Abstract
In vivo delivery of large RNA molecules has significant implications for novel gene therapy, biologics delivery, and vaccine applications. We have developed cationic nanolipoprotein particles (NLPs) to enhance the complexation and delivery of large self-replicating mRNA (replicon) in vivo. NLPs are high-density lipoprotein (HDL) mimetics, comprised of discoidal lipid bilayer stabilized by apolipoproteins, that are readily functionalized to provide a versatile delivery platform. Herein, we systematically screened NLP assembly with a wide range of lipidic and apolipoprotein constituents, using biophysical metrics to identify lead candidates for in vivo RNA delivery. NLPs formulated with cationic lipids successfully complexed with RNA replicons encoding luciferase, provided measurable protection from RNase degradation, and promoted replicon in vivo expression. The NLP complexation of the replicon and in vivo transfection efficiency were further enhanced by modulating the type and percentage of cationic lipid, the ratio of cationic NLP to replicon, and by incorporating additive molecules.
Keywords: cationic, nanolipoprotein particle, NLP, HDL, self-amplifying mRNA, replicon, in vivo delivery
Graphical Abstract
Cationic HDL mimetics, termed nanolipoprotein particles (NLPs), can bind and complex with large replicon RNA molecules. Tuning the NLP composition and NLP-to-RNA ratios enabled control over formulation charge and stability. NLP-mediated in vivo delivery enhanced expression encoded protein cargo, and provides a readily functionalized, versatile platform for in vivo delivery of mRNA constructs.
INTRODUCTION
Self-replicating mRNA constructs (also referred to as replicons) have recently garnered significant attention for therapeutic applications(1-4). Derived from the single-stranded RNA genome of alphaviruses (in particular Venezuela Equine Encephalitis Virus, VEEV), replicons are able to self-replicate in the cytoplasm of host cells by importing the necessary transcriptional machinery for effective propagation(5, 6). In lieu of structural viral proteins, genes encoding the desired proteins are engineered into the replicon. As the mRNA itself is actively amplified, the protein of interest can often express at a high level, for extended periods of time (16 to 60 days, depending on the formulation)(4, 7). With respect to vaccines, replicons are anticipated to provide a number of benefits over protein-based vaccines due to their self-replicating abilities, highly efficient protein synthesis, and enhanced immunogenicity potential(8). In addition, replicons can be rapidly constructed with specific mutations of interest, are easily produced, and are more potent than pDNA(1). The administration of these larger nucleic acid constructs, however, is often limited by significant obstacles for in vivo delivery(9).
Several non-viral replicon delivery methods have shown promise for vaccine development, including lipid nanoparticle formulations (LNPs) and cationic nanoemulsions (CNEs) (7, 10, 11). LNPs are functionalized liposomes that encapsulate the large mRNA molecules (~150nm diameter, z-average)(7), while the cationic nanoemulsions are spherical nano-droplets (~100nm diameter, z-average)(10) that provide a charged surface to which the mRNA can bind. These synthetic lipid-based systems have demonstrated efficiency as in vivo replicon delivery platforms and provide a number of key benefits relative to viral vectors, including enhanced safety profiles and relative ease in manufacturing. Our focus was on smaller nanoparticle platforms (e.g. below 40 nm diameter) that could be used to associate with and condense large mRNA molecules, effectively decorating the mRNA with multiple nanoparticles to package and potentially protect the mRNA. In this study, we evaluated the ability of biocompatible nanoparticles, analogous to high density lipoproteins (HDLs), to effectively package replicon RNA for in vivo delivery. Nanolipoprotein particles (NLPs), commonly referred to as nanodiscs or reconstituted high-density lipoproteins (rHDLs), exhibit a unique structure and marked versatility to complex with other biological entities, differentiating it from other nanoparticle formulations(12, 13). NLPs are discoidal nanoparticles with tunable diameters between 6 and 25nm, featuring a lipid bilayer supported by an apolipoprotein belt at the bilayer periphery(12, 14) (Figure 1). The self-assembly of NLPs is highly versatile and can be used to prepare particles with a wide range of lipid, apolipoprotein derivates, and lipidic/amphipathic components. Judicious use of starting components and ratios provides a facile means of tailoring NLP size, stability, and functionality(15). NLPs also provide several advantages for in vivo drug delivery, including rapid cellular uptake, high biocompatibility, while exhibiting no overt toxicity(12). Importantly, NLPs have significantly greater surface area than other lipidic nanoparticles, such as liposomes(16), and have been used successfully for biomolecule formulations and delivery(17-20). Recent work has also focused on tailoring NLPs for modulating the host immune system, either by stimulating the innate immune response or by eliciting a vaccine-based humoral immune response through co-localized delivery of both antigen and adjuvant molecules(14, 18, 21-23). While cationic rHDLs have been shown to enhance delivery of short oligonucleotides in vitro(17), a systematic study exploring and analyzing various zwitterionic bulk lipids, cationic lipids, and scaffold proteins for use in nucleic acid delivery in vivo has not been reported. As such, NLPs provide a number of possible benefits as a delivery platform for mRNA molecules: 1) facile tailoring of NLP physicochemical parameters (e.g. charge, charge density, size) provides a rich pool of candidates to enhance in vivo delivery, 2) increased surface area and smaller size could more effectively complex with and protect mRNA molecules, 3) controlled versatility in incorporating additional functional elements, e.g. adjuvants for vaccine applications, and 4) can be complexed with mRNA immediately prior to administration without further purification, providing a versatile point-of-care delivery option for mRNA therapeutics. These possible advantages merited an in-depth analysis of how NLPs can be formulated to package replicon mRNA molecules and evaluate their ability to mediate in vivo delivery to promote gene expression.
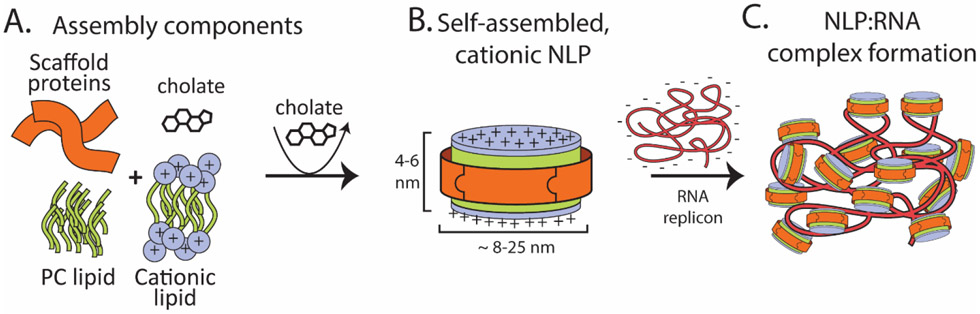
Figure 1.
Schematic of NLP assembly and RNA complex formation. (A) Purified starting components (apolipoprotein scaffold protein, zwitterionic phosphatidylcholine lipids, cationic lipids) were solubilized in sodium cholate. (B) Incubation with bio-beads removed cholate and initiated self-assembly of cationic NLPs. (C) Cationic NLPs incubated with replicon RNA form complexes used for further characterization and in vivo efficacy assessments.
