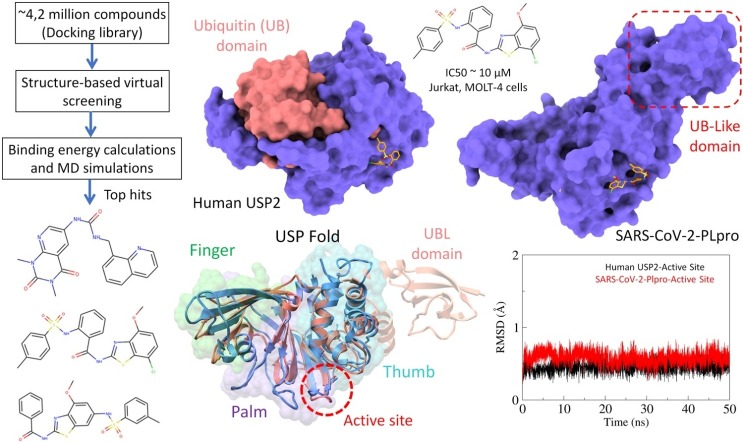
Graphical abstract
Keywords: Deubiquitination, Leukemia, Ubiquitin-specific protease 2 (USP2), SARS-CoV-2 papain-like protease (PLpro), COVID-19
Highlights
-
•
SARS-CoV-2 papain-like protease (PLpro) and human ubiquitin carboxyl-terminal hydrolase 2 (USP2) share strikingly similar structural scaffold and conserved catalytic triad.
-
•
Inhibition of Jurkat and MOLT-4 cell-growth in low micromolar range (≈10 μM) was achieved by a new human USP2 inhibitor (Z93).
-
•
This is the first account in the on-going COVID-19 outbreak where we purposed the identification of SARS-CoV-2 PLpro inhibitor with a proof of human USP2 inhibition.
Abstract
Human ubiquitin carboxyl-terminal hydrolase-2 (USP2) inhibitors, such as thiopurine analogs, have been reported to inhibit SARS-CoV papain-like proteases (PLpro). The PLpro have significant functional implications in the innate immune response during SARS-CoV-2 infection and considered an important antiviral target. Both proteases share strikingly similar USP fold with right-handed thumb–palm–fingers structural scaffold and conserved catalytic triad Cys-His-Asp/Asn. In this urgency situation of COVID-19 outbreak, there is a lack of in-vitro facilities readily available to test SARS-CoV-2 inhibitors in whole-cell assays. Therefore, we adopted an alternate route to identify potential USP2 inhibitor through integrated in-silico efforts. After an extensive virtual screening protocol, the best compounds were selected and tested. The compound Z93 showed significant IC50 value against Jurkat (9.67 μM) and MOTL-4 cells (11.8 μM). The binding mode of Z93 was extensively analyzed through molecular docking, followed by MD simulations, and molecular interactions were compared with SARS-CoV-2. The relative binding poses of Z93 fitted well in the binding site of both proteases and showed consensus π-π stacking and H-bond interactions with histidine and aspartate/asparagine residues of the catalytic triad. These results led us to speculate that compound Z93 might be the first potential chemical lead against SARS-CoV-2 PLpro, which warrants in-vitro evaluations.
1. Introduction
The recently emerged severe acute respiratory syndrome (SARS-CoV-2) created a pandemic and COVID-19 outbreak resulted in more than 0.87 million deaths worldwide (Feldmann and Geisbert, 2011), there is still no therapeutics clinically approved against SARS-CoV-2. Although phenomenal efforts are underway particularly in preclinical and clinical trials (Ahn et al., 2020, Liu et al., 2020, Rosenbaum, 2020, Sanders et al., 2020, Wang et al., 2020a) including nine existing protease inhibitors (Yamamoto et al., 2020). Following this, several other FDA-approved drugs are also under investigation, e.g., remdesivir and chloroquine (Wang et al., 2020b). Moreover, ivermectin, a broad spectrum anti-parasitic agent have presented remarkable in vitro activity (∼5000-fold reduction) (Caly et al., 2020). Others include favipiravir, nitazoxanide and nafamostat (Wang et al., 2020b).
Papain-like protease (PLpro) produced by coronaviruses plays a requisite role in viral replication and pathogenesis mainly by converting viral polyprotein into a functional replicase complex (Shin et al., 2020). It is primarily considered as a cysteine protease with multifunctional protease activity having tendency to hydrolyze different types of ubiquitin including diubiquitin, polyubiquitin and synthetic ubiquitin peptide substrates (Chen et al., 2009). Along with deubiquitination activity, it also possesses deISGylation activity which is believed to facilitate viral invasion (Barretto et al., 2005). Thus, PLpro inhibitors could provide a route towards SARS-CoV2 treatment.
The design and development of PLpro inhibitors is believed to be a daunting task due to a number of reasons including lack of specificity and toxicity resulted by the covalent interaction of free cysteine moieties (Ratia et al., 2008). After the first SARS outbreak (2002–2004) some studies have exhibited that human ubiquitin carboxyl-terminal hydrolase 2 (USP2) antagonists can inhibit SARS-CoV by inhibiting viral PLpro (Chen et al., 2009, Cheng et al., 2015, Chou et al., 2008, Chuang et al., 2018). Deubiquitinating peptidases (DUBs) are a broad class of proteases that dissociate ubiquitin from proteins and disrupt proteasomal and lysosomal degradation of its substrate proteins. Many members of DUB have found to play their role in the progression of different cancers (Young et al., 2019). For instance, USP2 is specific deubiquitinase of a proto-oncoprotein cyclin D1 and stabilizes it via restriction of ubiquitin-dependent degradation (Shan et al., 2009). This affirms that the type of cancers which depend on cyclin D1 for their growth, e.g., T-cell associated ALL (T-ALL) (Aref et al., 2006), can be suppressed by using USP2 inhibitors.
The active site of human USP2 shares a similar architecture with SARS PLpro which belongs to USP class (Báez-Santos et al., 2015, Lindner et al., 2005, Ratia et al., 2006). The PLpro together with 3C-like protease (3CLpro) proteolytically cleave the coronaviral polyprotein and release non-structural proteins. As compared to SARS-CoV-2 3CLpro, the PLpro has deubiquitinating activity and directs important implications on viral replication and pathogenesis (Barretto et al., 2005), therefore considered an important target against SARS-CoV-2. The overall structural analysis revealed a similar active site of SARS-CoV-2 PLpro, including the signature catalytic triad (Cys-His-Asp/Asn) located at the interface of the thumb and palm sub-domains (Chou et al., 2014). Various studies have reported the efficacy of USP2 potent inhibitors including thiopurine analogs on SARS (Chen et al., 2009, Chou et al., 2008) and MERS (Cheng et al., 2015) which were found to interact with the signature catalytic triad (IC50 values ranges from 5 to 21.6 μM).
