Abstract
Background
Lysin motif-containing proteins (LYP), which act as pattern-recognition receptors, play central roles in growth, node formation, and responses to biotic stresses. The sequence of Chinese white pear genome (cv. ‘Dangshansuli’) along with the seven other species of Rosaceae has already been reported. Although, in these fruit crops, there is still a lack of clarity regarding the LYP family genes and their evolutionary history.
Results
In the existing study, eight Rosaceae species i.e., Pyrus communis, Prunus persica, Fragaria vesca, Pyrus bretschneideri, Prunus avium, Prunus mume, Rubus occidentalis, and Malus × domestica were evaluated. Here, we determined a total of 124 LYP genes from the underlined Rosaceae species. While eighteen of the genes were from Chinese white pear, named as PbrLYPs. According to the LYPs structural characteristics and their phylogenetic analysis, those genes were classified into eight groups (group LYK1, LYK2, LYK3, LYK4/5, LYM1/3, LYM2, NFP, and WAKL). Dispersed duplication and whole-genome duplication (WGD) were found to be the most contributing factors of LYP family expansion in the Rosaceae species. More than half of the duplicated PbrLYP gene pairs were dated back to the ancient WGD (~ 140 million years ago (MYA)), and PbrLYP genes have experienced long-term purifying selection. The transcriptomic results indicated that the PbrLYP genes expression was tissue-specific. Most PbrLYP genes showed differential expression in leaves under fungal pathogen infection with two of them located in the plasmalemma.
Conclusion
A comprehensive analysis identified 124 LYP genes in eight Rosaceae species. Our findings have provided insights into the functions and characteristics of the Rosaceae LYP genes and a guide for the identification of other candidate LYPs for further genetic improvements for pathogen-resistance in higher plants.
Keywords: Chinese white pear, Lysin motif containing protein, Comparative analysis, Gene family, Evolution, Fungal pathogen resistance
Background
In contrast to mammals, plants have no sophisticated mobile defender cells or a somatic adaptive immune system. Plants have been developed their survival strategies, depending on the innate immunity along with signals arising from the site of infection via pathogen co-evolution [1, 2]. Similar to other organisms, plants can recognize PAMPs via recruiting plasmalemma localized pattern-recognition receptors (PRRs) to initiate immune reactions, such as PAMP-triggered immunity (PTI) responses [3]. Upon the perception between ectodomain and corresponding ligand, the cytoplasmic kinase domain (KD) of PRRs could transmit the signal to downstream and activate defense responses, such as reactive oxygen species production (ROS), phytoalexins, accumulation of callose, as well the stimulation of MAPK (Mitogen-activated protein kinase) pathways and the pathogenesis-related (PR) proteins expression.
Plant PRRs could be divided in two clusters: Receptor-like kinases (RLKs), which contain an extracellular sensor domain, a transmembrane domain and an intracellular domain with homology to protein kinases, involved in signal transduction; Receptor-like proteins (RLPs), which are similar to RLKs but lack intracellular region [4]. RLK/Ps are firstly reported in animals, but the gene number is particularly expanded in plants [5]. As a plant specific PRR family, the functions of lysin motif (LysM) containing proteins (LYPs) in fungal and bacterial microbe perceptions have been well studied in rice and Arabidopsis. LYPs are common in land plants and may have evolved before land colonization and symbiosis with mycorrhiza as a signaling module [6, 7], and most of LYPs that have been characterized were related to the perception of N-acetyl glucosamine containing molecules and/or to be involved in plant-microbe interaction pathway including activating of defense responses and establishment of root endosymbioses. For example, the Arabidopsis genome encodes five LysM-RLKs, and three of them participate in chitin signaling with chitin affinity: AtCERK1 or LYK1, LYK4 and LYK5 [7–12]. AtCERK1 is essential for chitin signaling pathway in Arabidopsis by forming hetero-oligomeric complexes with LYK5 to initiate downstream PTI, and LYK4 is also involved in that pathway having functions partly redundant with LYK5. While OsCEBiP, the main chitin binding protein in rice, recruits OsCERK1 to activate the chitin-triggered immune responses [13–15]. In addition to activating innate immunity, LYPs in legumes are essential receptors for the perception of nodulation factors (NFs) released by rhizobia and the establishment of nitrogen fixing symbiosis [16–21].
Few LYPs have been previously reported in fruit trees including apple (MdCERK1 and MdCERK1–2). MdCERK1, the ortholog of AtCERK1, has been shown to directly bind chitin and to be involved in transcriptional responses to pathogen infection of a soilborne pathogen Rhizoctonia solani [22]. MdCERK1–2 is also involved in the anti-fungal defense responses as a PRR and significantly upregulated after Botryosphaeria dothidea infection [23]. However, for other therophyte and perennial species in the Rosaceae, members of the LYP gene family involved in fungal pathogen perception and their evolutionary history are poorly defined.
In this study, we identified the Rosaceae LYP genes at the genome-wide scale by employing bioinformatics and publicly available data, and analyzed part of their functions in pear. We annotated full-length LYP genes in pear and other Rosaceae species, investigated their subcellular localization, and analyzed their expression patterns in different pear tissue types. We investigated the expression of PbrLYPs in response to the infection by a fungal pathogen Botryosphaeria dothidea, and provided a relatively complete profile of the LYP gene family in the Rosaceae. The genetic structure, evolutionary analysis, and experimental data of LYPs provide potential candidate LYPs for the future genetic modifications of pathogen-resistance in Rosaceae fruit crops and other higher plants.
Results
Identification and classification of LYP genes in the Rosaceae
To identify the members of LYP gene family in the genus Rosaceae, HMM search was performed using both the HMM profile (PF01476) and a self-built HMM model against the whole-genome protein sequences of each species. A total of 141 LYP genes were identified from eight investigated Rosaceae species. After removing redundant and incomplete gene sequences, the longest transcript of the same gene was retained. Subsequently, the NCBI Batch CD-Search was used to further confirm the presence of a LysM domain (Table 1 and Fig. S3a). Finally, we identified 124 LYP genes in eight Rosaceae species, including 18 genes in Chinese white pear, 14 in European pear, 21 in apple, 15 in peach, 13 in strawberry, 16 in Mei (Japanese apricot), 14 in sweet cherry and 13 in black raspberry. The PbrLYP genes showed a random distribution on eight of the 17 chromosomes and three unanchored scaffolds (scaffolds681.0, scaffolds831.0, and scaffolds897.0) in pear (Supplementary Fig. S1).
Table 1.
Classification of LYP genes in eight Rosaceae species
| Group name | Chinese white pear | European pear | Apple | Strawberry | Peach | Mei | Sweet cherry | Black rasberry | |
|---|---|---|---|---|---|---|---|---|---|
| (Number of genes) | Pbr | Pco | Md | Fv | Ppe | Pm | Pav | Ro | |
| LYK1 | LYK1a | Pbr000107.1 | PCP017929.1 | MD05G1351500 | FvH4_3g01490.1 | ppa017142m | Pm009697 | Pav_sc0000824.1_g060.1.mk | Ro03_G13508 |
| (22) | LYK1b1 | Pbr005151.1 | PCP003883.1 | MD09G1111800 | FvH4_6g42640.1 | ppa003023m | Pm015128 | Pav_sc0001077.1_g520.1.mk | Ro06_G06708 |
| LYK1b2 | Pbr005152.1 | PCP008886.1 | MD17G1102100 | Pm015129 | Ro06_G17308 | ||||
| LYK1b3 | Pbr021830.1 | ||||||||
| LYK2 | LYK2 | Pbr014439.4 | PCP023229.1 | MD17G1014100 | FvH4_6g53170.1 | ppa024632m | Pm016182 | Ro06_G03822 | |
| (7) | |||||||||
| LYK3 | LYK3a1 | Pbr019107.1 | PCP003907.1 | MD09G1202500 | FvH4_6g32430.1 | ppa002771m | Pm013417 | Pav_sc0000759.1_g060.1.mk | Ro06_G20150 |
| (21) | LYK3a2 | Pbr034737.1 | PCP003981.1 | MD17G1183700 | Pav_sc0002893.1_g120.1.mk | ||||
| LYK3b1 | Pbr036151.1 | PCP000376.1 | MD16G1098600 | ppa019853m | Pm006733 | Pav_sc0006018.1_g500.1.mk | Ro04_G02801 | ||
| LYK3b2 | PCP002145.1 | MD06G1169200 | |||||||
| LYK4/5 | LYK4/5a1 | Pbr022856.1 | MD02G1156000 | FvH4_1g14190.1 | ppa002539m | Pm022870 | Ro05_G22711 | ||
| (24) | LYK4/5a2 | FvH4_5g26710.1 | ppa015910m | Pm026556 | |||||
| LYK4/5a3 | FvH4_5g26740.1 | ||||||||
| LYK4/5b | Pbr022855.1 | FvH4_1g14260.1 | ppa016660m | Pm026555 | Pav_sc0000348.1_g1000.1.mk | Ro01_G00968 | |||
| LYK4/5c1 | PCP028343.1 | MD00G1205900 | FvH4_3g31710.1 | ppa027141m | Pm001054 | Pav_sc0000480.1_g310.1.mk | Ro03_G07424 | ||
| LYK4/5c2 | MD11G1140300 | ||||||||
| NFP | NFP1 | Pbr014715.1 | MD02G1156500 | FvH4_1g14270.1 | ppa002872m | Pm009175 | Pav_sc0000998.1_g140.1.mk | Ro01_G00967 | |
| (12) | NFP2 | Pbr022854.1 | MD13G1203700 | ppa002949m | Pm026553 | Pav_sc0000348.1_g980.1.mk | |||
| WAKL | WAKL1 | Pbr035317.1 | PCP000222.1 | MD04G1238700 | ppa002979m | Pm003663 | Pav_sc0000744.1_g060.1.mk | ||
| (11) | WAKL2 | PCP038026.1 | MD04G1238900 | Pav_sc0004456.1_g300.1.br | |||||
| WAKL3 | MD04G1239400 | ||||||||
| WAKL4 | MD04G1239000 | ||||||||
| LYM1/3 | LYM1/3–1 | Pbr002167.1 | PCP020213.1 | MD06G1215700 | FvH4_2g26750.1 | ppa006147m | Pm004817 | Pav_sc0000195.1_g1050.1.mk | Ro02_G01432 |
| (18) | LYM1/3–2 | Pbr020280.1 | PCP035131.1 | MD08G1165500 | FvH4_5g12220.1 | ppa017855m | Pm025193 | Pav_sc0000383.1_g420.1.mk | Ro05_G16294 |
| LYM1/3–3 | Pbr040557.1 | MD15G1351200 | |||||||
| LYM2 | LYM2–1 | Pbr016347.1 | PCP000950.1 | MD00G1083800 | FvH4_2g16390.1 | ppa008092m | Pm020791 | Pav_sc0000377.1_g320.1.mk | Ro02_G18370 |
| (9) | LYM2–2 | Pbr039238.2 | |||||||
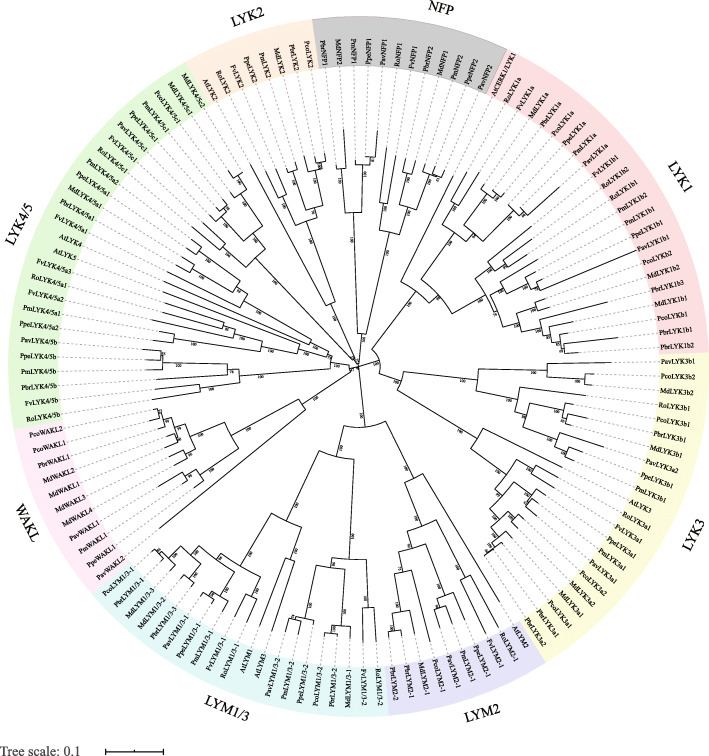
Phylogenetic analyses of the LYP protein sequences were performed in order to classify the LYP genes and investigate their evolutionary relationships. The phylogenetic tree showed that the LYP genes are separated into eight well-supported clades. According to the name of the best hit gene in Arabidopsis, these subfamilies were named LYK1–3, LYK4/5, LYM1/3, LYM2, NFP (Nod factor perception protein), and WAKL (Wall associated kinase-like) (Fig. 1). The subfamily classification and corresponding names of LYPs are shown in Table 1. Although, the best local BLASTP hit gene of most LYPs in NFP clade were AtLYK4 or AtLYK5, their best NCBI BLASTP results were categorized as NFP (data not shown). Notably, the numbers of genes in the LYK1, LYK3 and LYK4/5 clades were more than that of others, suggesting these three subgroups may have undergone the subfamily specific expansion.