Here, we discuss a method whereby we rapidly synthesize and screen for stable NLP constructs containing different cationic lipids formulated with RNA replicon. Moreover, we demonstrate that RNA replicon formulated with cationic lipid charged NLPs is a promising platform for in vivo transfection and gene delivery applications. Our NLP formulations tightly bind to RNA replicon, forming macromolecular complexes driven by electrostatic interactions. NLP:replicon complexes were characterized using gel electrophoresis, zeta potential analysis, and dynamic light scattering. Subsequently, the NLP:replicon complex was screened for in vivo activity. Further optimization of NLP formulations by incorporating glycerol monooleate (GMO) enhanced in vivo activity, as 1 μg of NLP formulated replicon resulted in comparable levels of luciferase protein expression as with 30 μg of unformulated replicon.
METHODS
Materials
Lipids were purchased from Avanti Polar Lipids: 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), didodecyldimethylammonium bromide (DDAB), 3ß-[N-(N’,N’-dimethylaminoethane)-carbamoyl]cholesterol hydrochloride (Chol), 1,2-dimyristoyl-sn-glycero-3-ethylphosphocholine (DMEPC), 1,2-dioleoyl-sn-glycero-3-ethylphosphocholine (DOEPC), dimyristoyltrimethylaminopropane (DMTAP), 1,2-dioleoyl-3-trimethylammoniumpropane (DODAP), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE), 1,2-dioleoyl-3-trimethylammoniumpropane (DOTAP), 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), N1-[2-((1S)-1-[(3-aminopropyl)amino]-4-[di(3-amino-propyl)amino]butylcarboxamido)ethyl]-3,4-di[oleyloxy]-benzamide (MVL-5). Glyceryl monooleate (GMO) was purchased from Nu-Chek (Elysian, MN). All other chemicals were purchase from Sigma-Aldrich (St. Louis, MO) or as indicated in the methods below.
Production and Purification of Apolipoproteins
Two different forms of apolipoproteins were produced and purified. The expression plasmid to produce murine apoE4-22k (apoE) was kindly provided by Dr. Karl Weisgraber. ApoE4-22k is the N-terminal 22 kDa fragment of murine apolipoprotein E4, cloned into pET32b vector to have a 6xHis- and thioredoxin fusion tag. The apoE4-22k was expressed and purified using a similar protocol as previously described(24, 25).
Murine apoA1-Δ49 (apoA1) was cloned into expression vector pET11 and was engineered to include a TEV-cleavable N-terminal His-tag for purification. The expression and purification were also described previously(26), with slight modifications(27).
NLP Synthesis
NLPs were synthesized according to a previously described in vitro assembly method with slight modifications(12). Additional details can be found in the Supplementary Materials.
Preparation of replicon RNA
The replicon vector is based on the current investigational new drug (IND) VEE virus vaccine (TC-83)(28). The red firefly (rFF) luciferase gene was PCR-amplified from a commercial DNA plasmid (ThermoFisher), and cloned as an AscI fragment into the replicon plasmid as previously described(29). The orientation of the gene was determined by restriction digest analysis and clones in the sense orientation were selected and sequence verified. The replicon construct was 9420 bases in length. Replicon RNA was prepared as described previously(30). Briefly, capped replicon RNAs were in vitro transcribed using a T7 RiboMax kit (Promega, Madison WI) following the manufacturer’s instructions, supplemented with 7.5 mM CAP analog (Promega, Madison, WI), from NotI linearized replicon plasmid. RNAs were purified using RNEasy purification columns (Qiagen, Valencia, CA) following the manufacturer’s instructions. RNA was aliquoted and stored at −80 °C until utilized.
Preparation of NLP:RNA formulations
Replicons were formulated with NLPs at various NLP:replicon molar ratios at RNA concentrations between 2 and 600 μg/ml. Formulations were always prepared by adding NLPs to the replicon, which was pre-diluted in PBS. Formulations were incubated at room temperature for at least 5 minutes prior to analysis or use. NLP:replicon ratios were based on molar ratios, assuming a replicon MW of 2,900 kDa. NLP MWs were based on assuming 2 scaffolds per apoA1 NLP(31) and 5 per apoE NLP(25), with apolipoprotein molecular weights of 21.1 and 23.3 kDa, respectively. Nitrogen/phosphate (N/P) ratios were calculated using the number of negatively charged phosphates in the replicon (9420 bases = 9420 negative charges (P)). Cationic charges (N) were based on the percentage of cationic lipids, assuming 160 and 850 total lipid molecules per apoA1-and apoE-based NLP, respectively.
Agarose gel electrophoresis
Agarose gels were freshly prepared with 1% bacto-agar and 1X PBS pH 7.2. Before pouring into chamber, ethidium bromide solution was added at a final concentration of 0.5 μg/ml. NLP:replicon complexes were formulated at molar ratios ranging from 15:1 – 500:1 (NLP-to-replicon). Reactions were incubated for 5 minutes at room temperature before being combined with gel loading dye and ran at 400 mA for 30 minutes with 1x PBS pH 7.2 as running buffer. Unformulated RNA was also loaded as a control. Gel was imaged at 600 nm for 2 minutes with LiCor Odyssey FC system.
RNase protection assay
Unformulated RNA and NLP:replicon complexes were incubated with 3.8 mAU of RNase A (Life Technologies) per μg RNA for 1 minute at room temperature. RNase was then inactivated with the addition of 2 AU of Anti-RNase (Life Technologies) per 1 ng RNase at room temperature for 30 minutes. Subsequently, 1% Triton was added to each sample to disrupt NLP and release RNA. A 1% agarose gel in tris-acetic acid-EDTA (TAE) buffer was prepared, loaded (0.1 μg RNA per lane), and run at 90V for 30 minutes. The gel was stained with SYBR Gold (Invitrogen; 1:1000 dilution in water) for 1 hour while shaking. Gels was then directly imaged with a Kodak Gel Logic 200 using UV illumination.
Dye exclusion analysis
NLP:replicon complexes were formulated at indicated molar ratios. In short, PBS, RNA, then NLP were mixed together and allowed to incubate at room temperature for at least 5 minutes. 0.1 μg RNA was added to each vial. Dye intercalation was visualized via Quant-iT Ribogreen RNA Reagent and Kit (Invitrogen) according to manufacturer’s instructions.
RT-PCR quantitation
Unformulated RNA and NLP:replicon complexes were prepared as described in “RNase protection assay” for RNase and Anti-RNase treatment. QuantiTect Probe RT-PCR kit (Qiagen) was used to run samples, adding 1% Triton to the Master Mix. 5 ng RNA was loaded per PCR well. PCR was run at 30 °C for 30 minutes, 95 °C for 15 minutes, then [95 °C for 15 seconds, 60 °C for 30 seconds) X 30 cycles] (7900 HT Fast Real-Time PCR Machine; Applied Biosystems/Life Technologies).
Zeta potential analysis
The Zeta potential and particle size of the NLP and NLP:replicon complexes were measured using a Zetasizer Nano ZS 90 (Malvern Instruments, Orsay, France). NLP or NLP:replicon samples in PBS were diluted 25-fold in water (final total salt concentration was 6mM). Zeta potential and size distribution were analyzed at 25°C according to the manufacturer’s instructions. Particle sizes are reported as Number Mean (d.nm).