Even with the therapeutic advancement, there is a dearth of in vitro facilities available to test SARS-CoV-2 inhibitors in whole-cell assays. Therefore, an alternate route might be the selective inhibition of USP2 by small molecules which may plausibly lead to the identification of SARS-CoV-2 PLpro inhibitors due to structurally conserved similar active site fold. Based on these assumptions, the present study focuses on the identification of potential USP2 inhibitor through in-depth structure-based virtual screening workflow and later repurposed against SARS-CoV-2 PLpro. The computational methods already discovered potential antiviral compounds against a wide range of viruses including influenza (Du et al., 2012), Ebola (Madrid et al., 2013, Mirza and Ikram, 2016, Shurtleff et al., 2012), Dengue (Leela et al., 2016, Luzhkov et al., 2013, Wang et al., 2009, Zhou et al., 2008, Parida et al., 2014, Tahir ul Qamar et al., 2016), Zika Virus (Tahir ul Qamar et al., 2016, Nitsche, 2018, Pattnaik et al., 2018) and recently emerged CoVs (Ramajayam et al., 2010, Wang et al., 2017, Wu et al., 2020, Tahir ul Qamar et al., 2020). Moreover, the significance of molecular dynamics (MD) simulations has been potentially aided in antiviral drug discovery (Luzhkov et al., 2013, Hou and Yu, 2007, Tu et al., 2017, Anusuya and Gromiha, 2017, Guan et al., 2017, Bhakat et al., 2014, Speelman et al., 2001, Mottin et al., 2017, Zhang et al., 2017). The computational work in the present study was carried out in sequential way followed by molecular dynamics (MD) simulation and interaction energetics using AMBER simulation package. The identified compounds were purchased and tested on Jurkat and MOLT-4 cells against T-ALL. The tested compounds with significant activity were repurposed against SARS-CoV-2 PLpro and binding mode was extensively analyzed to examine the molecular interactions at SARS-CoV-2 PLpro subsites, including a catalytic triad site and binding site in the cleft leading to the active site (Ghosh et al., 2010). To the best of our knowledge, the utilized method is the first study reported in on-going COVID-19 outbreak where we propose SARS-CoV-2 PLpro inhibitor with a proof of human USP2 inhibition based on highly similar structural attributes.
2. Material and methods
2.1. Data collection and docking structure preparation
For the structure-based study, the co-crystalized structure of human USP2 complex with 6-thioguanine (5XU8) was utilized for docking. The structure was prepared accordingly, as described previously (Ikram et al., 2019, Mirza et al., 2019). Briefly, all heteroatoms were removed, hydrogens and charges were added into USP2. The structure was minimized for the 1000 steepest descent steps at root means square gradient of 0.02 having an update interval of 10 and using the AMBER ff14SB force field. A chemical library of ∼4.2 million molecules was retrieved from ZINC. The library was reduced after applying various drug-like parameter, which mainly includes the Lipinski’s Rule of five (with 2 violations), oral bioavailability rule including: rotatable bonds ≤10 and TPSA ≤ 140 Å2. The resulted dataset was utilized for structure-based virtual screening workflow.
2.2. Virtual screening workflow
Before subsequent docking runs, the docking grid was defined by selecting the specific area around the catalytic triad residues (Cys276, His557, and Asn574). The filtered database was finally uploaded into the Mcule drug discovery platform and AutoDock Vina was utilized to screen the docking library (Kiss et al., 2012). Diversity filter was also applied in workflow, which reduced the docking library by removing the closest analogs and maximized the coverage of chemical space to identify active scaffolds. Autodock Vina was selected among the top-ranked scoring functions in terms of docking power and screening test according to CASF (Comparative Assessment of Scoring Functions) benchmark 2016 (Su et al., 2018). The compounds were ranked accordingly on docking score which is based on the Vina empirical scoring function that approximates the ligand binding affinity in kcal/mol. The top hits were analysed for ADMET criteria and top hits were analysed in Chimera in terms of binding poses and interactions with the catalytic triad.
2.3. Molecular dynamics simulations and energy calculations
The overall stability of the best complexes was analysed over a period of 20 ns using AMBER 18 simulation package (Case et al., 2018). We used the same MD simulation protocol as described previously (Mirza et al., 2019, Mirza et al., 2016). Briefly, the Antechamber was utilized to generate the general AMBER force field (GAFF) parameters for the studied ligands. After a stepwise minimization, heating and equilibration in explicit solvent environment (TIP3), a production run of 20 ns was performed at standard pressure (p =1.0 atm) and temperature (T =300 K). The binding free energies (ΔGtol) of USP2 complexed with screened compounds were calculated using the MM-PB(GB)SA method of AMBER 18 using the following equations.
| ΔEMM = ΔEint + ΔEele + ΔEvdw |
| ΔGsol = ΔGp + ΔGnp |
| ΔGtotal = ΔEMM + ΔGsol |
| ΔGbind (MM-PB(GB)SA) = ΔEMM + ΔGsol – TΔS |
Where, ΔEMM is further divided into internal energy (ΔEint), electrostatic energy (ΔEele), and van der Waals energy (ΔEvdw), and the polar (ΔGp) and non-polar (ΔGnp) energy components contributed to total solvation free energy (ΔGsol). ΔGbind is the free energy of binding evaluated after entropic calculations (-TΔS), for both MM-GBSA and MM-PBSA methods.
The compounds with significant interaction profile were purchased based on availability and price.
2.4. Binding mode analysis on SARS-CoV-2 PLpro
Based on the similarity shared by USP2 and PLpro supported by various studies (Chen et al., 2009, Cheng et al., 2015, Chou et al., 2008, Chuang et al., 2018), we redocked the tested compounds on recently resolved crystal structure of SARS-CoV-2 PLpro (6W9C) and compared the molecular interactions with USP2/ligand complex (Ghosh et al., 2010). For molecular docking, docking grid was setup around the catalytic triad (Cys112, His272 and Asp286). The same MD simulation protocol was utilized for SARS-CoV-2 PLpro complexes. To further rationalize the binding of best compound with the SARS-CoV-2, the molecular interactions were analyzed on the binding site (site 2) adjacent to catalytic triad as reported recently (PDB ID: 7JIW) in complex with compound 3 (5-acrylamide-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide) (Osipiuk et al., 2020) and compared the binding affinities with the co-crystalized inhibitor.