Fig. 1.
Phylogenetic analysis of LYP genes in Rosaceae and Arabidopsis. The tree was formed via MEGA 7.0 with the NJ method and 1000 bootstraping replicates. The proteins were assembled into eight groups. Different background colors indicate the different groups of the LYP proteins
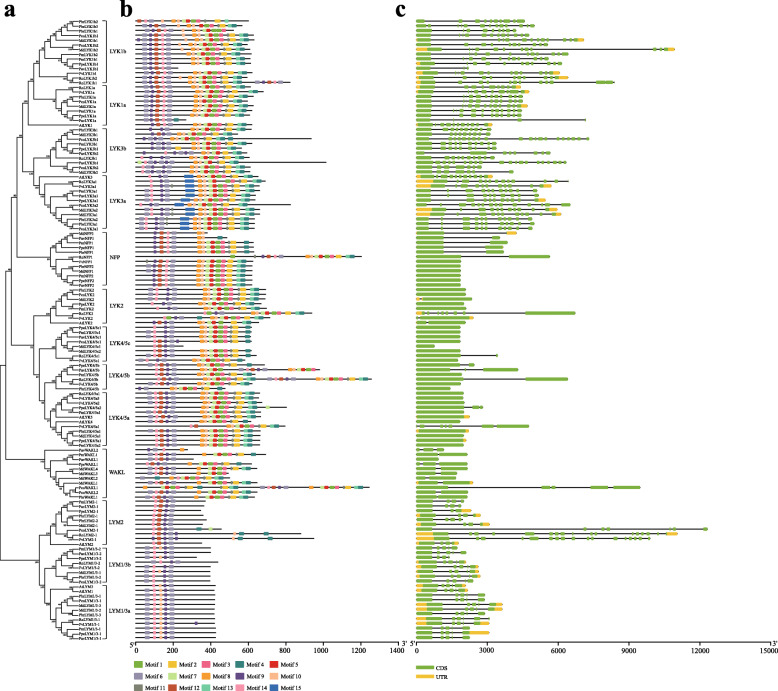
To explore the structural diversity of Rosaceae LYP genes, an exon distribution analysis was performed (Fig. 2). The results showed that these Rosaceae LYP subgroups displayed different exon abundance and the numbers of exons in each gene in the same subgroup were similar, supporting the phylogenetic classification of the LYP genes (Fig. 2a, c). However, among the Rosaceae, the number of exons in subgroups LYK1 and LYK3 was much higher compared to others, about 11 on average. These results were consistent with Arabidopsis. Exon number were relatively conserved in subgroups LYM1/3 and LYM2, at about 4 to 5 (Table 2 and Supplementary Table S1). These results were well consistent with previous reports about the conserved exon number of different LYP types. Type I LYP genes contained was up to 10 exons (group LYK1 and LYK3), type II LYP contained approximately 5 (group LYM1/3 and LYM2), and of type III contained approximately 2 (group LYK2, LYK4/5, NFP and WAKL) [10, 24–26]. The conserved and specific exon numbers of certain LYP type may have been due to similar replication events, implying that the different LYP type genes originated through the evolutionary path separate from genes in other types.
Fig. 2.
Structural and motif analysis of LYP genes. a Subgroup classification. Neighbor-joining phylogenetic tree was generated among 132 LYP genes with MEGA7. The subgroup names were labeled accordingly. b Motif analysis. Fifteen distinct motifs were determined with MEME suite and the representation of each motif was carried out with a different color. c Gene structural analysis. The exon sizes are comparable to their sequence length
Table 2.
Characteristics of the LYP proteins
| Gene name | Coding sequence length (bp) | Pprotein length (aa) | MW | PI | GRAVY | Extron number | PDB domain 4EBZ | CDD domain LysM | Best hit gene in Arabidopsis | Corresponding gene name in Arabidopsis |
|---|---|---|---|---|---|---|---|---|---|---|
| AtCERK1/LYK1 | 1854 | 618aa | 67.32 kDa | 6.79 | −0.012* | 12 | D | D | AT3G21630.1 | LYK1 |
| AtLYK2 | 1965 | 655aa | 73.17 kDa | 6.18 | −0.291* | 2 | D | D | AT3G01840.1 | LYK2 |
| AtLYK3 | 1956 | 652aa | 71.45 kDa | 6.28 | −0.004* | 11 | D | D | AT1G51940.1 | LYK3 |
| AtLYK4 | 1839 | 613aa | 66.63 kDa | 5.13 | −0.057* | 1 | D | D | AT2G23770.1 | LYK4 |
| AtLYK5 | 1995 | 665aa | 72.58 kDa | 5.84 | −0.117* | 1 | D | D | AT2G33580.1 | LYK5 |
| AtLYM1 | 1251 | 417aa | 43.49 kDa | 4.65 | 0.293 | 5 | D | D | AT1G21880.2 | LYM1 |
| AtLYM2 | 1053 | 351aa | 37.75 kDa | 6.23 | 0.04 | 4 | D | D | AT2G17120.1 | LYM2 |
| AtLYM3 | 1272 | 424aa | 44.16 kDa | 4.77 | 0.243 | 5 | D | D | AT1G77630.1 | LYM3 |
| FvLYK1a | 2037 | 679aa | 75.29 kDa | 6.26 | −0.054* | 10 | D | D | AT3G21630.1 | LYK1 |
| FvLYK1b1 | 1860 | 620aa | 68.52 kDa | 4.94 | −0.2* | 12 | D | AT3G21630.1 | LYK1 | |
| FvLYK2 | 2142 | 714aa | 78.52 kDa | 6.84 | −0.03* | 2 | D | D | AT3G01840.1 | LYK2 |
| FvLYK3a1 | 1974 | 658aa | 72.71 kDa | 6.51 | −0.047* | 11 | D | D | AT1G51940.1 | LYK3 |
| FvLYK4/5a1 | 2385 | 795aa | 87.54 kDa | 6.82 | 0.021 | 7 | D | AT2G23770.1 | LYK4 | |
| FvLYK4/5a2 | 2016 | 672aa | 73.26 kDa | 6.23 | −0.113* | 1 | D | D | AT2G33580.1 | LYK5 |
| FvLYK4/5a3 | 1965 | 655aa | 70.90 kDa | 6.14 | −0.138* | 1 | D | D | AT2G33580.1 | LYK5 |
| FvLYK4/5b | 1860 | 620aa | 69.09 kDa | 6.72 | 0.04 | 1 | D | D | AT2G23770.1 | LYK4 |
| FvLYK4/5c1 | 1743 | 581aa | 64.21 kDa | 6.39 | 0.041 | 1 | D | D | AT2G23770.1 | LYK4 |
| FvLYM1/3–1 | 1257 | 419aa | 43.65 kDa | 4.98 | 0.268 | 5 | D | D | AT1G21880.2 | LYM1 |
| FvLYM1/3–2 | 1191 | 397aa | 42.60 kDa | 6.32 | 0.181 | 5 | D | D | AT1G21880.2 | LYM1 |
| FvLYM2–1 | 2847 | 949aa | 101.63 kDa | 7.62* | 0.323 | 17 | D | D | AT4G38380.1 | MATE efflux family protein |
| FvNFP1 | 1872 | 624aa | 68.95 kDa | 8.05* | 0.105 | 1 | D | D | AT2G33580.1 | LYK5 |
| MdLYK1a | 1878 | 626aa | 68.98 kDa | 6.37 | −0.112* | 12 | D | AT3G21630.1 | LYK1 | |
| MdLYK1b1 | 1890 | 630aa | 69.54 kDa | 5.03 | −0.09* | 12 | D | AT3G21630.1 | LYK1 | |
| MdLYK1b2 | 1824 | 608aa | 66.72 kDa | 5.38 | −0.12* | 12 | D | D | AT3G21630.1 | LYK1 |
| MdLYK2 | 2067 | 689aa | 75.24 kDa | 6.92 | −0.069* | 1 | D | D | AT3G01840.1 | LYK2 |
| MdLYK3a1 | 1980 | 660aa | 72.94 kDa | 7.13* | −0.066* | 11 | D | D | AT1G51940.1 | LYK3 |
| MdLYK3a2 | 1980 | 660aa | 72.65 kDa | 6.27 | −0.056* | 11 | D | D | AT1G51940.1 | LYK3 |
| MdLYK3b1 | 1629 | 543aa | 60.77 kDa | 5.73 | −0.109* | 9 | D | D | AT1G51940.1 | LYK3 |
| MdLYK3b2 | 1824 | 608aa | 68.76 kDa | 6.18 | −0.267* | 7 | D | D | AT1G51940.1 | LYK3 |
| MdLYK4/5a1 | 1986 | 662aa | 72.19 kDa | 4.87 | −0.074* | 1 | D | D | AT2G23770.1 | LYK4 |
| MdLYK4/5c1 | 756 | 252aa | 27.80 kDa | 4.43 | 0.04 | 1 | D | D | AT2G23770.1 | LYK4 |
| MdLYK4/5c2 | 1845 | 615aa | 68.29 kDa | 6.71 | 0.002 | 1 | D | D | AT2G23770.1 | LYK4 |
| MdLYM1/3–1 | 1185 | 395aa | 42.24 kDa | 6.37 | 0.255 | 5 | D | D | AT1G21880.2 | LYM1 |
| MdLYM1/3–2 | 1254 | 418aa | 43.49 kDa | 4.7 | 0.368 | 5 | D | D | AT1G21880.2 | LYM1 |
| MdLYM1/3–3 | 1260 | 420aa | 43.85 kDa | 5.99 | 0.374 | 5 | D | D | AT1G21880.2 | LYM1 |
| MdLYM2–1 | 1068 | 356aa | 38.31 kDa | 7.25* | 0.09 | 4 | D | D | AT2G17120.1 | LYM2 |
| MdNFP1 | 1860 | 620aa | 69.00 kDa | 6.98 | 0.017 | 1 | D | D | AT2G33580.1 | LYK5 |
| MdNFP2 | 1143 | 381aa | 41.79 kDa | 8.29* | −0.006* | 2 | D | D | AT2G23770.1 | LYK4 |
| MdWAKL1 | 1938 | 646aa | 72.01 kDa | 5.94 | −0.121* | 2 | D | D | AT1G16120.1 | WAKL1 |
| MdWAKL2 | 1500 | 500aa | 56.17 kDa | 6.83 | −0.207* | 2 | D | D | AT1G16130.1 | WAKL2 |
| MdWAKL3 | 1479 | 493aa | 54.68 kDa | 7.8* | 0.006 | 2 | D | D | AT1G16120.1 | WAKL1 |
| MdWAKL4 | 1935 | 645aa | 71.87 kDa | 6.32 | −0.103* | 2 | D | D | AT1G16150.1 | WAKL4 |
| PavLYK1a | 801 | 267aa | 30.13 kDa | 7.74* | −0.049* | 4 | D | AT3G21630.1 | LYK1 | |
| PavLYK1b1 | 675 | 225aa | 25.03 kDa | 6.73 | −0.144* | 3 | D | D | AT3G21630.1 | LYK1 |
| PavLYK3a1 | 1914 | 638aa | 70.56 kDa | 6.56 | −0.003* | 10 | D | D | AT1G51940.1 | LYK3 |
| PavLYK3a2 | 1776 | 592aa | 66.34 kDa | 4.99 | −0.107* | 8 | D | D | AT1G51940.1 | LYK3 |
| PavLYK3b1 | 3042 | 1014aa | 115.03 kDa | 6.74 | −0.214* | 20 | D | D | AT1G60780.1 | HAPLESS 13 |
| PavLYK4/5b | 2940 | 980aa | 108.71 kDa | 7.2* | −0.106* | 3 | D | D | AT2G23770.1 | LYK4 |
| PavLYK4/5c1 | 1836 | 612aa | 68.08 kDa | 6.1 | 0.011 | 1 | D | D | AT2G23770.1 | LYK4 |
| PavLYM1/3–1 | 1275 | 425aa | 44.24 kDa | 6.32 | 0.324 | 5 | D | D | AT1G21880.2 | LYM1 |
