Abstract
McrBC is a conserved modification-dependent restriction system that in Escherichia coli specifically targets foreign DNA containing methylated cytosines. Crystallographic data show that the N-terminal domain of Escherichia coli McrB binds substrates via a base flipping mechanism. This region is poorly conserved among the plethora of McrB homologs, suggesting that other species may use alternative binding strategies and/or recognize different targets. Here we present the crystal structure of the N-terminal domain from Stayphlothermus marinus McrB (Sm3–180) at 1.92 Å, which adopts a PUA-like EVE fold that is closely related to the YTH and ASCH RNA binding domains. Unlike most PUA-like domains, Sm3–180 binds DNA and can associate with different modified substrates. We find the canonical ‘aromatic cage’ binding pocket that confers specificity for methylated bases in other EVE/YTH domains is degenerate and occluded in Sm3–180, which may contribute to its promiscuity in target recognition. Further structural comparison between different PUA-like domains identifies motifs and conformational variations that correlate with the preference for binding either DNA or RNA. Together these data have important implications for PUA-like domain specificity and suggest a broader biological versatility for the McrBC family than previously described.
Keywords: restriction nuclease, McrB, PUA domain, aromatic cage, RNA binding, DNA binding
Introduction
Restriction modification systems (RMS) are conserved defense systems that protect bacteria against viral bacteriophage (phage) infection (Labrie et al., 2010). RMS contain a site-specific DNA binding module, endonuclease core, and associated methyltransferase to protect the host genome (Tock and Dryden, 2005). As phages incorporated modifications into their genomes to evade RMS, bacteria in turn evolved modification-dependent restriction systems (MDRS) – that specifically target and cleave methylated and/or glucosylated DNA – to restore the balance in the ongoing arms race for survival (Loenen and Raleigh, 2014). These systems define the epigenetic landscape of bacterial populations (Ishikawa et al., 2010) and complement CRISPR systems as essential barriers to foreign invaders (Dupuis et al., 2013, Bernheim and Sorek, 2020).
McrBC is a highly conserved, two-component MDRS consisting of the McrB and McrC proteins. Escherichia coli (Ec) McrB contains an N-terminal DNA binding domain that recognizes 4-methyl-, 5-methyl-, or 5-hydroxymethylcytosines (Raleigh and Wilson, 1986, Sutherland et al., 1992, Krüger et al., 1995, Gast et al., 1997) and a C-terminal AAA+ domain that hydrolyzes GTP and facilitates nucleotide-dependent oligomerization into hexamers (Panne et al., 2001, Nirwan et al., 2019a, Nirwan et al., 2019b). EcMcrC contains a C-terminal PD-(D/E)XK nuclease domain but cannot bind DNA on its own (Pieper and Pingoud, 2002). To exert its function, EcMcrC associates with the EcMcrB oligomer (Pieper and Pingoud, 2002) and inserts a stalk domain into the central pore of the hexameric ring (Nirwan et al., 2019b). This interaction stimulates the GTP hydrolysis and subsequent DNA translocation in vitro (Pieper et al., 1999, Panne et al., 1999). Collision of two McrBC complexes is thought to trigger DNA cleavage on both strands near the modified sites (Steward et al., 2000; Pieper et al., 2002). These mechanochemical properties are reminiscent of type I and type III RMS, which bind DNA at non-modified sites separated by up to thousands of base pairs and use ATP hydrolysis to power long-range translocation (type I) or sliding/diffusion (type III) events through which cleavage occurs either by collision or stalling (Dryden et al., 2001; Gupta et al. 2015; Schwarz et al., 2013; Ahmad et al., 2018). It remains to be seen whether these structural and mechanistic features are conserved in other organisms beyond E. coli.
Crystallographic studies have shown that EcMcrB binds modified DNA via a base-flipping mechanism (Sukackaite et al., 2012, Zagorskaitė et al, 2018). The N-terminal domain of EcMcrB, however, is poorly conserved among the wide array of McrBC homologs, suggesting that other species may use different mechanisms for substrate binding and/or may preferentially target other sequences and modifications (Hosford et al., 2020). This remains a largely unexplored area of study. Here we present the crystal structure of the N-terminal domain from Staphylothermus marinus McrB (Sm3–180) at 1.92 Å, which adopts a PseudoUridine synthase and Archaeosine transglycosylase (PUA)-like EVE domain fold that is prevalent among prokaryotic RNA binding proteins and shares homology with eukaryotic YTH/ASCH family proteins. Sm3–180, however, binds DNA and can associate with different modified substrates. We find the canonical ‘aromatic cage’ binding pocket responsible for specifically recognizing modified bases in other PUA-like domains is degenerate and occluded in Sm3–180, which may contribute to its promiscuity in target recognition. Further structural comparison identifies other motifs and conformational variations in different PUA-like domains that correlate with individual preferences for binding RNA or DNA. Together these data have important implications for PUA-like domain specificity and suggest new mechanistic possibilities for McrBC enzymes, underscoring the modular nature of these nuclease complexes.
Materials and methods
Cloning, expression and purification of SmMcrB 3–180 constructs
DNA encoding the Staphylothermus marinus F1 McrB protein (DOE IMG/M ID 640109242; Chen et al., 2017) was codon optimized for E. coli expression and synthesized commercially by Integrated DNA Technologies (IDT), Inc. DNA encoding the N-terminal domain (Sm3–180) was amplified by PCR and cloned into pCAV4, a modified T7 expression vector that introduced an N-terminal 6xHis-NusA tag followed by a Hrv3C protease site upstream of the inserted sequence. Selenomethionine-labeled (SeMet) Sm3–180 was transformed into BL21(DE3) cells, grown at 37°C in minimal media, and expressed in the absence of auxotrophs as described previously (Van Duyne et al., 1993). Native Sm3–180 was transformed into BL21(DE3) cells, grown at 37°C in Terrific Broth to an OD600 of 1.0, and then induced with 0.3 mM IPTG overnight at 19°C. All cells were harvested, washed with nickel load buffer (20 mM HEPES pH 7.5, 500 mM NaCl, 30 mM imidazole, 5% glycerol (v:v), and 5 mM β-mercaptoethanol), and pelleted a second time. Pellets were typically flash frozen in liquid nitrogen and stored at −80°C.Thawed pellets from 500 ml cultures were resuspended in 30 ml of nickel load buffer supplemented with 10 mM PMSF, 5 mg DNAse I (Roche), 1 mM MgCl2, and a complete protease inhibitor cocktail tablet (Roche). Lysozyme was added to 1 mg/ml and the mixture was incubated for 15 minutes rocking at 4°C. Cells were disrupted by sonication and the lysate was cleared of debris by centrifugation at 13 000 rpm (19 685 g) for 30 minutes at 4°C. For native and SeMet Sm3–180, the supernatant was filtered, loaded onto a 5 ml HiTrap chelating column charged with NiSO4 and then washed with nickel load buffer. Sm3–180 was eluted with an imidazole gradient from 30 mM to 1 M. Hrv3C protease was added to pooled fractions and dialyzed overnight at 4°C into SP loading buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA, 5% glycerol (v:v), and 5 mM DTT). The sample was applied to a 5 ml HiTrap SP HP column equilibrated with SP loading buffer and then washed with SP loading buffer. Sm3–180 was eluted with a NaCl gradient from 50 mM to 1 M. Pooled fractions were subjected to a 30 kDa Millipore centrifugal concentrator, flow through collected, and concentrated on a 10 kDa centrifugal concentrator. The concentrated protein was further purified by size exclusion chromatography (SEC) using a Superdex 75 10/30 pg column and was exchanged into a final buffer of 20 mM HEPES pH 7.5, 150 mM KCl, 5 mM MgCl2, and 1 mM DTT (5 mM for SeMet labelled) during SEC and concentrated to 5–40 mg/ml. Protein concentration was determined by SDS-PAGE with BSA standards
Cloning, expression, and purification of HsYTHDC1
DNA encoding the human (Homo sapiens, Hs) YTHDC1 YTH domain (residues 344–509) was codon optimized for E. coli expression and synthesized commercially by Integrated DNA Technologies (IDT). Synthesized DNA was cloned into pET15bP, a modified pET15b (Novagen) plasmid in which an Hrv3C protease site (LEVLFQGP) replaces the thrombin site after the N-terminal 6xHis tag. HsYTHDC1 344–509 was transformed into BL21(DE3) cells and expressed in the same manner as Sm3–180. Cells were harvested, washed with nickel load buffer, pelleted, and flash frozen in liquid nitrogen for storage at −80°C. His-tagged HsYTHDC1 344–509 was purified from 500-mL cultures were thawed and resuspended in 30-ml of nickel load buffer supplemented with 10 mM PMSF, 5 mg of DNAse, 5 mM MgCl2, and a complete protease inhibitor cocktail tablet. Lysozyme was added to 1 mg/ml and the cells were sonicated and pelleted as described above. The clarified supernatant was filtered, loaded onto a 5-ml HiTrap chelating column charged with NiSO4, washed with nickel load buffer, and eluted with an imidazole gradient from 30 mM to 1 M. Pooled fractions were concentrated and further purified by SEC using a Superdex 75 10/300 GL column. HsYTHDC1 344–509 was exchanged into a final buffer of 20 mM HEPES pH 7.5, 150 mM KCl, 5 mM MgCl2, and 1 mM DTT during SEC and concentrated to 5–30 mg/ml.