In vivo assessment of luciferase expression
All in vivo experiments were conducted after review and approval by the Institutional Animal Care and Use Committee (IACUC) at Lawrence Livermore National Laboratory. Female BALB/c mice (Harlan), aged 5-8 weeks and weighing about 20 g, were used for all in vivo imaging studies. Mice were bilaterally injected intramuscularly at the midline of each hindlimb quadricep with 50 μL of unformulated RNA or NLP formulated RNA complexes. All test materials were formulated in PBS and filter-sterilized. Four days after injection, luciferase expression levels were assessed using an IVIS Spectrum instrument (Perkin Elmer) at the University of California, Davis, Center for Molecular and Genomic Imaging (CMGI). Five minutes before imaging, mice were injected intraperitoneally with 0.15 mg/g of luciferin solution (Caliper Lifesciences). Mice were subsequently anesthetized [2% (vol/vol) isoflurane in oxygen] and positioned in the IVIS instrument to image from both dorsal and ventral positions. Relative luciferase expression was based on quantifying the total flux (photons per second) in a pre-defined area (region of interest was consistent for all analyses). The expression level of luciferase for each mouse is reported as the highest total flux value from one of the limbs and positions. All acquisition and imaging parameters were kept constant across experiments (e.g. exposure time, binning, field-of-view). Error bars represent SD. Statistical analyses was performed using GraphPad PRISM 7 software. Significance analyses for in vivo luciferase expression was performed using ANOVA followed by Kruskal-Wallis or Tukey’s post hoc test for multiple comparisons.
RESULTS
In order to evaluate the ability of NLPs to deliver large RNA molecules in vivo, it was first necessary to demonstrate that the NLPs could be prepared with a variety of cationic lipids and cationic lipid densities. As such, NLPs were assembled with different cationic lipids at varying molar ratios to alter the cationic charge on the lipid bilayer surface (Figure 1). The ability to form NLPs with a variety of lipid constituents was screened with truncated versions of two mouse apolipoproteins, ApoA1-Δ49 and ApoE4-22k, as candidate scaffold proteins for the NLP synthesis (referred to as apoA1 and apoE, respectively). ApoE4-22k, unlike the apoE2 and apoE3 isoforms, does not contain any cysteine residues, thus simplifying and increasing efficiency of recombinant protein expression, purification, and NLP assembly. NLP formation with cationic lipids was assessed using three common zwitterionic bulk lipids: DOPC, POPC, and DMPC, which have previously been shown to result in stable NLP formation(18, 32, 33). We tested a wide range of cationic lipids at increasing molar percentages of total lipid to test the compatibility with apoA1 and apoE scaffold proteins and zwitterionic lipids (Supplemental Table 1). NLPs were assembled with a range of different lipid and protein ratios and were analyzed by size exclusion chromatography (SEC) for proper nanoparticle formation(32-34). The SEC chromatograms from the high-throughput NLP synthesis were assessed for peak height, elution time (representing the size of NLP formed), and homogeneity of the NLP population, as well as the presence of larger lipidic aggregates and unincorporated apolipoprotein. The type of cationic lipid has a profound impact on NLP formation. For example, as shown in Figure 2A, NLPs prepared with apoA1, DMPC, and increasing molar percentages of DDAB were stable up to 20% DDAB , while 40% DDAB formulations produce a heterogeneous mixture of large NLPs and aggregates. DMPC-based apoE NLPs readily accommodate up to 30% DODAP, and only exhibit a mild increase in heterogeneity in NLP size at 40% DODAP (Figure 2B). Similar assessments were made for DMPC-based NLPs prepared with a series of cationic lipids, and the results are summarized in Figure 2C. DMEPC and DMTAP are among the most tolerated cationic lipids and can comprise up to 40% of the total lipid content. Other cationic lipids, such as DOTAP, DOTMA and MVL-5, are well tolerated at up to 20% of the total lipid content (Figure 2C). Furthermore, in most cases both apoA1 and apoE can successfully form cationic NLPs (Figure 2C), whereby apoA1 NLPs can typically accommodate higher percentages of cationic lipid relative to apoE, although some exceptions to this trend were observed (e.g. DODAP). For apoE-based NLPs, Bulk DMPC lipid tended to tolerate higher percentages of cationic lipids as compared to bulk POPC or DOPC (Supplemental Figure S1).
Figure 2.
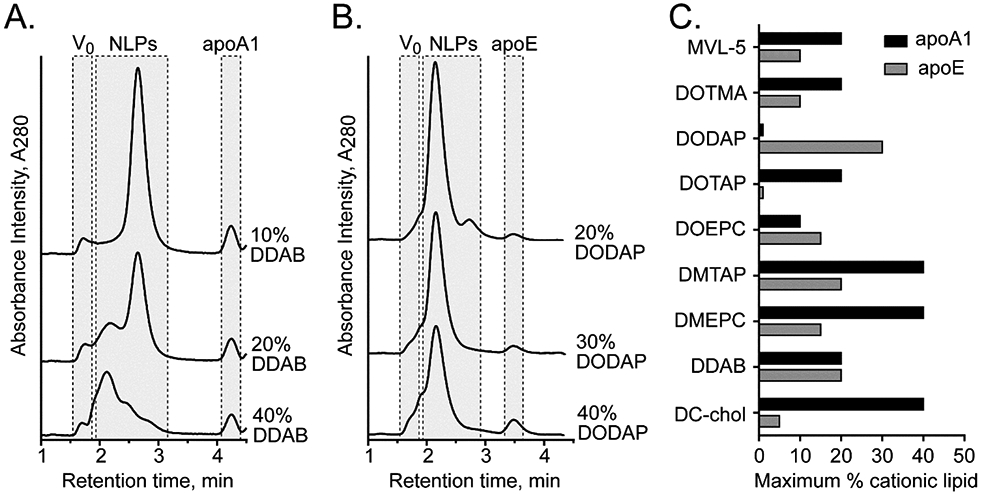
High-throughput screening of cationic NLP assembly. (A, B) Representative SEC chromatograms of cationic NLPs prepared with (A) apoA1 and DMPC, assembled at increasing mol% of DDAB, and (B) apoE and DMPC, assembled with increasing mol% of DODAP. Elution regions corresponding to column void volume (V0), NLP species (NLP), and unincorporated free apolipoprotein (apo) are indicated. (C) Summary graph of the maximal cationic lipid content (% of total lipid during assembly) that successfully formed DMPC-based NLPs prepared with apoA1 (black bars) or apoE (gray bars).