| PavLYM1/3–2 | 1194 | 398aa | 42.74 kDa | 6.02 | 0.095 | 5 | D | D | AT1G21880.2 | LYM1 |
| PavLYM2–1 | 1089 | 363aa | 38.78 kDa | 7.7* | 0.108 | 4 | D | D | AT2G17120.1 | LYM2 |
| PavNFP1 | 1455 | 485aa | 53.72 kDa | 8.02* | −0.094* | 2 | D | D | AT2G23770.1 | LYK4 |
| PavNFP2 | 1857 | 619aa | 68.76 kDa | 8.13* | 0.012 | 1 | D | D | AT2G33580.1 | LYK5 |
| PavWAKL1 | 921 | 307aa | 33.36 kDa | 7.97* | 0.097 | 1 | D | D | AT2G33580.1 | LYK5 |
| PavWAKL2 | 828 | 276aa | 30.46 kDa | 8.75* | −0.103* | 3 | D | D | AT2G23770.1 | LYK4 |
| PbrLYK1a | 1878 | 626aa | 68.85 kDa | 6.31 | −0.088* | 12 | D | AT3G21630.1 | LYK1 | |
| PbrLYK1b1 | 1563 | 521aa | 57.39 kDa | 7.51* | −0.188* | 9 | D | D | AT3G21630.1 | LYK1 |
| PbrLYK1b2 | 1800 | 600aa | 65.81 kDa | 5.97 | −0.137* | 10 | D | D | AT3G21630.1 | LYK1 |
| PbrLYK1b3 | 1698 | 566aa | 61.85 kDa | 8.5* | −0.199* | 13 | D | AT3G21630.1 | LYK1 | |
| PbrLYK2 | 2076 | 692aa | 75.39 kDa | 6.74 | −0.084* | 1 | D | D | AT3G01840.1 | LYK2 |
| PbrLYK3a1 | 1896 | 632aa | 69.70 kDa | 6.73 | −0.106* | 11 | D | D | AT1G51940.1 | LYK3 |
| PbrLYK3a2 | 1896 | 632aa | 69.64 kDa | 6.85 | −0.108* | 11 | D | D | AT1G51940.1 | LYK3 |
| PbrLYK3b1 | 1845 | 615aa | 68.86 kDa | 6.43 | −0.12* | 10 | D | D | AT1G51940.1 | LYK3 |
| PbrLYK4/5a1 | 1989 | 663aa | 72.36 kDa | 5.07 | −0.057* | 1 | D | D | AT2G23770.1 | LYK4 |
| PbrLYK4/5b | 1428 | 476aa | 52.71 kDa | 7.34* | −0.05* | 1 | D | AT2G23770.1 | LYK4 | |
| PbrLYM1/3–1 | 1263 | 421aa | 43.73 kDa | 5.99 | 0.343 | 5 | D | D | AT1G21880.2 | LYM1 |
| PbrLYM1/3–2 | 1188 | 396aa | 42.09 kDa | 6.48 | 0.222 | 5 | D | D | AT1G21880.2 | LYM1 |
| PbrLYM1/3–3 | 1254 | 418aa | 43.48 kDa | 5.53 | 0.361 | 5 | D | D | AT1G21880.2 | LYM1 |
| PbrLYM2–1 | 1077 | 359aa | 38.51 kDa | 7.29* | 0.149 | 4 | D | D | AT2G17120.1 | LYM2 |
| PbrLYM2–2 | 1134 | 378aa | 40.55 kDa | 8.24* | 0.165 | 4 | D | D | AT2G17120.1 | LYM2 |
| PbrNFP1 | 1887 | 629aa | 69.18 kDa | 6.51 | −0.066* | 2 | D | D | AT2G23770.1 | LYK4 |
| PbrNFP2 | 1860 | 620aa | 69.04 kDa | 8.3* | 0.011 | 1 | D | D | AT2G33580.1 | LYK5 |
| PbrWAKL1 | 1938 | 646aa | 72.04 kDa | 5.97 | −0.148* | 2 | D | D | AT1G16130.1 | WAKL2 |
| PcoLYK1a | 1785 | 595aa | 65.41 kDa | 5.93 | −0.09* | 11 | D | AT3G21630.1 | LYK1 | |
| PcoLYK2 | 2076 | 692aa | 75.44 kDa | 6.81 | −0.088* | 1 | D | D | AT3G01840.1 | LYK2 |
| PcoLYK3a1 | 1893 | 631aa | 69.78 kDa | 6.73 | −0.122* | 11 | D | D | AT1G51940.1 | LYK3 |
| PcoLYK3a2 | 2472 | 824aa | 90.91 kDa | 6.64 | −0.179* | 14 | D | D | AT1G51940.1 | LYK3 |
| PcoLYK3b1 | 2805 | 935aa | 106.20 kDa | 5.58 | −0.211* | 20 | D | D | AT1G60780.1 | HAPLESS 13 |
| PcoLYK3b2 | 1830 | 610aa | 68.88 kDa | 5.85 | −0.242* | 7 | D | D | AT1G51940.1 | LYK3 |
| PcoLYK4/5c1 | 1845 | 615aa | 68.30 kDa | 6.5 | 0.015 | 1 | D | D | AT2G23770.1 | LYK4 |
| PcoLYKb1 | 1881 | 627aa | 68.55 kDa | 5.4 | −0.124* | 12 | D | D | AT3G21630.1 | LYK1 |
| PcoLYKb2 | 1782 | 594aa | 65.09 kDa | 5.51 | −0.111* | 10 | D | D | AT3G21630.1 | LYK1 |
| PcoLYM1/3–1 | 1260 | 420aa | 43.74 kDa | 6.32 | 0.335 | 5 | D | D | AT1G21880.2 | LYM1 |
| PcoLYM1/3–2 | 1185 | 395aa | 42.11 kDa | 6.48 | 0.199 | 5 | D | D | AT1G21880.2 | LYM1 |
| PcoLYM2–1 | 1368 | 456aa | 48.61 kDa | 4.67 | 0.295 | 7 | D | D | AT2G17120.1 | LYM2 |
| PcoWAKL1 | 3732 | 1244aa | 138.78 kDa | 5.9 | −0.202* | 5 | D | D | AT1G16130.1 | WAKL2 |
| PcoWAKL2 | 1938 | 646aa | 72.10 kDa | 6.07 | −0.164* | 2 | D | D | AT1G16130.1 | WAKL2 |
| PmLYK1a | 1857 | 619aa | 68.14 kDa | 6.37 | −0.009* | 12 | D | AT3G21630.1 | LYK1 | |
| PmLYK1b1 | 1827 | 609aa | 66.66 kDa | 6.43 | −0.106* | 12 | D | D | AT3G21630.1 | LYK1 |
| PmLYK1b2 | 1845 | 615aa | 67.51 kDa | 5.38 | −0.085* | 12 | D | D | AT3G21630.1 | LYK1 |
| PmLYK2 | 2088 | 696aa | 76.31 kDa | 7.03* | −0.086* | 1 | D | D | AT3G01840.1 | LYK2 |
| PmLYK3a1 | 1980 | 660aa | 72.77 kDa | 6.31 | −0.052* | 11 | D | D | AT1G51940.1 | LYK3 |
| PmLYK3b1 | 1854 | 618aa | 69.19 kDa | 5.9 | −0.092* | 10 | D | D | AT1G51940.1 | LYK3 |
| PmLYK4/5a1 | 1995 | 665aa | 72.37 kDa | 6.91 | −0.076* | 1 | D | D | AT2G33580.1 | LYK5 |
| PmLYK4/5a2 | 1980 | 660aa | 72.78 kDa | 5.24 | −0.13* | 1 | D | D | AT2G23770.1 | LYK4 |
| PmLYK4/5b | 1908 | 636aa | 70.84 kDa | 5.62 | −0.021* | 1 | D | AT2G23770.1 | LYK4 | |
| PmLYK4/5c1 | 1845 | 615aa | 68.61 kDa | 6.63 | 0.028 | 1 | D | D | AT2G23770.1 | LYK4 |
| PmLYM1/3–1 | 1275 | 425aa | 44.56 kDa | 5.98 | 0.309 | 5 | D | D | AT1G21880.2 | LYM1 |
| PmLYM1/3–2 | 1194 | 398aa | 42.71 kDa | 5.82 | 0.124 | 5 | D | D | AT1G21880.2 | LYM1 |
| PmLYM2–1 | 1110 | 370aa | 39.57 kDa | 6.47 | 0.131 | 4 | D | D | AT2G17120.1 | LYM2 |
| PmNFP1 | 1878 | 626aa | 69.89 kDa | 6.92 | −0.098* | 2 | D | D | AT2G23770.1 | LYK4 |
| PmNFP2 | 1854 | 618aa | 68.99 kDa | 8.19* | −0.007* | 1 | D | D | AT2G33580.1 | LYK5 |
| PmWAKL1 | 2076 | 692aa | 77.10 kDa | 7.28* | −0.133* | 2 | D | D | AT1G16130.1 | WAKL2 |
| PpeLYK1a | 1821 | 607aa | 66.68 kDa | 6.27 | 0.018 | 10 | D | AT3G21630.1 | LYK1 | |
| PpeLYK1b1 | 1836 | 612aa | 66.63 kDa | 6.36 | −0.135* | 12 | D | D | AT3G21630.1 | LYK1 |
| PpeLYK2 | 2004 | 668aa | 73.24 kDa | 6.77 | −0.174* | 1 | D | D | AT3G01840.1 | LYK2 |
| PpeLYK3a1 | 1908 | 636aa | 70.35 kDa | 6.67 | −0.021* | 10 | D | D | AT1G51940.1 | LYK3 |
| PpeLYK3b1 | 1689 | 563aa | 62.86 kDa | 6.04 | −0.109* | 9 | D | D | AT1G51940.1 | LYK3 |
| PpeLYK4/5a1 | 1983 | 661aa | 72.33 kDa | 4.98 | −0.125* | 1 | D | D | AT2G23770.1 | LYK4 |
| PpeLYK4/5a2 | 2409 | 803aa | 87.75 kDa | 7.1* | −0.091* | 3 | D | D | AT2G33580.1 | LYK5 |
| PpeLYK4/5b | 2058 | 686aa | 76.08 kDa | 5.71 | −0.062* | 2 | D | D | AT2G33580.1 | LYK5 |
| PpeLYK4/5c1 | 1848 | 616aa | 68.58 kDa | 6.9 | 0.014 | 1 | D | D | AT2G23770.1 | LYK4 |
| PpeLYM1/3–1 | 1278 | 426aa | 44.26 kDa | 5.98 | 0.324 | 5 | D | D | AT1G21880.2 | LYM1 |
| PpeLYM1/3–2 | 972 | 324aa | 34.79 kDa | 6.62 | 0.171 | 4 | D | D | AT1G21880.1 | LYM1 |
| PpeLYM2–1 | 1038 | 346aa | 36.84 kDa | 7.13* | 0.125 | 3 | D | D | AT2G17120.1 | LYM2 |
| PpeNFP1 | 1881 | 627aa | 69.66 kDa | 7.26* | −0.081* | 2 | D | D | AT2G23770.1 | LYK4 |
| PpeNFP2 | 1860 | 620aa | 68.93 kDa | 8.35* | −0.009* | 1 | D | D | AT2G33580.1 | LYK5 |
| PpeWAKL1 | 1848 | 616aa | 68.83 kDa | 7.02* | −0.19* | 3 | D | D | AT1G16130.1 | WAKL2 |
| RoLYK1a | 1833 | 611aa | 67.26 kDa | 6.13 | 0.013 | 9 | D | D | AT3G21630.1 | LYK1 |
| RoLYK1b1 | 2466 | 822aa | 90.99 kDa | 5.32 | −0.044* | 13 | D | D | AT3G21630.1 | LYK1 |
| RoLYK1b2 | 1779 | 593aa | 65.27 kDa | 5.96 | −0.027* | 10 | D | D | AT3G21630.1 | LYK1 |
| RoLYK2 | 2814 | 938aa | 102.59 kDa | 6.65 | −0.03* | 8 | D | D | AT3G01840.1 | LYK2 |
| RoLYK3a1 | 2070 | 690aa | 76.39 kDa | 6.5 | −0.029* | 13 | D | D | AT1G51940.1 | LYK3 |