2.5. In vitro cytotoxicity
2.5.1. Materials
Jurkat and MOLT-4 cells were used to inspect the cytotoxicity of the selected compound against T-ALL. The test compound and cisplatin were purchased from MolPort (Life Chemicals Inc.). Cell culture media, fetal calf serum (FCS), antibiotic/antimycotic, MTT (3-[4-C-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) and accompanying materials were purchased from Thermo Fisher Scientific, Inc, (Waltham, MA, USA).
2.5.2. MTT assay
The cells were cultured in RPMI-1640 medium accompanied with 10 % heat-inactivated FCS, 100 μg/ml streptomycin and 100 U/mL penicillin and incubated in a humidified atmosphere at 37 °C and 5% CO2. The cell suspension was adjusted to a final concentration of 3 × 105 cells/mL and 100 μL cell suspension dispensed per well. The plate was incubated at 37 °C for 12 h in 5% CO2 incubator. The stock solutions (10 mM) of the test compound and cisplatin were prepared in DMSO and the test concentrations were prepared by diluting the stocks with the growth media. The test dilutions (100 μL) were added to the wells giving final concentrations of 1, 5, 10, 20 and 40 μM. MTT reagent (20 μL, 5 mg/ml in PBS) was added to each microwell after 48 h, followed by incubation for 5 h. The contents were solubilized with 100 μL 10 % sodium dodecylsulfate in 10 mM HCl. The absorbance was measured after 12 h with the help of BioRad PR 4100 microplate reader at a wavelength of 570 nm. Percentage viability was calculated by using following relation, and dose-response curves were plotted. The results represent mean ± SD of six readings; three determinations of two independent experiments (Ikram et al., 2019, Papa et al., 2008).
Where, As, Ab and Ac are absorbance of sample, blank and control, respectively.
3. Results
3.1. Virtual screening workflow validation using set of known USP2 binders
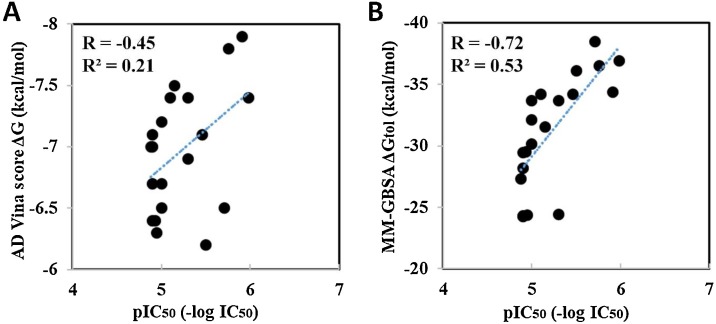
To validate the docking protocol, a total of 12 reported USP2 inhibitors were subjected to an established virtual screening pipeline. The IC50 values of inhibitors against USP2 were converted to pIC50 values (-log of IC50) and plotted against the AD Vina docking scores and MMGBSA total binding free energy scores calculated by Amber-mmgbsa method. The corresponding values are listed in Table S1 and correlation plot between pIC50 values and docking scores is displayed in Fig. 1 . The AD Vina docking scores showed weak correlation with the experimental pIC50 values (R 2 = 0.21), whereas MMGBSA scores showed better correlation (R 2 = 0.53). Here, we speculated that the weak correlation between AD Vina docking scores and experimental pIC50 values might be indorsed by side chains conformational flexibility of binding site residues, which were adjusted during MD simulations in explicit solvent environment. Hence, the MMGBSA total binding free energy showed better correlation with experimental values, as it incorporates all electrostatic and van der Waals (vdW) interactions. Overall, combining AD Vina followed by MMGBSA calculations suggested a reliable approach to select the compounds against USP2 (together with expected false positives and negatives).
Fig. 1.
Correlation between the AutoDock Vina docking score and IC50 for 20 known human-USP2 binders. (A) Plot of AD Vina docking score against pIC50. (B) Plot of MMGBSA value (total binding free energy calculated after 5 ns MD simulations) against pIC50.
3.2. Virtual screening
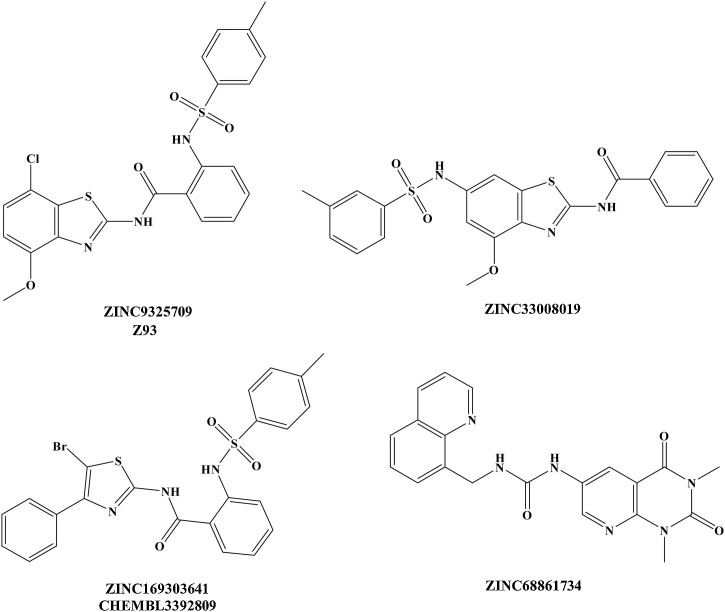
To identify potential inhibitors against UPS2, structure-based virtual screening was employed. A substantial number of compounds were eliminated through several ADMET filters, which include drug-like and toxicity assessment. A total of 52 compounds were selected based on the AD Vina score from a known USP2 binder (CHEMBL3392809, AD Vina score: -7.3 kcal/mol) as cut-off value. Among these hits, each compound was investigated for molecular interactions with signature catalytic triad (Cys276, His557, and Asn574), which further reduced the list to 14 compounds. The final selection of compounds was made after extensive MD simulation analysis of these 14 complexes which included, 1) constant interaction with any of the catalytic triad residues, 2) lowest MMGBSA total binding free energy values and 3) stable conformation of ligand over a period of 20 ns (Figure S1). A total of 3 compounds were finally selected and only one compound (ZINC9325709 denoted as Z93) was purchased for biological evaluations based on the specific scaffold, its availability and price ( Fig. 2 and Table S2). The detailed ADMET properties are tabulated in (Table S3).
Fig. 2.