To evaluate the ability of the cationic NLPs to associate and complex with large RNA molecules, we conducted a detailed analysis using a representative cationic NLP formulated with the cationic lipid MVL-5. The RNA molecule used for these experiments was a replicon encoding the bioluminescent protein firefly luciferase (9,420 bases in length). First, gel electrophoresis was used as a qualitative measure of association between NLP and replicon. The complexation of cationic NLPs with the replicon was expected to result in a mobility shift of the RNA molecules through the agarose gel by impeding the migration of the anionic RNA molecule to the cathode due to both an increase in RNA complex size as well as an attenuation of the RNA negative charge by the cationic NLPs. A representative demonstration of this is shown in Figure 3A. replicon incubated with non-cationic NLPs (denoted as 0% MVL-5) has little to no impact on replicon migration through the gel relative to replicon alone, regardless of NLP:replicon ratio. However, NLPs formulated with 1 mol% cationic lipid MVL-5 exhibited a dose-dependent effect on replicon migration: as the NLP:replicon molar ratio was increased, the migration of the RNA relative to replicon alone was reduced. NLPs incorporating 2% and 5% MVL-5 exhibited identical trends in reducing RNA mobility. In some cases, the RNA failed to migrate out of the well, suggesting either full charge attenuation or the formation of significantly large structures impeding penetration into the gel matrix. These association trends are seen across NLPs formed with different cationic lipids, apolipoproteins, cationic lipid percentages, and NLP:replicon ratios (Supplemental Figures S2-S3). We also demonstrated that these NLP:replicon complexes are stable when stored at 4 °C over a span of two weeks (Supplemental Figure S4), whereas cationic NLPs alone are stable for at least 50 days under these same conditions (Supplemental Figure S5).
Figure 3.
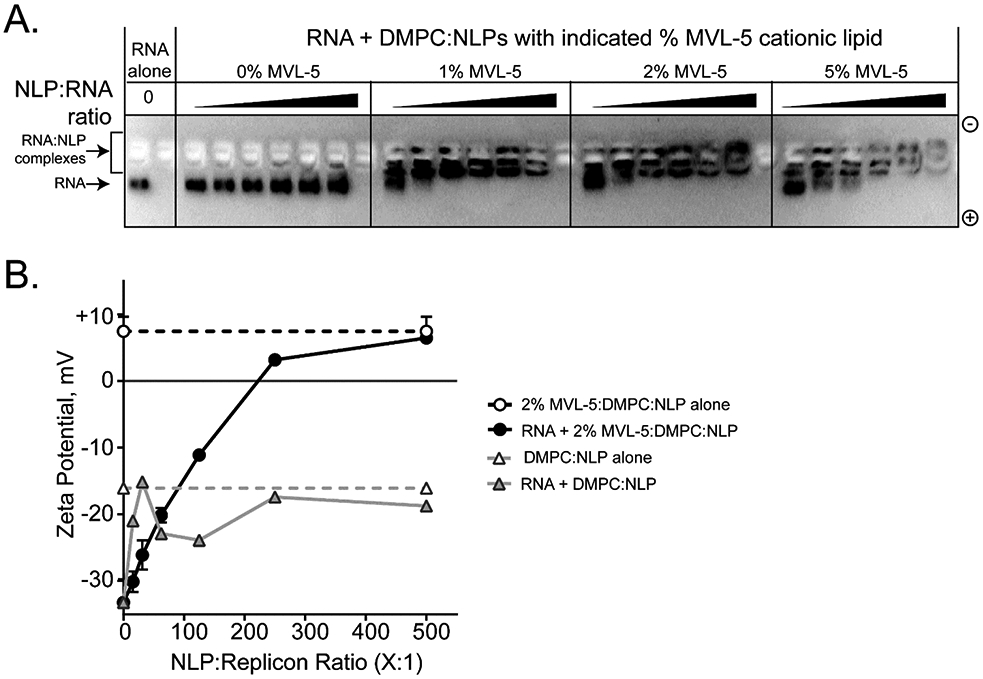
Cationic NLPs form complexes with replicon RNA. (A) Agarose gel electrophoresis demonstrates interaction between replicon RNA and cationic NLPs. In this representative example, apoA1 NLPs prepared with increasing percentages of MVL-5 (0, 1, 2, and 5%) were incubated with replicon RNA (NLP:RNA ratios = 15:1, 31:1, 62.5:1, 125:1, 250:1, and 500:1) for 30 minutes prior to agarose gel electrophoresis. Decreased RNA mobility was observed at higher NLP:RNA ratios. Non-cationic NLPs (0% MVL-5) had no effect on RNA mobility. Gels were stained with SybrGold to visualize RNA. (B) Zeta potential was characterized for representative NLP:replicon formulations. RNA complexes with cationic NLPs (circles, black trace; apoA1, DMPC, 2% MVL-5) trend toward increasingly positive zeta potential at increasing NLP:RNA ratios. Non-cationic NLPs (triangles, gray trace; apoA1, DMPC, 0% MVL-5) do not alter the intrinsically negative zeta potential of the RNA replicon. Images are representative of 3 independent experiments.
The cationic MVL-5- NLPs were further characterized to determine change in zeta potential of RNA replicon upon NLP binding at increasing molar ratios (Figure 3B). Unformulated RNA has a zeta potential of approximately −30 mV (ranges between −20 mV to −40 mV), whereas the cationic MVL-5 based NLPs exhibit a zeta potential of +10 mV. Cationic NLPs were formulated with RNA replicon at molar ratios ranging from 15:1 to 500:1 (NLP:replicon). At increasing cationic NLP to RNA ratios, the zeta potential of the complex became more positive, transitioning from negative to positive at around 200:1 ratio (NLP:replicon). Depending on the type of cationic NLP, the transition point from negative to positive zeta potential occurred at molar ratios between 125.5:1 and 500:1 (NLP:replicon) (Supplemental Figure S6A). When mixed with non-cationic NLPs (DMPC only), no trend in association between replicon and NLP is observed. An analysis of the zeta potential distribution further demonstrates the direct association between charged NLP and replicon. Cationic NLPs form a single complex with replicon, and these complex increase in zeta potential as the ratio of NLP increases, whereas two distinct populations are observed when the non-cationic NLPs are mixed with replicon, irrespective of ratio (Supplemental Figure S6B). These observations further demonstrate that cationic NLPs form stable interactions with RNA that are driven by electrostatic interactions.
The particle size of the complex was measured using dynamic light scattering (DLS) (Supplemental Tables 2 and 3). The hydrodynamic diameters of apoA1-based cationic NLPs were between 12-25 nm, whereas the apoE based cationic NLPs were slightly larger, between 30-40 nm. The replicon alone has a size of approximately 90 nm under our experimental conditions. The NLP:replicon complex size ranged between 60 nm and 900 nm, depending on the type of cationic NLP and NLP:replicon ratios. In general, apoA1 cationic NLPs formed larger structures than apoE NLPs when complexed with RNA replicon. This may indicate structural differences in the complex formation between apoA1 and apoE cationic NLP:replicon complexes and may be driven in part by the significantly larger surface area of apoE-based NLPs.
One key requirement for an effective RNA delivery formulation is the ability to protect RNA from RNase digestion. We demonstrated that NLP association stabilized the RNA and significantly reduced degradation by RNase as ratios of NLP-to-RNA increased, whereas unformulated RNA is completely degraded (Supplemental Figure S7). This protective effect is also demonstrated using an RT-PCR assay that measures integrity of the replicon RNA (Supplemental Figure S8). In addition to RNase protection, the RNA replicon also becomes less accessible to small dye molecules when associated with NLP (Supplemental Figure S9).