| RoLYK3b1 | 1815 | 605aa | 67.14 kDa | 5.94 | 0.012 | 10 | D | D | AT1G51940.1 | LYK3 |
| RoLYK4/5a1 | 1980 | 660aa | 71.61 kDa | 6.28 | −0.099* | 1 | D | D | AT2G33580.1 | LYK5 |
| RoLYK4/5b | 3765 | 1255aa | 139.28 kDa | 7.69* | −0.105* | 2 | D | D | AT2G23770.1 | LYK4 |
| RoLYK4/5c1 | 1926 | 642aa | 71.38 kDa | 6.76 | −0.004* | 2 | D | D | AT2G23770.1 | LYK4 |
| RoLYM1/3–1 | 1260 | 420aa | 43.57 kDa | 4.67 | 0.262 | 6 | D | D | AT1G21880.2 | LYM1 |
| RoLYM1/3–2 | 1311 | 437aa | 47.21 kDa | 7.97* | 0.03 | 5 | D | D | AT1G21880.2 | LYM1 |
| RoLYM2–1 | 2637 | 879aa | 95.20 kDa | 8.24* | 0.244 | 17 | D | D | AT4G38380.1 | MATE efflux family protein |
| RoNFP1 | 3606 | 1202aa | 135.40 kDa | 8.21* | −0.118* | 2 | D | D | AT5G66631.1 | Tetratricopeptide repeat like superfamily protein |
The basic information and characteristics of LYP proteins and encoding genes were shown
MW: Protein Molecular Weight. PI: Isoelectric point. GRAVY: Grand average of hydropathy. 4EBZ: Crystal structure of the LysMs in the ectodomain of AtCERK1 in PDB databases (https://www.rcsb.org/). CDD domain LysM: The domain of LysM or LysM superfamily in CDD v3.18. Best hit gene in Arabidopsis: The gene ID with the highest score in BLASTP result
The symbol * indicate the values more than 7 or less than 0 and the letter D indicate the corresponding domain could be detected
Features of the LYPs in the Rosaceae
The characteristics of the LYPs and their coding genes are shown in Table 2. The lengths of the LYPs protein sequences ranged from 225 to 1255 amino acids and the molecular weights were 25.03 to 139.28 kD. Protein isoelectric points (PI) ranged from 4.43 to 8.75, with the majority lower than 7 (Table 2). The highest number of exons in pear LYPs was found in the LYK1 and LYK3 subgroups. A similar trend was also observed in the other five Rosaceae species (European pear, apple, peach, sweet cherry and Mei) and Arabidopsis (Table 2), confirming that the genes in LYK1 and LYK3 groups have undergone specific evolutionary events as type I LYPs. However, the highest exon numbers in strawberry and black raspberry were detected in the LYM2 subgroup, suggesting that these species may have experienced an unknown evolutionary process or some specific selection forces. The grand average of hydropathy (GRAVY) for most LYK proteins in pear was positive, while that of LYMs was negative. The GRAVY of NFP and WAKL subgroups was random. A similar trend for all subgroups was also observed in the other Rosaceae species. These results indicated that similar to Arabidopsis, most of the LYK proteins are hydrophobic and all LYM proteins are hydrophilic in the LYP gene family (Table 2).
Synteny analysis of LYPs
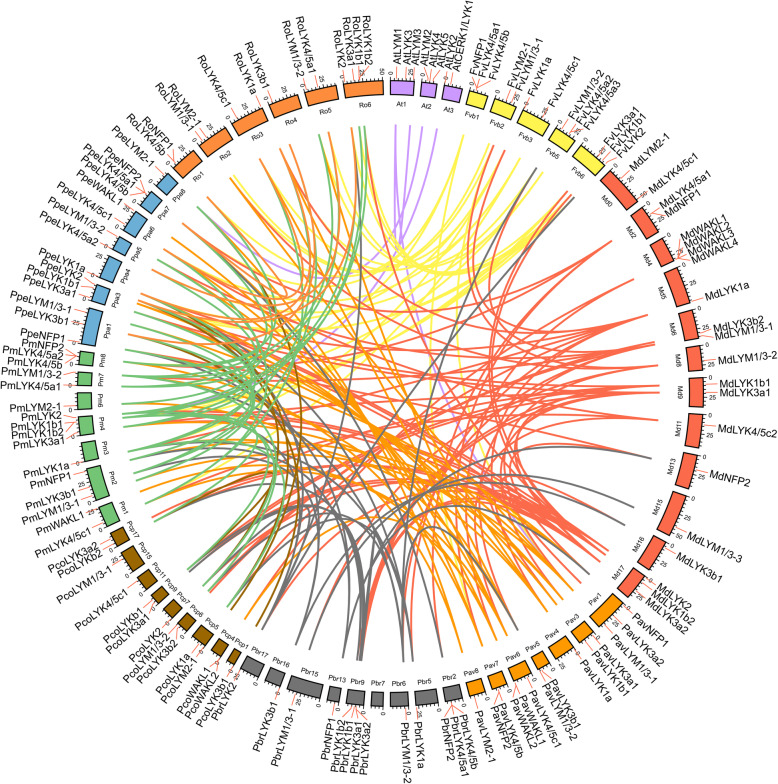
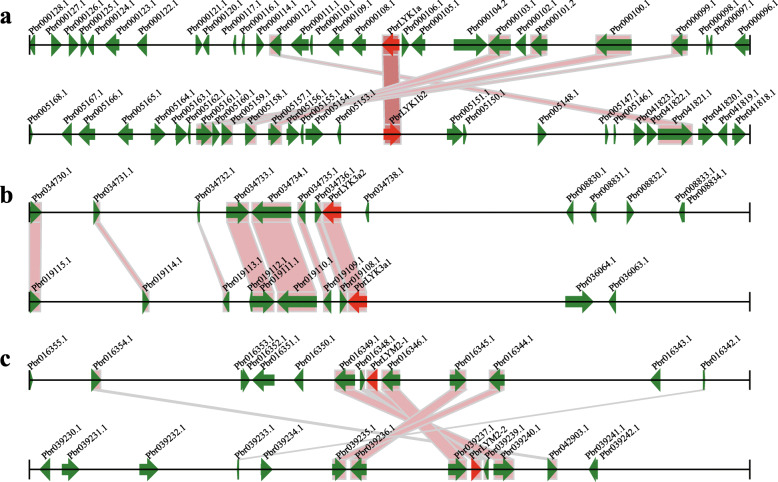
The gene duplication events, such as tandem duplication, the whole genome duplication (WGD)/segmental duplication, and transposition events are the contributing factors in gene family development that impact the protein-coding gene family’s evolution [27]. By MCScanX package, we detected the events duplication related to the LYP gene family, and assigned each of LYP genes to one of the five various types of duplication: WGD/segmental, singleton, proximal, dispersed, or tandem. In Arabidopsis, only two LYP genes duplicated during the WGD/segmental event, while the others originated from a dispersed duplication. Unlike in Arabidopsis, the five duplication types were all detected in the Rosaceae driving the expansion of the LYP genes (Table 3 and Supplementary Table S2). WGD occurred in all the Rosaceae species studied, with 38.9% of LYP genes in Chinese white pear and 57.2% in apple retained and duplicated from WGD/segmental events. However, the percentage of genes retained following dispersed duplication in peach (53.3%), strawberry (46.2%), Mei (56.3%), sweet cherry (57.1%), and black raspberry (69.2%) was higher than that in apple (19%). Peach, strawberry, Mei, sweet cherry, and black raspberry did experience a WGD from the time of their divergence from pear and apple. Hence, these species may have experienced more genome rearrangements and gene losses during the long-term evolution in the absence of WGD, resulting in the larger ratios of dispersed genes. Although pear and apple have undergone the same recent WGD event, Chinese white pear and European pear showed a higher percentage of dispersed LYP genes (38.9 and 50%, respectively) compared to apple. This may be due to the differences in the ratio of self-incompatibility and the domestication process between pear and apple. However, proximal duplication events of LYP genes were only detected in apple (14.3%), strawberry (23.2%), peach (6.7%), Mei (6.2%), and sweet cherry (14.3%) as depicted in Table 3. The obtained data suggested that WGD and dispersed gene duplication have an effective contribution to the development of LYP gene family, belong to Rosaceae. To reveal the LYP genes (belong to Rosaceae) evolutionary routes that made them the most diverse, here, we evaluated both intra- and intergenomic synteny analyses to identify conservation chromosome blocks within Chinese white pear and among eight Rosaceae species and Arabidopsis. The landscape of inter-species orthologous LYP gene pairs among Rosaceae species and Arabidopsis presented in Fig. 3 and their chromosomal distribution was random. In the Chinese white pear genome, 7 conserved syntenic blocks containing PbrLYPs were detected, including most of WGD/segmental type LYP gene pairs (PbrLYK1a-PbrLYK1b2, PbrLYM2–1-PbrLYM2–2, and PbrLYK3a1-PbrLYK3a2) (Fig. 4). The timing of the WGD/segmental duplication events could be estimated by the Ks value (synonymous substitutions per site) [28]. Based on previous reports, the Ks values, show that the genome of apple and pear have undergone two genome-wide duplication events: the ancient WGD from γ triplication (Ks ~ 1.6) and a recent WGD (Ks ~ 0.2) [29] in the apple genome, as well the ancient WGD (Ks ~ 1.5–1.8) that took place ~ 140 MYA [30] and the recent WGD (Ks ~ 0.15–0.3) occurred at 30–45 MYA [31] in pear. Hence, Ks values were used to estimate the time for the gene duplication events among the PbLYP gene family members. The Ks values suggest that most PbrLYP genes were duplicated from around the time of the ancient WGD event, while some originated from the recent WGD (Table 4). The Ka/Ks ratio represents the selection intensity and direction. The Ka/Ks value of one showed neutral evolution, positive selection when the Ka/Ks value is greater than one, and purifying selection when the Ka/Ks value is lower than one [32]. Our results showed all Ka/Ks ratios of the PbrLYP gene pairs were lower than one, demonstrating, PbrLYPs primarily evolved under purifying selection (Table 4).