Chemical structure of selected compounds from in silico screen. CHEMBL3392809, potent USP2 inhibitor is included as reference.
3.3. Biological evaluation
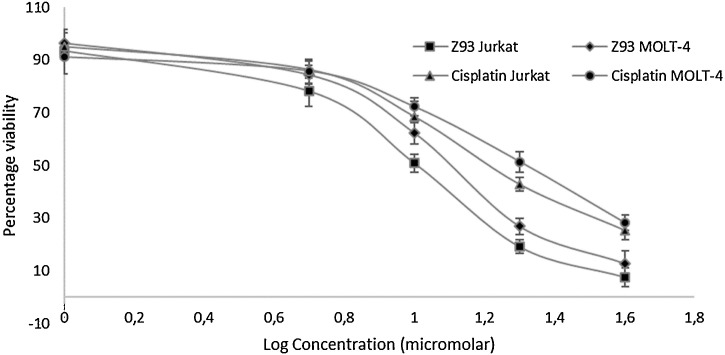
Jurkat and MOLT-4 cell lines were used to examine cytotoxicity of selected USP2 inhibitor (Z93) against T-ALL while cisplatin was used as a control. Both selected cell lines display versatile characteristics to study cytotoxicity of drugs against T-ALL. The dose-response curve is presented in Fig. 3 . The IC50 value of Z93 against Jurkat and MOTL-4 cells were 9.67 and 11.8 μM, respectively, after 48 h of exposure. Under same conditions cisplatin depicted 16.88 and 20.01 μM IC50 values against Jurkat and MOLT-4, respectively.
Fig. 3.
Dose-response curve for Z93 and cisplatin against Jurkat and MOLT-4 cell lines.
3.4. Conserved active site insights of USP2 and SARS-CoV-2 PLpro
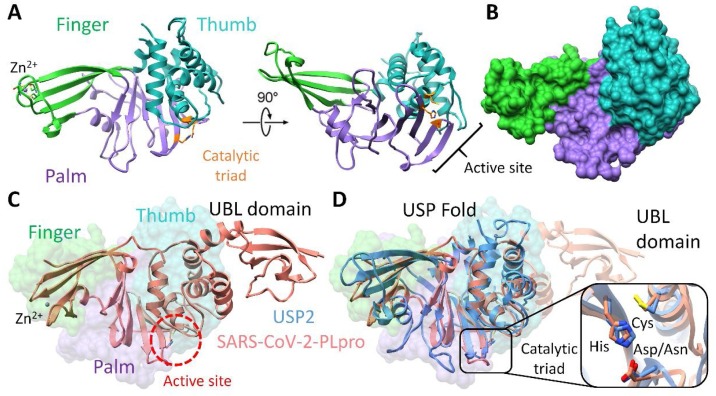
Recent studies have determined the crystal structure of MERS, SARS-CoV and CoV-2 PLpro, which corroborated the similar palm-thumb-fingers structural scaffold and conserved catalytic triad Cys-His-Asn/Asp (Báez-Santos et al., 2015, Chou et al., 2014, Lei et al., 2014). This arrangement is similar to USPs, albeit the low sequence identities (Hilgenfeld, 2014, Mielech et al., 2014). The structural relationship between the CoV PLpros and the catalytic core domain of the USPs have also been identified in previously studies (Lindner et al., 2005, Sulea et al., 2006, Sulea et al., 2005). Briefly, the larger catalytic domain of SARS-CoV-2 holds three characteristic domains. Similar to USP2, the palm subdomain is consisted of 6 β-strands which contains a canonical cysteine protease conserved catalytic triad (Cys111, His272, and Asp286) present at the interface of thumb and palm subdomains. Moreover, the conformationally identical catalytic site formed by two β-hairpins of palm domain, first coordinated by β3 and β4 and second coordinated by β5 and β6 showed an RMSD value 0.8 Å and 0.9 Å with USP2, respectively. An additional identical feature of conserve active site is evident from an important β-turn/loop formed by β3 and β4 that closes upon inhibitor binding (Osipiuk et al., 2020). In addition to conserved catalytic Cys111, the zinc binding motif in thumb subdomain is coordinated by four conserved cysteine residues (Cys189, 192, 224, and 226 of SARS-CoV-2 PLpro), located on two loops of two β-hairpins identical to USP2 (Cys425, 428, 477, and 479). The zinc binding has a prime importance in structure integrity and protease activity (Barretto et al., 2005, Ratia et al., 2006). After superimposition, the corresponding domain architecture of both PLpro displayed strikingly similar representation of conserved USP fold and orientation of catalytic triad residues (Fig. 4 A-D).
Fig. 4.
Molecular modelling of human USP2 and SARS-CoV-2 papain-like protease (PLpro). (A) Overall structure of the USP12 comprises of finger (green), palm (purple) and thumb (sea green) domains. The catalytic centre (Cys box) is shown in orange between the palm and thumb domains. (B) Surface representation of the structure of the human USP2. (C) The SARS-CoV-2 PLpro monomer (PDB: 6W9C) consists of four domains: beginning from N- to the C-terminus, the extended UBL, the thumb, palm and fingers domain. The active site is circled red. (D) An overlay of SARS-CoV-2 PLpro (salmon) with USP2 (cornflower blue) displaying the conserved USP fold. The catalytic triad residues are magnified.
3.5. Binding mode of Z93 with USP2 and SARS-CoV-2 PLpro
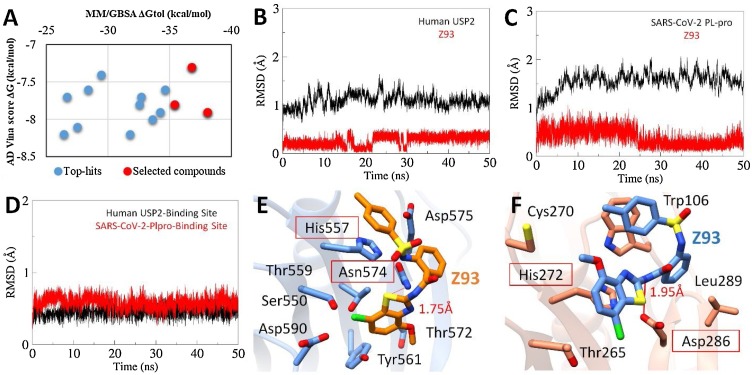
The structural findings led us to speculate that identified Z93, a potential USP2 inhibitor might be able to inhibit the SARS-CoV-2 PLpro because both proteases share a similar catalytic triad. Overall, the integrated virtual screening identified 14 hits which were reduced to 3 after analysing the binding free energies (Fig. 5 A).