To study the in vivo transfection efficiency of our NLP formulations, RNA replicons encoding a firefly luciferase reporter gene were administered to BALB/c mice as a single bilateral dose injected intramuscularly (i.m.) at the midline of each hindlimb quadricep, as is routine for the administration of self-amplifying mRNA constructs(7, 10, 35) None of the mice exhibited negative side-effects from the injection, based on physiological cues. Animals were imaged 4 days after replicon administration by whole body luminescence imaging (representative images shown in Supplemental Figure S10). Typical formulations consisted of 1 μg RNA replicon complexed with various NLP formulations, and were compared to control formulations (PBS sham or unformulated replicon, 1 μg or 30 μg) (Figure 4). To study the ability of NLPs enhancing replicon delivery and protein expression, parameters including cationic lipid constituents, cationic lipid percentages (up to 40%), and NLP:replicon formulation ratios were assessed. From the control groups, the average total flux resulting from injection of 30 μg non-formulated RNA was about 4-fold higher than that from injection of 1 μg. Replicons formulated with NLPs prepared with either apoA1 or apoE (Figure 4) exhibited a wide range of in vivo bioluminescence intensities, from no luciferase expression (Figure 4; apoE, 12.5% MVL-5, 50:1 NLP:replicon) to levels comparable to 30 μg of unformulated replicon (Figure 4b; apoE, DODAP 30%, 12.5:1 NLP:replicon). While the majority of formulations did not enhance mean total flux over unformulated RNA, a handful of NLP/RNA formulations demonstrated enhanced luciferase expression. In particular, replicons formulated with apoA1 20% DMTAP NLPs (250:1 NLP:replicon ratio) and apoE 30% DODAP NLPs (12.5:1 NLP:replicon ratio) exhibited the greatest apparent increases in expression compared to unformulated RNA (1 μg), although these differences were not statistically significant. As the toxicity of cationic moieties is well documented(36), we assessed the in vitro cytotoxicity of our top cationic NLP formulations in cultured HEK 293 T cells (Supplemental Figure S11)(35). Cationic NLPs (apoA1 20% DMTAP and apoE 30% DODAP NLPs) are well tolerated with no statistically significant effect on cell viability relative to a PBS vehicle control.
Figure 4.
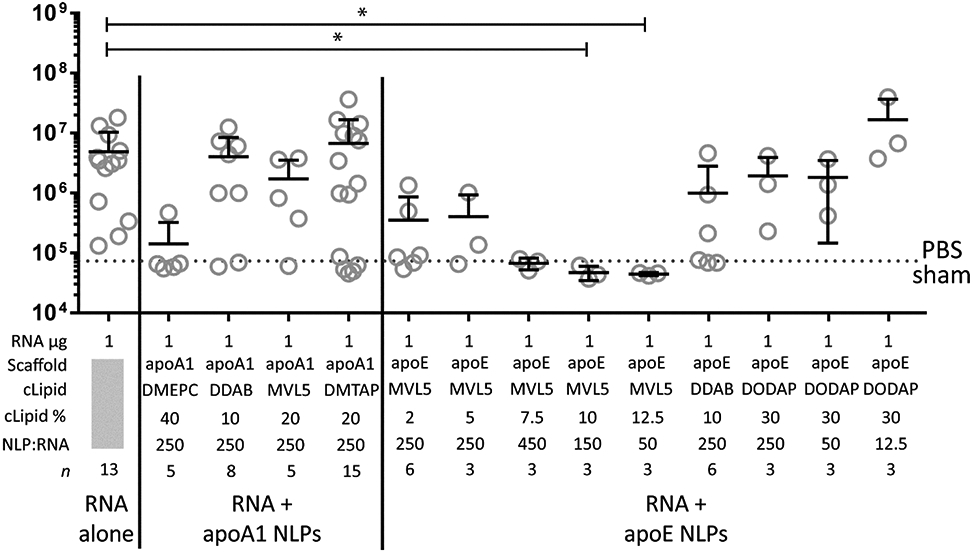
Cationic NLPs enhance in vivo replicon activity. In vivo luminescence in groups of female BALB/c mice injected i.m. with PBS (sham, dotted line), unformulated RNA (1 μg and 30 μg)and RNA (1 μg) formulated with NLPs prepared with apoA or apoE scaffold proteins. RNA amount, scaffold protein, cationic lipid (cLipid), cationic lipid percentage (cLipid%), ratio of NLP -to-replicon, and experimental replicates (n) are indicated. Mean and S.D. are shown. * = p < 0.05 by ANOVA followed by Kruskal-Wallis test for multiple comparisons.
Our initial efficacy screen provided a number of candidate formulations that merited additional optimization to enhance the overall in vivo gene expression from the RNA replicon. We picked the top performing apoA1 and apoE NLP formulations to see if we could further enhance the in vivo delivery and expression of the replicon: apoA1 20% DMTAP NLPs (250:1 NLP:replicon ratio) and apoE 30% DODAP NLPs (12.5:1 NLP:replicon ratio), respectively. To explore further optimization of the NLP formulations we included GMO. Recent reports have identified the ability GMO, in conjunction with cationic lipids, to enhance the efficacy or RNA delivery to cells(37, 38). In part, the ability of GMO to induce a cubic phase in lipidic membranes is believed to promote endosomal escape through enhanced pore formation, thus improving the delivery of RNA molecules to the cytosol. We hypothesized that a similar approach may enhance the efficacy of our cationic NLPs to deliver the replicons in vivo. NLPs were readily assembled with GMO: both apoA1 20% DMTAP and apoE 30% DODAP identified in our initial in vivo screen were assembled with 20% GMO with no discernable effect on NLP size or homogeneity (Figure 5A). When incubated with replicon, NLPs containing GMO exhibited similar binding characteristics to replicon as non-GMO formulations, based on the shift in zeta potential of the NLP:replicon complex at increasing formulation ratios (Figure 5B).
Figure 5.
