Abstract
Chromosomal inversions are among the primary drivers of genome structure evolution in a wide range of natural populations. Although there is an impressive array of theory and empirical analyses that have identified conditions under which inversions can be positively selected, comparatively little data are available on the fitness impacts of these genome structural rearrangements themselves. Because inversion breakpoints can disrupt functional elements and alter chromatin domains, the precise positioning of an inversion’s breakpoints can strongly affect its fitness. Here, we compared the fine-scale distribution of low-frequency inversion breakpoints with those of high-frequency inversions and inversions that have gone to fixation between Drosophila species. We identified a number of differences among frequency classes that may influence inversion fitness. In particular, breakpoints that are proximal to insulator elements, generate large tandem duplications, and minimize impacts on gene coding spans which are more prevalent in high-frequency and fixed inversions than in rare inversions. The data suggest that natural selection acts to preserve both genes and larger cis-regulatory networks in the occurrence and spread of rearrangements. These factors may act to limit the availability of high-fitness arrangements when suppressed recombination is favorable.
Keywords: polymorphic inversion, insulator, Drosophila melanogaster, position effect
Introduction
Chromosomal inversions, which are large genomic regions that are generated by double-strand breakage and repair in reverse orientation, are widespread in many natural populations. These rearrangements have a long history of study in Drosophila species (Sturtevant 1917; Dobzhansky 1962). The primary theories explaining the prevalence of inversions in natural populations are that suppressed recombination over the inverted region is favored by natural selection (Sturtevant and Beadle 1936; Mukai et al. 1971; Kirkpatrick and Barton 2006; Corbett-Detig and Hartl 2012; Langley et al. 2012; Kapun, Schmidt, et al. 2016; Fuller et al. 2019). Alleles contained in inversions can interact epistatically or additively to maintain a complex polygenic phenotype such as body size, stress resistance, fecundity, and lifespan (Hoffmann et al. 2004; Hoffmann and Rieseberg 2008; Kirkpatrick 2010). Inversions that suppress recombination between alleles that contribute to a beneficial phenotype can be selected for. Biogeographic data support this hypothesis; natural populations of Drosophila melanogaster maintain inversion frequency clines strongly correlated with climatic clines (Mettler et al. 1977; Knibb 1982; Rane et al. 2015; Kapun, Fabian, et al. 2016; Kapun, Schmidt, et al. 2016; Simões and Pascual 2018). Furthermore, an ever-expanding set of taxa appear to contain polymorphic inversions that are associated with adaptive phenotypes (Butlin et al. 1982; Huynh et al. 2011; Oneal et al. 2014). It is increasingly accepted that a major source of positive selection on chromosomal inversions is the maintenance of linkage among alleles that are favorable in similar contexts.
Whereas the potential fitness benefits of maintaining linkage among synergistic alleles are well established, the impacts of inversion breakpoints on the individuals that carry them are not well understood. Nonetheless, these impacts are likely to play an important role in shaping evolutionary outcomes for new arrangements. An inversion breakpoint that disrupts a key gene sequence could result in the death or sterility of the individual that carries it, preventing the inversion from reaching polymorphic frequencies in natural populations. Accumulated evidence is consistent with the idea that an inversion’s breakpoint positions might have large impacts on its fitness. The distribution of polymorphic inversion breakpoints along the genome is not random (Tonzetich et al. 1988; Pevzner and Tesler 2003; González et al. 2007; Calvete et al. 2012; Puerma et al. 2014, 2016b; Orengo et al. 2015). In fact, many apparently independently formed inversions seem to precisely share breakpoint locations (Pevzner and Tesler 2003; González et al. 2007; Puerma et al. 2014; Corbett-Detig et al. 2019). Even when inversion breakpoints are not precisely reused at the molecular level, their broad-scale distributions across the genome are nonuniform (Pevzner and Tesler 2003; Ranz et al. 2007). Though this pattern is well established, the factors underlying breakpoint localization and the fitness of new arrangements are poorly understood.
There are two mechanisms that shape the fine-scale distribution of inversion breakpoints. First, mutational biases are factors that affect the probability that an inversion breakpoint occurs at a specific genomic location (Tonzetich et al. 1988; Pevzner and Tesler 2003; Calvete et al. 2012; Guillén and Ruiz 2012). In many species inversions occur through ectopic recombination between repetitive sequences, an example of a mutational bias (Guillén and Ruiz 2012), though this is relatively rare in the melanogaster subgroup (Ranz et al. 2007; Corbett-Detig and Hartl 2012). Additionally, some evidence indicates that physical instability due to unstable secondary structure or local chromatin environment may also bias breakpoint localization (Falk et al. 2010). Second, specific breakpoint positions can affect the fitness of a new arrangement. These “position effects” have been identified in a variety of organisms (Frischer et al. 1986; Lakich et al. 1993; Hough et al. 1998; Puig et al. 2004; Castermans et al. 2007). Deleterious effects associated with breakpoint positions could hypothetically be as large as positive impacts from the maintenance of allele complexes contributing to polygenic traits. Deleterious position effects are therefore expected to limit the number of individual inversions that could evolve and maintain polygenic phenotypes in the population.
There are several specific factors that could influence the fitness of inversion breakpoints. First, disruption of gene sequence and enhancer–promoter interactions can cause mRNA truncation, chimeric transcripts, or misregulation of genes overlapping and near to breakpoints (Frischer et al. 1986; Castermans et al. 2007; Ren and Dixon 2015; Lupiáñez et al. 2016). Previous work has found that common inversions in D. melanogaster and fixed inversions in Drosophila pseudoobscura are less likely to disrupt gene coding sequences that would be expected under a random breakpoint model (Corbett-Detig and Hartl 2012; Fuller et al. 2017), possibly indicating that natural selection acts against inversions which disrupt gene sequences. However, a mutational bias that preferentially creates breakpoints in intergenic regions is also consistent with these findings. Inversions in the D. melanogaster species group tend to create inverted duplications of sequence at their breakpoints in the repair process, which can preserve copies of disrupted sequence (Ranz et al. 2007; Puerma et al. 2016b). Duplication size may therefore also influence the fitness of an inversion breakpoint because large duplications can avoid disrupting individual genes. A study in Anopheles gambiae has shown that the inversion 2L+a is likely viable because it preserves functional copies of disrupted genes through this mechanism (Sharakhov et al. 2006). Location in respect to gene sequence and duplication size should both contribute to the fitness of a new inversion arrangement.
Factors related to gene regulation may also impact the fitness of newly formed arrangements. These include topologically associated domains (TADs), chromatin state, and the locations of insulator elements. TADs are genomic features that appear in HiC proximity ligation mappings (Lieberman-Aiden et al. 2009) which reflect the physical folding and arrangement of the genome (Sexton et al. 2012; Jost et al. 2014; Lupiáñez et al. 2016). Disruption of these domains may alter local gene expression (Lupiáñez et al. 2016). Chromatin marks often determine local expression and repressive chromatin is capable of suppressing nearby gene activity when translocated (Cryderman et al. 1998). Boundaries between domains are often associated with insulator elements in D. melanogaster (Sexton et al. 2012). Insulators limit the influence of repressive chromatin marks and block ectopic enhancer activity, and could therefore act as a compensatory mechanism to maintain native regulatory environments (Sigrist and Pirrotta 1997; Gaszner and Felsenfeld 2006; Bushey et al. 2008; Yang and Corces 2012). We hypothesize that high-fitness inversions disrupt local gene regulation less than would be expected by chance, by avoiding disrupting crucial domains or by colocalization with insulator elements.