Fig. 5.
(A) The energy plot of top hits between corresponding AD Vina docking score and MMGBSA total binding free energy calculated after a production run of 20 ns MD simulations). The selected compounds are highlighted red. Root-mean-square-deviation of USP2 (B) and SARS-CoV-2 PLpro (C) backbone are estimated over a period of 20 ns with bound Z93 ligand (red). (D) RMSD plot of binding site residues of corresponding proteases. The MD simulated binding modes of Z93 (sticks representation) inside the active site of USP2 (E) and SARS-CoV-2 PLpro (F).
To gain further insight into the binding mode, we performed molecular docking followed by MD simulations against both PLpro and most favourable binding conformation of active compounds, Z93 was obtained from the largest cluster after 20 ns production run through clustering analysis. The RMSD plot indicated the stable backbone conformation of binding site residues with docked Z93 (Fig. 5B). Overall, both complexes fluctuated in the start and remained stable for last 20 ns indicated that Z93 adopted more favourable conformation (Fig. 5 C and D). Despite the similar docked sites on both PLpro, the binding poses were found different. More insights revealed the consensus binding interactions of Z93 with the catalytic triad in both PLpro. In UPS2, the major interaction was established due to stable stacking interaction between terminal benzene of Z93 and His557 of catalytic triad. Moreover, the benzothiazole moiety of Z93, apart from establishing a H-bond with the side chain nitrogen atom of Asn574 also formed extensive hydrophobic interactions with Thr572, Thr559, Ser550 and Tyr561 ( Fig. 5E). In relation to SARS-CoV-2 PLpro, the Z93 also found interacted with the catalytic triad where the benzothiazole moiety was positioned to make direct stacking interaction with His272, and terminal benzene was positioned to form stacking interaction with Trp106 (Fig. 5F). Additionally, the nitrogen atom next to benzothiazole established H-bond with the sidechain oxygen atom of conserved Asp286 of catalytic triad. The predicted binding site was close to that found for SARS-CoV PLpro (Chou et al., 2008).
3.6. MM/PB(GB)SA free binding energy calculations at both sites
The binding free energy calculations by MM/GBSA and MM/PBSA are both end-point methods, which represent more physically meaningful depiction than docking scoring functions. These methods have been widely used in the discovery of potential antiviral small molecules (Chen et al., 2016, Hou et al., 2011a, Sirin et al., 2014, Srivastava and Sastry, 2012). The absolute energy of binding (ΔGbind) of Z93 was estimated at the catalytic site of USP2 and SARS-CoV-2 PLpro. Moreover, ΔGbind of Z93 was also determined at site 2 located in close proximity of catalytic triad, together with co-crystallized SARS-CoV-2 PLpro inhibitor (Compound 3) as a positive control of site 2 (Osipiuk et al., 2020). The values were predicted through mechanics/Poisson–Boltzmann (generalized born) surface area (MM/PB(GB)SA) method from 100 snapshots extracted from the complete 50 ns trajectory. During this energy calculations, we also incorporated a more computationally expensive entropic contributions (-TΔS) in ligand binding, which is reported to give improved accuracy (Hou et al., 2011b) and play essential role in protein-ligand interactions (Sun et al., 2018). The overall energy contributions are tabulated in Table 1 .
Table 1.
Binding free energy calculations by MM-PB(GB)SA method.
| Energy component | Catalytic Site |
Site 2 |
||
|---|---|---|---|---|
| Human-UPS2/Z93 (kcal/mol) | SARS-CoV-2 PLpro/Z93 (kcal/mol) | Z93 (kcal/mol) | Compound 3a (kcal/mol) | |
| MM (Gas term) | ||||
| ΔEvdw | −36.4 | −39.45 | −43.41 | −41.33 |
| ΔEele | −22.36 | −19.45 | −13.71 | −12.55 |
| ΔEMM | −58.76 | −58.9 | −57.12 | −53.88 |
| (-)TΔS | 24.4 | 25.9 | 22.2 | 21.09 |
| PBSA (solvation term) | ||||
| ΔGp(PBSA) | 33.4 | 34.14 | 33.04 | 32.45 |
| ΔGnp(PBSA) | −7.91 | −7.8 | −6.23 | −7.22 |
| ΔGsol(PBSA) | 25.49 | 26.34 | 26.81 | 25.23 |
| GBSA (solvation term) | ||||
| ΔGp(GBSA) | 30.25 | 31.87 | 31.14 | 29.42 |
| ΔGnp(GBSA) | −7.81 | −8.02 | −6.74 | −6.48 |
| ΔGsol(GBSA) | 22.44 | 23.85 | 24.4 | 22.94 |
| Binding free energy | ||||
| ΔGbind(MM/PBSA) | −8.87 | −6.66 | −8.11 | −7.56 |
| ΔGbind(MM/GBSA) | −11.92 | −9.15 | −10.52 | −9.85 |
Compound 3 is reported as potent inhibitor co-crystalized with SARS-CoV-2 PLpro (PDB ID: 7JIW) (Osipiuk et al., 2020).
At catalytic site, the findings from MM/PB(GB)SA calculations displayed van der Waals (ΔEvdW) interactions majorly contributed in USP2/Z93 and SARS-CoV-2 PLpro/Z93 complex stabilization with a value of -36.4 kcal/mol and -39.45 kcal/mol respectively. These favorable ΔEvdW was evident from strong stacking interactions with relatively less electrostatic contributions, which were predicted to be -22.36 kcal/mol and -18.45 kcal/mol in USP2/Z93 and SARS-CoV-2 PLpro/Z93 complex respectively. Together with the solvation effect in Z93/USP2 complex (ΔGsol(PBSA) = 25.49; ΔGsol(GBSA) =22.44 kcal/mol), and Z93/SARS-CoV-2 PLpro (ΔGsol(PBSA) = 26.34; ΔGsol(GBSA) =23.85 kcal/mol) and incorporation of entropic terms, the absolute ΔGbind was account for -8.87 (Z93 complexed with USP2) and -6.66 (Z93 complexed with SARS-CoV-2) as determined through MM-PBSA (ΔGbind(MM/PBSA)) approach and values of 11.92 and -9.15 kcal/mol, respectively as determined from the MM-GBSA (ΔGbind(MM/PBSA)) approach.
At site 2, a similar trend was examined, where Z93 mainly interacted through van der Waals interactions and these findings were in agreement with the co-crystalized compound 3 (Osipiuk et al., 2020). With the solvation and entropic contributions, the overall obtained ΔGbind of Z93 at site 2 revealed a slightly stronger value by MM/PBSA (-8.11 kcal/mol) and MM/GBSA (-10.52 kcal/mol) methods and compared to co-crystalized compound 3 (Table 1).