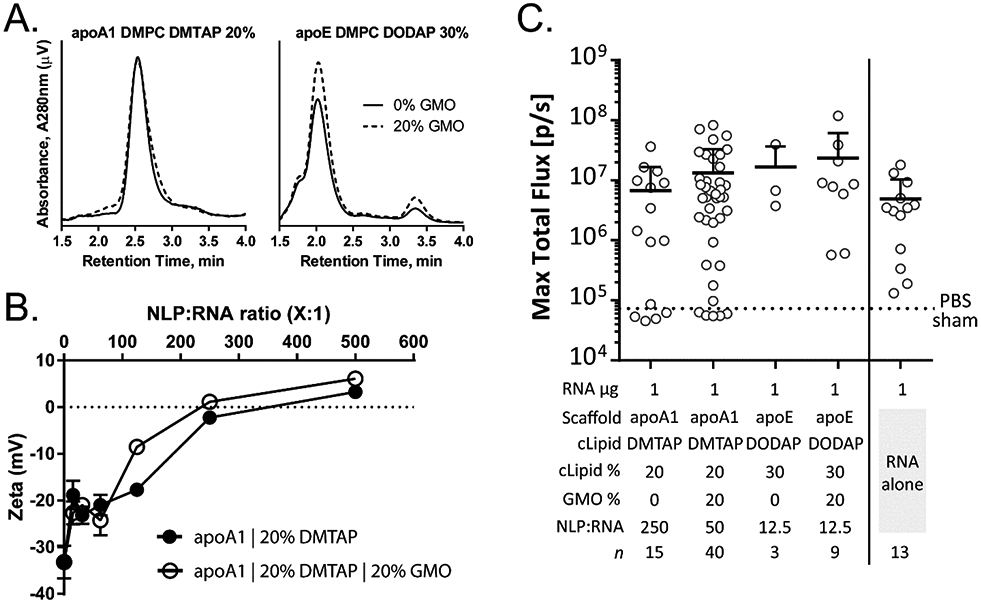
GMO enhances efficacy of cationic NLP:replicon formulations. (A) Analytical SEC of cationic NLPs supplemented with 20% GMO has no effect on NLP purity, size, or homogeneity. This trend is observed with cationic NLPs prepared with apoA1 NLPs containing the cationic DMTAP lipid (left) or apoE NLPs containing the cationic DODAP lipid (right). (B) apoA1 NLPs with DMTAP lipid exhibit only slight differences in complex formation and charge attenuation with and without 20% GMO. (C) In vivo luminescence of RNA (1 μg) formulated with cationic NLPs assembled with indicated cationic lipids ± 20% GMO, or unformulated RNA (1 and 30 μg). Each symbol represents a single animal. The scaffold protein, cationic lipid (cLipid), cationic lipid percentage (cLipid%), GMO percentage (GMO%), ratio of NLP-to-replicon, and experimental replicates (n) are indicated. Mean and S.D. are shown (upper bound only). Significance analysis was performed using ANOVA followed by Tukey’s post hoc test for multiple comparisons. No statistical significance was observed between the groups.
GMO-based cationic NLPs exhibited enhancement in replicon gene expression. Mirroring the experimental approach described above, GMO-containing cationic NLP compositions and replicon formulations were assessed for luciferase expression in vivo (Figure 5C). Enhanced mean total flux is observed with both the apoA1 20% DMTAP and apoE 30% DODAP formulations in the presence of GMO. Interestingly, the optimal NLP:replicon ratio for apoA1 20% DMTAP NLPs decreased from 250:1 to 50:1 in the presence of GMO. Again, while the gene expression differences were not statistically significant between the groups (even between the RNA-alone controls), the NLP-mediated delivery of the replicon trended toward enhanced expression, demonstrating that the NLP platform is amenable to in vivo mRNA delivery.
DISCUSSION
Self-amplifying mRNA molecules, or replicons, demonstrate great potential for in vivo therapeutic gene expression(39, 40). Developing novel delivery technologies to enhance or tune the in vivo delivery and gene expression of these self-amplifying mRNAs is of great interest to the research community. The NLP platform has been successfully implemented for a wide range of therapeutic applications, and, when coupled with replicon therapeutics, could provide synergistic therapeutic effects. As a first step towards developing NLPs for replicon in vivo delivery, we evaluated NLPs in the current study to establish if they are able to associate with replicons, and if these constructs can prevent RNA degradation by RNase. Since it has been reported that the in vivo and in vitro delivery of therapeutics require very different optimization in formulations, we decided to directly test and optimize the NLP:replicon formulations in vivo(41).
We demonstrated that cationic NLPs can be readily synthesized with a wide range of different types of cationic lipids, spanning incorporation percentages of 1-40% (contingent on lipid identity). Cationic NLPs are able to associate with replicon RNA, driven by charge complementarity between the cationic headgroups and the anionic replicon RNA backbone. In addition, we demonstrated that a number of cationic NLPs are able to facilitate the in vivo protein expression of the gene encoded by the replicon. How NLPs help enhance in vivo replicon transfection is an intriguing question. Naturally, HDL is known to trigger endocytosis upon attaching to the surface of various cell types(42, 43). Our previous studies demonstrated that NLPs exhibited significant cellular uptake of small cargo molecules conjugated to the NLPs(12, 18). However, the exact mechanism by which NLP:replicon complexes achieve cellular uptake will require in-depth in vitro experiments, which are currently ongoing.
One key advantage of our NLP-based formulations is the relatively low amount of cationic lipid required for efficacy. We compared cationic lipid usage between our NLP formulations and other effective systems by calculating the nitrogen/phosphate (N/P) ratio of our formulations, based on the number of moles of protonatable nitrogen in the cationic molecules to the number of moles of phosphate present in the RNA molecules. The N/P ratio of two “gold standard” formulations, cationic lipid nanoparticles and cationic nanoemulsions, are 8:1(7) and 7:1(10), respectively. The N/P ratios of NLP:replicon however, are often below 1:1 (Supplemental Table 4; Supplemental Figure S12). For instance, the N/P ratios of the two best performing formulations, apoA1 20% DMTAP (at a 50:1 NLP:replicon ratio) and apoE 30% DODAP (at a 12.5:1 NLP:replicon ratio) are only 0.17:1 and 0.34:1, respectively. While significant cytotoxicity has been observed with other delivery platforms utilizing similar cationic lipids(36, 44, 45), we hypothesized that the low N/P ratios achievable with our NLP platform would translate significantly greater biocompatibility. Indeed, no cytotoxic effects were observed in vitro with our top performing cationic NLPs (Supplemental Figure S11). The reason behind the low N/P ratio may lay in the unique structure of the NLP:replicon complex, whereby the small size and high surface are of the NLPs (relative to LNPs and CNEs) may effective form an NLP shell around the replicon (see schematic in Figure 1). We show that the NLP association provides measurable protection to RNA against RNase digestion, and the level of protection is proportional to the NLP:replicon ratio (Supplemental Figures S7 and S8), indicating that a higher coverage of replicon by NLP leads to better protection. Based on this model, the biophysical and biological properties of the complex are dictated by the components of NLP, including scaffold protein, lipids type, charge density, as well as the size of NLP and the molar ratio of NLP to RNA.
Both apoA1 and apoE apolipoproteins spontaneously form discoidal-shaped NLPs with DMPC lipids(32). However, apoA1 NLPs have a considerably smaller diameter than apoE NLPs(32, 33). This difference has direct impact on the number of positive charges on each NLP, as well as how NLPs interact with RNA molecule. For example, when looking at the difference in zeta potential between the respective replicon formulations, the apoE 10% DDAB formulation is about +14 mV, whereas the apoA1 10% DDAB formulation is about −12mV both at 250:1 ratio (Supplemental Figure S6). This large contrast in overall charge of the NLP:replicon complex may have impact on the overall structure and in vivo stability as well as transfection efficiency.