Comparisons among fixed, high- and low-frequency inversions can reveal the impact of natural selection on chromosomal inversion breakpoints (Cáceres et al. 1997; Corbett-Detig 2016). Because they have persisted and spread within natural populations, we expect both high-population frequency and ancestrally fixed chromosomal inversion breakpoints to show a biased distribution of features consistent with higher fitness. Conversely, low-frequency inversions, often identified in only a single individual within a population, are most likely recently arisen arrangements. The low-frequency inversions’ breakpoint distribution should therefore primarily reflect mutational biases. By examining the distributions of fixed, high-frequency, and low-frequency inversion breakpoints, we can identify the factors that shape the fitness of newly arisen arrangements.
We leverage population-resequencing data sets from >1,000 D. melanogaster isolates to detect and de novo assemble both breakpoints of 18 rare naturally occurring inversions. We compare these “rare” inversion breakpoints to known high-frequency inversion breakpoints in D. melanogaster (Corbett-Detig and Hartl 2012) as well as a set of fixed inversion breakpoints between species in the Melanogaster subgroup (Ranz et al. 2007). By comparing rare, common, and fixed inversion breakpoints, we find evidence supporting the idea that both mutational biases and natural selection play important roles in shaping the fine-scale distribution of inversion breakpoints in natural populations.
Materials and Methods
Defining Inversion Categories
In our analysis, we define three classes of inversion population frequency. Previous work in D. melanogaster has typically referred to four categories of inversion, “common cosmopolitan,” “rare cosmopolitan,” “recurrent endemic,” and “unique endemic” (Mettler et al. 1977; Krimbas and Powell 1992). The latter half of each of these terms refers to the geographic distribution of the inversion. As long as an inversion reached high frequency in any population, it has not been strongly impacted by negative selection. We label these high-frequency inversions “common” inversions. We use “rare” to refer to inversions which were found in only single samples (with the exception of In(2R)Mal, which is present in three samples studied here). The distribution of rare inversions, while possibly containing high-fitness inversions that could eventually spread to high frequencies, are likely to primarily reflect mutational biases in their overall breakpoint distribution. To summarize, “common cosmopolitan,” “rare cosmopolitan,” and “recurrent endemic” will all fall under our label “common,” whereas we refer to “unique endemic” as “rare” inversions, similarly to the analysis in Corbett-Detig (2016).
The third class in our framework, “fixed” inversions, are inversions that have gone to fixation within one lineage during divergence of the D. melanogaster subgroup (Ranz et al. 2007). Originally all fixed inversions occurred as unique events in a Drosophila ancestor. They subsequently spread until they reached fixation in populations ancestral to contemporary species in the melanogaster subgroup. These fixed inversions were discovered by comparing the locations of homologous sequences in the genomes of between D. melanogaster and its relatives (Lemeunier and Ashburner 1976) and have been molecularly characterized previously (Ranz et al. 2007). It is important to note that the vast majority of these fixed inversions occurred on the Drosophila yakuba branch and not in a direct D. melanogaster ancestor (Krimbas and Powell 1992; Ranz et al. 2007). The reference genome of D. melanogaster should therefore generally reflect the ancestral state and the genetic background on which these inversions originated rather than a derived state evolved after fixation. Common and rare inversions annotated here occurred in contemporary D. melanogaster populations and thus in the absence of additional changes unrelated to genome structure, on a similar genetic background to that on which the D. yakuba inversions were fixed. The functional annotations used here are also based on the D. melanogaster standard arrangement, meaning these annotations should represent the genetic background of all three inversion frequency categories.
Short-Read Alignment
We obtained short-read data as fastq files from the Sequence Read Archive. All short-read data are described in Lack et al. (2016) and was originally produced in Pool et al. (2012), Lack et al. (2015), Mackay et al. (2012), Kao et al. (2015), and Grenier et al. (2015). We aligned the short-read data using bwa v0.7.15 using the “mem” function and default parameters (Li 2013). All postprocessing (sorting, conversion to BAM format, and filtering) was performed in SAMtools v1.3.1 (Li et al. 2009). We filtered these BAM files to include only those alignments with a minimum mapping quality of 20 or more.
Rare Breakpoint Identification
As in previous works that characterized structural variation using short-insert paired-end Illumina libraries (Cridland and Thornton 2010; Rogers et al. 2014; Corbett-Detig et al. 2019), we first identified aberrantly mapped read “clusters.” Briefly, here, a cluster is defined as three or more read pairs that align in the same orientation (for inversions, this is either both forward-mapping or both reverse-mapping) and for which all reads at one edge of the cluster map to within 1 kb of all other reads in the cluster. We considered only aberrant clusters where both ends mapped to the same chromosome arm as the vast majority of inversions in Drosophila are paracentric (Krimbas and Powell 1992). We required that all read pairs included in a cluster map a minimum of 500 kb apart. We then retained only those potential inversions for which we recovered both forward- and reverse-mapping clusters there were within 100 kb of one another. The choice of a maximum distance between possible breakpoint coordinates was included to reduce the possible rates of false-positives and because none of the known inversions whose breakpoints have previously been characterized included a duplicated region of 100 kb or more (Ranz et al. 2007; Corbett-Detig and Hartl 2012). When breakpoint assemblies existed in very close proximity or appeared to delete short sequences, we set the duplication size to 1 base. We further filtered all breakpoint assemblies that overlapped annotation transposable elements as these are the primary source of aberrantly mapping read clusters in previous works (Corbett-Detig and Hartl 2012).
As an additional check for the accuracy of our newly discovered breakpoints, we compared our distribution of rare breakpoints to the known cytogenetic distribution and found no chromosomal or by-region differences (P = 0.7, χ2 test; cytogenetic data from Corbett-Detig (2016) who summarized Krimbas and Powell (1992)). The short insert size from previous sequencing experiments ranged from ∼200 to ∼600 bp, which may have led to a nontrivial false-negative rate of breakpoint discovery particularly if the breakpoints contain repetitive elements or other large DNA insertions. However, we do not expect that these potential false-negatives will bias our downstream analyses, and all previously characterized inversion breakpoints in the Melanogaster species complex occurred in unique sequences (Ranz et al. 2007; Corbett-Detig and Hartl 2012). All software used to perform these analyses is available from the github repositories associated with this project. Specifically, scripts used for breakpoint detection and assembly are in https://github.com/dliang5/breakpoint-assembly (last accessed May 26, 2020).
De Novo Rare Breakpoint Assembly
For each putative inversion, we then extracted all reads for which either pair mapped to within 5 kb of the predicted breakpoint position. We converted all fastq read files to fasta and qual files as is required by Phrap, and we assembled each using otherwise default parameters but including the “-vector_bound 0 -forcelevel 10” command line options (Corbett-Detig and Hartl 2012; Rogers et al. 2014). We then used BLAST to align the resulting de novo assembled contigs to the D. melanogaster reference genome to identify the contig that overlapped the predicted breakpoint using the flybase BLAST tool (https://flybase.org/blast/, last accessed May 26, 2020). We retained only inversions for which we could de novo assemble contigs overlapping both breakpoints, and we further discarded any contigs where the sequence intervening two distant genomic regions contained sequence with homology to known transposable elements. All of the assembled breakpoint sequences are available in supplementary file S1, Supplementary Material online. Assembly scripts are available from https://github.com/dliang5/breakpoint-assembly (last accessed May 26, 2020).
Overlapping Inversions and In(2R)Mal
We also attempted to find sets of overlapping inversions. Briefly, for overlapping inversions, where one inversion arises on a background that contains another inversion with one breakpoint inside and one outside of the inverted region, the breakpoint-spanning read clusters should be largely the same as inversions that arose on a standard arrangement chromosome. However, the key difference is that rather than pairs of forward- and reverse-mapping read clusters, we expect to observe two distantly mapping read clusters in the reverse–forward and forward–reverse arrangements. We applied this approach for the 17 rare inversions that we initially discovered as well as to all samples that contained common inversions that are known from previous work (Corbett-Detig and Hartl 2012; Lack et al. 2015). We found only one such overlapping rare inversion, which is consistent with the known segregation distorter-associated chromosomal inversion In(2R)Mal, which is composed of two overlapping inversions (Presgraves et al. 2009). In our analysis here, we treat these overlapping inversions as independent, but our results are qualitatively unaffected if we simply exclude the second inversion.