4. Discussion
Ubiquitin carboxyl-terminal hydrolase 2 (USP2) is a highly conserved protein present in most eukaryotes. It is found to be involved in intracellular proteasomal degradation, cell-cycle regulation, and stress response. The upregulation of USP2 results in the elevated concentration levels of deubiquitinated substrates, for example, cyclin D1, MDM2, fatty acid synthase, and Aurora-A (Chuang et al., 2018). The overexpression of USP2 has been found involved in progression and metastasis of several cancers, e.g., triple negative breast cancer, colorectal cancer and non-Hodgkin's lymphoma (Qu et al., 2015). Most of the proteasome inhibitors usually pose extensive off-target effects and an alternative approach of USP2 inhibition can selectively target proteasomal degradation of its multiple oncogenic substrates with least effect on non-cancerous cells (Halford, 2012, Davis et al., 2016).
The inhibition of USP2 antagonizes ubiquitin-dependent degradation of cyclin D1 resulting in its stabilization. The cells involving cyclin D1 independent cell-cycle progression, for example, normal human fibroblasts or cancer cells that don’t show cyclin D1 expression are not affected by the inactivation of USP2. These evidences suggest that inhibition of USP2 in cancer cells which express cyclin D1 for their growth could induce growth suppression (Shan et al., 2009). MOLT-4 and jurkat cells were selected in this study to examine the effect of proposed USP2 inhibitor (Z93). These cell lines make use of cyclin D1 during progression and could serve as a potential model to study USP2 inhibition. The low micromolar IC50 values of Z93 against both the cell lines suggest its promising selectivity and warrants its further investigations.
The proposed USP2 inhibitor identified in this study (Z93) has close structural similarity with an already reported potent USP2 inhibitor ML364, with both sharing 2-((4-methylphenyl)sulfonamido)benzamide residue. ML364 induces cyclin D1 degradation resulting in cell-cycle arrest, indicating key role in progression of non-Hodgkin's lymphoma and colorectal cancer (Davis et al., 2016). Cyclin D1 is necessarily involved in cell-cycle progression from G1 to S phase and its overexpression is evident in many human cancers (Fernandes et al., 2018). It is found to be upregulated in most of the ALL patient while its expression is not affected in AML patients as compared to the normal controls (Aref et al., 2006). Reduced survival rates have been observed in ALL patients with higher expression of cyclin D1, suggesting its involvement in the progression of aggressive forms of ALL (Fernandes et al., 2018). These evidences clearly suggest that the inhibition of USP2 could serve as a promising strategy in treating cancers with cyclin D1 expression.
6-Mercaptopurine (6 M P) and 6-thioguanine (6 TG) has been reported effective against the treatment of various cancers including leukemia. These both compounds are hypoxanthine-guanine phosphoribosyltransferase inhibitors and possess purine residue (Pieters et al., 1992). Another potential effect of these compounds have been reported against MERS-CoV and SARS-CoV PLpro (Cheng et al., 2015). Several other purine derivatives have also been reported as antiviral compounds; these include famciclovir, acyclovir, valacyclovir, ganciclovir, and penciclovir. These drugs convert to their triphosphate analogues in the body and inhibit viral DNA polymerase. These drugs have been reported effective against hepatitis B, herpes simplex, varicella zoster, and cytomegalovirus (Thomasy and Maggs, 2016, Seley-Radtke and Yates, 2018). It was evident from our virtual screening outcomes that three out of four top hits against PLpro possessed thiazole moiety and two of the top hits possessed benzo[d]thiazole moiety which closely resembles 6 M P and 6 TG. These results further strengthened our hypothesis that 6 M P and 6 TG type of USP2 inhibitors can also inhibit viral PLpro.
The benzothiazole moiety of Z93 resembles 6 M P and 6 TG purine, having [4.3.0] bicyclic system. In another study, indole based 3-chymotrypsin-like protease (3CLpro) inhibitor of SARS-CoV have been reported with low micromolar range (Turlington et al., 2013). These analogies strongly potentiate the hypothesis that Z93 can inhibit SARS-CoV-2 by antagonizing its PLpro. The modelling studies indicated the consensus molecular interactions governed by Z93 with both proteases. There was no major structural deviation observed over a period of 20 ns in both docked complexes. With both proteases, Z93 interacted with the His and Asp/Asn of catalytic triad in a similar way, albeit with different binding mode and revealed favourable total binding free energies. The Z93 bound conformation was found more stable with SARS-CoV-2 PLpro due to an additional staking interaction with Trp106, therefore showed more favourable vdW energy contributions, as compared to Z93/USP2. Conclusively, based on the in vitro and repurposed modelling analysis, it is entirely possible to develop selective, non-covalent inhibitors against SARS-CoV-2 PLpro by using structure-based approach against USPs involved in disease processes. At site 2, Asp165 is reported as highly conserved among USP family of DUBs (Quesada et al., 2004) and among most coronaviral PLpros (Barretto et al., 2005, Ratia et al., 2006). Several studies of USPs reported that this aspartate establish H-bonds with ubiquitin molecules, thus considered to be vital for ligand stabilization (Hu et al., 2002, Hu et al., 2005, Renatus et al., 2006). The combination of molecular docking and MM/PB(GB)SA has proven a promising approach in in defining the correct binding poses (Hou et al., 2011a, Sirin et al., 2014, Rastelli et al., 2010) and ranking of virtual hits based on binding affinities (Thompson et al., 2008, Zhang et al., 2014, Greenidge et al., 2014). Further evidence from binding free energy calculations by MM/PB(GB)SA method with the incorporation of entropy effect expounded the binding potential and contributions of major energy components participated by Z93. Apart from molecular mechanics force field, entropic contributions also play an essential role in protein-ligand interactions (Sun et al., 2018).
Although, our study lacks the binding assay, the significance of this study basically lies in the concept to facilitate the efforts towards COVID-19 treatment via bypassing the extensive viral enzymatic inhibition assays, which have been reported in previous studies. This concept will also provide a comprehensive starting point with the availability of already available potent USP2 inhibitors.