Table 3.
Numbers of LYP genes from different origins in Arabidopsis and Roseceae genomes
| Species | No. of total LYP genes | No. of LYP genes from different origins (percentage) | ||||
|---|---|---|---|---|---|---|
| Singleton | Dispersed | Proximal | Tandem | WGD/segmental | ||
| Arabidopsis | 8 | 0 (0) | 6 (75) | 0 (0) | 0 (0) | 2 (25) |
| Chinese white pear | 18 | 0 (0) | 7 (39) | 0 (0) | 4 (22) | 7 (39) |
| European pear | 14 | 0 (0) | 7 (50) | 0 (0) | 2 (14) | 5 (36) |
| Apple | 21 | 0 (0) | 4 (19) | 3 (14) | 2 (10) | 12 (57) |
| Strawberry | 13 | 0 (0) | 6 (46) | 3 (23) | 2 (15) | 2 (15) |
| Peach | 15 | 0 (0) | 8 (53) | 1 (7) | 4 (27) | 2 (13) |
| Mei | 16 | 0 (0) | 9 (56) | 1 (6) | 4 (25) | 2 (13) |
| Sweet cherry | 14 | 0 (0) | 8 (57) | 2 (14) | 2 (14) | 2 (14) |
| Black rasberry | 13 | 0 (0) | 9 (69) | 0 (0) | 2 (15) | 2 (15) |
Note: The table shows the total numbers of LYP genes and the numbers of genes from each kind of duplication events in Arabidopsis and eight Rosaceae species
Fig. 3.
Distribution and collinearity of the LYP genes. Red lines along the circumference of the circle mark the gene positions. The lines in different colors inside the circle indicate collinearity relationships among the genes from Arabidopsis and eight Rosaceae species
Fig. 4.
Segmental duplications between PbrLYPs. This figure depicts a stretch of nucleotides, comprising 100 kb on each side flanking the LYP genes. Pairs of homologous gene are linked with bands. The black horizontal line showed the chromosome segment, and the green broad line containing arrowhead indicates the gene along with its transcriptional orientation. The text beside the gene is the gene ID. The LYP genes are indicated in red, and other genes indicated in green. a PbrLYK1a-PbrLYK1b2, b PbrLYK3a1-PbrLYK3a2, c PbrLYM2–1-PbrLYM2–2
Table 4.
The duplicate mode and estimation of the absolute date for large-scale duplication events in Chines white pear
| Duplicated pair | Duplicated mode | Number of conserved flanking protein-coding genes | Mean Ka | Mean Ks | Date (million years ago) | Ka/Ks | |
|---|---|---|---|---|---|---|---|
| LYK1 | PbrLYK1a — PbrLYK1b2 | WGD/segmental | 6 | 0.322 | 2.047 | 684.62 | 0.16 |
| PbrLYK1a — PbrLYK1b3 | Dispersed | \ | 0.356 | 2.093 | 700.28 | 0.17 | |
| PbrLYK1b1 — PbrLYK1b3 | Dispersed | \ | 0.111 | 0.183 | 61.35 | 0.61 | |
| PbrLYK1b2 — PbrLYK1b1 | Tandem | \ | 0.090 | 0.173 | 57.72 | 0.52 | |
| PbrLYK1b2 — PbrLYK3a2 | Dispersed | \ | 0.675 | 2.305 | 771.03 | 0.29 | |
| PbrLYK1b3 — PbrLYK3a2 | Dispersed | \ | 0.729 | 1.828 | 611.36 | 0.40 | |
| LYK2 | PbrLYK2 — PbrLYK4/5a1 | Dispersed | \ | 0.802 | \ | \ | \ |
| LYK3 | PbrLYK3a1 — PbrLYK3b1 | Dispersed | \ | 0.642 | 2.259 | 755.67 | 0.28 |
| PbrLYK3a2 — PbrLYK3a1 | WGD/segmental | 8 | 0.001 | 0.021 | 6.90 | 0.07 | |
| PbrLYK3a2 — PbrLYK3b1 | Dispersed | \ | 0.638 | 2.073 | 693.36 | 0.31 | |
| LYK4/5 | PbrLYK4/5b — PbrLYK1a | Dispersed | \ | 0.870 | \ | \ | \ |
| PbrLYK4/5b — PbrLYK4/5a1 | Tandem | \ | 0.572 | 2.616 | 875.16 | 0.22 | |
| LYM1/3 | PbrLYM1/3–1 — PbrLYM1/3–2 | Dispersed | \ | 0.605 | 2.786 | 931.98 | 0.22 |
| PbrLYM1/3–2 — PbrLYM1/3–3 | Dispersed | \ | 0.588 | 2.000 | 668.94 | 0.29 | |
| LYM2 | PbrLYM2–1 — PbrLYM1/3–2 | Dispersed | \ | 0.735 | 1.290 | 431.63 | 0.57 |
| PbrLYM2–1 — PbrLYM2–2 | WGD/segmental | 8 | 0.009 | 0.011 | 3.78 | 0.77 | |
| PbrLYM2–2 — PbrLYM1/3–3 | Dispersed | \ | 0.755 | 1.529 | 511.53 | 0.49 | |
| NFP | PbrNFP1 — PbrNFP2 | Dispersed | \ | 0.584 | \ | \ | \ |
| PbrNFP2 — PbrLYK4/5a1 | Dispersed | \ | 0.765 | \ | \ | \ | |
| PbrNFP2 — PbrLYK4/5b | Tandem | \ | 0.708 | 2.581 | 863.31 | 0.27 |
Conserved motif analysis of the LYP gene family in Rosaceae species
The types and composition of inner motifs primarily determine the protein function. To further identify motif construction of the LYP gene family in the Rosaceae, the online MEME program was used in this study to detect motif patterns of LYPs. Fifteen conserved motifs with low E values were recognized (Fig. 2b). The number of motifs in LYPs were varied and there were distinctive differences in motif composition between LYM type LYP proteins and other types. The details of each motif and the motif patterns of each subgroup are shown in Supplementary Figs. S2 and S3, respectively. Among Rosaceae LYPs, pattern [#6,14,12,9,6] was shown in almost LYPs as the conserved motifs for the LYP family. However, the pattern [#8,10,7,5,1,3,2,13,4] only could be detected in the KD sequence of LYK type proteins. Without KD, the characteristic motifs were on the N-terminal of LYMs, such as common pattern [#6,14,12,9,6] for LYM1/3 s and partially incomplete pattern [#14,12,9,6] for LYM2s.
Each subfamily had its own relatively certain motif composition with significant differences between LYM and other types of LYPs (Supplementary Fig. S3b), indicating that LYPs are relatively conserved in their evolutionary history and the division among groups may have occurred at an early period. Previous reports have shown that AtLYM2 and AtLYK1/4/5 were all involved in the chitin signal pathway and played a role as core participators or co-receptors to mediate the signaling for their chitin binding ability of the second LysM on the ectodomain [7, 8, 10–13]. In this work, 3 of the 7 conserved residues for chitin binding on that LysM domain were detected on motif #12 (Supplementary Figs. S2 and S3b). This conserved motif was only detected in the extracellular domains of LYK1, LYK4/5, NFP, WAKL, and LYM type groups, which is indicative of inner links between the chitin affinity and the presence of motif #12 and that the evolution between each subgroup may not be completely independent. Interestingly, as the unique motif in LYP family, motif #15 was only detected in LYK3a subfamily (Supplementary Fig. S3b). This indicates that the conserved motif #15 may be related to the opposite function of AtLYK3, as a negative regulator in chitin-induced immunoreactions. Our results suggest that the occurrence of motif #12 and #15 in the ectodomain of Rosaceae LYPs may be related to the chitin affinity and the negative regulation of defensive responses to fungal pathogens, respectively.
Expression levels of the PbrLYPs
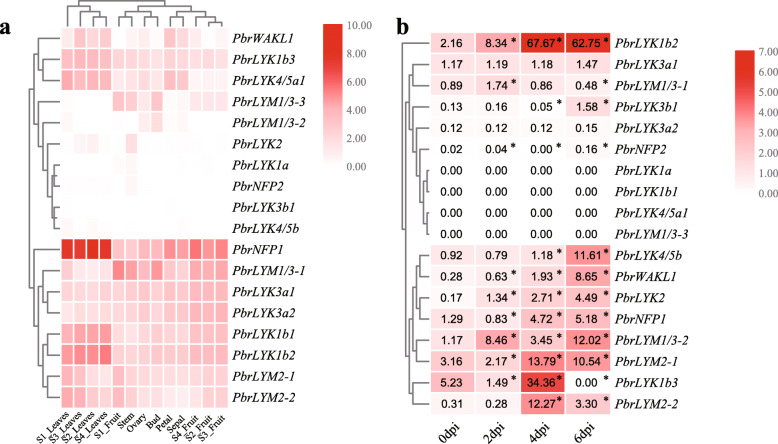
Previous transcriptome analysis of Chinese white pear revealed tissue-specific expression patterns in petal, sepal, ovary, stem, bud, leaves and fruit [33, 34]. The results indicated that the background expression of most PbrLYP genes was rarely detected, however other genes were primarily expressed in fruit and leaves (Fig. 5a). For example, PbrLYK3a1 and PbrLYK3a2 were mainly expressed in fruit, petal, sepal and ovary, while PbrLYK1b3, PbrLYK1b1, PbrLYK1b2, PbrLYK4/5a1 and PbrNFP1 showed preferential expression levels in leaves. However, PbrLYM1/3–1 showed highest expression in fruit, stem, and bud, but relatively low expression in leaves.
Fig. 5.
Expression pattern analyses of the PbrLYPs in Chinese white pear. a Expression analyses of the 18 PbrLYPs using previously published transcriptome data in different tissues and stages of Chinese white pear. b Expression analyses of the PbrLYP genes in leaves by qRT-PCR after B. dothidea infection treatments. The pear actin was used as an internal reference for the normalization. Asterisks indicated a significant difference in statistics compared with 0dpi at the indicated time points (* P < 0.05)
To verify whether PbrLYPs participate in the defense response against Botryosphaeria dothidea (B. dothidea) pathogen infection, a fungal pathogen that can cause the ring rot disease in apple and pear, we performed an infection treatment experiment with 6-weeks-old pear seedlings. The qRT-PCR (quantitative real-time PCR) results indicated that most of PbrLYPs were up-regulated by the infection of B. dothidea, with the peak expression occurring at 4 or 6 dpi (Fig. 5b). For example, at 4 dpi, the relative expression of PbrLYK1b2, LYM2–1 and LYM2–2 was significantly higher than controls at the highest expression. At 6 dpi, the expression levels of PbrLYK1b2, PbrLYK3b1, PbrLYK4/5b and PbrWAKL1 were still relatively higher than control, as well the peak levels of PbrNFP2, PbrLYK2, PbrNFP1 and PbrLYM1/3–2. The results indicated that these differentially expressed genes may participate in the defense reactions. However, the expression of PbrLYK3a1, PbrLYK3a2, PbrLYK1a, PbrLYK1b1, PbrLYK4/5a-1 and PbrLYM1/3–3 showed no significant change following infection in Chinese white pear. Furthermore, the expression of PbrNFP1, PbrLYK1b2, PbrWAKL1 and PbrLYM2–2 was also significantly up-regulated in Qiuzi pear induced by the pathogen infection. On the contrary, the expression levels of PbrLYM2–1, PbrLYK3a2 and PbrLYM1/3–1 were down-regulated after the infection (Supplementary Fig. S4).