Crystallization, X-ray data collection, and structure determination
SeMet Sm3–180 was crystallized by sitting drop vapor diffusion in 0.1 M BisTris Propane pH 7.5, 0.2 M Na2SO4, and 23% PEG3350 with a drop size of 2 μL and reservoir volume of 65 μL. The reservoir was supplemented with 5 mM DTT immediately prior to setting up the drop. Crystals appeared within 2–8 days at 20°C and were of the space group P43212 with unit cell dimensions a = 62.41 Å, b = 62.41 Å, c = 118.63 Å and α = 90.00°, β = 90.00°, γ = 90.00°. Samples were cryoprotected with Parabar 10312 and frozen in liquid nitrogen. Single-wavelength anomalous diffraction (SAD) data were collected remotely on the tuneable NE-CAT 24-ID-C beamline at the Advanced Photon Source at the selenium edge energy at 12.663 kEv (Table 1). Data were integrated and scaled using XDS (Kabsch, 2010) and AIMLESS (Evans, 2006) via the NE-CAT RAPD pipeline. Heavy atom sites were located using SHELX (Sheldrick, 2008) and phasing, density modification, and initial model building was carried out using the Autobuild routines of the PHENIX package (Adams et al., 2010). Further model building and refinement was carried out manually in COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2010) respectively. The final model was refined to 1.92 Å resolution with Rwork/Rfree = 0.1964 / 0.2356 (Supplementary Table 1) and contained one molecule in the asymmetric unit: chain A, 3–180. All structural models were rendered with Pymol (http://www.pymol.org) and surface electrostatics were calculated with APBS (Jurrus et al., 2018).
Table 1.
Dissociation constants from filter binding and MST experiments.
| Method | Construct | DNA or RNA | Kd (μM) | Error (%) |
|---|---|---|---|---|
| Filter binding | HsYTHDC1 YTH | m6A RNA 7mer | 0.68 | 1.29 |
| HsYTHDC1 YTH | nmA RNA 7mer | ND | ND | |
| Sm3–180 | m6A RNA 7mer | ND | ND | |
| Sm3–180 | nm dsDNA | 0.07 | 1.85 | |
| Sm3–180 | m6A dsDNA | 0.07 | 1.55 | |
| Sm3–180 | m5C dsDNA | 0.11 | 2.23 | |
| MST | Sm3–180 | Rm6A ds DNA | 0.93 | 18.19 |
| Sm3–180 | RA ds DNA | 0.31 | 2.07 | |
| Sm3–180 | Rm5C ds DNA | 0.20 | 3.04 | |
| Sm3–180 | RC ds DNA | 1.67 | 14.22 | |
| Sm3–180 | Ym6A ds DNA | 0.22 | 0.95 | |
| Sm3–180 | YA ds DNA | 0.53 | 1.73 | |
| Sm3–180 | Ym5C ds DNA | 0.73 | 10.73 | |
| Sm3–180 | YC ds DNA | 0.09 | 1.48 | |
| Sm3–180 | Rm6A ss DNA | 0.64 | 5.94 | |
| Sm3–180 | RA ss DNA | 0.86 | 7.24 | |
| Sm3–180 | Rm5C ss DNA | 0.68 | 7.32 | |
| Sm3–180 | RC ss DNA | 0.46 | 4.79 | |
| Sm3–180 | Ym6A ss DNA | 0.31 | 0.70 | |
| Sm3–180 | YA ss DNA | 0.50 | 1.55 | |
| Sm3–180 | Ym5C ss DNA | 0.76 | 7.85 | |
| Sm3–180 | YC ss DNA | 1.06 | 15.53 | |
| Sm3–180 Y164A | Rm6A ds DNA | 0.34 | 7.52 | |
| Sm3–180 Y164A | RA ds DNA | 0.59 | 3.33 | |
| Sm3–180 Y164A | Rm5C ds DNA | 0.16 | 2.10 | |
| Sm3–180 Y164A | RC ds DNA | 1.65 | 21.75 | |
| Sm3–180 Y164A | Ym6A ds DNA | 0.42 | 3.03 | |
| Sm3–180 Y164A | YA ds DNA | 1.00 | 7.45 | |
| Sm3–180 Y164A | Ym5C ds DNA | 0.40 | 3.53 | |
| Sm3–180 Y164A | YC ds DNA | 0.19 | 4.59 | |
| Sm3–180 Y164A | Rm6A ss DNA | 0.72 | 11.55 | |
| Sm3–180 Y164A | RA ss DNA | 0.12 | 1.34 | |
| Sm3–180 Y164A | Rm5C ss DNA | 0.16 | 1.98 | |
| Sm3–180 Y164A | RC ss DNA | 1.31 | 19.49 | |
| Sm3–180 Y164A | Ym6A ss DNA | 0.65 | 1.66 | |
| Sm3–180 Y164A | YA ss DNA | 0.97 | 5.75 | |
| Sm3–180 Y164A | Ym5C ss DNA | 1.47 | 17.96 | |
| Sm3–180 Y164A | YC ss DNA | 1.37 | 18.37 |
‘ss’ and ‘ds’ denote ‘single-stranded’ and ‘double-stranded’. The 6-FAM-labeled upper strand oligos associated with each substrate were used to assess ssDNA binding in the MST experiments. See Supplementary Table 2 for oligonucleotide sequences. Binding isotherms used to determine Kd values are shown in Figs. 4 (filter binding) and 5 (MST). ‘ND’ signifies ‘not determined’ due to incomplete saturation within the data acquisition range.
Preparation of oligonucleotide substrates
DNA (Integrated DNA technologies, IDT) and RNA (Dharmacon) were purchased commercially as lyophilized, single-stranded oligonucleotides. For filter binding, all oligonucleotides were resuspended to 1 mM in 10 mM Tris-HCl and 1 mM EDTA and stored at −20°C until needed. Single-stranded oligonucleotides were 5’ end-labeled with [γ32P]ATP using polynucleotide kinase (New England Biolabs) and then purified on P-30 spin columns (BioRad) to remove unincorporated label. Duplex substrates were prepared by heating equimolar concentrations of complementary strands to 95°C for 5 minutes and then cooling to room temperature overnight. Single stranded DNA was removed using an S-300 spin column (GE). Three duplex substrates – 5mC dsDNA (5mC DNA US and 5mC DNA LS), m6A dsDNA (m6A DNA US and m6A DNA LS), and nm dsDNA (nm DNA US and nm DNA LS) – were prepared. Two single-stranded substrates – m6A ssRNA 7mer and nmA ssRNA 7mer – were left unannealed and used accordingly. For microscale thermophoresis, all oligonucleotides were resuspended in molecular grade HEPES pH 7.5 at a concentration of 200 μM and stored at −20°C until needed. Eight duplex substrates were prepared as described above by annealing 6-FAM-labeled upper strand oligonucleotides to unlabeled, complementary lower strand oligonucleotides: Rm6A ds DNA (Rm6A DNA US and Rm6A DNA LS), RA ds DNA (RA DNA US and RA DNA LS), Ym6A ds DNA (Ym6A DNA US and Ym6A DNA LS), YA ds DNA (YA DNA US and YA DNA LS), Rm5C ds DNA (Rm5C DNA US and Rm5C DNA LS), RC ds DNA (RC DNA US and RC DNA LS), Ym5C ds DNA (Ym5C DNA US and Ym5C DNA LS), YC ds DNA (YC DNA US and YC DNA LS). The unannealed, 6-FAM-labeled upper strand oligonucleotides were used as single-stranded DNA substrates. See Supplementary Table 2 for oligonucleotide sequences.
Filter binding assays
The standard buffer for the DNA and RNA binding assays contained 25 mM MES (pH 6.5), 2.0 mM MgCl2, 0.1 mM DTT, 0.01 mM EDTA, and 40 μg/mL BSA. Binding was performed with purified SmMcrB 3–180 and the HsYTHDC1 YTH domain at 30°C for 10 min in a 30 μL reaction mixture containing 14.5 nM unlabeled DNA and 0.5 nM γ32P-labelled DNA. Samples were filtered through KOH-treated nitrocellulose filters (Whatman Protran BA 85, 0.45 μM) using a Hoefer FH225V filtration device for approximately 1 min. Filters were subsequently analyzed by scintillation counting on a 2910TR digital, liquid scintillation counter (PerkinElmer). All measured values represent the average of at least three independent experiments (mean ± SD) and were compared with a negative control to determine fraction bound. Binding constants were determined by nonlinear curve fitting using Kaleidagraph (Synergy Software) and defined as the concentration of the protein at which 50% of the labeled DNA substrate is retained. Calculated Kd values are listed in Table 1.