Authors contributions
M.U.M and M.F conceived and designed the experiments. M.U.M. performed the in-silico studies. M.U.M. S.A. and I.A performed experiments. M.F supervised the study. M.U.M. wrote the main manuscript text. S.A, I.A and M.F critically reviewed the manuscript. All the authors approved the final manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The authors are grateful to Rega Institute for Medical Research and Department of Chemistry, Malaya University (Grant Number: IIRG003A-2019) for providing necessary facilities to accomplish this study.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2020.107376.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Ahn D.-G. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30(3):313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anusuya S., Gromiha M.M. Quercetin derivatives as non-nucleoside inhibitors for dengue polymerase: molecular docking, molecular dynamics simulation, and binding free energy calculation. J. Biomol. Struct. Dyn. 2017;35(13):2895–2909. doi: 10.1080/07391102.2016.1234416. [DOI] [PubMed] [Google Scholar]
- Aref S. Cyclin D1 expression in acute leukemia. Hematology. 2006;11(1):31–34. doi: 10.1080/10245330500322321. [DOI] [PubMed] [Google Scholar]
- Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat S., Martin A.J., Soliman M.E. An integrated molecular dynamics, principal component analysis and residue interaction network approach reveals the impact of M184V mutation on HIV reverse transcriptase resistance to lamivudine. Mol. Biosyst. 2014;10(8):2215–2228. doi: 10.1039/c4mb00253a. [DOI] [PubMed] [Google Scholar]
- Caly L. The FDA-approved Drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D.A. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- Chen X., Chou C.-Y., Chang G.-G. Thiopurine analogue inhibitors of severe acute respiratory syndrome-coronavirus papain-like protease, a deubiquitinating and deISGylating enzyme. Antivir. Chem. Chemother. 2009;19(4):151–156. doi: 10.1177/095632020901900402. [DOI] [PubMed] [Google Scholar]
- Chen F. Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein–protein binding free energies and re-rank binding poses generated by protein–protein docking. J. Chem. Soc. Faraday Trans. 2016;18(32):22129–22139. doi: 10.1039/c6cp03670h. [DOI] [PubMed] [Google Scholar]
- Cheng K.-W. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.-Y. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75(8):1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.Y. Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease. Acta Crystallogr. D Biol. Crystallogr. 2014;70(2):572–581. doi: 10.1107/S1399004713031040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang S.-J. 6-Thioguanine is a noncompetitive and slow binding inhibitor of human deubiquitinating protease USP2. Sci. Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-21476-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.I. Small molecule inhibition of the ubiquitin-specific protease USP2 accelerates cyclin D1 degradation and leads to cell cycle arrest in colorectal cancer and mantle cell lymphoma models. J. Biol. Chem. 2016;291(47):24628–24640. doi: 10.1074/jbc.M116.738567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Cross T.A., Zhou H.-X. Recent progress in structure-based anti-influenza drug design. Drug Discov. Today. 2012;17(19-20):1111–1120. doi: 10.1016/j.drudis.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H., Geisbert T.W. Ebola haemorrhagic fever. Lancet. 2011;377(9768):849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J.C. Increased levels of cyclin D1 negatively impacts on acute lymphoblastic leukemia overall survival. Appl. Cancer Res. 2018;38(1):7. [Google Scholar]
- Ghosh A.K. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein− ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53(13):4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenidge P.A. Improving docking results via reranking of ensembles of ligand poses in multiple X-ray protein conformations with MM-GBSA. J. Chem. Inf. Model. 2014;54(10):2697–2717. doi: 10.1021/ci5003735. [DOI] [PubMed] [Google Scholar]
- Guan S. Exploration of binding and inhibition mechanism of a small molecule inhibitor of influenza virus H1N1 hemagglutinin by molecular dynamics simulation. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-03719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford B. From discovery to drug. Chem. Eng. News. 2012;90(35):34–35. [Google Scholar]
- Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T., Yu R. Molecular dynamics and free energy studies on the wild-type and double mutant HIV-1 protease complexed with amprenavir and two amprenavir-related inhibitors: mechanism for binding and drug resistance. J. Med. Chem. 2007;50(6):1177–1188. doi: 10.1021/jm0609162. [DOI] [PubMed] [Google Scholar]
- Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M. Structure and mechanisms of the proteasome‐associated deubiquitinating enzyme USP14. EMBO J. 2005;24(21):3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram N. Inhibition of oncogenic kinases: an in vitro validated computational approach identified potential multi-target anticancer compounds. Biomolecules. 2019;9(4) doi: 10.3390/biom9040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss R., Sandor M., Szalai F.A. http://Mcule. com: a public web service for drug discovery. J. Cheminform. 2012;4(S1):P17. [Google Scholar]
- Leela S.L. Drug repurposing of minocycline against dengue virus infection. Biochem. Biophys. Res. Commun. 2016;478(1):410–416. doi: 10.1016/j.bbrc.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Lei J. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res. 2014;109:72–82. doi: 10.1016/j.antiviral.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79(24):15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. ACS Publications; 2020. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhkov V. Evaluation of adamantane derivatives as inhibitors of dengue virus mRNA cap methyltransferase by docking and molecular dynamics simulations. Mol. Inform. 2013;32(2):155–164. doi: 10.1002/minf.201200107. [DOI] [PubMed] [Google Scholar]
- Madrid P.B. A systematic screen of FDA-approved drugs for inhibitors of biological threat agents. PLoS One. 2013;8(4):e60579. doi: 10.1371/journal.pone.0060579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech A.M. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res. 2014;194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M.U., Ikram N. Integrated computational approach for virtual hit identification against ebola viral proteins VP35 and VP40. Int. J. Mol. Sci. 2016;17(11) doi: 10.3390/ijms17111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M.U. Towards peptide vaccines against Zika virus: immunoinformatics combined with molecular dynamics simulations to predict antigenic epitopes of Zika viral proteins. Sci. Rep. 2016;6:37313. doi: 10.1038/srep37313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M.U. In silico structural elucidation of RNA-dependent RNA polymerase towards the identification of potential crimean-congo hemorrhagic fever virus inhibitors. Sci. Rep. 2019;9(1):1–18. doi: 10.1038/s41598-019-43129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottin M. Molecular dynamics simulations of Zika virus NS3 helicase: insights into RNA binding site activity. Biochem. Biophys. Res. Commun. 2017 doi: 10.1016/j.bbrc.2017.03.070. [DOI] [PubMed] [Google Scholar]
