et al. 2016) to link the contigs.
Genome Annotation
Three different approaches, such as ab initio prediction, protein homology, and RNA-seq-based annotation, were carried out to predict the gene structure. For protein homology–based approaches, the reference protein sequences from NCBI database of goat (Capra hircus) and cattle (Bos taurus) were downloaded. Additionally, proteins sequences of NCBI refseq database of domestic sheep (Oar_rambouillet_v1.0) and reviewed uniprot sequences of mammals were also downloaded. All these proteins sequences were aligned against the snow sheep genome using Spaln2, verion 2.4.0 (Iwata and Gotoh 2012). For RNA-seq-based annotation, the RNA-seq reads of five different tissues (cerebrum, heart, large intestine, liver, and skin), which consisted of paired-end as well as single-end reads, were filtering using trim_galore, version 1.4 (Martin 2011). Subsequently, all the filtered reads were de novo assembled using Trinity, version 2.10.0 (Grabherr et al. 2011; parameters: -no_normalize_reads –min_kmer_cov 1 -SS_lib_type F). To reduce the redundancy of the assembled transcripts, we used the “tr2aacds” tool as implemented in the EvidentialGene (Gilbert 2019) pipeline. In the next step, Program to Assemble Spliced Alignments (PASA), version 2.0.0 (Haas et al. 2003), was used to obtain the refined gene models from the assembled transcripts. The ab initio prediction of gene structure was carried out SNAP, version 2006-07-28 (Korf 2004), Augustus, version 3.3.3 (Stanke et al. 2006), and GeneId, version 1.4 (Parra et al. 2000) using the parameters either trained for human or mammals. Hisat2 with default parameters was used to align the RNA-seq reads against the snow sheep genome. Later, these aligned reads were assembled into gene models using Stringtie, version 1.3.6 (Pertea et al. 2015). Subsequently, gene models predicted by all the above approaches were provided as inputs for the EvidenceModeler, version 1.1.1 tool (Haas et al. 2008), to generate the nonredundant sets of gene structure. The final set of high-quality protein-coding sequences was also prepared based on the following criteria: 1) a gene should be supported by all the three methods of ab initio predictions or 2) a gene should at least be supported either by transcript-based evidence or by protein homology-based evidence. The amino acid sequences were functionally annotated using emapper, version 2.0.1.4 (Huerta-Cepas et al. 2017), based on eggNOG orthology data (Huerta-Cepas et al. 2018). For this purpose, sequence searchers were performed using DIAMOND (Buchfink et al. 2015). Additionally, functional annotation of protein sequences was also carried out using InterProScan 5.36.75.0 database (Jones et al. 2014). Both these procedures also assigned the gene ontology terms associated with the protein function.
Identification of Noncoding RNAs
Various noncoding RNAs were also predicted using combination of different tools and Rfam database. To identify, cytoplasmic transfer (t)RNA gene, tRNAscan-SE, version 2.0.5 (Chan et al. 2019), was used with the default settings. Further, tRNAs were filtered based on the following criteria: 1) it overlapped with the Short Interspersed Nuclear Elements identified by Repeatmasker, 2) it was identified as pseudogene, and 3) it had mismatched isotypes. Further, miRNAs, SnRNAs, SnoRNAs, and rRNA were annotated by searching the Rfam database (Griffiths-Jones et al. 2005, release 14.1) with Infernal, version 1.1.3 (Nawrocki and Eddy 2013). We also annotated rRNA genes using RNAmmer, vesion 1.2 (Lagesen et al. 2007). Subsequently, we filtered out rRNAs identified using Rfam database that had no overlap with rRNAs identified using RNAmmer.