Microscale thermophoresis
MST experiments were conducted using a Monolith NT.115 instrument (NanoTemper Technologies) equipped with red and blue filters. Protein was two-fold serially diluted 16 times and mixed with 40 nm of each 6-FAM-labeled oligo. Buffer conditions contained 20 mM HEPES pH 7.5, 150 mM KCl, 5 mM MgCl2, 1mM DTT and 0.05% TWEEN-20. Samples were incubated at room temperature in the dark for 30 minutes before being loaded into standard capillaries (NanoTemper Technologies), tested with 40% excitation power, medium MST power, and measured using M.O. Control software (NanoTemper Technologies). Data were analyzed using M.O. Affinity Analysis software (version 2.3, Nanotemper Technologies) to determine the normalized fluorescence (Fnom) at each concentration. Fnorm is calculated by dividing Fhot (average fluorescence value in the heated state) by Fcold (average fluorescence value measured in the cold state before the IR laser is turned on) and plotted as parts per thousand (%). Fnorm values from at least three independently pipetted measurements were averaged (mean ± standard deviation) and plotted against the respective concentration of Sm 3–180 (wildtype and Y164A mutant) to obtain a binding isotherm for each substrate (Fig. 5). Binding constants (Kd) were determined by nonlinear curve fitting using Kaleidagraph (Synergy Software) (Table 1). All experiments were carried out using multiple, independently purified batches of protein.
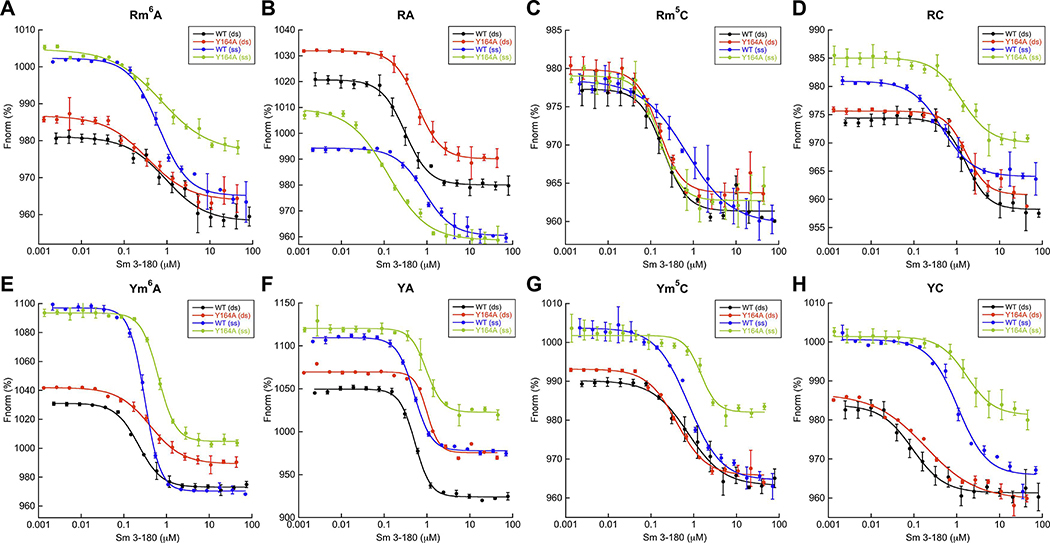
Figure 5. MST confirms SmMcrB DNA binding promiscuity.
All data points represent average of three independent experiments (mean ± standard deviation) using multiple, independently purified batches of protein. Fnorm is calculated by dividing Fhot (average fluorescence value in the heated state) by Fcold (average fluorescence value measured in the cold state before the IR laser is turned on) and plotted as parts per thousand (%). Binding constants were determined by nonlinear curve fitting using Kaleidagraph (Synergy Software). Calculated Kd values are listed Table 1. Abbreviations are as follows: ds, double stranded; nm, non-methylated; ss, single stranded; m6A, 6-methyladenine-modified; m5C, 5-methylcytosine-modified. Binding isotherms with the Rm6A (A), RA (B), Rm5C (C), RC (D), Ym6A (E), YA (F), Ym5C (G), and YC (H) substrates are colored as follows: wildtype (WT) + dsDNA, black; wildtype + ssDNA, blue; Y164A + dsDNA, red; Y164A + ssDNA, green. Substrate sequences are shown in Supplementary Table 2. The 6-FAM-labeled upper strand oligos associated with each substrate were used to assess ssDNA binding.
Results
SmMcrB 3–180 adopts an EVE domain fold
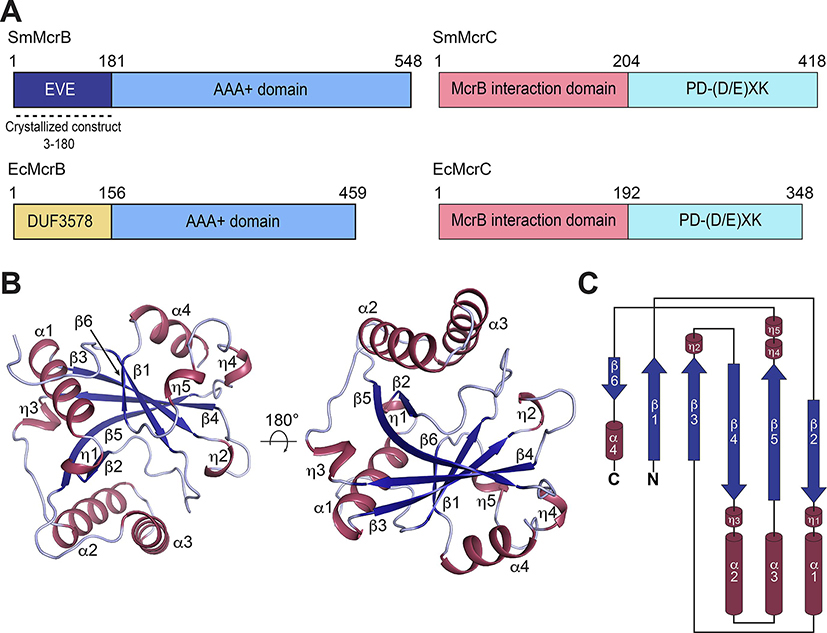
Previous biochemical and structural studies established that the EcMcrB N-terminal domain (residues 1–155) recognizes DNA containing methylated cytosines (5mC) (Sutherland et al., 1992, Krüger et al., 1995, Gast et al., 1997) via a base flipping mechanism (Sukackaite et al., 2012, Zagorskaitė et al., 2018). This domain, however, is only conserved in a handful of McrB homologs (Hosford et al., 2020), suggesting other species use different strategies for substrate binding and/or may preferentially target other sequences and modifications. To test this hypothesis and explore the evolutionary diversity of this family, we screened divergent McrB homologs containing unique N-terminal sequences and identified the N-terminal domain from Staphylothermus marinus McrB (Sm3–180; Fig. 1A) as a suitable candidate for structural and biochemical characterization. This construct is thermally stable, could be expressed in E. coli and purified to homogeneity in milligram quantities, and readily crystallized by sitting drop vapor diffusion. The C-terminal AAA+ domain of SmMcrB and the accompanying SmMcrC nuclease share identifiable homology with their E. coli counter parts (Fig. 1A), suggesting that only the putative substrate binding module is distinct in this species while the motor and cleavage machinery remain unaltered.
Figure 1. Structure and topology of Sm3–180.
A. Domain architecture of SmMcrBC and EcMcrBC. Dashed line denotes crystallized construct (residues 3–180). B. Cartoon representations of SmMcrB 3–180 in two orientations. Helices and β-strands are colored raspberry and blue respectively. C. Topology diagram of SmMcrB 3–180.
Recombinant selenomethionine-labeled Sm3–180 yielded crystals of the space group P43 21 2 with 1 molecule in the asymmetric unit. The structure was solved by single wavelength anomalous diffraction (SAD) phasing (Hendrickson, 2014) and the final model was refined to 1.92Å resolution with Rwork and Rfree values of 0.1964 and 0.2356 (Supplementary Table 1).
