Abstract
Early detection and diagnosis are critical factors to control the COVID-19 spreading. A number of deep learning-based methodologies have been recently proposed for COVID-19 screening in CT scans as a tool to automate and help with the diagnosis. These approaches, however, suffer from at least one of the following problems: (i) they treat each CT scan slice independently and (ii) the methods are trained and tested with sets of images from the same dataset. Treating the slices independently means that the same patient may appear in the training and test sets at the same time which may produce misleading results. It also raises the question of whether the scans from the same patient should be evaluated as a group or not. Moreover, using a single dataset raises concerns about the generalization of the methods. Different datasets tend to present images of varying quality which may come from different types of CT machines reflecting the conditions of the countries and cities from where they come from. In order to address these two problems, in this work, we propose an Efficient Deep Learning Technique for the screening of COVID-19 with a voting-based approach. In this approach, the images from a given patient are classified as group in a voting system. The approach is tested in the two biggest datasets of COVID-19 CT analysis with a patient-based split. A cross dataset study is also presented to assess the robustness of the models in a more realistic scenario in which data comes from different distributions. The cross-dataset analysis has shown that the generalization power of deep learning models is far from acceptable for the task since accuracy drops from 87.68% to 56.16% on the best evaluation scenario. These results highlighted that the methods that aim at COVID-19 detection in CT-images have to improve significantly to be considered as a clinical option and larger and more diverse datasets are needed to evaluate the methods in a realistic scenario.
Keywords: COVID-19, Deep learning, EfficientNet, Pneumonia, Chest radiography
Highlights
-
•
Voting-based approach extending the EfficientNet Family of deep artificial neural networks.
-
•
A cross-dataset study with the two largest public datasets available.
-
•
A specific data augmentation process and transfer learning for such task.
-
•
A high-quality yet compact deep-learnign model for the screening of COVID-19 in CT scans.
-
•
Highest accuracy (and F1-measure) to date on COVID-CT dataset and SARS-CoV-2 CT-scan dataset.
1. Introduction
In March 2020, the World Health Organization (WHO) officially declared the outbreak of COVID-19, the disease caused by SARS-CoV-2, a pandemic. COVID-19 is highly infectious and can potentially evolve to fatal acute respiratory distress syndrome (ARDS). Early detection and diagnosis is a critical factor to control the COVID-19 spreading. The most common screening method to detect it is the reverse-transcription polymerase chain reaction (RT-PCR) testing. However, it is a laborious method and some studies reported its low sensitivity in early stages [1].
Chest scans such as X-rays and Computer tomography (CT) scans have been used to identify morphological patterns of lung lesions linked to the COVID-19. However, the accuracy of the diagnosis of COVID-19 by Chest scans strongly depends on experts [2] and Deep learning techniques have been studied as a tool to automate and help with the diagnosis [[3], [4], [5], [6], [7], [8]].
A computed tomography scan, or CT scan, produces detailed images of organs, bones, soft tissues and blood vessels. CT images allow physicians to identify internal structures and see their shape, size, density and texture. Different from conventional X-Rays, CT scans produce a set of slices of a given region of the body without overlaying the different body structures. Thus, CT scans give a much more detailed picture of the patient's condition than the conventional X-Rays. This detailed information can be used to determine whether there is a medical problem as well as the extent and exact location of the problem. For these reasons, a number of deep learning based methodologies have been recently proposed for COVID-19 screening in CT scans [[9], [10], [11], [12], [13], [14]].
The main bottleneck for the realization of a study such as the ones cited above is the lack of good quality comprehensive data sets. Possibly the first attempt to create such a data set was the so-called COVID-CT dataset [15] which consists of images mined from research papers. Different versions of this dataset were used in Refs. [[9], [10], [11], [12]]. For its most updated version, the highest reported accuracy, F1-score, and AUC were 86%, 85%, and 94% [9], respectively. More recently, Soares et al. [14] made another set of CT scans publicly available. It consists of 2482 CT scans taken from hospitals in the city of Sāo Paulo, Brazil. They have reported an accuracy, sensitivity, and positive predictive value of 97.38%, 95.53%, and 99.16%, respectively.
These two datasets are, to date, the biggest publicly available datasets. It can be seen that the difference in the best results obtained in each of them is significant which raises two questions: (i) Are the discrepancies in the results due to the differences in the datasets? (ii) Does a model trained in one dataset have good performance when tested with the other? This work aims to answer these two questions.
Another drawback of the best performing techniques is their immense number of parameters which directly influence their footprint and latency. Improving these two metrics allows the model to be more easily embedded in mobile applications and to be less of a burden on the server if provided as a web-service receiving an enormous number of requests per second. In addition, having a more compact baseline model allows the exploitation of higher resolution inputs without making the computational cost prohibitively high. Broadly speaking, the computational cost is an important factor in the accessibility and availability of the technology to the public.