Genome Version, Insulator, and Gene Annotations
All our analyses are based on alignments to D. melanogaster genome version 6.26 (Hoskins et al. 2015). We obtained genome annotation data including gene locations from flybase. We treated long noncoding RNAs as genes for our purposes, as they perform essential functions and can be disrupted in the same way as protein-coding genes. We obtained insulator-binding site positions from Nègre et al. (2010, accession GSE16245). As necessary, we converted the coordinates of genomic features from genome version 5 to 6 using the flybase coordinate batch conversion tool (https://flybase.org/convert/coordinates, last accessed May 26, 2020).
Selection of Public Data Sets for Topological Domains and Chromatin Marks
We obtained TAD data including annotations of chromatin state from Sexton et al. (2012). This data set is composed of domains detected by genome-wide chromosome conformation capture sequencing, HiC, on early stage embryos, and annotated with an epigenetic state using a clustering method applied to another source of linear epigenomic data (Sexton et al. 2012). Their annotations include four categories: “active,” “null,” “PcG” (polycomb), and “HP1” (centromeric heterochromatin). For the sake of consistency, we refer to Sexton et al.’s “null” domains as “inactive.” Early stage embryos are likely to be the environment in which any regulatory disruption induced by inversions is most deleterious given the sensitive nature of development, which makes this a promising source of context for our analysis of inversion frequency. This data set also allows us to separately analyze breakpoint occurrence within TADs and chromatin states in tandem, because they are derived from the same source. It should be noted, however, that the annotations of these TADs are relatively coarse and may not reflect the more local environment of an inversion breakpoint.
We therefore performed a second analysis on finer scales using the data set of Kharchenko et al. (2011, accession GSE25321). This data set in its raw form consists of short spans marked with one of a set of chromatin markers, in both a nine-state model and a 30-state model. As we desired a representation of the local chromatin environment around inversion breakpoints, we chose to bin the nine-state representation into total counts of bases assigned to a state of the given type over windows of 10 kb. About 10 kb was selected based on the average heterogeneity of the windows; we wanted our window size to be as small as possible but for most windows to contain at least one region with an annotated chromatin state. This yielded a distribution of values for each window which represented the overall enrichment of each state in each 10-kb span. As we lacked statistical power to evaluate these mark types individually with our relatively small inversion breakpoint data sets, we further assigned each 10-kb window an activity state based on the majority of present marks. Windows in which the vast majority of sites were assigned states one through five, annotated by Kharchenko et al. (2011) as being various components of genes including promoters, exons, and introns, were designated “active.” Windows where states six through nine, which include PcG, HP1, and other heterochromatic marks, were most prominent, were designated “inactive.” Windows in which both groups each constituted at least 5% of all marks were designated “mixed.” This yields an alternative representation of chromatin environments surrounding inversion breakpoints that is much finer-grained than the annotations of Sexton et al. (2012).
We compared this representation to Sexton et al.’s annotated chromatin states as an additional check for the validity of our approach. We found that 10-kb windows located within each annotated TAD generally aligned with the annotation of that TAD, but that substantial heterogeneity of chromatin marks exists within each TAD span (supplementary fig. S3, Supplementary Material online). For example, ∼19% of windows within TADs annotated as “active” are enriched for chromatin state 9, which is associated with extended silenced regions, and conversely 26% of windows within TADs annotated as inactive are enriched for chromatin state 2, which is associated with the active transcription. This indicates that one cannot be treated as a direct substitute for the other.
As a final check on the validity of the domains obtained from Sexton et al. (2012), we obtained polytene domain data from Eagen et al. (2015), repeated our analysis, and found them to be generally consistent with our conclusions. These results may be found in supplementary text S1, Supplementary Material online.
Permutations and Statistical Tests
To compare inversion breakpoint positions to a randomized distribution, permutations for all categories of inversions (rare, common, and fixed) were performed with 1,000 iterations of a group of randomly located breakpoints, holding the inversion number, duplication lengths, and chromosome arms constant. Specifically, for each inversion breakpoint, 1,000 starting positions were chosen from a uniform distribution between the start of that chromosome arm and the end minus the length of the duplication—that is, from the entire set of possible points for that size of breakpoint. Random breakpoints were located independently for most tests, as most values were calculated for each breakpoint individually rather than the inversion as a whole. The exception is the chromatin-blending test, in which we additionally controlled for inversion lengths to account for the role of inversion length in biasing pairs of chromatin environments. Features of the genome at each of these breakpoints were recorded as our expected value for the random distribution of breakpoints.
Tests were divided by the nature of the factor. For factors that are a discrete numerical value for each break, such as distance to an element or length of a duplication, P values were calculated as percentiles of real values within a large set of random distributions. Tests between categories of the distance-based factors and the duplication length test were performed distribution to distribution with pairwise Mann–Whitney rank-sum tests.
For categorical values, such as disrupting a gene span or not, rates of category occurrence were calculated for 1,000 permutations. We define disruptions of genes and other elements as both forward and reverse single-strand breaks occurring within a single-annotated functional element. It is important to note that our method of defining disruption is likely to overestimate the proportion of fixed inversion breakpoints that truly disrupt genic sequences. Ranz et al.’s (2007) method to identify sequences duplicated by the original break relies on sequence homology, and in fixed inversions divergence of noncoding sequences can interfere with the precise identification of breakpoint regions. For example, if the original duplicated region includes a gene coding span and some noncoding bases, a complete gene copy will be produced along with a partial duplication. Over time, the noncoding region will tend to accumulate more mutations than the intact gene copy. In this case, coordinates obtained from BLAST alignments may not detect the homology between the noncoding regions and instead only yield apparent homology from duplication within the conserved gene span. This would be counted as a gene disruption event by our analysis. This bias will tend to make our analysis conservative with respect to identifying the impacts of natural selection, because breakpoints are more likely to be identified within coding regions and because we should tend to underestimate the sizes of breakpoint-adjacent duplicated regions after sequence homology has decreased. All scripts used to produce the results of the permutation tests described above are available from the github repository associated with this project https://github.com/jmcbroome/breakpoint_analysis (last accessed May 26, 2020).
Lethal and Sterile Phenotype Analysis
Additionally, we obtained phenotype data from Flybase using the query builder (https://flybase.org/cgi-bin/qb.pl, last accessed May 26, 2020) to get the IDs of all genes which have lethal phenotypes and sterile phenotypes. These data were incorporated into the gene disruption analysis and we sought evidence of difference in disruption rates between genes annotated with these phenotypes and the overall set of annotated genes. Supplementary table S2, Supplementary Material online, contains the set of inversion breakpoints which appear to disrupt these genes.
Results and Discussion
Common and Fixed Inversion Breakpoints
Common and fixed inversion breakpoints have been characterized extensively in D. melanogaster and in the Melanogaster species complex in previous works. We obtained the breakpoint locations for nine common inversions from Corbett-Detig and Hartl (2012) and Lack et al. (2016). We note that although population frequencies and geographic ranges vary among common inversions (Krimbas and Powell 1992; Corbett-Detig and Hartl 2012; Lack et al. 2016), each has reached frequencies of at least 10% within local subpopulations and all have been observed in several geographically widespread populations, suggesting that their breakpoints do not cause strong deleterious fitness consequences. From Ranz et al. (2007), we obtained the breakpoint positions of 26 inversions that have fixed in a lineage since the common ancestor of the Melanogaster species complex. To confirm that the breakpoint-adjacent regions have not been modified or updated in the more recent genome assemblies for either D. melanogaster or D. yakuba, we extracted each surrounding 100-kb region from the genome that contains the ancestral arrangement and used BLAST to align these to the genome containing the derived rearrangement. We recorded the most breakpoint proximal high quality, that is, BLAST score >50, sequence alignment as the putative location of the inversion breakpoint.