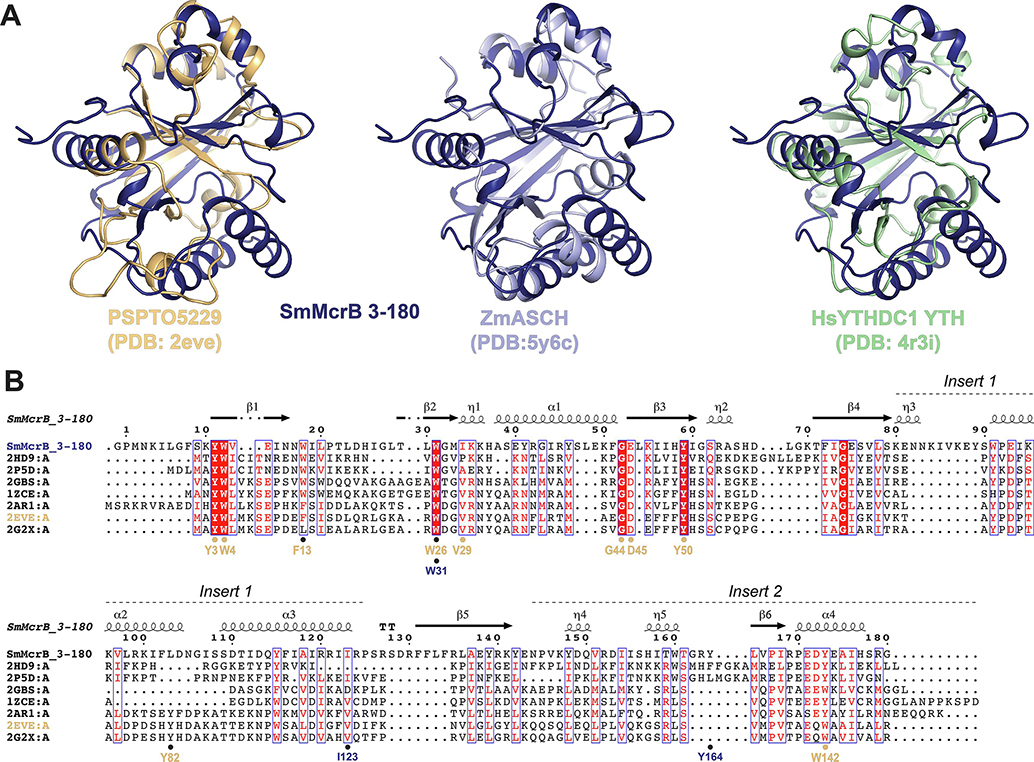
Sm3–180 is composed of a central six-stranded pseudobarrel, connected with an intricate network of extended loops and α-helical inserts (Fig. 1B). The strands are ordered β6–1-3–4-5–2 with each oriented in an antiparallel configuration except β1 and β3, which are parallel (Fig. 1B). Helix α1 lies between β2-β3 while helices α2 and α3 are inserted in tandem between β4-β5 (Fig. 1C). A fourth helix, α4, follows β6 at the C-terminal end of the domain (Fig.1C). Attempts to superimpose Sm3–180 with N-terminal DNA binding domain of EcMcrB (pfam: DUF3578, PDB: 3SSD) failed to yield a consistent structural alignment, suggesting their structural topologies differ significantly. Analysis via the Dali server (Holm and Rosenström, 2010) instead revealed that Sm3–180 shares homology with PUA-like domains, which at their core contain a five-stranded pseudobarrel architecture with an intervening α-helix often localized between β1 and β2 (Supplementary Fig. 1). Structural superposition with representative Dali hits like the PSPTO5229 EVE domain (PDB: 2eve, Z-score = 9.8, # structurally equivalent Cα (lali) = 129 of 149, RMSD (of structurally equivalent Cα) = 2.9Å), the Zymomonas mobilis (Zm) ASCH domain (PDB: 5y6c, Z-score = 10.9, lali = 128 of 142, RMSD = 2.6Å), and YTHDC1 YTH domain (PDB: 4r3i, Z-score = 9.1, lali = 116 of 164, RMSD = 2.6Å) confirms an overall structural similarity with this superfamily (Fig. 2A, Supplementary Fig. 1).
Figure 2. Sm3–180 adopts an EVE domain fold.
A. Structural superpositions of SmMcrB 3–180 with PSPTO5229 (PDB: 2eve), ZmASCH (PDB: 5y6c), and HsYTHDC1 YTH (PDB: 4r3i). B. Structure based sequence alignment of SmMcrB 3–180 with EVE domain homologs. Secondary structure of SmMcrB 3–180 is mapped above alignment. Aromatic cage residues for Sm3–180 (dark blue) and the PSPTO5229 EVE domain (orange) are marked below. Sequence shading indicates conservation: white text on red background, 100% conserved; boxed red text on white background, 70% conserved. Sequence labeling associated with the listed PDB codes is as follows: 2HD9:A, PH1033 from Pyrococcus horikoshii OT3; 2P5D:A, MJECL36 from Methanocaldococcus jannaschii DSM 2661; 2GBS:A, Rpa0253 from Rhodopseudomonas palustris; 1ZCE:A, Atu2648 from Agrobacterium tumefaciens; 2AR1:A, Hypothetical protein from Leishmania major; 2EVE:A, PSPTO5229 from Pseudomonas syringae; 2G2X:A, Q88CH6 from Pseudomonas putida.
PUA-like domains can be further subdivided by the presence of topological embellishments to the core fold (Iyer et al., 2006, Perez-Arellano, 2007, Bertonati, et al. 2009). ASCH domains, for example, contain a ~40 residue insert between β4 and β5 (Insert 1) that features numerous short helices (Supplemental Fig. 1, yellow). EVE and YTH domains both exhibit a shortened Insert 1 along with a C-terminal extension (Insert 2) characterized by helices of variable length and an additional strand (β6) that pairs with β1 in an anti-parallel arrangement (Supplementary Fig. 1, green). EVE domains are further distinguished at the sequence level by a set of conserved residues – Y3, W4, W26, V29, G44, D45, Y50, and W142 in the prototypical PSPTO5229 (Fig. 2B, orange) – that are absent in other PUA family members (Bertonati, et al. 2009). Canonical PUA domains lack Insert 2 but contain a topologically distinct sixth strand (Insert 0) that lies between β2 and β3 and pairs with β1 in an antiparallel manner (Supplementary Fig. 1, red). SET and RING finger-associated (SRA) domains, in contrast, are comprised of eight β-strands (Iyer et al., 2011) with β3, β6, β7, β1/β8, and β5 forming the PUA core and α1/β2 spatially aligning with Insert 2 (Supplementary Fig. 1). We designate Sm3–180 as an EVE domain based on the presence of Inserts 1 and 2 and conservation of the characteristic EVE residues distributed throughout the fold (Fig. 2B, Supplementary Fig. 1).
SmMcrB binds DNA
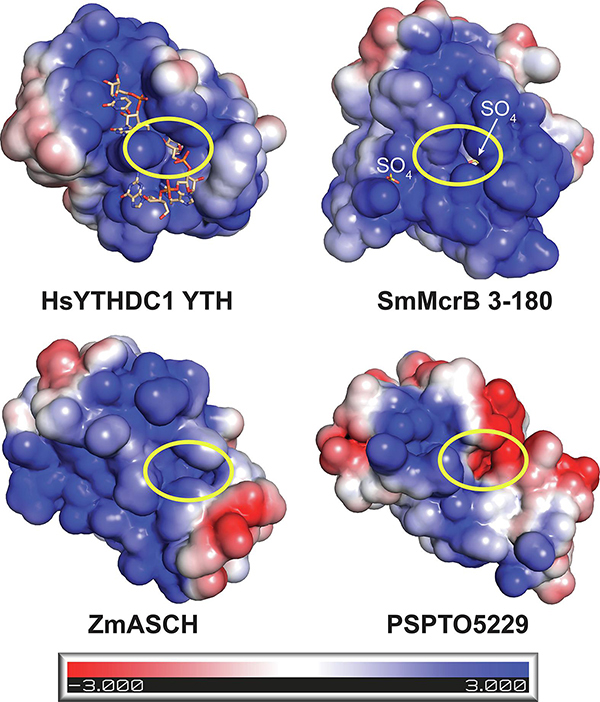
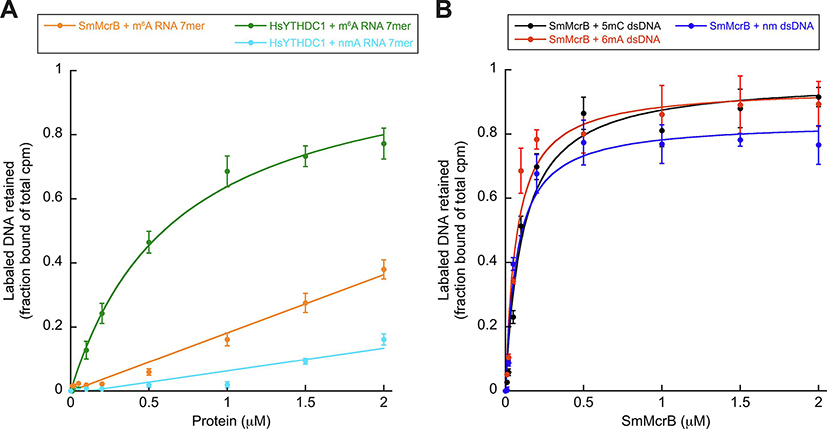
Many PUA-like domains bind RNA and contain a swath of positively charged residues on one face that forms a cleft for the negatively charged phosphate backbone (Fig. 3). Two sulfate ions from the crystallization buffer are bound in this cleft and localized along the positively charged electrostatic surface of Sm3–180 (Fig. 3). EVE domains remain largely uncharacterized and minimal biochemical data exists supporting their RNA binding capacity. YTH domains, however, have been extensively studied and shown to associate preferentially with short, methylated RNAs containing a primary consensus site of G(m6A)C (Li et al., 2014, Luo and Tong, 2014, Theler et al., 2014, Zhu et al., 2014, Xu et al., 2014, Xu et al. 2015, Wang et al., 2016). To determine whether SmMcrB shares this RNA binding activity, we analyzed Sm3–180’s association with methylated RNA containing the YTH consensus sequence via filter binding (Fig. 4A, Supplementary Table 2). Sm3–180 shows weak affinity for this m6A RNA at a level where a Kd for binding could not be calculated (Fig. 4A, orange; Table 1). The human (Hs) YTHDC1 YTH domain, in contrast, specifically binds this m6A-modified RNA with a Kd of ~680 nM and does not bind a non-methylated version of the same substrate (Fig. 4A, green and cyan; Table 1).