Results and Discussion
Genome Assembly and Quality Assessment
A total of 56 Gb data in 17 million PromethION sequences (fig. 1B) with a read N50 of 7,029 was used to assemble the snow sheep genome. The genome assembly (fig. 1C) resulted in 7,373 contigs and was estimated to be 2.62 Gb in length with L50 (length at N50) and L90 (length at N90) of 1.76 Mb (N50 = 438) and 323.32 kb (N90 = 1,674), respectively. The size of the assembly comparable with domestic sheep (O. aries, Oar 4.1, ∼2.61 Gb) but it is smaller than goat genome assembly (Capra hircus, ARS1, 2.92 Gb). The completeness of the assembly was assessed using BUSCO tool (version 3.1.0); the single-copy orthologs set in mammalian lineage were searched against the assembled genome of snow sheep using BUSCO tool (version 3.1.0). The results (supplementary table S1, Supplementary Material online) indicated that the assembly covered 3,947 (∼97%) of the total 4,104 orthologs. Of these 3,947 genes, 3,762 (∼92%) were completely covered in the assembled genome, whereas only about 3% were assumed to be missing in the assembly. Additionally, the assessment of the base content also indicated that GC content of the assembly was 42.12%, which was comparable with that of domestic sheep (41.9%) and goat (41.5%).
Repeat Annotation
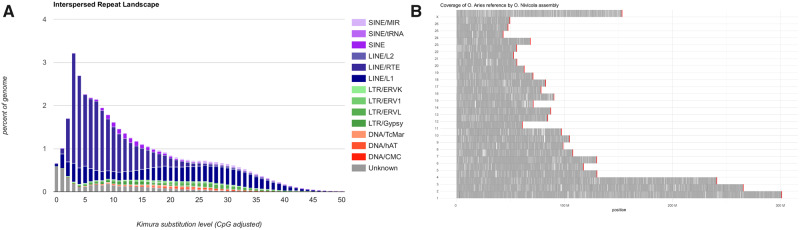
RepeatModeler identified 605 repeat consensus sequences in snow sheep genome. Among these, long interspersed nuclear elements (LINE) had highest number of consensus followed by long terminal repeats consensus. Subsequently, RepeatMasker procedure (supplementary table S2, Supplementary Material online) soft-masked about 1,087 Mb (∼41.5%) sequences of the genome. An interspersed repeat landscape, to study the divergence of transposable element classes, was produced for the snow sheep assembly using the scripts calcDivergenceFromAlign.pl and createRepeatLandscape.pl as provided with RepeatMasker package. The resulting landscape fig. 2A) identified LINE repeat families as the most abundant, constituting about 30% of the snow sheep genome (supplementary table S2, Supplementary Material online). The lowest substitution level in some LINE L1 copies suggested that these are the youngest repeat elements and are still probably expanding and diversifying in snow sheep genome.
Fig. 2.
(A) Interspersed repeat landscape of snow sheep genome. (B) Coverage of Ovis aries reference (Oar_rambouillet_v1.0) by snow sheep genome assembly. The x axis represents the position, and y axis represents choromosome number of O. aries reference. The red bars represent the size of the chromosomes of O. aries reference.
RNA-Based Scaffolding and Quality Assessment
RNA-based scaffolding approach (fig. 1C) reduced the number of contigs from 7,373 to 7,157 and it also increased L50 from ∼1.76 Mb to ∼2.01 Mb and L90 from ∼323 kb to ∼346 kb. After carrying out RNA-based scaffolding, we aligned the Illumina paired-end sequences, of the same individual from which the assembly was generated, to the assembly using bwa-mem, and the result suggested that about 99.42% reads were mapped and 97.6% were aligned as proper pairs. In another approach, to assess the completeness of the snow sheep assembly, we aligned the snow sheep assembly against the latest domestic sheep assembly (Oar_rambouillet_v1.0) available in NCBI genome database. The results (fig. 2B) suggested that scaffolds of the snow sheep assembly covered the entire genome of domestic sheep assembly.