Rare Inversion Breakpoints Discovered
We realigned all sequence data from over 1,000 D. melanogaster natural isolates that have been sequenced previously using paired-end sequencing methods (Langley 2012; Mackay et al. 2012; Pool et al. 2012; Grenier 2015; Kao et al. 2015; Lack et al. 2015; summarized in detail in Lack et al. [2016] ). We identified 5,318 short-read clusters that corresponded to possible inversion breakpoints that are a minimum of 1 Mb from each other and for which we found both forward- and reverse-mapping read clusters (supplementary fig. S1, Supplementary Material online). That is, for a given inversion relative to the reference genome, we expect to find a cluster of read pairs where both maps in the “forward” orientation and another cluster where each pair of reads both map in the “reverse” orientation (Corbett-Detig and Hartl 2012; see Materials and Methods). We also searched for overlapping inversions using a slight modification of this approach (see Materials and Methods). To be as conservative as possible with our analysis, we retained only the set for which we recovered and successfully de novo assembled both breakpoints for a given inversion. Additionally, we removed any putative breakpoint-spanning contig that mapped with high confidence to multiple locations in the D. melanogaster reference genome. We ultimately retained 18 rare inversions. Three of our candidate rare inversions are corroborated by previous cytological evidence (Presgraves et al. 2009; Huang et al. 2014). Similarly, previous molecular evidence (Grenier et al. 2015) supports the identified breakpoints of another chromosomal inversion. The breakpoints of our putative rare inversions do not show unusual genetic distances from other samples isolated from the same populations, suggesting that these are relatively recent events and not older inversions that have recently gone to lower frequencies (supplementary text S2 and table S1, Supplementary Material online).
The genomic and population distributions of candidate rare inversions are largely consistent with our expectations based on extensive cytological work. First, our estimated rate of occurrence of rare inversions, 1.6% per genome, is within the range of estimates from cytological data across diverse populations 0.47–2.71% (Krimbas and Powell 1992; Aulard et al. 2002). Furthermore, we found no rare inversions on the X chromosome, which contains very few chromosomal inversions in natural populations of this species (Krimbas and Powell 1992; Aulard et al. 2002). However, because we conservatively required that both breakpoints are detected from discordant short-read alignments and completely assembled de novo, and because we excluded any breakpoints that contained homology to annotated transposable elements, it is possible that our approach has underestimated the prevalence of rare inversions in these data sets. It is also possible that a portion of the rare inversions may be false-positives owing to the challenges of short-read based de novo assembly and interpretation. Nonetheless, as an additional check to ensure the robustness of our results, we repeated all of our analyses on the subset of rare inversions which have been cytologically or molecularly characterized or are very simple in their breakpoint structures and found no major differences between data sets (supplementary text S3, Supplementary Material online).
Inversion Breakpoints Could Truncate Coding Sequences
Inversions can strongly disrupt sequences at their breakpoints (fig. 1). This has multiple classes of potential negative consequences, including the truncation of gene spans and the creation or alteration of enhancer–gene interactions (Frischer et al. 1986; Castermans et al. 2007; Ren and Dixon 2015; Lupiáñez et al. 2016). We investigated interactions with gene spans to test the hypothesis that higher frequency inversions are more likely to exhibit features which reduce large-scale disruptions of local functional elements. For each category, we calculated the percentile of the count of disrupting breakpoints against the permuted distribution, where low percentiles correspond to less disruption than expected. All three inversion frequency categories disrupt annotated gene spans less often than the random expectation (rare P = 0.0415, common P = 0.0055, fixed P < 0.001, permutation test). The proportion of gene-disrupting inversions is inversely correlated to population frequency category (44% of rare inversion breakpoints, 28% of common inversion breakpoints, 24% of fixed inversion breakpoints).
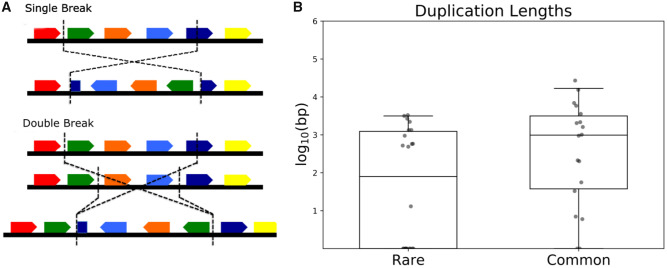
Fig. 1.
—(A) Staggered breakpoints generate duplications that might suppress the impacts of sequence disruption. In the single break, the dark blue gene span is divided into half in the inverted line with no functional copies remaining. In the double break, the dark blue gene span is duplicated and a functional copy remains to the right of the break region. (B) Common inversion breakpoints exhibit longer duplications. The boxplots represent duplication lengths of each inversion class. Note that the y axis is in logarithmic scale, and all short deletions were set to length 1.
Our results are consistent with gene disruption being negatively selected after inversion formation. We note here that the baseline rate of disruption is still relatively high even in the most conservative category, at 24% of fixed inversion breakpoints. Nonetheless, for reasons described above (see Materials and Methods), this should be considered a conservative upper bound on the rate of gene disruption in the fixed inversion class. In all cases of putative disruption, the D. yakuba genome contains an intact ortholog; this indicates that if breakpoints occurred within an annotated gene, they rarely completely disrupt the coding sequence or that secondary sequence evolution can suppress the deleterious effects. All putatively disrupting breakpoints within the fixed inversion class lie within 1,000 bases of the start or the end of the disrupted gene (supplementary table S3, Supplementary Material online). The trend across categories indicates that there is a negative association between population frequency and the occurrence of inversion breakpoints within gene sequences in our data.
We also note that rare inversions appear to disrupt genes less often than expected by chance. This could be explained by the critical nature of many genes to survival. At a minimum, each inversion must not be lethal for us to discover it. The preservation of gene spans by rare inversions may also be explained by a mutational bias of chromatin state or basepair composition favoring intergenic regions, reducing gene disruption rates below random expectations. As a final possible explanation, we note that because many of the samples used in this work were inbred, either intentionally or passively as isofemale lines, inversions that induce recessive strongly deleterious fitness effects might still be exposed to selection and purged from the line prior to sequencing.
We further investigated the possible fitness impacts of disrupted gene sequences by examining the subset of disrupted genes that are annotated as having lethal or sterile alleles. We expect to observe a reduction in the rates that inversion breakpoints interrupt genes with lethal alleles and sterile alleles owing to the importance for organism survival and reproduction. In applying a similar permutation test as above, but instead asking if inversion breakpoints are less likely than expected by chance to disrupt essential genes specifically, we do not find a significant decrease in the rate of essential gene disruptions compared with genes overall (supplementary table S4, Supplementary Material online). We note that only one gene with an annotated sterile phenotype was disrupted among all inversion breakpoints considered here. However, we still failed to reject the null model possibly due to a general paucity of known sterility-inducing genes compared with unannotated genes.
Furthermore, it is possible that a significant portion of genes remains functional despite the presence of both breaks within the annotated span. For example, the common inversion In(X)A disrupts a gene with annotated lethal alleles. The disrupted gene, NFAT, encodes an important transcription factor (Keyser et al. 2007). The inversion breakpoint is very near the 5′ start of the gene, where some annotated transposable element insertions have produced viable alleles (Bellen et al. 2011). It is possible that the breakpoint does not actually render the gene nonfunctional and is therefore not lethal. Further functional work will be needed to understand the specific effects of localized gene disruption on individual phenotypes.