Figure 3. Surface electrostatics define the nucleic acid binding surfaces of PUA-like domains.
Electrostatic surfaces of HsYTHDC1 YTH domain with bound 5’-GG(m6A)CU-3’ ssRNA (PDB: 4r3i), SmMcrB 3–180, ZmASCH (PDB: 5y6c), and PSPTO5229 (PDB: 2eve). Bound RNA and sulfate ions are shown in stick representation and colored wheat and yellow respectively. Yellow circle shows location of the aromatic cage binding pocket. Scale bar indicates electrostatic surface coloring from −3 KbT/ec to +3 KbT/ec.
Figure 4. SmMcrB preferentially binds DNA and is promiscuous.
All data points represent average of three independent experiments (mean ± standard deviation) using multiple, independently purified batches of protein. Binding constants were determined by nonlinear curve fitting using Kaleidagraph (Synergy Software) and defined as the concentration of the protein at which 50% of the labeled DNA substrate is retained. Substrate sequences and calculated Kd values are listed in Supplementary Table 2 and Table 1 respectively. Abbreviations are as follows: ds, double stranded; nm, non-methylated; m6A, 6-methyladenine- modified; m5C, 5-methylcytosine-modified. A. Filter binding analysis of Sm3–180 and HsYTHDC1 YTH domain interactions with RNA containing the YTH consensus sequence. B. Filter binding analysis of Sm3–180 interactions with dsDNA substrates.
Emerging data suggest that some PUA family members can instead target DNA. Biochemical characterization of PUA-like domains in bacterial HNH and PD-(D/E)XK nucleases showed that those most similar to EVE and YTH domains (e.g. TspA15I) target DNA containing 5-hydroxy-methylcytosines (m5hC) (Lutz et al., 2019). Mass spectrometry screening similarly showed that the EVE domain-containing human protein THYN1/Thy28 binds m5hC modifications in DNA (Spruijt et al., 2013). In vitro analysis of a putative Zymomonas mobilis (Zm) ASCH protein revealed both DNA and RNA binding capabilities (Kim et al., 2017). We recently demonstrated that the N-terminal domain of Thermococcus gammatolerans (Tg) McrB adopts a YTH domain fold that preferentially associates with m6A-modified DNA (Hosford et al., 2020). Based on these observations, we therefore tested whether Sm3–180 could interact with methylated (m6A or m5C) and/or non-methylated DNA substrates using filter binding (Fig. 4B, Supplementary Table 2). Sm3–180 binds double stranded (ds) DNA with strong affinity on the order of ~70–100 nM, but in this sequence context does not discriminate between different modifications (Fig. 4B, black, red, and blue; Table 1).
To examine Sm3–180 DNA binding by an orthogonal method and avoid potential artifacts, we generated a second set of substrates with an alternative nucleotide sequence (Supplementary Table 2) and measured binding affinities by microscale thermophoresis (MST) (Fig. 5). Substrates directly compared m5C and m6A modifications to their respective unmodified bases within the same sequence context. Because EcMcrBC preferentially binds methylated cytosines where the adjacent nucleotide upstream is a purine (RmC) (Raleigh and Wilson, 1986, Sutherland et al., 1992), we also included substrate variants to evaluate the effects of having a purine (R) versus a pyrimidine (Y) immediately adjacent to each modified site (e.g. Rm6A versus RA versus Ym6A versus YA) (Supplementary Table 2). Sm3–180 binds these dsDNA substrates with affinities ranging from 90 nM (for YC dsDNA) up to 1.67 μM (for RC dsDNA) (Fig. 5, Table 1). We observe no specific preference for a particular modification in this sequence context nor any clear trend with regard to how the adjacent nucleotide modulates binding. For example, the Rm5C and Ym6A dsDNA substrates both bind on the order of ~200 nM while unmodified RA dsDNA binds at 310 nM. We also tested Sm3–180’s interaction with single stranded (ss) DNA (Fig. 5). While we observe a nearly 10-fold tighter association of YC dsDNA versus ssDNA, the measured Kd values for binding other ssDNA substrates all are within a factor of two relative to their dsDNA counterparts (Fig. 5, Table 1), suggesting a general lack of ds/ss discrimination within this sequence context. Taken together, these data suggest that Sm3–180 is an EVE domain that binds DNA.
SmMcrB contains a degenerate aromatic cage
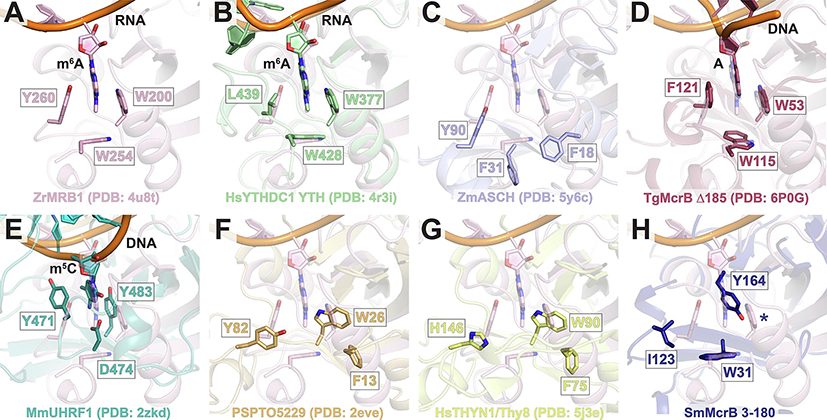
We next examined available structural models in greater detail to identify features that could explain Sm3–180’s unique substrate binding profile. A defining feature of many PUA-like domains is the presence of a conserved hydrophobic pocket – colloquially termed the ‘aromatic cage’ – that sits at the base of the positively charged cleft (Fig. 3, yellow circles) and plays a critical role in substrate binding and discrimination. The canonical arrangement of this cage is exemplified in the Zygosaccharomyces rouxii (Zr) MRB1 protein, where three aromatic residues (W200, W254, and Y260) are arranged to stabilize the m6A base through a combination of hydrophobic interactions and π-π stacking (Fig. 6A). The HsYTHDC1 YTH domain achieves the same interactions with modified RNA but substitutes a leucine residue (L439) for one of the cage walls (Fig. 6B). The apo Zm ASCH domain also contains a set of spatially conserved aromatic residues (Y90, F31, and F18), but these adopt a different conformation likely owing to the absence of a bound RNA substrate (Fig. 6C). TgMcrB’s YTH domain uses a canonical cage to recognize methylated adenines that are flipped out of the DNA duplex (Fig. 6D). SRA domains in eukaryotic proteins and bacterial modification-dependent restriction enzymes similarly combine base flipping and π-stacking to recognize methylated cytosines (Hashimoto et al., 2008, Avvakumov et al., 2008, Arita et al., 2008, Qian et al., 2008, Horton et al., 2014, Kazrani et al., 2014, Horton et al., 2014b, Shao et al., 2014, Horton et al., 2014c, Kisiala et al., 2018), albeit with a different pocket (Fig. 6E). Structural superposition reveals a distinct, shared arrangement of aromatic residues in the PSPTO5229 and HsTHYN1/Thy28 EVE domains (Fig. 6F and G), which completely reorganizes the putative binding pocket and may reflect unique features of this branch of the PUA superfamily. In contrast to all of these, Sm3–180 contains a degenerate cage with only two hydrophobic side chains (W31 and I123; Fig. 6H). An additional tyrosine (Y164) occludes the top of the pocket in a configuration that would prohibit modified bases from accessing the cage cavity (Fig. 6H).
Figure 6. SmMcrB contains a degenerate aromatic cage.
A. Zoomed view of ‘canonical’ aromatic cage from ZrMRB1 with bound m6A RNA (pink). Cage residues are shown as sticks and labeled. View is centered around the m6A-modified base that is flipped into the cage pocket. B-H. Structural superposition ZrMRB1 YTH domain with HsYTHDC1 YTH domain (B, green), ZmASCH (C, light blue), TgMcrB N-terminal YTH domain (D, raspberry); MmUHRF1 SRA domain (E, teal), PSPTO5229 EVE domain (F, orange), HsTHYN1/Thy28 EVE domain (G, yellow), and SmMcrB 3–180 (G, dark blue). Panels are oriented to illustrate the positions of the cage residues from each structure (sticks) relative to the ZrMRB1 aromatic cage. Where applicable, bound nucleic acid substrates with bases flipped into the cage pocket are shown (B, D, and E). Blue asterisk in H denotes absence of third hydrophobic cage residue in Sm3–180.