Subcellular localization of the PbrLYPs
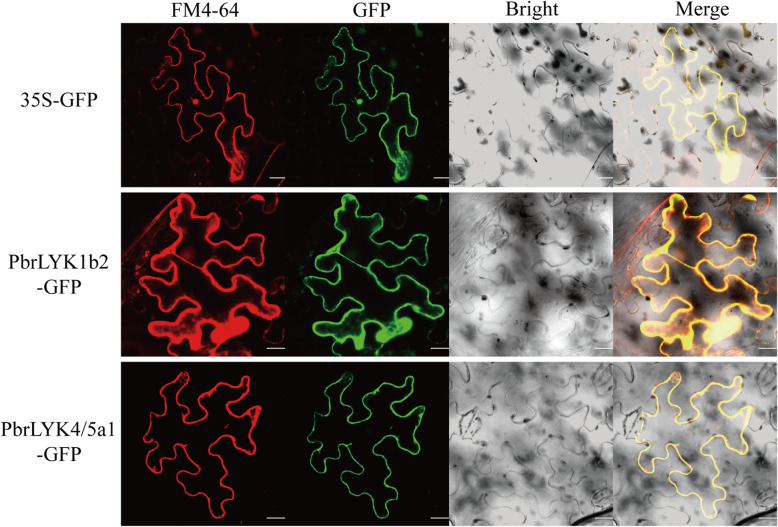
PRRs are primarily located in the plasma membrane and are in direct contact with the ligand. To verify whether LYP proteins were also present on the plasma membrane in the Rosaceae and had the potential to act as PRRs, we first performed structural analyses of PbrLYP proteins using the TMHMM online software. The sequence analysis showed that, except for LYM type LYPs, all PbrLYPs contained a transmembrane (TM) region (Supplementary Additional file 3), demonstrating that they can also be localized in the membrane. Considering the effect of signal peptides (SP) on subcellular localization, we selected PbrLYK1b2 and PbrLYK4/5a1 to verify the localization of PbrLYPs. The open reading frame of each gene was cloned from pear branches and PbrLYP-35S-GFP fusion proteins or control (35S-GFP alone) were transformed separately into Nb leaves. Based on fluorescence microscopy, using the control plasmid, the green fluorescence was found to be scattered in the overall cell. However, PbrLYK1b2-GFP and PbrLYK4/5a1-GFP containing vectors showed the green fluorescence only in the cell membrane, as depicted in Fig. 6. Therefore, all PbrLYPs with SP and TM seems to have the potential to act as PRRs.
Fig. 6.
Subcellular localization of PbrLYPs protein. The two fusion proteins and 35S-GFP as the control were expressed transiently in Nb leaves in an independent manner and observed via confocal microscope. The merged images include the membrane dye FM4–64 red fluorescence channel (first panels) and green fluorescence channel (second panels). The analogous bright-field images are represented on the right. Bar = 20 μm
Discussion
The LysM-containing proteins have been primarily implicated in the PTI immune processes including the early stage of node formation and pathogen perception by direct or indirect PAMP binding. As crucial components of the chitin receptor complex, some members of the LYP gene family have been extensively studied in model plants, such as rice and Arabidopsis. However, there have been few such efforts to annotate the LYP gene family in pear. In this study, we identified 124 LYP genes among eight Rosaceae species. In Chinese white pear, 18 LYP genes were identified compared to the 21 LYP members in apple, while the other Rosaceae species contained 13 to 16 LYPs. The number of LYP genes in the Rosaceae was much higher than those reported in the model eudicot Arabidopsis thaliana. In other words, the expansion of LYP family genes in Rosaceae species appeared like a common event. The larger gene numbers in certain LYK1, LYK3 and LYK4/5 groups suggested that these groups may play diverse roles in the adaptive evolution of Rosaceae species to environmental stresses.
The gene duplication analysis showed that the expansion of LYP genes in Chinese white pear and apple was primarily due to WGD/segmental events, along with dispersed duplication as the major expansion driving force for LYPs in the other six Rosaceae species. According to the widely- spanning Ks values, many large-scale duplication events were detected at the ancient stage (Ks values of 12 of 20 duplicated gene pairs were around 1.290 ~ 2.786) in Chinese white pear (Table 4). The results suggested that the selection of the function of perception and defense response to chitin was beginning at a very early stage and continuing up to now. These functions are fundamental and vital for plant survival. The LYPs in LYK1/3/4/5 and LYM2 groups were reported to be closely related to chitin signaling [7, 8, 10–13]. In this study, six out of seven WGD/segmental-type PbrLYPs were detected in LYK1, LYK3 and LYM2 groups, suggesting that the evolution of chitin response was mainly derived from the WGD/segmental events and remained in Chinese white pear. The Ka/Ks ratios of all duplicated PbrLYP pairs were less than one, which implied that the PbrLYPs are undergoing purifying selection and they seem to be necessary for adaptation to the current environment in their evolutionary history.
Phylogenetic analysis classified the Rosaceae LYPs into eight subgroups, which suggested that the evolution of different subfamilies was relatively independent. Analysis of the gene structure and protein motif showed the high similarity of the motif composition and exon-intron architecture within each subgroup also confirming independent evolution (Fig. 2b, c). The above results suggested that the genes in the same clade may have similar evolutionary histories and may perform a similar function. As shown in the gene structure analyses, subfamilies LYK1 and LYK3 contained the highest number of exons in LYKs, indicating that intronization in the exons of the genes (in these groups) might have happened. The number of exons also has a key contribution to their divergent functionality in various tissues, organs, or growth periods. A similar case was also found in the LYM2 group in the LYMs in European pear, strawberry, and black raspberry.
According to the previous works about the evolution of the plant LYP gene family, the LYPs have evolved through local and segmental duplications and can be grouped into three main types: LYP-I (about 10 exons per gene and containing conserved KD), LYP-II (one to five exons per gene, lacking the KD), and LYP-III (one or two exons per gene, with a KD unlike that of LYP-I), likely arising from the fusion of other type LYP genes [16, 35–37]. The LYP-I type gene products are the main PRRs in each signaling pathway. LYP-II types are likely to not function as core receptor kinases, but form complexes with other LYPs, such as that AtLYK1 that could interact with AtLYM1/3 and AtLYK4/5 to mediate bacterial and fungal pathogen perception in Arabidopsis, respectively [7, 11, 12, 38]. The Rosaceae LYPs were well-matched to the characteristics in protein and gene structure of AtLYPs, and therefore may potentially have similar roles in signaling. With the higher number of genes and exons, the genes in the LYM2 group of sweet cherry and black raspberry and genes in LYK1 and LYK3 groups of other species seemed to have undergone stronger evolutionary selection and may be more diverse in function. In addition, we also investigated the conserved motifs of LYPs and determined the putative protein localization as well as their collinearity relationships. In total, 15 distinct conserved motifs among various LYP proteins were predicted by the MEME analysis. As shown above in Fig. 2b and Fig. S3b, motif patterns [#6,14,12,9,6] and [#8,10,7,5,1,3,2,13,4] might represent the functional motifs of ectodomain and intracellular kinase domain of LYPs, respectively. Meanwhile, a LYK3a-unique motif #15 was detected in the region of the juxtamembrane domain in the LYK3a protein group, where it could be regulated by phosphorylation to affect the activity of the kinase domain [39]. The AtLYK3 was also placed into LYK3a group with the motif #15. Therefore, it is reasonable to consider that the motif #15 was related to the negative regulatory functions of the genes in LYK3a. However, this question requires further research. The conservative residues for chitin binding were detected in the motif #12, and that motif was only found in LYK1/4/5, NFP, WAKL and LYM type groups (Fig. S3b), suggesting that the genes in those groups likely shared a common ancestor and had the similar functions at the ancient period in response to chitin. After a long period of evolution and selection, the duplicated Rosaceae LYPs remained in relatively large numbers, suggesting that the LYP genes were important for Rosaceae species in adaptation to the complex and changing environments.
The LYP gene family plays various important roles in growth and response to biotic stresses. For example, AtLYK1 encodes a plasma membrane-localized receptor kinase protein. AtCERK1 works as a receptor homodimer or the core element of the hetero-tetramer with AtLYK4/5 or AtLYM1/3. These complexes are involved in initiating PTI responses against the fungal or bacterial pathogen infection in Arabidopsis [7, 8, 10–12, 38]. Transcriptome data showed that in the common target tissues for pathogen infection, such as leaves and fruit, some PbrLYPs had relatively higher expression than in other tissues for host protection. Based on the expression patterns, these PbrLYPs may be regarded as putative defense-related genes at the background level. In China, fruit ring rot and stem wart diseases caused by pathogen B. dothidea occur in almost all pear-growing areas, and the target organs including pear fruit, stem, shoots and leaf [40]. Although the pathogenesis of B. dothidea was poorly understood, the previous work in apple had reported that a LysM-containing protein gene, MdCERK1–2, was involved in the anti-fungal defense responses as a PRR and significantly upregulated after B. dothidea infection [23]. In other word, the chitin signaling pathway was likely recruited during the infection of B. dothidea. To verify whether PbrLYPs were involved in the defense reaction, we performed an infection treatment and qRT-PCR analysis. Our qRT-PCR results indicated that some of PbrLYPs participated in the immune response to B. dothidea infection (Fig. 5 and Fig. S4). In addition, after the infection with B. dothidea, significantly increased relative expression of several putative defense-related genes was detected by qRT-PCR, including PbrLYK1b2, PbrNFP1, PbrWAKL1 and PbrLYM2–2, which is consistent with the case of MdCERK1–2 in apple [23]. It is interesting to note that, although the expression level of PbrLYK4/5b in Chinese white pear was strongly up-regulated by the fungal pathogen infection, it could not be detected in Qiuzi pear before or after infection. This may have been due to the relatively high expression levels of PbrLYK1b1, PbrLYK1b2 and PbrWAKL1 in Qiuzi pear compared to Chinese white pear. Therefore, if some of the LYPs were the core PRR of chitin perception complex and able to form a homodimer, like in Arabidopsis, then these extensively expressed proteins may perform full functions independently activate the chitin signaling pathway. Hence, there may be no need to recruit co-receptors like PbrLYK4/5b to form a recognition complex, possibly accounting for the high pathogen resistance of Qiuzi pear. This question requires further investigation to reveal the infection strategy of B. dothidea and the resistant mechanism of host pear plant. Furthermore, the subcellular localization analysis demonstrated that pathogen-inducible genes (PbrLYK1b2 and PbrLYK4/5b) were also located at the plasmalemma, suggesting a potential capacity for PbrLYKs to act as PRRs at the subcellular level. In addition to the expression analysis, these results were consistent with previous studies that have implicated LYP genes in biotic stress tolerance via chitin-binding chitin and activation of the downstream immune response as plasmalemma-located PRRs [7, 11, 12, 38].
However, further investigation will be required to determine whether the expansion of LYPs could provide more advanced pathogen detection model to increase the chances of surviving under the complex environmental changes and the receptor complex in Chinese white pear or other Rosaceae species similar to that in Arabidopsis or rice. The characterization of key elements and the composite pattern of these complexes was also crucial to the understanding of the functional mechanisms of LYPs in the Rosaceae.