Larger Inverted Duplications May Prevent Gene Disruption
Duplications that occur during inversion formation may maintain functional elements and suppress the local gene-interrupting effects of inversion breakpoints. Paired staggered double-strand breakage is the major mechanism by which inversions events occur in the D. melanogaster subgroup (Ranz et al. 2007; Puerma et al. 2016b). These breaks leave an overhang of sequence at each end of the putative inversion. After repair in inverted orientation, the result is inverted duplicated regions on either side of a new inversion with length equal to the overhang left over after the double-strand break (fig. 1). To guarantee disruption of a given functional element at the sequence level without creating a complete duplicate, both sides of a double-strand break must fall into that same functional element. Longer duplications are thus less likely to disrupt individual elements. Therefore, we hypothesized that selection will favor longer duplicated regions that minimize impacts on local sequence functions.
To test this, we first verified that tandem duplications of inversion breakpoints which do not disrupt genes are longer than those that do (P = 0.0096, Mann–Whitney U test). Dividing the data by frequency category, we found that common polymorphic inversions have significantly longer duplications than rare inversions (fig. 1, P = 0.0095, Mann–Whitney U test). We did not include fixed inversion breakpoints, as secondary sequence evolution and gaps between synteny blocks made determination of exact original duplication length inaccurate and likely an underestimate. These results are consistent with the idea that long duplications act as a compensatory mechanism for otherwise negative position effects by preserving intact functional elements or by maintaining proximity among functional elements within duplicated regions.
Formally, our analysis is consistent with higher relative fitness of inversions with longer inverted repeats, but does not necessarily require deleterious effects at single breakpoints. It is also possible that inversions are positively selected when they contain larger breakpoint-adjacent duplications because of positive effects associated with gene duplications or chimeric gene products (Puerma et al. 2016a, 2016b). However, given that microsynteny is largely maintained over evolution and given that the Drosophila genome contains a high density of functional elements, we favor our hypothesis that larger repeats can be favored by natural selection because they can avoid disrupting functional elements.
Inversions Could Alter Local Regulatory Environments
Impacts on gene regulation in the regions surrounding inversion breakpoints are also likely to be an important determinant of inversion fitness. By translocating large sections of the genome, inversions can reshape local regulatory environments and interfere with nuclear structures. They can separate enhancers from their gene targets, bring chromatin marks of varying kinds into close proximity, and alter the content and size of local regulatory domains. Translocations of repressive chromatin marks can lead to the silencing of nearby genes, such as in the phenomenon of position-effect variegation, which is variable silencing of a gene near a translocated section of heterochromatin (Eissenberg et al. 1992; Cryderman et al. 1998; Puig et al. 2004; Vogel et al. 2009; Shatskikh et al. 2018). Chromatin environments also guide the activity of different double-strand break repair mechanisms including nonhomologous end joining, which may serve as a mutational bias in the occurrence of inversions (Lemaître and Soutoglou 2014; Marnef et al. 2017). We investigated the occurrence of inversions in different chromatin domains, hypothesizing that both mutational biases and selective pressures may influence breakpoints within these domains.
We examined patterns related to chromatin states and marks at two resolutions. The coarser resolution is the level of TADs. TADs are often highly conserved and associated with coordinated gene regulatory blocks (Cavalli and Misteli 2013). In D. melanogaster, TADs have been identified through high-resolution chromatin conformation capture, or HiC, sequencing and found to contain distinct chromatin states (Sexton et al. 2012). Although any inversion whose breakpoints occur within these domains can and does alter relative TAD boundary positions, inversions with breakpoints that capture boundary elements within associated duplicated regions might form entirely new boundaries and TADs by duplicating those boundary elements. We hypothesized that inversion breakpoints would be less likely to duplicate boundary elements at higher population frequencies, as the formation or division of TADs may be more deleterious than resizing them.
Only two polymorphic inversion breakpoints, one rare and one from the common inversion In(X)A, could have duplicated a boundary element annotated by Sexton et al. (2012). This occurs less often than we would expect by chance for both categories (rare P = 0.02275, rare and common combined P = 0.001, permutation test). As common inversions have a modest sample size (n = 9), no level of boundary duplication for them alone is statistically significant. The low rates of boundary duplication are relatively invariant across frequency categories, so we speculate that a mutational bias may protect boundary regions from breakage. This could occur through a concentration of bound proteins in boundary regions (Sexton et al. 2012). Alternatively, it may be extremely deleterious to duplicate boundary regions, purging these inversions from our rare inversion data set as well as from inversions at higher frequencies.
We also discovered an enrichment of inversion breakpoints within TADs marked with active chromatin by Sexton et al. (2012) (rare P = 0.003, common P = 0.055, fixed P < 0.001, all categories P < 0.001, permutation test). As part of our hypothesis that mixing chromatin states is deleterious, we investigated correlations between domain annotations at either end of an inversion—that is, whether the identity of the domain at one inversion breakpoint is correlated with the domain type at the other inversion breakpoint. There does not appear to be any enrichment for particular combinations of chromatin environments around each breakpoint in our inversions (P = 0.36, χ2 test), suggesting that the biased distribution of inversion breakpoint chromatin domains is driven by a marginal increase of breakpoints within active regions rather than a pairwise effect of breakpoint-adjacent chromatin domains. The increased rate of breakpoints in active regions is consistent with a mutational bias, as it is relatively invariant across frequency categories. This bias could be due to a difference in the rate of occurrence of double-strand breaks in open chromatin, or it could be due to a difference in the accuracy and efficiency of double-stranded break repair in these active environments (Marnef et al. 2017).
Because this coarse representation may not fully represent the role of chromatin environment on inversion occurrence, we additionally examined enrichment of chromatin marks at a finer scale. Kharchenko et al. (2011) created a genome-wide data set representing local chromatin mark enrichment, represented as a computationally derived nine-state model with high resolution. We binned this data into windows of 10 kb and examined the regions immediately surrounding each inversion breakpoint for the presence of Kharchenko’s chromatin states, assigning each window to a general state of “active,” “inactive,” or “mixed” (see Materials and Methods). We found that the enrichment of inversion breakpoints in active regions was not replicated in this finer-scale data set (P = 0.59, permutation test; fig. 2). The enrichment may not exist at these scales because the majority of windows designated “active” contain genes and breakpoints are less likely to occur within genes than in intergenic regions (see above).
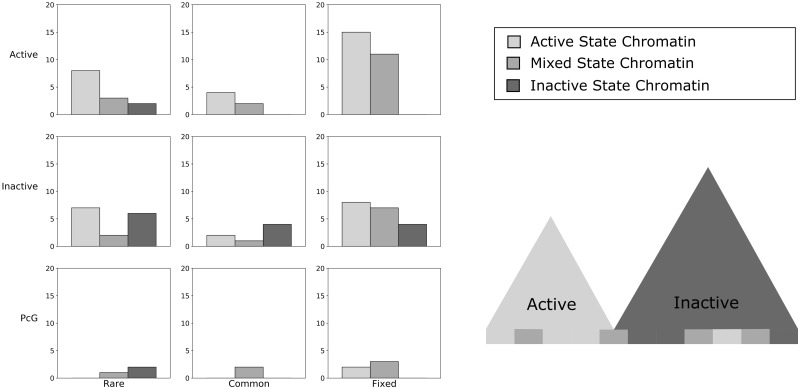
Fig. 2.
—Chromatin states around inversion breakpoints are active and heterogeneous. Inversion breakpoints are grouped by frequency into columns, then by domain annotation from Sexton et al. (2012) into rows. Each group is further subdivided into three bars, representing the overall states assigned to the 10-kb chromatin windows around each break in that combination of categories (i.e., using the Kharchenko et al. [2011] data set for chromatin state assignment, see Materials and Methods). The y axis of each plot is the count of inversion breakpoints which exist in that combination of states and frequencies. The plot below the legend displays the intuitive relationship between these two levels of annotation; that is, the larger domain annotations (triangles) contain a heterogeneous but biased set of local chromatin states (bars).