While NLP formation and association with replicon was observed across a wide range of cationic lipids, in vivo protein expression data strongly favored DMTAP- and DODAP-based cationic NLPs over others (Figure 4 and Supplemental Table 4). Meanwhile, some NLP formulations resulted in no protein expression, a significantly worse outcome than unformulated replicon. This is exemplified by NLPs with MVL-5 ratios above 7.5% (Figure 4). We expect there may be different mechanisms at play here, inhibiting either the delivery, endosomal release, or complex dissociation, and the mechanisms are likely dependent on the individual cationic lipid used. For example, a previous study suggested that the shorter alkyl chain of DMTAP in comparison to other cationic lipids, like DOTAP and DOTMA, allows the lipid:nucleotide complex to transfect cells more efficiently in vivo(46). Also, the similar alkyl chain between DMPC and DMTAP helps stabilize the bilayer inside NLP, therefore increasing the stability of the NLP:replicon complex. In the case of DDAB, it has been known that DDAB is a potent immunogenic adjuvant that facilitates interactions between an antigen and host cells(47). On the other hand, each MVL-5 molecule has 5 positive charges, while all other cationic lipids tested in this study featured a single positive charge. The increased charge density at the NLP surface may result in tightly bound lipid:RNA complex, impeding the full release of RNA and protein translation. In addition to the type of lipid, the percentage of charged lipid in the NLP and the NLP to replicon ratio both have an impact on the overall charge and charge density of the complex. Interestingly, the choice of NLP scaffold seems to perform best when paired with a specific cationic lipid, as apoA1 with DMTAP or apoE with DODAP. Future studies assessing the structure of the NLP:replicon complexes will help elucidate the role of different cationic lipids in stabilizing RNA replicon in both extracellular and intracellular environments and may help reveal how that impacts in vivo transfection efficiency.
In addition to enhancing RNA in vivo delivery, the defined structure and the versatile synthesis of NLP also make it an ideal platform for particle functionalization(15). Functional moieties can be readily coupled onto NLPs through hydrophobic interaction(20), chemical conjugation(48) or chelation(24, 25). Such functional moieties can include targeting ligand, therapeutic molecules, imaging agents, or even functional proteins(49). Recent work in our lab has successfully tailored NLPs for modulating the host immune system through co-localized delivery of both antigen and adjuvant molecules(14, 18, 24, 50, 51). The success of NLP-assisted replicon in vivo delivery provides a new way of designing the next generation of nucleic acid delivery systems.
In conclusion, we have demonstrated that cationic NLPs successfully promote in vivo delivery of large RNA molecules. Using cationic lipids, we can formulate reproducibly homogeneous NLP populations for consistent transfection in vivo. The synthesis of cationic charged NLPs and the association with RNA replicon are efficient, biocompatible, and versatile. In the future, we can easily implement additional features for gene therapy and vaccine development, such as adding adjuvants, functional polymers, and membrane-associated proteins for targeted in vivo delivery(13, 22, 52, 53).
Supplementary Material
ACKNOWLEDGEMENTS
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory (LLNL) under Contract DE-AC52-07NA27344 (LLNL-JRNL-747570). Funding was provided by LLNL LDRD award 20-ERD-004 and a Work for Others Agreement from Synthetic Genomics Vaccine Inc. (La Jolla, CA) to LLNL. Authors are employed by or have received funding from Synthetic Genomics Vaccine Inc. We kindly thank the University of California Davis Center for Molecular and Genomic Imaging for training on the IVIS imaging system.
Footnotes
Synthetic Genomics Vaccine Inc. (La Jolla, CA), provided funding under a Work for Others Agreement to LLNL. Authors are employed by or have received funding from Synthetic Genomics Vaccine Inc.
REFERENCES
- 1.Ljungberg K, Liljestrom P. Self-replicating alphavirus RNA vaccines. Expert review of vaccines. 2015;14(2):177–94. [DOI] [PubMed] [Google Scholar]
- 2.McCullough KC, Milona P, Thomann-Harwood L, Demoulins T, Englezou P, Suter R, et al. Self-Amplifying Replicon RNA Vaccine Delivery to Dendritic Cells by Synthetic Nanoparticles. Vaccines (Basel). 2014;2(4):735–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DY, Atasheva S, McAuley AJ, Plante JA, Frolova EI, Beasley DW, et al. Enhancement of protein expression by alphavirus replicons by designing self-replicating subgenomic RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(29):10708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel AB, Lambert L, Kinnear E, Busse D, Erbar S, Reuter KC, et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol Ther. 2018;26(2):446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, et al. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Virol. 2003;77(19):10394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayner JO, Dryga SA, Kamrud KI. Alphavirus vectors and vaccination. Rev Med Virol. 2002;12(5):279–96. [DOI] [PubMed] [Google Scholar]
- 7.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(36):14604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vander Veen RL, Harris DL, Kamrud KI. Alphavirus replicon vaccines. Animal health research reviews / Conference of Research Workers in Animal Diseases. 2012;13(1):1–9. [DOI] [PubMed] [Google Scholar]
- 9.Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brito LA, Chan M, Shaw CA, Hekele A, Carsillo T, Schaefer M, et al. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol Ther. 2014;22(12):2118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajj KA, Whitehead KA. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nature Reviews Materials. 2017;2(10). [Google Scholar]
- 12.Fischer NO, Weilhammer DR, Dunkle A, Thomas C, Hwang M, Corzett M, et al. Evaluation of nanolipoprotein particles (NLPs) as an in vivo delivery platform. PloS one. 2014;9(3):e93342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He W, Luo J, Bourguet F, Xing L, Yi SK, Gao T, et al. Controlling the diameter, monodispersity, and solubility of ApoA1 nanolipoprotein particles using telodendrimer chemistry. Protein science : a publication of the Protein Society. 2013;22(8):1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer NO, Rasley A, Corzett M, Hwang MH, Hoeprich PD, Blanchette CD. Colocalized delivery of adjuvant and antigen using nanolipoprotein particles enhances the immune response to recombinant antigens. Journal of the American Chemical Society. 2013;135(6):2044–7. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore SF, He W, Rasley A, Fischer NO. Strategies for Functionalizing Lipoprotein-Based Nanoparticles Control of Amphiphile Self-Assembling at the Molecular Level: Supra-Molecular Assemblies with Tuned Physicochemical Properties for Delivery Applications. ACS Symposium Series. 1271: American Chemical Society; 2017. p. 131–50. [Google Scholar]