To probe the function of Y164 in substrate discrimination, we mutated this side chain to alanine (Y164A) and used MST to assess how this change alters DNA binding affinity and selectivity (Fig. 5). This substitution has limited effects on dsDNA binding, with the majority of substrates showing less than a two-fold change relative to wildtype (WT) (Table 1). The Rm5C and RC substrates, for instance, bind to both constructs with the nearly same affinity (Rm5C: 200 nM for WT versus 160 nM for Y164A; RC: 1.67 μM for WT versus 1.65 μM for Y164A). Interestingly, the Y164A mutation increased the affinity for the single stranded RA and Rm5C substrates by 7-fold and 4-fold, respectively. Potential exposure of the cage pocket via this perturbation, however, does not impart selectivity for a specific base and/or modification, implying that Sm3–180’s degenerate cage lacks the capacity for stringent discrimination and/or that DNA binds differently than has been observed in other previously characterized YTH and SRA domains.
Structural and conformational variations that correlate with PUA-like domain specificity
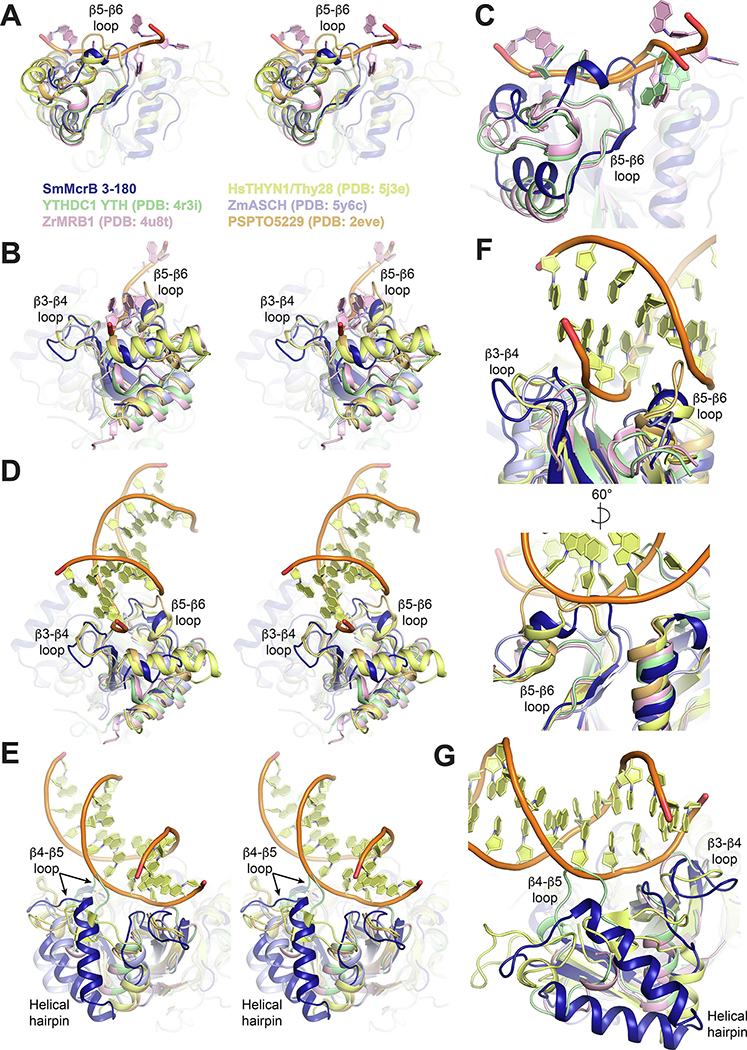
Sm3–180’s degenerate cage and preference for DNA suggest that other structural motifs may contribute its substrate specificity. Superpositions of PUA-like domains with RNA-bound YTH domain structures and m5C DNA-bound HsTHYN1/Thy28 show the β3-β4, β4-β5, and β5-β6 loops adopt different conformations that specifically correlate with either DNA or RNA binding (Fig. 7). In YTH domains that preferentially bind RNA, the β5-β6 loop is pulled down in a bent conformation that accommodates a tight association of the single stranded substrate while the β3-β4 loop tilts toward the phosphate backbone to provide additional stabilization (Fig. 7A–C). The β5-β6 loop is flipped up in Sm3–180, which would cause significant clashing with the phosphate backbone and a number of bases (Fig. 7C, dark blue). As similar upward flipped conformation is observed for the β5-β6 loop in the PSPTO5229 EVE domain and HsTHYN1/Thy28, resulting in even more pronounced clashing (Fig. 7A–B, orange and yellow). The β3-β4 loop is longer in each of these structures, including Sm3–180, and angles away from the RNA binding cleft.
Figure 7. Structural and conformational variations that correlate with PUA-like domain specificity.
Superposition of PUA-like domains with coloring as follows: SmMcrB 3–180, dark blue; YTHDC1, light green; ZrMRB1, light pink; HsTHYN1/Thy28, yellow; ZmASCH, light blue; PSPTO5229, light orange. Bound RNA and DNA substrates in each domain are colored according to the same scheme when shown. β3-β4, β4-β5, β5-β6 loops and the helical hairpin inserts are labeled where applicable. A,B. Side (A) and end on (B) stereo views of superposition with ZrMRB1 RNA substrate shown. C. Zoomed view of β5-β6 loop interactions with RNA. D,E. End on (D) and side (E) stereo views of superposition with HsTHYN1 DNA shown. F. Zoomed view of β3-β4 and β5-β6 interactions with DNA. G. Zoomed view of β3-β4 and β4-β5 loop interactions with DNA.
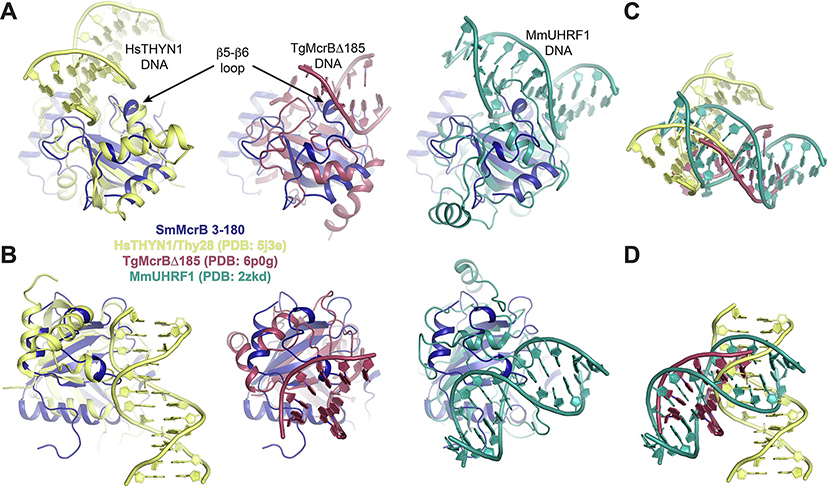
While these structural differences are incompatible with RNA binding, they are necessary to allow HsTHYN1/Thy28 to bind double stranded DNA (Fig. 7D–G, yellow). Here the β5-β6 packs into the minor groove of the duplex as the shifted β3-β4 loop sandwiches one strand from the outside (Fig. 7D and E, yellow). The shared conformational arrangement of these loops in Sm3–180 would permit DNA binding in the same orientation without any steric hindrance (Figs. 7D and E and 8A and B, dark blue). Superposition of Sm3–180 with the TgMcrB YTH domain or mouse (Mm) UHRF1 SRA domain, however, yields multiple points of collision with each bound DNA substrate, owing to the conformation of the β5-β6 loop (Fig. 8A and B). The radically different binding orientations in these structures (Fig. 8C and D) is likely coupled to whether base-flipping is explicitly used (TgMcrB and MmUHRF1) or not (HsTHYN1/Thy28).
Figure 8. The DNA-binding orientation in HsYTHN1/Thy28 is most compatible with the conformational organization of Sm 3–180 structural elements.
A,B. Superposition of of SmMcrB 3–180 (dark blue) with the DNA-bound structures of HsTHYN1/Thy28 (yellow; EVE domain), TgMcrBΔ185 (raspberry; YTH domain), and MmUHRF1 (teal; SRA domain) in two orientations. Arrows denote the position of the β5-β6 loop. DNA substrates are labeled and colored to match their respective structures. Structural alignments were guided by topological comparison shown in Supplementary Fig. 1. C,D. Comparison of isolated substrates from the superpositions in A and B illustrating the distinct orientation of DNA in HsTHYN1 relative to TgMcrB and MmUHR1.
We also note that the corresponding β4-β5 loop adopts a kinked conformation in the HsYTHDC1 YTH domain that collides with the DNA backbone, further highlighting the conformational differences between PUA-like domains evolved for RNA binding versus DNA binding (Fig. 7E,G). Sm3–180 and ZmASCH both contain helical hairpins that are inserted into this loop (Figs. 1C and 7G), which likely constrain the conformation such that it is compatible with DNA binding (Fig. 4A) (Kim et al., 2017). Together these structural comparisons reveal different conformational variations that correlate with specificity.