We discovered an enrichment of mixed chromatin activity states (i.e., both active and inactive states) in windows around fixed inversion breakpoints (P < 0.001, permutation test) and a decreased occurrence of fixed inversion breakpoints within windows containing only inactive states (P = 0.0075, permutation test; fig. 2). Common inversions show a similar pattern, though with no significant association between inversion breakpoints and windows containing only active chromatin states, but with a significant depletion of breakpoints in windows designated inactive (P = 0.026, permutation test). Rare inversion breakpoints appear randomly distributed with regards to their fine-scale chromatin environments (fig. 2). It is possible that the common inversions would reflect this same pattern with a larger sample size, but we are limited by the scarcity of high-frequency inversions. We additionally explored whether there is a correlation between local chromatin windows between two breakpoints of each inversion, but found no statistical enrichment (P = 0.25, χ2 test). Overall these results suggest that high-frequency breakpoints tend to occur on epigenetic boundaries within regions of the genome that contain some active chromatin.
Insulator Elements Maintain Boundaries of Local Regulatory Environments
We discovered an enrichment for inversions within active domains, including inversions which occur between pairs of active and inactive domains. This led us to ask whether we can identify a candidate compensatory mechanism that might suppress the disruption of local regulatory environments by the translocation of chromatin-defining elements. Insulator elements are key to the structure and function of genome regulatory networks, serving as structural anchor points, physical blockers of enhancer interactions, and boundary elements between TADs or chromatin compartments (Chung et al. 1993; Roseman et al. 1993; Nègre et al. 2010; Sexton et al. 2012). Insulators may reduce or prevent the effects of repressive chromatin on local gene activity after translocation (Sigrist and Pirrotta 1997; Gaszner and Felsenfeld 2006; Bushey et al. 2008; Yang and Corces 2012). In fact, these elements have been previously shown to be associated with fixed structural rearrangement breakpoints that alter local synteny, which includes inversion breakpoints (Nègre et al. 2010).
Strong association with these elements may act as a compensatory mechanism that preserves local chromatin state and thereby allows inversions to occur between heterochromatic and euchromatic regions while minimizing negative consequences (fig. 3A). Insulator element-binding sites are strongly associated with local active and mixed chromatin window states in our data (active P = 4.7e-24, mixed P = 2.7e-8, Fisher’s exact test) and correspondingly rare in windows annotated as inactive (P = 2.7e-14, Fisher’s exact test). This further supports that these insulator elements represent epigenetic boundaries, and a strong association with these insulator elements may explain the enrichment of mixed chromatin window states around higher frequency inversions in our data.
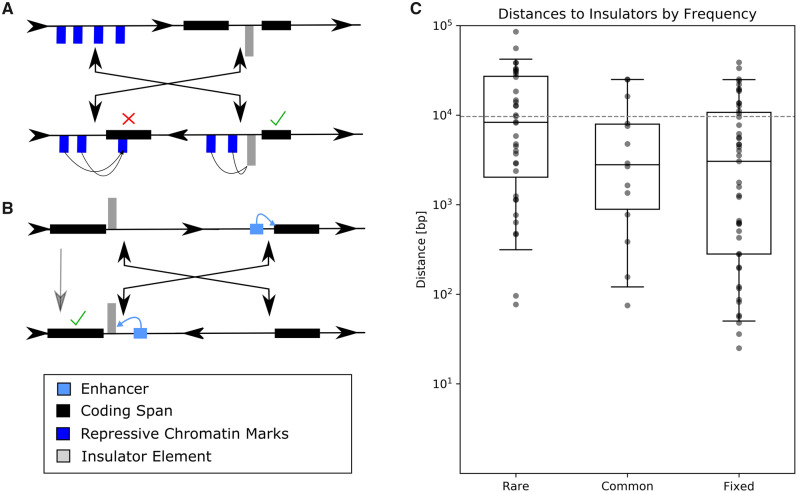
Fig. 3.
—(A) Insulators could prevent chromatin repression across inversion breakpoints. This pair of hypothetical inversions has four breakpoints that do not disrupt gene spans. The left inversion breakpoint is in a repressive gene-free chromatin landscape indicated by blue chromatin marks, whereas the other is between two genes separated by an insulator element. After inversion, repressive chromatin marks are relocated to be adjacent to one of the genes, but their repressive effect is blocked by the insulator. The check mark indicates normal gene expression, whereas the cross indicates disrupted gene expression. (B) Insulators block enhancer activity. This simple example displays how an inversion may translocate an enhancer into the proximity of a new gene. In this example, the enhancer’s activity is blocked by the presence of an insulator, in gray. (C) Higher population frequency correlates with insulator proximity. The distributions of distances are from breakpoints to the nearest insulator element. Note that the y axis is a logarithmic scale. The dashed gray line is the expected median distance of a random breakpoint model.
Association with insulator elements may also prevent ectopic enhancer activity, or the activation of genes other than the target gene by an enhancer (fig. 3A). Previous studies have shown developmental disorders can occur in mammals from inversion rearrangements with no additional mutation via ectopic enhancer activity (Ren and Dixon 2015; Lupiáñez et al. 2016). Notably, recent data suggest inversion breakpoints are rarely associated with local perturbed expression in Drosophila (Fuller et al. 2016; Lavington and Kern 2017; Said et al. 2018; Ghavi-Helm et al. 2019). However, there still appears to be some cases of ectopic enhancer/promoter interactions as in subdued and Dscam4 (Ghavi-Helm et al. 2019). This suggests that new ectopic interaction may be promoted by inversions but that these interactions rarely impact overall expression levels.
Our data show that inversion breakpoints are significantly closer to insulator elements than would be expected for randomly distributed breakpoints (rare P = 0.0446, common P < 0.001, fixed P < 0.001, permutation test, fig. 3). Common inversion breakpoints are significantly closer to insulators than are rare inversions breakpoints (P = 0.0373, Mann–Whitney U test), fixed and common inversion breakpoints are not statistically different (P = 0.312, Mann–Whitney U test) and fixed inversion breakpoints are significantly closer to insulator elements than are rare inversion breakpoints (P = 0.00242, Mann–Whitney U test). We asked whether insulators are found within or outside the duplicated regions and found no evidence for any directionality to the association (supplementary fig. S2, Supplementary Material online). We additionally note that insulator-binding sites are much more common in active TADs (supplementary fig. S3, Supplementary Material online), and that correspondingly inversions in active regions are somewhat more closely associated with insulators across frequency categories (P = 0.075, Mann–Whitney U test).
Because inversion breakpoints tend to occur in active domains, the enrichment for proximity to insulator elements might result from the increased density of insulator elements within those regions and not for selection for proximity to insulators per se. We saw a bias toward breakpoints situated in active domains, particularly among fixed inversions. We therefore tested for an association between fixed inversion breakpoints that occur within inactive domains and proximity to insulator elements. After controlling for the domain type, the close proximity of insulators persists, suggesting the observation is partially independent of the correlation between active domains and insulators (P = 0.03, Mann–Whitney U test). We thus conclude that this is not likely to be a strong confounding factor for our insulator association results. Conversely, if proximity to insulators is beneficial for any reason, this may explain the enrichment of mixed chromatin states around high-frequency inversions.