- Nitsche C. Dengue and Zika: Control and Antiviral Treatment Strategies. Springer; 2018. Strategies towards protease inhibitors for emerging flaviviruses; pp. 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipiuk J. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. bioRxiv. 2020 doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa V. Proapoptotic activity and chemosensitizing effect of the novel Akt inhibitor perifosine in acute myelogenous leukemia cells. Leukemia. 2008;22(1):147–160. doi: 10.1038/sj.leu.2404980. [DOI] [PubMed] [Google Scholar]
- Parida P. Potential of plant alkaloids as dengue ns3 protease inhibitors: molecular docking and simulation approach. Bangladesh J. Pharmacol. 2014;9(3):262–267. [Google Scholar]
- Pattnaik A. Discovery of a non-nucleoside RNA polymerase inhibitor for blocking Zika virus replication through in silico screening. Antiviral Res. 2018;151:78–86. doi: 10.1016/j.antiviral.2017.12.016. [DOI] [PubMed] [Google Scholar]
- Pieters R. Hypoxanthine‐guanine phosphoribosyl‐transferase in childhood leukemia: relation with immunophenotype, in vitro drug resistance and clinical prognosis. Int. J. Cancer. 1992;51(2):213–217. doi: 10.1002/ijc.2910510208. [DOI] [PubMed] [Google Scholar]
- Qu Q. USP2 promotes cell migration and invasion in triple negative breast cancer cell lines. Tumor Biol. 2015;36(7):5415–5423. doi: 10.1007/s13277-015-3207-7. [DOI] [PubMed] [Google Scholar]
- Quesada V. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004;314(1):54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- Ramajayam R. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010;20(12):3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli G. Fast and accurate predictions of binding free energies using MM‐PBSA and MM‐GBSA. J. Comput. Chem. 2010;31(4):797–810. doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- Ratia K. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. 2008;105(42):16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renatus M. Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure. 2006;14(8):1293–1302. doi: 10.1016/j.str.2006.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum L. Facing Covid-19 in Italy—ethics, logistics, and therapeutics on the epidemic’s front line. N. Engl. J. Med. 2020;382(20):1873–1875. doi: 10.1056/NEJMp2005492. [DOI] [PubMed] [Google Scholar]
- Sanders J.M. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. Jama. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Seley-Radtke K.L., Yates M.K. The evolution of nucleoside analogue antivirals: a review for chemists and non-chemists. Part 1: early structural modifications to the nucleoside scaffold. Antiviral Res. 2018;154:66–86. doi: 10.1016/j.antiviral.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J., Zhao W., Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell. 2009;36(3):469–476. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020:1–10. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff A.C. Therapeutics for filovirus infection: traditional approaches and progress towards in silico drug design. Expert Opin. Drug Discov. 2012;7(10):935–954. doi: 10.1517/17460441.2012.714364. [DOI] [PubMed] [Google Scholar]
- Sirin S. A computational approach to enzyme design: predicting ω-aminotransferase catalytic activity using docking and MM-GBSA scoring. J. Chem. Inf. Model. 2014;54(8):2334–2346. doi: 10.1021/ci5002185. [DOI] [PubMed] [Google Scholar]
- Speelman B., Brooks B.R., Post C.B. Molecular dynamics simulations of human rhinovirus and an antiviral compound. Biophys. J. 2001;80(1):121–129. doi: 10.1016/S0006-3495(01)75999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava H.K., Sastry G.N. Molecular dynamics investigation on a series of HIV protease inhibitors: assessing the performance of MM-PBSA and MM-GBSA approaches. J. Chem. Inf. Model. 2012;52(11):3088–3098. doi: 10.1021/ci300385h. [DOI] [PubMed] [Google Scholar]
- Su M. Comparative assessment of scoring functions: the CASF-2016 update. J. Chem. Inf. Model. 2018;59(2):895–913. doi: 10.1021/acs.jcim.8b00545. [DOI] [PubMed] [Google Scholar]
- Sulea T. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 2005;79(7):4550–4551. doi: 10.1128/JVI.79.7.4550-4551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulea T. Binding site‐based classification of coronaviral papain‐like proteases. Proteins Struct. Funct. Bioinform. 2006;62(3):760–775. doi: 10.1002/prot.20802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. J. Chem. Soc. Faraday Trans. 2018;20(21):14450–14460. doi: 10.1039/c7cp07623a. [DOI] [PubMed] [Google Scholar]
- Tahir ul Qamar M. Discovery of novel dengue NS2B/NS3 protease inhibitors using pharmacophore modeling and molecular docking based virtual screening of the zinc database. Int. J. Pharmacol. 2016;12:621–632. [Google Scholar]
- Tahir ul Qamar M. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasy S.M., Maggs D.J. A review of antiviral drugs and other compounds with activity against feline herpesvirus type 1. Vet. Ophthalmol. 2016;19:119–130. doi: 10.1111/vop.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.C., Humblet C., Joseph-McCarthy D. Investigation of MM-PBSA rescoring of docking poses. J. Chem. Inf. Model. 2008;48(5):1081–1091. doi: 10.1021/ci700470c. [DOI] [PubMed] [Google Scholar]
- Tu J. Exploring the binding mechanism of Heteroaryldihydropyrimidines and Hepatitis B Virus capsid combined 3D-QSAR and molecular dynamics. Antiviral Res. 2017;137:151–164. doi: 10.1016/j.antiviral.2016.11.026. [DOI] [PubMed] [Google Scholar]
- Turlington M. Discovery of N-(benzo [1, 2, 3] triazol-1-yl)-N-(benzyl) acetamido) phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23(22):6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.Y. A small-molecule dengue virus entry inhibitor. Antimicrob. Agents Chemother. 2009;53(5):1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Discovery of unsymmetrical aromatic disulfides as novel inhibitors of SARS-CoV main protease: chemical synthesis, biological evaluation, molecular docking and 3D-QSAR study. Eur. J. Med. Chem. 2017;137:450–461. doi: 10.1016/j.ejmech.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-s. A review of the 2019 Novel Coronavirus (COVID-19) based on current evidence. Int. J. Antimicrob. Agents. 2020:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020:1–3. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv. 2020 [Google Scholar]
- Young M.-J. The role of ubiquitin-specific peptidases in cancer progression. J. Biomed. Sci. 2019;26(1):42. doi: 10.1186/s12929-019-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wong S.E., Lightstone F.C. ACS Publications; 2014. Toward Fully Automated High Performance Computing Drug Discovery: a Massively Parallel Virtual Screening Pipeline for Docking and Molecular mechanics/generalized Born Surface Area Rescoring to Improve Enrichment. [DOI] [PubMed] [Google Scholar]
- Zhang C. Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design. Biochem. Biophys. Res. Commun. 2017;492(4):624–630. doi: 10.1016/j.bbrc.2016.11.098. [DOI] [PubMed] [Google Scholar]
- Zhou Z. Antiviral compounds discovered by virtual screening of small− molecule libraries against dengue virus e protein. ACS Chem. Biol. 2008;3(12):765–775. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.