Discussion
Here we showed that the N-terminal domain of SmMcrB is an EVE domain. Although EVE domains are distinct among PUA-like domains, the overall topological similarities to YTH, ASCH, and SRA domains allow for structural and functional comparisons. YTH domains typically bind m6A modified RNA containing the consensus sequence G(m6A)C, with the conserved aromatic cage residues serving as the primary determinant of substrate recognition and discrimination (Luo and Tang, 2014, Xu et al., 2014, Theler et al., 2014; Xu et al., 2015) (Fig. 6A and B). Our data show that Sm3–180 does not bind a short RNA containing the YTH consensus sequence but does bind DNA (Figs. 4 and 5, Table 1). While Sm3–180 can associate with different modified substrates, the strength and selectivity of this binding varies depending on the sequence context. Structural superposition reveals that the Sm3–180 aromatic cage is degenerate with only two hydrophobic side chains present instead of three. The missing residue would form one of the walls of the cage, opposite I123 (Fig. 6H, asterisk). Without this residue, Sm3–180 is unable to form π-stacking interactions that stabilize a base within this pocket to allow for recognition. Additionally, the orientation of the Y164 side chain at the top of the Sm3–180 cage occludes the pocket and would prevent base flipping as a means of distinguishing between different modified substrates. While both of these features may contribute to Sm3–180’s substrate binding promiscuity, the degenerate nature of the cage appears to be more critical in this regard. The Y164A mutation, which presumably exposes the pocket cavity, fails to restore specificity for any singular modification and instead enhances binding to some ssDNA substrates. Structure based sequence alignment indicates that Y164 localizes to a region in Insert 2 that is not strictly conserved among other EVE domains (Fig. 2B). This could represent a unique evolutionary adaptation that distinguishes Sm3–180 from other PUA-like domains and biases the substrate preference toward a specific mode of dsDNA binding (Fig. 8).
The putative cage residues in the PSPTO5229 and HsTHYN1/Thy28 EVE domains are conserved in only a subset of other EVE domains (Fig. 2B) and form a unique pocket that is distinct from that of YTH domains, SRA domains, and Sm3–180 (Fig. 6). Though previously proposed to play a role in specificity (Bertonati et al., 2009, Lutz et al., 2019), these side chains do not directly contact the m5C modification in the HsTHYN1/Thy28-DNA complex (Figs. 7 and 8, yellow). They do, however, serve as a binding site for small molecule ligands like 3-(N-morpholino)propanesulfonic acid and glycerol in other EVE domain structures (Arakaki et al., 2006, Sugahara et al., 2008, Bertonati et al., 2009) and may yet be important for base recognition.
Our structural comparisons suggest that the β3-β4, β4-β5, and β5-β6 loops contribute to specificity, as different conformations of these segments appear to be more compatible with either DNA or RNA binding (Fig. 7). The position of these loops in the HsYTHDC1 YTH domain, for example, stabilizes the bound RNA substrate but would clash with a double stranded substrate (Fig. 7, light green). We see the opposite effect in the DNA-bound HsTHYN1/Thy28 structure (Fig. 7D–G, yellow). Sm3–180’s loop conformations similarly favor DNA (Fig. 7), in agreement with our binding experiments. The β5-β6 loop in particular constrains the approach of all nucleic acid substrates, preventing single-stranded substrates from associating as in YTH domains (Fig 7A–C) and limiting the possible orientations of dsDNA (Fig. 8). Our modeling specifically predicts Sm3–180 could bind dsDNA like HsTHYN1/Thy28 without any steric hindrance. It remains to be seen exactly how ssDNA associates, but could be largely driven by electrostatic interactions along Sm3–180’s expanded basic surface (Fig. 3). Given the small sample size of PUA domain structures with bound substrates and the lack of available binding data for different family members, we cannot rule out that additional conformational changes may occur when these modules are presented with different types of nucleic acids. This is likely true for ZmASCH, which has been shown to bind both DNA and RNA with high affinity (Kim et al., 2017). Collectively, our findings highlight the plasticity of the PUA family core fold and could serve as an important predictive diagnostic to guide the biochemical characterization of new PUA domain-containing proteins following structure determination.
Sm3–180 constitutes the substrate binding domain of Staphylothermus marinus McrB but, despite its ability to DNA, does not show a clear preference for a specific modification within the sequence contexts we examined (Fig. 4B and 5). This is in stark contrast to the N-terminal domains of EcMcrB, which strictly binds DNA containing methylated cytosines (Sutherland et al., 1992; Krüger et al., 1995; Gast et al., 1997, Zagorskaitė et al., 2018) and TgMcrB, which targets m6A DNA via a YTH domain (Hosford et al., 2020). Both of these homologs flip the modified base out of the duplex, where it is then recognized via a constrained pocket that is finely tuned for specificity (Sukackaite et al., 2012, Zagorskaitė et al., 2018, Hosford et al., 2020). As noted above, the degenerate nature of the Sm3–180 aromatic cage would diminish the effectiveness of this strategy and could explain, at least partially, the discrepancy in binding preference. Alternatively, the apparent lack of specificity could arise from sequence-specific binding interactions that occur in close proximity to the modified site and are absent from our substrates. For EcMcrBC, the presence of a purine immediately adjacent to a methylated cytosine dramatically enhances binding (Raleigh and Wilson, 1986, Sutherland et al., 1992). In other restriction enzymes, the sequence context is more stringent and can have a much greater influence on modulating the affinity for a specific modified base (Cohen-Karni et al., 2011; Horton et al., 2012; Horton et al., 2014a; Horton et al., 2014c; Sasnauskas et al., 2015; Slyvka et al., 2019). Recent biochemical characterization of novel HNH and PD-(D/E)XK nucleases containing PUA-like domains showed that subtle binding differences in vitro translated to significant functional differences in vivo with respect to restriction/defense activities (Lutz et al., 2019). Finding the proper sequence context, if it exists, could therefore radically change the specificity profile of our construct.
From our data, we also cannot rule out the possibility that SmMcrBC binds unmodified DNA site-specifically as is customary in other restriction modification systems. The Sm mcrB (Smar_0573) and mcrC (Smar_0572) genes appear in tandem like other McrBC homologs but are situated in a larger gene cluster downstream from a putative DNA methyltransferase (Sm_0576), which could potentially protect the host from sequence-specific targeting. Ultimately, a more detailed and rigorous exploration of the sequence context and modification requirements will be necessary to define SmMcrB specificity fully, both in the isolated domain and in the context of the reconstituted restriction complex.
More overt differences among McrBC homologs reflect distinct evolutionary pressures, such as attack by lytic bacteriophages with different genomic content and modifications (Weigele and Raleigh, 2016). Distantly related McrB homologs like LlaJI, LlaI, and BsuMI target DNA site-specifically – a direct consequence of the unmodified viruses they provide protection against (O’Sullivan et al., 1995; Ohshima et al., 2002; O’Driscoll et al., 2006). We previously showed that N-terminal domain of LlaJI.R1 from Helicobacter pylori adopts a B3 domain fold to recognize DNA independent of modifications (Hosford and Chappie, 2018). Collectively these observations suggest an emerging theme in which bacteria have adapted a conserved set of core machinery – the GTP-specific AAA+ motor of McrB and the associated McrC nuclease – to different biological contexts through the incorporation of alternative N-terminal binding domains. Although EcMcrBC is historically described as a prototypical MDRS (Loenen and Raleigh, 2014), it appears members of the McrBC superfamily can be classified more broadly as modular nucleases and can be tuned for different substrates.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant GM120242 (to J.S.C.) and is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines under general user proposal GUP51113 (PI: J.S.C), which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The NanoTemper Monolith NT.115 instrument used for MST binding experiments was funded by an equipment supplement from the National Institutes of Health (GM120242-02S1 to J.S.C). J.S.C. is a Meinig Family Investigator in the Life Sciences. We thank the NE-CAT beamline staff for assistance with remote X-ray data collection.
Footnotes
Conflict of Interest
The authors declare that they have no competing financial interests.