Although these results are consistent with the idea that insulators are a compensation mechanism preventing misexpression, our speculation is not directly proven here. A recent work discovered a lack of gene expression differences over and around inversion breakpoint regions in balancer chromosomes (Ghavi-Helm et al. 2019). Because they are maintained as heterozygotes, balancer chromosomes are shielded from selection in homozygotes in a similar fashion to rare inversions. We investigated proximity to insulator elements as a potential cause for this lack of misexpression associated with the balancer chromosome inversion breakpoints but found no association (P = 0.57, Mann–Whitney). The result challenges our specific hypothesis because Ghavi-Helm et al. (2019) found minimal differential gene expression around balancer inversion breakpoints despite the lack of insulator association. The association between proximity to insulators and population frequencies is robust in our data, but Ghavi-Helm et al.’s results suggest it may not be necessary to prevent disruption of gene expression. A potential explanation may be that insulators affect more subtle gene expression phenotypes. For example, if the presence of insulator elements decreases the variance in expression across cells or facilitates coordinated timing of expression during development, this might not be reflected in mean expression values obtained via bulk RNA-seq. Regardless of the mechanism, our results suggest that inversions, and possibly synteny changes in general, are more fit when associated with insulator elements in the D. melanogaster genome.
Cross-Feature Analysis
Each of the features that we examined in this work do not exist in isolation, and it is possible that interactions among features also impact the fitness of new arrangements. Our primary hypothesis for insulator association is that it compensates for the negative effects of other features; for example, mixing chromatin may be permissible when insulator elements near the inversion breakpoint suppress expression modifying effects (see above). Therefore, we performed several cross analyses between the features explored here both without condition and conditioning on population frequencies of inversions studied in this work. The results of the cross-feature analysis are described in supplementary text S4, Supplementary Material online. We discovered a few obvious associations between features, such as gene disruption and active chromatin, which is expected given that active chromatin is typically associated with genes. We discovered no feature correlations based on population frequency, though this may be due to the modest sample sizes, which likely limited our power to detect interaction effects among features considered here.
Despite our lack of statistical power, we do observe individual cases where a combination of features may mitigate negative fitness effects. For example, in the common polymorphic inversion In(2L)t, the first breakpoint is in a region containing active chromatin states, whereas the second is in a region that contains inactive states. The active region breakpoint is <1 kb from an insulator-binding motif; it may be that insulator binding at this site limits the influence of repressive chromatin on the other side of the breakpoint. The common polymorphic inversions In(2R)NS, In(3L)P, and In(3R)K are similarly arranged, with breakpoints occurring in regions of active and inactive chromatin marks and with an insulator-binding site very near to the active region breakpoint. Consistent with this idea, Said et al. (2018) recently showed that breakpoint-adjacent genes in In(2L)t and In(3R)K are expressed at similar levels between standard and inverted arrangements. These breakpoint arrangements of common inversions are therefore qualitatively consistent with the idea that insulator elements could suppress deleterious consequences of mixing chromatin states.
Conclusion
In this work, we present evidence that mutational biases and natural selection have played a role in shaping the fine-scale distribution of chromosomal inversion breakpoints in the D. melanogaster subgroup. Natural selection likely plays a role in maintaining gene sequences, as we found that high-frequency inversions are less likely to disrupt gene coding spans and that they produce correspondingly longer tandem duplications. We also identified two levels of association between chromatin activity and inversion frequency—there appears to be a mutational bias toward occurrence in active regions of the genome, but inversions also tend to occur in areas with locally mixed chromatin states. We found evidence explaining this second pattern consistent with natural selection for the association of inversion breakpoints with insulator elements, which are in turn strongly associated with these mixed chromatin states. Our analyses therefore clarify the mutational context and fitness impacts of novel chromosomal inversions in natural populations and guide future research into specific fitness and gene regulatory impacts of chromosomal inversions.
A stronger understanding of the factors underlying the distribution of polymorphic inversions is requisite for the study of their evolutionary impact. Although it is possible that breakpoint effects sometimes increase the fitness of new arrangements, such as by creating new expression patterns of transposed loci or chimeric transcripts (Puerma et al. 2016a, 2016b), our data are consistent with the conclusion that selection acts against breakpoints that disrupt functional elements within breakpoint regions, consistent with studies in other Drosophila species (Fuller et al. 2017). Factors that mitigate fitness costs determine what parts of the genome can tolerate polymorphic inversions that maintain complex phenotypes. Variations in these factors may also explain differences in the formation and role of inversions across species. Before an inversion can be selected for recombination suppression or other features it must form in a genomic context where the presence of inversion breakpoints is not immediately and strongly detrimental. Given two additional features of chromosomal inversions, 1) the de novo mutation rate is likely very low (Krimbas and Powell 1992) and 2) the conditions for a new arrangement to be favored by natural selection are sometimes restrictive (Hoffmann et al. 2004; Kirkpatrick and Barton 2006; Charlesworth and Barton 2018), the impacts of fine-scale inversion breakpoint positions on the fitness of new arrangements suggest that the availability of suitable, high-fitness arrangements may often be rate-limiting for adaptive evolution when suppressed recombination is favorable.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors give their thanks to three anonymous reviewers, Kirk Lohmueller, Stephen Schaeffer, and Zach Fuller, for their detailed suggestions in article preparation and Iskander Said for his assistance collecting the low-frequency inversion data. This work was supported by the National Institutes of Health (R35GM128932) and an Alfred P. Sloan Fellowship to R.C.-D. During this work, J.M. was supported by an NIH training award (T32HG008345).
Literature Cited
- Aulard S, David J, Lemeunier F.. 2002. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet Res. 79(1):49–63. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, et al. 2011. The Drosophila Gene Disruption Project: progress using transposons with distinctive site specificities. Genetics 188(3):731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG.. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 32(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK, Read IL, Day TH.. 1982. The effects of a chromosomal inversion on adult size and male mating success in the seaweed fly, Coelopa frigida. Heredity 49(1):51–62. [Google Scholar]
- Cáceres M, Barbadilla A, Ruiz A.. 1997. Inversion length and breakpoint distribution in the Drosophila buzzatii species complex: is inversion length a selected trait? Evolution 51(4):1149–1155. [DOI] [PubMed] [Google Scholar]
- Calvete O, González J, Betrán E, Ruiz A.. 2012. Segmental duplication, microinversion, and gene loss associated with a complex inversion breakpoint region in Drosophila. Mol Biol Evol. 29(7):1875–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castermans D, et al. 2007. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur J Hum Genet. 15(4):422–431. [DOI] [PubMed] [Google Scholar]
- Cavalli G, Misteli T.. 2013. Functional implications of genome topology. Nat Struct Mol Biol. 20(3):290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Barton NH.. 2018. The spread of an inversion with migration and selection. Genetics 208(1):377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G.. 1993. A 5’ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74(3):505–514. [DOI] [PubMed] [Google Scholar]
- Corbett-Detig RB. 2016. Selection on Inversion Breakpoints Favors Proximity to Pairing Sensitive Sites in Drosophila melanogaster. Genetics 204(1):259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, et al. 2019. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics 213(4):1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL.. 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8(12):e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Thornton KR.. 2010. Validation of rearrangement break points identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol Evol. 2:83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman DE, Cuaycong MH, Elgin SCR, Wallrath LL.. 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107(5):277–285. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1962. Rigid vs. flexible chromosomal polymorphisms in Drosophila. Am Nat. 96(891):321–328. [Google Scholar]
- Eagen KP, Hartl TA, Kornberg RD.. 2015. Stable Chromosome Condensation Revealed by Chromosome Conformation Capture. Cell 163(4):934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T.. 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131(2):345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M, Lukasova E, Kozubek S.. 2010. Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat Res. 704(1–3):88–100. [DOI] [PubMed] [Google Scholar]