Conclusions
One hundred twenty-four full-length LYP genes were determined in the eight genomes of Rosaceae, along with the 18 LYP genes of the Chinese white pear genome. Based on the protein sequences and CDS structural characteristics, comparison with Arabidopsis homologs, and phylogenetic analysis, the LYP genes were classified into eight groups i.e., LYK1, LYK2, LYK3, LYK4/5, LYM1/3, LYM2, NFP, and WAKL, with groups LYK1 and LYK3 possibly having higher functional diversity. According to the analysis of collinearity, the ancient and recent WGD and dispersed duplication might have a role in the evolution of the LYP gene family, associated with apple and Chinese white pear. The LYP family genes were found to be greatly influenced via evolutionary negative selection. qRT-PCR revealed that LYP genes might have a vital role against the fungal pathogenesis. The underlined collected data establish a foundation for advanced studies to evaluate the complexity of LYP gene family in the Rosaceae.
Methods
Determination of LYP genes in Chinese white pear and other species of Rosaceae
For the determination of the LYP genes in pear and other species of Rosaceae, several databases were employed. To acquire LYP family genes, we used the following strategy: The genome sequences of eight species belong to Rosaceae were downloaded from each genome project (Supplementary Table S3). Subsequently, we built a Hidden Markov Model (HMM) with the extracellular domain sequences of 12 well-studied LYP proteins (AtLYK1–5, AtLYM1–3, OsCERK1, OsCEBiP, OsLYP4 and OsLYP6, the accession numbers and extracellular domain sequences were shown in Table S5) [41], using the HMMER3 software package [42, 43], and downloaded the seed file of Lysin Motif domain (PF01476) from the Pfam database (http://pfam.xfam.org/). The sequences of eight Arabidopsis proteins and four rice proteins were acquired from TAIR (https://www.arabidopsis.org/) and NCBI (https://www.ncbi.nlm.nih.gov/), respectively. Then HMM searches with PF01476 and self-build model were independently conducted for the local protein databases of eight species of Rosaceae via HMMER3 with E-values <1e− 10. Furthermore, two resulting gene lists were intersected and the protein sequences were detected by the NCBI Batch CD-Search tools (Batch CD-Search: https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) based on CDD v3.18 and SMART v6.0 databases for the validation of the existence of the LysM domain. The sequences of proteins with E-values greater than 1e− 6 or without a LysM domain were deleted. The relevant accession numbers of LYP genes were shown in Table 1.
Structure and conserved motif analysis of the LYP genes
The Gene Structure Display Server (GSDS 2.0) (http://gsds.cbi.pku.edu.cn/) was used to analyze the structures of the LYP genes by aligning the cDNA sequences with their corresponding genomic DNA sequences. Conserved motif analysis of LYP proteins was performed by online Multiple Expectation Maximization for Motif Elicitation (MEME) (http://meme.nbcr.net/meme/cgibin/meme.cgi) [44]. Maximum number parameter of motifs was seted as 15.
Phylogenetic analysis
The construction of phylogenetic trees was carried out with Neighbor-Joining (NJ) and a bootstrap of 1000 in MEGA7.0 (http:// www.megasoftware.net/) [45]. The p-distance was used and the optional parameters for pairwise deletion were considered.
Chromosomal localization and synteny analysis
Genome annotation files of Arabidopsis and eight Rosaceae species were downloaded from TAIR and each genome project (Supplementary Table S3). The same procedure used in the PGDD (http://chibba.agtec.uga.edu/duplication/) [46] was performed to analyze the synteny among the LYPs. Primarily, for the investigation of considerable pairs of the homologous gene, the local all-vs-all BLASTP searches among Arabidopsis and eight species belong to Rosaceae genomes were conducted (E < 1e− 10). Afterward, MCScanX was employed for the determination of syntenic gene pairs with the BLASTP result and gene location information used as input files [47]. The downstream analysis tool (duplicate_gene_classifier) in the MCScanX package was employed for the identification of tandem, proximal dispersed, and segmental/whole-genome duplications (WGD) of LYP family genes. The results were visualized using circos-0.69 software [48]. The Ka and Ks values were analyzed via KaKs_Calculator 2.0 [49]. For the estimation of the date of segmental duplication events, the succeeding pairs of homologous genes within 100 Kb on all sides of the LYP genes, considered for the mean Ks calculation.
Subcellular localization of the PbrLYPs
The amplification of total-length CDS of the PbrLYK1b2 and PbrLYK4/5a1 was carried out via PCR, respectively. Purified products were subcloned directionally into the modified pCAMBIA1300-GFP vector (Clontech, Beijing, China), and resulted in PbrLYK1b2-GFP and PbrLYK4/5a1-GFP. Primers assisting gene cloning and vectors construction, depicted in added Table S4. The agrobacterium carrying above products were transformed into 4-week-old Nb leaves, respectively, as the method reported previously with slight modification [50]. Images were obtained via the Zeiss LSM Image Browser (Zeiss LSM 780, Germany). The independent assays were conducted at a minimum of thrice for each gene. The empty vector pCAMBIA1300-GFP was used as control.
Infection treatment and quantitative real-time PCR
Chinese white pear (Dangshansuli, Pyrus bretschneideri Rehd.) and “Qiuzi” pear (Pyrus ussuriensis Maxim) seeds were obtained from the pear germplasm orchard of the Center of Pear Engineering Technology Research situated at Hushu in Nanjing and were allowed to grow in soil pots in a maintained environment (2:1 light/ dark period, 25 °C) in the phytotron. Sixty leaves of each kind of pears were harvested from 15 six-week-old seedlings and placed on the sterile water wetted filter paper in a petri dish overnight. Then the 5-day-old fresh B. dothidea mycelia, which grown in the PDA plat, were stuck to the paraxial surface of leaves to perform infection. The infected leaves were cryopreserved with liquid nitrogen at 0 dpi (day post infection), 2dpi, 4dpi, and 6dpi. Total RNA extraction and the synthesis of cDNA were according to the instructions of RNA kit (Tiangen, Beijing, China) and PrimeScript RT reagent Kit (Trans Gen). Specialized primers of the constitutive TUB and PbrLYP genes were designed via NCBI online tool Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi? LINK_LOC=BlastHome) with the Specificity Parameters Organism option set as Pyrus bretschneideri (taxid:225117) (Supplementary Table S4). The specificity of each primer pairs was verified by the online program Primer search-Paired against the pear genome. The qRT-PCR assays were conducted with three technical copies. QRT-PCR reactions (20 μl per hole) were performed as previously reported [51]. The expression was evaluated for each sample via the 2−ΔΔCt method, and Duncan’s multiple range test was conducted. A P-value of less than 0.05 was the considerable variation and indicated with asterisks. The reported RNA-seq data was processed for the evaluation of the expression patterns of PbrLYPs (obtained from the NCBI bioproject PRJNA563942 and PRJNA498777) [33, 34], the differentially expressed genes were identified with |log2FC| > 1. The heatmaps were drawn in TBtools v0.666 [52].
Supplementary information
Additional file 1: Table S1. Exon number statistical analysis of LYP genes. Table S2. Duplication type of LYP genes in Arabidopsis and eight Rosaceae species. Table S3. Genome information of eight Rosaceae species. Table S4. Primers of PbrLYPs for qRT-PCR and vector construction. Table S5. Extracellular domain sequences of 12 LYP proteins from Arabidopsis and rice.
Additional file 2: Fig. S1. Gene position of PbrLYPs. The characters in blue indicate the chromosome or scaffold number. The height of columns in red indicate the length of chromosome. The number on the right side of the chromosome indicate the start position of each gene. Fig. S2 15 MEME motifs of LYPs. Over-represented motifs in Arabidopsis and the eight Rosaceae species were identified using the MEME tool. The stack’s height indicates the level of sequence conservation. The heights of the residues within the stack indicate the relative frequencies of each residue at that position. The star symbols under Motif 12 indicate the conservative positions for chitin binding. Fig. S3. Schematic diagram and distribution of conserved motifs among the LYP proteins of Arabidopsis and eight Rosaceae species. a: The schematic diagram of conserved motifs among the LYP proteins detected by NCBI Batch CD-Search. 4EBZ: the ectodomain of AtCERK1 in PDB databases (https://www.rcsb.org/). b: The distribution of conserved motifs among the LYP proteins detected by MEME. The red and black squares represent non- and conservative MEME domains in the subfamily. The blue square represents the unique motif in LYK3a subgroup. Fig. S4 qRT-PCR analyses of the PbrLYP genes in Qiuzi pear leaves after B. dothidea infection. The pear actin was used as internal reference for the normalization. Asterisks indicated significant difference in statistics compared with 0dpi at the indicated time points (* P < 0.05).
Additional file 3. Transmembrane regions prediction of AtLYPs and PbrLYPs.
Acknowledgements
We would like to thank Xun Sun from Nanjing agricultural university to provide us with pathogens and associate professor Xin Qiao for revising the manuscript. Bioinformatic analysis was supported by the Bioinformatics Center of Nanjing Agricultural University.
Abbreviations
- LYP
Lysin motif containing protein
- LYK
LysM receptor kinase
- LYM
LysM-containing glycosylphosphatidylinositol-anchored protein
- LysM
Lysin motif
- RLK
Receptor-like kinase
- NFP
Nod factor perception protein
- WAKL
Wall associated kinase-like
- WGD
Whole-genome duplication
- MYA
Million years ago
- PAMP
Pathogen-associated molecular pattern
- PRR
Pattern-recognition receptor
- PTI
PAMP-triggered immunity
- EF-Tu
Elongation factor Tu
- flg22
Bacterial flagellin epitope
- FLS2
Flagellin sensing 2
- KD
Kinase domain
- kd
Dissociation constant
- ROS
Reactive oxygen species
- MAPK
Mitogen-activated protein kinase
- PR
Pathogenesis-related
- CEBiP
Chitin elicitor binding protein
- CERK
Chitin elicitor receptor kinase
- PI
Protein isoelectric point
- GRAVY
Grand average of hydropathy
- Ka
Nonsynonymous substitutions per site
- Ks
Synonymous substitutions per site
- B. dothidea
Botryosphaeria dothidea
- qRT-PCR
Quantitative real-time Polymerase Chain Reaction
- CDS
Coding sequence
- Nb
Nicotiana benthamiana
- dpi
Day post infection
Authors’ contributions
QC designed and carried out the experiments and wrote the manuscript. QL contributed in bioinformatics analysis supports. XQ and HY directed and revised the manuscript. All authors read, reviewed and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31830081, 31772276, 31471839), Earmarked Fund for China Agriculture Research System (CARS-28). The funders were not involved in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The relevant accession numbers of all LYP genes from eight used Rosaceae species were obtained from each genome annotation files and shown in Table 1: All needed genome sequences and genome annotation files of Chinese white pear, Arabidopsis and Mei were obtained from the Nanjing Agricultural University pear genome project website (http://peargenome.njau.edu.cn), The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and a clone of the genome data of the Prunus mume genome project website from GitHub (https://github.com/lileiting/prunusmumegenome), respectively; all needed genome sequences and genome annotation files of other six used Rosaceae species were downloaded from the Genome Database for Rosaceae (European pear: ftp://ftp.bioinfo.wsu.edu/species/Pyrus_communis/Pcommunis-draft_genome.v1.0/; Apple: ftp://ftp.bioinfo.wsu.edu/species/Malus_x_domestica/Malus_x_domestica-genome_GDDH13_v1.1; Strawberry: ftp://ftp.bioinfo.wsu.edu/species/Fragaria_vesca/Fvesca-genome.v4.0.a2/; Peach: ftp://ftp.bioinfo.wsu.edu/species/Prunus_persica/Prunus_persica-genome.v2.0.a1/; Sweet cherry: ftp://ftp.bioinfo.wsu.edu/species/Prunus_avium/Prunus_avium-genome.v1.0.a1; Black rasberry: ftp://ftp.bioinfo.wsu.edu/species/Rubus_occidentalis/Rubus_occidentalis-genome.v3.0/). The sequences of the 12 well-studied LYP proteins (AtLYK1–5, AtLYM1–3, OsCERK1, OsCEBiP, OsLYP4 and OsLYP6) were acquired from TAIR and NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/), respectively, and the accession numbers of them were listed in Table S5. The seed file of Lysin Motif domain (PF01476) were downloaded from the Pfam database (http://pfam.xfam.org/). The transcriptome raw reads from different pear tissue and stage have been deposited at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA563942 and PRJNA498777.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-07032-9.