- 16.Murakami T Phospholipid nanodisc engineering for drug delivery systems. Biotechnol J. 2012;7(6):762–7. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh M, Ren G, Simonsen JB, Ryan RO. Cationic lipid nanodisks as an siRNA delivery vehicle. Biochem Cell Biol. 2014;92(3):200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weilhammer DR, Blanchette CD, Fischer NO, Alam S, Loots GG, Corzett M, et al. The use of nanolipoprotein particles to enhance the immunostimulatory properties of innate immune agonists against lethal influenza challenge. Biomaterials. 2013;34(38):10305–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer NO, Blanchette C, Rasley A. Enhancing the efficacy of innate immune agonists: could nanolipoprotein particles hold the key? Nanomedicine. 2014;9(3):369–72. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh M, Singh AT, Xu W, Sulchek T, Gordon LI, Ryan RO. Curcumin nanodisks: formulation and characterization. Nanomedicine. 2011;7(2):162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman MA, Cappuccio JA, Blanchette CD, Gao T, Arroyo ES, Hinz AK, et al. Expression and Association of the Yersinia pestis Translocon Proteins, YopB and YopD, Are Facilitated by Nanolipoprotein Particles. PloS one. 2016;11(3):e0150166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He W, Scharadin TM, Saldana M, Gellner C, Hoang-Phou S, Takanishi C, et al. Cell-free expression of functional receptor tyrosine kinases. Sci Rep. 2015;5:12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater. 2017;16(4):489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer NO, Infante E, Ishikawa T, Blanchette CD, Bourne N, Hoeprich PD, et al. Conjugation to nickel-chelating nanolipoprotein particles increases the potency and efficacy of subunit vaccines to prevent West Nile encephalitis. Bioconjugate chemistry. 2010;21(6):1018–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchette CD, Fischer NO, Corzett M, Bench G, Hoeprich PD. Kinetic analysis of his-tagged protein binding to nickel-chelating nanolipoprotein particles. Bioconjugate chemistry. 2010;21(7):1321–30. [DOI] [PubMed] [Google Scholar]
- 26.Cappuccio JA, Blanchette CD, Sulchek TA, Arroyo ES, Kralj JM, Hinz AK, et al. Cell-free co-expression of functional membrane proteins and apolipoprotein, forming soluble nanolipoprotein particles. Molecular & cellular proteomics : MCP; 2008;7(11):2246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2002;2(8):853–6. [Google Scholar]
- 28.Kinney RM, Johnson BJ, Welch JB, Tsuchiya KR, Trent DW. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170(1):19–30. [DOI] [PubMed] [Google Scholar]
- 29.Hooper JW, Custer DM, Thompson E. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology. 2003;306(1):181–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamrud KI, Custer M, Dudek JM, Owens G, Alterson KD, Lee JS, et al. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology. 2007;360(2):376–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson WS, Thompson TB. The Structure of Apolipoprotein A-I in High Density Lipoproteins. Journal of Biological Chemistry. 2007;282(31):22249–53. [DOI] [PubMed] [Google Scholar]
- 32.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, et al. Different apolipoproteins impact nanolipoprotein particle formation. Journal of the American Chemical Society. 2007;129(46):14348–54. [DOI] [PubMed] [Google Scholar]
- 33.Fischer NO, Blanchette CD, Segelke BW, Corzett M, Chromy BA, Kuhn EA, et al. Isolation, characterization, and stability of discretely-sized nanolipoprotein particles assembled with apolipophorin-III. PloS one. 2010;5(7):e11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchette CD, Segelke BW, Fischer N, Corzett MH, Kuhn EA, Cappuccio JA, et al. Characterization and purification of polydisperse reconstituted lipoproteins and nanolipoprotein particles. International journal of molecular sciences. 2009;10(7):2958–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blakney AK, McKay PF, Christensen D, Yus BI, Aldon Y, Follmann F, et al. Effects of cationic adjuvant formulation particle type, fluidity and immunomodulators on delivery and immunogenicity of saRNA. Journal of Controlled Release. 2019;304:65–74. [DOI] [PubMed] [Google Scholar]
- 36.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114(1):100–9. [DOI] [PubMed] [Google Scholar]
- 37.Leal C, Ewert KK, Shirazi RS, Bouxsein NF, Safinya CR. Nanogyroids incorporating multivalent lipids: enhanced membrane charge density and pore forming ability for gene silencing. Langmuir. 2011;27(12):7691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leal C, Bouxsein NF, Ewert KK, Safinya CR. Highly efficient gene silencing activity of siRNA embedded in a nanostructured gyroid cubic lipid matrix. Journal of the American Chemical Society. 2010;132(47):16841–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brito LA, Kommareddy S, Maione D, Uematsu Y, Giovani C, Berlanda Scorza F, et al. Self-amplifying mRNA vaccines. Adv Genet. 2015;89:179–233. [DOI] [PubMed] [Google Scholar]
- 40.Brazzoli M, Magini D, Bonci A, Buccato S, Giovani C, Kratzer R, et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J Virol. 2016;90(1):332–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan S, Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Therapy. 2017;24:133. [DOI] [PubMed] [Google Scholar]
- 42.Azuma Y, Takada M, Shin HW, Kioka N, Nakayama K, Ueda K. Retroendocytosis pathway of ABCA1/apoA-I contributes to HDL formation. Genes to cells : devoted to molecular & cellular mechanisms. 2009;14(2):191–204. [DOI] [PubMed] [Google Scholar]
- 43.Lorenzi I, von Eckardstein A, Cavelier C, Radosavljevic S, Rohrer L. Apolipoprotein A-I but not high-density lipoproteins are internalised by RAW macrophages: roles of ATP-binding cassette transporter A1 and scavenger receptor BI. Journal of molecular medicine. 2008;86(2):171–83. [DOI] [PubMed] [Google Scholar]
- 44.Akhtar S, Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: potential impact on siRNA-mediated gene silencing activity and specificity. Adv Drug Deliv Rev. 2007;59(2-3):164–82. [DOI] [PubMed] [Google Scholar]
- 45.Kumar VV, Singh RS, Chaudhuri A. Cationic transfection lipids in gene therapy: successes, set-backs, challenges and promises. Curr Med Chem. 2003;10(14):1297–306. [DOI] [PubMed] [Google Scholar]
- 46.Chesnoy S, Huang L. Structure and function of lipid-DNA complexes for gene delivery. Annual review of biophysics and biomolecular structure. 2000;29:27–47. [DOI] [PubMed] [Google Scholar]
- 47.Hilgers LA, Snippe H. DDA as an immunological adjuvant. Res Immunol. 1992;143(5):494–503; discussion 74-6. [DOI] [PubMed] [Google Scholar]
- 48.Park HJ, Kuai R, Jeon EJ, Seo Y, Jung Y, Moon JJ, et al. High-density lipoprotein-mimicking nanodiscs carrying peptide for enhanced therapeutic angiogenesis in diabetic hindlimb ischemia. Biomaterials. 2018;161:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patriarchi T, Shen A, He W, Baikoghli M, Cheng RH, Xiang YK, et al. Nanodelivery of a functional membrane receptor to manipulate cellular phenotype. Sci Rep. 2018;8(1):3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He W, Felderman M, Evans AC, Geng J, Homan D, Bourguet F, et al. Cell-free production of a functional oligomeric form of a Chlamydia major outer-membrane protein (MOMP) for vaccine development. J Biol Chem. 2017;292(36):15121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weilhammer D, Dunkle AD, Blanchette CD, Fischer NO, Corzett M, Lehmann D, et al. Enhancement of antigen-specific CD4(+) and CD8(+) T cell responses using a self-assembled biologic nanolipoprotein particle vaccine. Vaccine. 2017;35(11):1475–81. [DOI] [PubMed] [Google Scholar]
- 52.Gao T, Blanchette CD, He W, Bourguet F, Ly S, Katzen F, et al. Characterizing diffusion dynamics of a membrane protein associated with nanolipoproteins using fluorescence correlation spectroscopy. Protein science : a publication of the Protein Society. 2011;20(2):437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao T, Petrlova J, He W, Huser T, Kudlick W, Voss J, et al. Characterization of de novo synthesized GPCRs supported in nanolipoprotein discs. PloS one. 2012;7(9):e44911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.