- Frischer LE, Hagen FS, Garber RL.. 1986. An inversion that disrupts the Antennapedia gene causes abnormal structure and localization of RNAs. Cell 47(6):1017–1023. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW.. 2016. Genomics of natural populations: how differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics 204(1):287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW.. 2017. Genomics of natural populations: evolutionary forces that establish and maintain gene arrangements in Drosophila pseudoobscura. Mol Ecol. 26(23):6539–6562. [DOI] [PubMed] [Google Scholar]
- Fuller ZL, Koury SA, Phadnis N, Schaeffer SW.. 2019. How chromosomal rearrangements shape adaptation and speciation: case studies in Drosophila pseudoobscura and its sibling species Drosophila persimilis. Mol Ecol. 28(6):1283–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G.. 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 7(9):703–713. [DOI] [PubMed] [Google Scholar]
- Ghavi-Helm Y, et al. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 51(8):1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Casals F, Ruiz A.. 2007. Testing chromosomal phylogenies and inversion breakpoint reuse in Drosophila. Genetics 175(1):167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier JK, et al. 2015. Global diversity lines–a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 (Bethesda) 5:593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén Y, Ruiz A.. 2012. Gene alterations at Drosophila inversion breakpoints provide prima facie evidence for natural selection as an explanation for rapid chromosomal evolution. BMC Genomics 13(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH.. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39(1):21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM, Weeks AR.. 2004. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 19(9):482–488. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, et al. 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25(3):445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough RB, Lengeling A, Bedian V, Lo C, Bucan M.. 1998. Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of kit expression. Proc Natl Acad Sci U S A. 95(23):13800–13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster genetic reference panel lines. Genome Res. 24(7):1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh LY, Maney DL, Thomas JW.. 2011. Chromosome-wide linkage disequilibrium caused by an inversion polymorphism in the white-throated sparrow (Zonotrichia albicollis). Heredity 106(4):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost D, Carrivain P, Cavalli G, Vaillant C.. 2014. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 42(15):9553–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Zubair A, Salomon MP, Nuzhdin SV, Campo D.. 2015. Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Mol Ecol. 24(7):1499–1509. [DOI] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, Flatt T.. 2016. Genomic evidence for adaptive inversion clines in Drosophila melanogaster. Mol Biol Evol. 33(5):1317–1336. [DOI] [PubMed] [Google Scholar]
- Kapun M, Schmidt C, Durmaz E, Schmidt PS, Flatt T.. 2016. Parallel effects of the inversion In(3R)Payne on body size across the North American and Australian clines in Drosophila melanogaster. J Evol Biol. 29(5):1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser P, Borge-Renberg K, Hultmark D.. 2007. The Drosophila NFAT homolog is involved in salt stress tolerance. Insect Biochem Mol Biol. 37(4):356–362. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471(7339):480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8(9):e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N.. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173(1):419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR. 1982. Chromosome inversion polymorphisms in Drosophila melanogaster II. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica 58(3):213–221. [Google Scholar]
- Krimbas CB, Powell JR.. 1992. Drosophila inversion polymorphism. CRC Press. [Google Scholar]
- Lack JB, et al. 2015. The Drosophila Genome Nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199(4):1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE.. 2016. A thousand fly genomes: an expanded Drosophila Genome Nexus. Mol Biol Evol. 33(12):3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakich D, Kazazian HH, Antonarakis SE, Gitschier J.. 1993. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet. 5(3):236–241. [DOI] [PubMed] [Google Scholar]
- Langley CH, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192(2):533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavington E, Kern AD.. 2017. The effect of common inversion polymorphisms In(2L)t and In(3R)Mo on patterns of transcriptional variation in Drosophila melanogaster. G3 (Bethesda) 7:3659–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C, Soutoglou E.. 2014. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair 19:163–168. [DOI] [PubMed] [Google Scholar]
- Lemeunier F, Ashburner MA.. 1976. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 193(1112):275–294. [DOI] [PubMed] [Google Scholar]
- Li H, . et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv:1303.3997 [q-Bio]. [Google Scholar]
- Lieberman-Aiden E, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326(5950):289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Spielmann M, Mundlos S.. 2016. Breaking TADs: how alterations of chromatin domains result in disease. Trends Genet. 32(4):225–237. [DOI] [PubMed] [Google Scholar]
- Mackay TFC, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Cohen S, Legube G.. 2017. Transcription-coupled DNA double-strand break repair: active genes need special care. J Mol Biol. 429(9):1277–1288. [DOI] [PubMed] [Google Scholar]
- Mettler LE, Voelker RA, Mukai T.. 1977. Inversion clines in populations of Drosophila melanogaster. Genetics 87(1):169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T, Mettler LE, Chigusa SI.. 1971. Linkage disequilibrium in a local population of Drosophila melanogaster. Proc Natl Acad Sci U S A. 68(5):1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, et al. 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6(1):e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oneal E, Lowry DB, Wright KM, Zhu Z, Willis JH.. 2014. Divergent population structure and climate associations of a chromosomal inversion polymorphism across the Mimulus guttatus species complex. Mol Ecol. 23:2844–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo DJ, Puerma E, Papaceit M, Segarra C, Aguadé M.. 2015. A molecular perspective on a complex polymorphic inversion system with cytological evidence of multiply reused breakpoints. Heredity 114(6):610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P, Tesler G.. 2003. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci U S A. 100(13):7672–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, et al. 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8(12):e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Gérard PR, Cherukuri A, Lyttle TW.. 2009. Large-scale selective sweep among segregation distorter chromosomes in African populations of Drosophila melanogaster. PLoS Genet. 5(5):e1000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerma E, et al. 2014. Characterization of the breakpoints of a polymorphic inversion complex detects strict and broad breakpoint reuse at the molecular level. Mol Biol Evol. 31(9):2331–2341. [DOI] [PubMed] [Google Scholar]
- Puerma E, Orengo DJ, Aguadé M.. 2016. a. The origin of chromosomal inversions as a source of segmental duplications in the Sophophora subgenus of Drosophila. Sci Rep. 6(1):30715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerma E, Orengo DJ, Aguadé M.. 2016. b. Multiple and diverse structural changes affect the breakpoint regions of polymorphic inversions across the Drosophila genus. Sci Rep. 6(1):36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Cáceres M, Ruiz A.. 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc Natl Acad Sci U S A. 101(24):9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA.. 2015. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol Ecol. 24(10):2423–2432. [DOI] [PubMed] [Google Scholar]
- Ranz JM, et al. 2007. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 5(6):e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Dixon JR.. 2015. A CRISPR connection between chromatin topology and genetic disorders. Cell 161(5):955–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RL, et al. 2014. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol. 31(7):1750–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman RR, Pirrotta V, Geyer PK.. 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12(2):435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said I, et al. 2018. Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proc Natl Acad Sci U S A. 115(21):5492–5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, et al. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148(3):458–472. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, et al. 2006. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc Natl Acad Sci U S A. 103(16):6258–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatskikh AS, Olenkina OM, Solodovnikov AA, Lavrov SA.. 2018. Regulated gene expression as a tool for analysis of heterochromatin position effect in Drosophila. Biochemistry (Mosc). 83(5):542–551. [DOI] [PubMed] [Google Scholar]
- Sigrist CJA, Pirrotta V.. 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147(1):209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões P, Pascual M.. 2018. Patterns of geographic variation of thermal adapted candidate genes in Drosophila subobscura sex chromosome arrangements. BMC Evol Biol. 18(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH. 1917. Genetic factors affecting the strength of linkage in Drosophila. Proc Natl Acad Sci U S A. 3(9):555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, Beadle GW.. 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21(5):554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonzetich J, Lyttle TW, Carson HL.. 1988. Induced and natural break sites in the chromosomes of Hawaiian Drosophila. Proc Natl Acad Sci U S A. 85(5):1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, et al. 2009. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenet Chromatin. 2(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Corces VG.. 2012. Insulators, long-range interactions, and genome function. Curr Opin Genet Dev. 22(2):86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.