Genome Annotation and Quality Assessment
A total of 57,267 protein-coding sequences (supplementary table S3, Supplementary Material online) were identified using comprehensive gene prediction pipeline. After filtering (see Materials and Methods for detail), these were reduced to 20,202 high-quality protein sequences. Of these 20,202, 18,870 proteins were annotated using InterPro database, whereas 19,175 protein sequences were annotated using eggNOG orthology data. Of these 19,175 protein sequences, 18,919 proteins were mapped to mammalian orthologs gene families. Combining the results of InterProScan and emapper, a total of 19,240 (∼96%) protein sequences were annotated. To assess the quality of protein-coding gene set of the snow sheep assembly, BUSCO was run in the protein mode against the mammalian lineage. The results indicated that the annotation successfully captured about ∼81% complete and ∼14% fragmented BUSCOs, and only ∼5% of BUSCOs were missing from the predicted protein sequences. Moreover, DOGMA, version 3.0 (Dohmen et al. 2016) was also used to measure the completeness of a proteome. It measures the completeness of conserved protein domains identified in a given proteome by providing it as a percentage of a defined core set. The DOGMA analysis based on the domain annotation by PfamScan annotation revealed that about 90% of the total expected conserved domain arrangements were present in the proteome of snow sheep assembly.
Annotation of Noncoding RNAs
A total of 1,484 high-quality tRNAs were identified. Blast pairwise comparison of these tRNAs against that of the domestic sheep genome (downloaded from GtRNAdb 2.0, Chan and Lowe 2016) identified 444 tRNAs of snow sheep that had sequence identity >95% over aligned sequence length or more than 95% of query coverage per hsp (high-scoring segment pairs) with the tRNAs of domestic sheep (reference: Oar4.1, BlastN settings: -perc_identity 0.95 -qcov_hsp_perc 0.95 -max_target_seqs 1). In addition, a total of 243 rRNAs, 1,931 snRNAs, and 782 miRNAs were also identified (supplementary table S4, Supplementary Material online).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported in part by the Ministry of Science and Higher Education of the Russian Federation within theme (Grant No. 0445-2019-0024) as well as by the Russian Foundation of Basic Research (Grant No. 18-316-20009). This work was also supported by LMUexcellent funding of LMU Munich.
Author Contributions
I.M., G.B., and N.Z. conceived and guided the study. G.B., I.O., and V.B. obtained the samples. S.K. and H.B. led the genome and RNA sequencing. A.D. and I.O. provided important inputs in data analysis. M.U., A.H., and E.K. analyzed the data. M.U., A.H., E.K., and A.D. wrote the manuscript. All authors read and approved the final manuscript.
Data deposition: All the fastq files of genome sequences generated using Illumina sequencing technology and fast5 files generated using PromethION sequencing technology are uploaded on EMBL-ENA with project accession PRJEB38329. All the supporting files related to genome annotation can be downloaded using the following link: https://doi.org/10.6084/m9.figshare.12458111.
Literature Cited
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. doi: 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Chan PP, Lin BY, Mak AJ, Lowe TM.. 2019. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. bioRxiv. 614032.doi: 10.1101/614032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2016. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44(D1):D184–D189. doi: 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N.. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 162(1):156–159. doi: 10.1006/abio.1987.9999 [DOI] [PubMed] [Google Scholar]
- Deniskova TE, et al. 2016. Whole genome SNP scanning of snow sheep (Ovis nivicola). Dokl Biochem Biophys. 469(1):288–293. doi: 10.1134/s1607672916040141 [DOI] [PubMed] [Google Scholar]
- Deniskova TE, et al. 2018. Characteristics of the genetic structure of snow sheep (Ovis nivicola lydekkeri) of the Verkhoyansk Mountain chain. Russ J Genet. 54(3):328–334. doi: 10.1134/s1022795418030031 [DOI] [Google Scholar]
- Dohmen E, Kremer LPM, Bornberg-Bauer E, Kemena C.. 2016. DOGMA: domain-based transcriptome and proteome quality assessment. Bioinformatics 32(17):2577–2581. doi: 10.1093/bioinformatics/btw231 [DOI] [PubMed] [Google Scholar]