Accession numbers
The atomic coordinates and structure factors for the N-terminal domain of Staphylothermus marinus McrB (residues 3–180) has been deposited in the Protein Data Bank (http://www.rcsb.org) under the PDB code 6N0S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. , 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Kulkarni M, Gopinath A, Saikrishnan K, 2018. Single-site DNA cleavage by Type III restriction endonuclease requires a site-bound enzyme and a trans-acting enzyme that are ATPase-activated. Nucleic Acids Res 46, 6229–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki T, Le Trong I, Phizicky E, Quartley E, DeTitta G, Luft J, Lauricella A, Anderson L, Kalyuzhniy O, Worthey E, Myler PJ, Kim D, Baker D, Hol WG, Merritt EA, 2006. Structure of Lmaj006129AAA, a hypothetical protein from Leishmania major. Acta Crystallogr. F 62, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M, 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455, 818–821. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S, 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455, 822–825. [DOI] [PubMed] [Google Scholar]
- Bertonati C, Punta M, Fischer M, Yachdav G, Forouhar F, Zhou W, et al. , 2009. Structural genomics reveals EVE as a new ASCH/PUA-related domain. Proteins 75, 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernheim A, Sorek R, 2020. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol 18, 113–119. [DOI] [PubMed] [Google Scholar]
- Chen IA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, et al. , 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45, D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Karni D, Xu D, Apone L, Fomenkov A, Sun Z, Davis PJ, Kinney SR, Yamada-Mabuchi M, Xu SY, Davis T, Pradhan S, Roberts RJ, Zheng Y, 2011. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. USA 108, 11040–11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden D, Murray NE, Rao D, 2001. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis MÈ, Villion M, Magadán AH, Moineau S, 2013. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun 4, 2087. [DOI] [PubMed] [Google Scholar]
- Gupta YK, Chan SH, Xu SY, Aggarwal AK, 2015. Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I. Nat. Commun 6, 7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X, 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Mabuchi MY, Cohen-Karni D, Zhang X, Griggs RM, Samaranayake M, Roberts RJ, Zheng Y, Cheng X, 2012. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res 40, 9763–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Nugent RL, Li. A, Mabuchi MY, Fomenkov A, Cohen-Karni D, Griggs RM, Zhang X, Wilson GG, Zheng Y, Xu SY, Cheng X, 2014a. Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI. Sci Rep 4, 4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Borgaro JG, Griggs RM, Quimby A, Guan S, Zhang X, Wilson GG, Zheng Y, Zhu Z, Cheng X, 2014b. Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA. Nucleic Acids Res 42, 7947–7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Wang H, Mabuchi MY, Zhang X, Roberts RJ, Zheng Y, Wilson GG, Cheng X, 2014c. Modification-dependent restriction endonuclease, MspJI, flips 5-methylcytosine out of the DNA helix. Nucleic Acids Res 42, 12092–12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosford CJ, Bui AQ, Chappie JS, 2020. The structure of the Thermococcus gammatolerans McrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs. J. Biol. Chem 295, 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Fukuda E, Kobayashi I, 2010. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res 17, 325–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, and Aravind L, 2006. The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism. Bioinformatics 22, 257–263. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L, 2011. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci 101, 25–104. [DOI] [PubMed] [Google Scholar]
- Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, et al. , 2018. Improvements to the APBS biomolecular solvation software suite. Protein Sci 27, 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, 2010. XDS. Acta Crystallogr D66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazrani AA, Kowalska M, Czapinska H, Bochtler M, 2014. Crystal structure of the 5hmC specific endonuclease PvuRts1I. Nucleic Acids Res 42, 5929–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BN, Shin M, Ha SC, Park SY, Seo PW, Hofmann A, et al. , 2017. Crystal structure of an ASCH protein from Zymomonas mobilis and its ribonuclease activity specific for single-stranded RNA. Scientific Reports 7, 12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisiala M, Copelas A, Czapinska H, Xu SY, Bochtler M, 2018. Crystal structure of the modification-dependent SRA-HNH endonuclease TagI. Nucleic Acids Res 46, 10489–10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger T, Wild C, Noyer-Weidner M, 1995. McrB: a prokaryotic protein specifically recognizing DNA containing modified cytosine residues. EMBO J 14, 2661–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S, 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol 8, 317–327. [DOI] [PubMed] [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y, 2014. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res 24,1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA, Raleigh EA, 2014. The other face of restriction: modification-dependent enzymes. Nucleic Acids Res 42, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Tong L, 2014. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA. 111,13834–13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz T, Flodman K, Copelas A, Czapinska H, Mabuchi M, Fomenkov A, He X, Bochtler M, Xu SY, 2019. A protein architecture guided screen for modification dependent restriction endonucleases. Nucleic Acids Res 47, 9761–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirwan N, Singh P, Mishra GG, Johnson CM, Szczelkun MD, Inoue K, Vinothkumar KR, Saikrishnan K, 2019a. Hexameric assembly of the AAA+ protein McrB is necessary for GTPase activity. Nucleic Acids Res 47, 868–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirwan N, Itoh Y, Singh P, Bandyopadhyay S, Vinothkumar KR, Amunts A, Saikrishnan K, 2019b. Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC. Nat. Commun 10, 3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll J, Heiter DF, Wilson GG, Fitzgerald GF, Roberts R, Van Sinderen D. 2006. A genetic dissection of the LlaJI restriction cassette reveals insights on a novel bacteriophage resistance system. BMC Microbiol. 6,40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan DJ, Zagula K, Klaenhammer TR, 1995. In vivo restriction by LlaI is encoded by three genes, arranged in an operon with llaIM, on the conjugative Lactococcus plasmid pTR2030. J. Bacteriol 177, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima H, Matsuoka S, Asai K, Sadaie Y, 2002. Molecular organization of intrinsic restriction and modification genes BsuM of Bacillus subtilis Marburg. J. Bacteriol 184, 381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM, 2008. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J. Biol. Chem 283, 34490–34494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Raleigh EA, Bickle TA, 1999. The McrBC endonuclease translocates DNA in a reaction dependent on GTP hydrolysis. J. Mol. Biol 290, 49–60. [DOI] [PubMed] [Google Scholar]
- Panne D, Müller SA, Wirtz S, Engel A, Bickle TA, 2001. The McrBC restriction endonuclease assembles into a ring structure in the presence of G nucleotides. EMBO J 20, 3210–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Arellano I, Gallego J, Cervera J, 2007. The PUA domain - a structural and functional overview. FEBS J 274, 4972–4984. [DOI] [PubMed] [Google Scholar]
- Pieper U, Schweitzer T, Groll DH, Gast FU, Pingoud A, 1999. The GTP-binding domain of McrB: more than just a variation on a common theme? J. Mol. Biol. 292, 547–556. [DOI] [PubMed] [Google Scholar]
- Pieper U, Pingoud A, 2002. A mutational analysis of the PD...D/EXK motif suggests that McrC harbors the catalytic center for DNA cleavage by the GTP-dependent restriction enzyme McrBC from Escherichia coli. Biochemistry 41,5236–5244. [DOI] [PubMed] [Google Scholar]
- Pieper U, Groll DH, Wünsch S, Gast FU, Speck C, Mücke N, et al. , 2002. The GTP-dependent restriction enzyme McrBC from Escherichia coli forms high-molecular mass complexes with DNA and produces a cleavage pattern with a characteristic 10-base pair repeat. Biochemistry 41, 5245–5254. [DOI] [PubMed] [Google Scholar]
- Raleigh EA, Wilson G, 1986. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 183, 9070–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasnauskas G, Zagorskaitė E, Kauneckaitė K, Tamulaitiene G, Siksnys V, 2015. Structure-guided sequence specificity engineering of the modification-dependent restriction endonuclease LpnPI. Nucleic Acids Res 43, 6144–6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Wang C, Zang J, 2014. Structural basis for the substrate selectivity of PvuRts1I, a 5-hydroxymethylcytosine DNA restriction endonuclease. Acta Crystallogr D70, 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM, 2008. A short history of SHELX. Acta Crystallogr A64,112–122. [DOI] [PubMed] [Google Scholar]
- Schwarz FW, Tóth J, van Aelst K, Cui G, Clausing S, Szczelkun MD, Seidel R, 2013. The helicase-like domains of type III restriction enzymes trigger long-range diffusion along DNA. Science 340, 353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slyvka A, Zagorskaitė E, Czapinska H, Sasnauskas G, Bochtler M, 2019. Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping. Nucleic Acids Res 47, 11943–11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Müller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M, 2013. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell, 152, 1146–1159. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Panne D, Bickle TA, Raleigh EA, 2000. Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J. Mol. Biol 298, 611–622. [DOI] [PubMed] [Google Scholar]
- Sugahara M, Asada Y, Morikawa Y, Kageyama Y, Kunishima N, 2008. Nucleant-mediated protein crystallization with the application of microporous synthetic zeolites. Acta Crystallogr D64, 686–695. [DOI] [PubMed] [Google Scholar]
- Sukackaite R, Grazulis S, Tamulaitis G, Siksnys V, 2012. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res 40, 7552–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland E, Coe L, Raleigh EA, 1992. McrBC: a multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol 225, 327–348. [DOI] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH, 2014. Solution structure of the YTH domain in complex with N6-methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 42, 13911–13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock MR, Dryden DT, 2005. The biology of restriction and anti-restriction. Curr. Opin. Microbiol 8, 466–472. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J, 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Weigele P, Raleigh EA, 2016. Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. Chem. Rev 116, 12655–12687. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhu Y, Bao H, Jiang Y, Xu C, Wu J, et al. , 2016. A novel RNA-binding mode of the YTH domain reveals the mechanism for recognition of determinant of selective removal by Mmi1. Nucleic Acids Res 44, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. , 2014. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol 10, 927–929. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J, 2015. Structural Basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem 290, 24902–24913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorskaitė E, Manakova E, Sasnauskas G, 2018. Recognition of modified cytosine variants by the DNA-binding domain of methyl-directed endonuclease McrBC. FEBS Lett 592, 3335–3345. [DOI] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, et al. , 2014. Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 24:1493–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.