References
- 1.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6(10):973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 2.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.00859. [DOI] [PubMed] [Google Scholar]
- 3.Dangl JL, Jones JDG. Defence Responses To Infection. Nature. 2001;411(June). 10.1038/nature05286. [DOI] [PubMed]
- 4.Buendia L, Girardin A, Wang T, Cottret L, Lefebvre B. LysM receptor-like kinase and lysM receptor-like protein families: an update on phylogeny and functional characterization. Front Plant Sci. 2018;871(October):1–25. doi: 10.3389/fpls.2018.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antolín-Llovera M, Ried MK, Binder A, Parniske M. Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol. 2012;50:451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- 6.Delaux PM, Radhakrishnan GV, Jayaraman D, et al. Algal ancestor of land plants was preadapted for symbiosis. Proc Natl Acad Sci U S A. 2015;112(43):13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue D-X, Li C-L, Xie Z-P, Staehelin C. LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J Exp Bot. 2019. 10.1093/jxb/erz313. [DOI] [PubMed]
- 8.Miya A, Albert P, Shinya T, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petutschnig EK, Jones AME, Serazetdinova L, Lipka U, Lipka V. The Lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285(37):28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan J, Zhang X-C, Neece D, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell Online. 2008;20(2):471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan J, Tanaka K, Zhang XC, et al. LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 2012;160(1):396–406. doi: 10.1104/pp.112.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Liang Y, Tanaka K, et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife. 2014;3:1–19. doi: 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kouzai Y, Nakajima K, Hayafune M, et al. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol Biol. 2014;84(4–5):519–528. doi: 10.1007/s11103-013-0149-6. [DOI] [PubMed] [Google Scholar]
- 14.Hayafune M, Berisio R, Marchetti R, et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci. 2014;111(3):E404–E413. doi: 10.1073/pnas.1312099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzai Y, Mochizuki S, Nakajima K, et al. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol Plant-Microbe Interact. 2014;27(9):975–982. doi: 10.1094/MPMI-03-14-0068-R. [DOI] [PubMed] [Google Scholar]
- 16.Arrighi J-F, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, et al. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 18.Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 19.Mulder L, Lefebvre B, Cullimore J, Imberty A. LysM domains of Medicago truncatula NFP protein involved in nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and nod factors. Glycobiol. 2006;16:801–809. doi: 10.1093/glycob/cwl006. [DOI] [PubMed] [Google Scholar]
- 20.Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, et al. LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Tian Y, Cong P, Zhu Y. Functional characterization of an apple (Malus x domestica) LysM domain receptor encoding gene for its role in defense response. Plant Sci. 2018;269:56–65. 10.1016/j.plantsci.2018.01.006. [DOI] [PubMed]
- 23.Chen Q, Dong C, Sun X, Zhang Y, Dai H, Bai S. Overexpression of an apple LysM- confers improved resistance to the pathogenic fungus, Alternaria alternata, in Nicotiana benthamiana. BMC Plant Biol. 2020;20:146. doi: 10.1186/s12870-020-02361-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KFX, Li WH. Comparative analysis of the receptor-like kinase family in Arabidopsis and Rice. Plant Cell. 2004;16(5):1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buendia L, Wang T, Girardin A, Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016;210(1):184–195. doi: 10.1111/nph.13753. [DOI] [PubMed] [Google Scholar]
- 26.Richards S, Rose LE. The evolutionary history of LysM-RLKs (LYKs/LYRs) in wild tomatoes. BMC Evol Biol. 2019;19(1):1–17. doi: 10.1186/s12862-019-1467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16(4):510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiao X, Li M, Li L, Yin H, Wu J, Zhang S. Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 2015;15(1):1–16. doi: 10.1186/s12870-014-0401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus × domestica Borkh.) Nat Genet. 2010;42(10):833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 30.Fawcett JA, Maere S, Van De Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci U S A. 2009;106(14):5737–42. 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed]
- 31.Wu J, Wang Z, Shi Z, et al. The genome of the pear (Pyrus bretschneideri Rehd.) Genome Res. 2013;23(2):396–408. doi: 10.1101/gr.144311.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. 10.1093/molbev/msm088. [DOI] [PubMed]
- 33.Li Q, Qiao X, Yin H, et al. Unbiased subgenome evolution following a recent whole-genome duplication in pear (Pyrus bretschneideri Rehd.). Hortic Res. 2019;6(1). 10.1038/s41438-018-0110-6. [DOI] [PMC free article] [PubMed]
- 34.Qiao X, Yin H, Li L, et al. Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri) Front Plant Sci. 2018;9(February):1–16. doi: 10.3389/fpls.2018.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang XC, Cannon SB, Stacey G. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 2009;9(1):1–13. doi: 10.1186/1471-2148-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang XC, Wu X, Findley S, et al. Molecular evolution of Lysin motif-type receptor-like kinases in plants. Plant Physiol. 2007;144(2):623–636. doi: 10.1104/pp.107.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lohmann GV, Shimoda Y, Nielsen MW, et al. Evolution and regulation of the lotus japonicus LysM receptor gene family. Mol Plant-Microbe Interact. 2010;23(4):510–521. doi: 10.1094/MPMI-23-4-0510. [DOI] [PubMed] [Google Scholar]
- 38.Willmann R, Lajunen HM, Erbs G, et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A. 2011;108(49):19824–19829. doi: 10.1073/pnas.1112862108/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1112862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Liu J, Wang J, et al. The juxtamembrane domains of Arabidopsis CERK1, BAK1, and FLS2 play a conserved role in chitin-induced signaling. J Integr Plant Biol. 2019:jipb.12847. 10.1111/jipb.12847. [DOI] [PubMed]
- 40.Zhai L, et al. Biological and Molecular Characterization of Four Botryosphaeria Species Isolated from Pear Plants Showing Stem Wart and Stem Canker in China. Plant Dis. 2014;98(6). 10.1094/PDIS-10-13-1060-RE. [DOI] [PubMed]
- 41.Liu B, Li JF, Ao Y, et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2012;24(8):3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7(10). 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed]
- 43.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(database):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(SUPPL. 1):281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed]
- 46.Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(D1):1152–1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang H, Wang X, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;(706):1944–54. 10.1101/gr.080978.108.1944. [DOI] [PMC free article] [PubMed]
- 48.Connors J, Krzywinski M, Schein J, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(604):1639–45. 10.1101/gr.092759.109.19. [DOI] [PMC free article] [PubMed]
- 49.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinforma. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Dong C, Zhang Y, Zhu J, Dai H, Bai S. An apple cyclic nucleotide-gated ion channel gene highly responsive to Botryosphaeria dothidea infection enhances the susceptibility of Nicotiana benthamiana to bacterial and fungal pathogens. Plant Sci. 2018;269(January):94–105. doi: 10.1016/j.plantsci.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Chen G, Li X, Qiao X, et al. Genome-wide survey and expression analysis of the SLAC/SLAH gene family in pear (Pyrus bretschneideri) and other members of the Rosaceae. Genomics. 2018;(June):0–1. 10.1016/j.ygeno.2018.07.004. [DOI] [PubMed]
- 52.Chen C, Chen H, Zhang Y, et al. TBtools - an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020:1–9. 10.1016/j.molp.2020.06.009. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Exon number statistical analysis of LYP genes. Table S2. Duplication type of LYP genes in Arabidopsis and eight Rosaceae species. Table S3. Genome information of eight Rosaceae species. Table S4. Primers of PbrLYPs for qRT-PCR and vector construction. Table S5. Extracellular domain sequences of 12 LYP proteins from Arabidopsis and rice.
Additional file 2: Fig. S1. Gene position of PbrLYPs. The characters in blue indicate the chromosome or scaffold number. The height of columns in red indicate the length of chromosome. The number on the right side of the chromosome indicate the start position of each gene. Fig. S2 15 MEME motifs of LYPs. Over-represented motifs in Arabidopsis and the eight Rosaceae species were identified using the MEME tool. The stack’s height indicates the level of sequence conservation. The heights of the residues within the stack indicate the relative frequencies of each residue at that position. The star symbols under Motif 12 indicate the conservative positions for chitin binding. Fig. S3. Schematic diagram and distribution of conserved motifs among the LYP proteins of Arabidopsis and eight Rosaceae species. a: The schematic diagram of conserved motifs among the LYP proteins detected by NCBI Batch CD-Search. 4EBZ: the ectodomain of AtCERK1 in PDB databases (https://www.rcsb.org/). b: The distribution of conserved motifs among the LYP proteins detected by MEME. The red and black squares represent non- and conservative MEME domains in the subfamily. The blue square represents the unique motif in LYK3a subgroup. Fig. S4 qRT-PCR analyses of the PbrLYP genes in Qiuzi pear leaves after B. dothidea infection. The pear actin was used as internal reference for the normalization. Asterisks indicated significant difference in statistics compared with 0dpi at the indicated time points (* P < 0.05).
Additional file 3. Transmembrane regions prediction of AtLYPs and PbrLYPs.
Data Availability Statement
The relevant accession numbers of all LYP genes from eight used Rosaceae species were obtained from each genome annotation files and shown in Table 1: All needed genome sequences and genome annotation files of Chinese white pear, Arabidopsis and Mei were obtained from the Nanjing Agricultural University pear genome project website (http://peargenome.njau.edu.cn), The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and a clone of the genome data of the Prunus mume genome project website from GitHub (https://github.com/lileiting/prunusmumegenome), respectively; all needed genome sequences and genome annotation files of other six used Rosaceae species were downloaded from the Genome Database for Rosaceae (European pear: ftp://ftp.bioinfo.wsu.edu/species/Pyrus_communis/Pcommunis-draft_genome.v1.0/; Apple: ftp://ftp.bioinfo.wsu.edu/species/Malus_x_domestica/Malus_x_domestica-genome_GDDH13_v1.1; Strawberry: ftp://ftp.bioinfo.wsu.edu/species/Fragaria_vesca/Fvesca-genome.v4.0.a2/; Peach: ftp://ftp.bioinfo.wsu.edu/species/Prunus_persica/Prunus_persica-genome.v2.0.a1/; Sweet cherry: ftp://ftp.bioinfo.wsu.edu/species/Prunus_avium/Prunus_avium-genome.v1.0.a1; Black rasberry: ftp://ftp.bioinfo.wsu.edu/species/Rubus_occidentalis/Rubus_occidentalis-genome.v3.0/). The sequences of the 12 well-studied LYP proteins (AtLYK1–5, AtLYM1–3, OsCERK1, OsCEBiP, OsLYP4 and OsLYP6) were acquired from TAIR and NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/), respectively, and the accession numbers of them were listed in Table S5. The seed file of Lysin Motif domain (PF01476) were downloaded from the Pfam database (http://pfam.xfam.org/). The transcriptome raw reads from different pear tissue and stage have been deposited at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA563942 and PRJNA498777.