- Dotsev AV, et al. 2018. Genome-wide SNP analysis unveils genetic structure and phylogeographic history of snow sheep (Ovis nivicola) populations inhabiting the Verkhoyansk Mountains and Momsky Ridge (northeastern Siberia). Ecol Evol. 8(16):8000–8010. doi: 10.1002/ece3.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotsev AV, et al. 2019. The first complete mitochondrial genomes of snow sheep (Ovis nivicola) and thinhorn sheep (Ovis dalli) and their phylogenetic implications for the genus Ovis. Mitochondrial DNA Part B. 4(1):1332–1333. doi: 10.1080/23802359.2018.1535849 [DOI] [Google Scholar]
- Gilbert DG. 2019. Genes of the pig, Sus scrofa, reconstructed with EvidentialGene. PeerJ. 7:e6374.doi: 10.7717/peerj.6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, et al. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33(Database issue):D121–D124. doi: 10.1093/nar/gki081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31(19):5654–5666. doi: 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9(1):R7. doi: 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 34(8):2115–2122. doi: 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47(D1):D309–D314. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Sheep Genomics Consortium, et al. 2010. The sheep genome reference sequence: a work in progress. Anim Genet. 41(5):449–453. doi: 10.1111/j.1365-2052.2010.02100.x [DOI] [PubMed] [Google Scholar]
- Iwata H, Gotoh O.. 2012. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40(20):e161. doi: 10.1093/nar/gks708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics (Oxford, England) 30(9):1236–1240. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 12(4):357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):59.doi: 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108. doi: 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Medvedev DG, et al. 2017. Genetic characteristics of Kodar snow sheep using SNP markers. Contemp Probl Ecol. 10(6):591–598. doi: 10.1134/s1995425517060099 [DOI] [Google Scholar]
- Miller JM, Moore SS, Stothard P, Liao X, Coltman DW.. 2015. Harnessing cross-species alignment to discover SNPs and generate a draft genome sequence of a bighorn sheep (Ovis canadensis). BMC Genomics. 16(1):397. doi: 10.1186/s12864-015-1618-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler CF, Hoffmann RS, Woolf A.. 1973. G-band patterns as chromosomal markers, and the interpretation of chromosomal evolution in wild sheep (Ovis). Experientia 29(1):117–119. doi: 10.1007/BF01913288 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR.. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29(22):2933–2935. doi: 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigó R.. 2000. GeneID in Drosophila. Genome Res. 10(4):511–515. doi: 10.1101/gr.10.4.511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, et al. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 33(3):290–295. doi: 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raul Valdez JB. 1982. The wild sheep of the world. Mesilla (New Mexico: ): Wild Sheep and Goat International. [Google Scholar]
- Rezaei HR, et al. 2010. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol Phylogenet Evol. 54(2):315–326. doi: 10.1016/j.ympev.2009.10.037 [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H.. 2019. Fast and accurate long-read assembly with wtdbg2. bioRxiv. doi: 10.1101/530972. [DOI] [PMC free article] [PubMed]
- Smit AFA, Hubley R.. 2008. –2015. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org. Accessed October 06, 2019.
- Smit AFA, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0. Seattle (WA: ): Institute for Systems Biology; Available from: http://www.repeatmasker.org. Accessed October 06, 2019. [Google Scholar]
- Stanke M, et al. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server):W435–W439. doi: 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–225. doi: 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2004. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. Chapter 4:Unit 4.10. doi: 10.1002/0471250953.bi0410s05 [DOI] [PubMed]
- Vaser R, Sovic I, Nagarajan N, Sikic M.. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27(5):737–746. doi: 10.1101/gr.214270.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. 2017. Draft genome of the Marco Polo Sheep (Ovis ammon polii). GigaScience. 6(12):1–17. doi: 10.1093/gigascience/gix106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SV, Zhuo L, Hahn MW.. 2016. AGOUTI: improving genome assembly and annotation using transcriptome data. GigaScience 5(1):31–31. doi: 10.1186/s13742-016-0136-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheleznov-Chukotskii NK. 1994. Ecology of snow sheep of Northern Asia. Moscow (Russia): Nauka Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.