Thus, the main goals of this work are: (i) to propose a high-quality yet compact deep-learnign model for the screening of COVID-19 in CT scans and (ii) to address, for the first time, the aforementioned questions regarding the two biggest datasets, and a (iii) proposal of a voting based evaluation approach.
To produce an efficient model we exploit and extend the EfficientNet Family of deep artificial neural networks along with a data augmentation process and transfer learning. Following previous evaluation protocols [9,14], state-of-the-art results are presented for the COVID-CT dataset (accuracy of 87.60%) and the SARS-CoV-2 CT-scan dataset (accuracy of 98.99%). The voting based approach showed promising results for the Covid-19 detection in CT images.
The remainder of this work is organized as follows. Section 2 present the details of COVID-CT [15] and SARS-CoV-2 CT-scan [14] datasets. The methodology is described in Section 3 and the experiments along with the results in Section 4. Finally, Section 5 presents the conclusion of this work.
2. Datasets
This section describes the two datasets considered in this work. To the best of our knowledge, these are the two largest public datasets to date.
2.1. SARS-CoV-2 CT-scan dataset
The SARS-CoV-2 CT-scan dataset [14] consists of 2482 CT scans from 120 patients, with 1252 CT scans of 60 patients infected by SARS-CoV-2 from males (32) and females (28), and 1230 CT scan images of 60 non-infected patients by SARS-CoV-2 from males (30) and females (30), but presenting other pulmonary diseases. Data was collected from hospitals of São Paulo, Brazil.
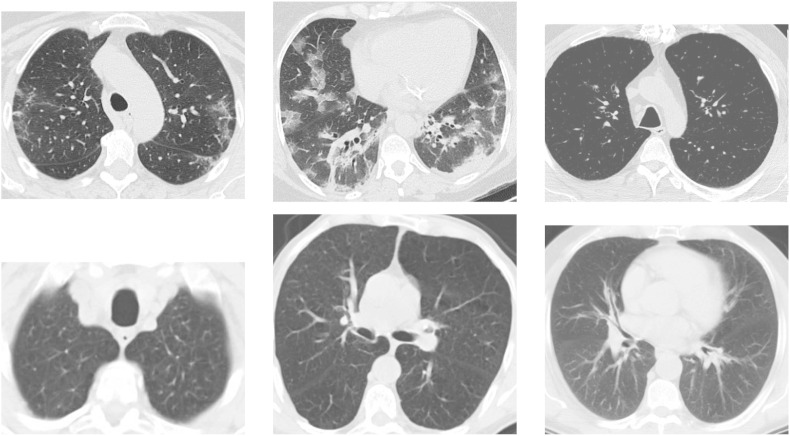
In this dataset the images consist of digital scans of the printed CT exams and they have no standard regarding image size (the dimensions of the smallest image in the dataset are while the largest images are ), Fig. 1 shows some examples.
Fig. 1.
Examples of CT images that are: positive for COVID-19 (top) and non-COVID-19 (bottom) from SARS-CoV-2 CT-scan dataset.
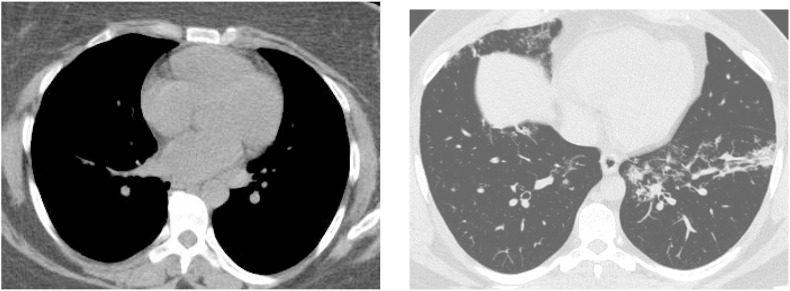
This dataset also lacks standardization regarding the contrast of the images, as can be seen in Fig. 2 .
Fig. 2.
Comparison among different contrast in images.
For method evaluation, the protocol presented in Ref. [14] proposes to randomly divide the dataset in training (80%) and test (20%) partitions. The dataset is available at https://www.kaggle.com/plameneduardo/sarscov2-ctscan-dataset.
2.2. COVID-CT dataset
To assemble the COVID-CT dataset [15], CT images of patients infected with COVID-19 were collected from scientific articles (pre-prints) deposited in the medRxiv and biRxiv repositories, from January 19 to March 25 and also some images were donated by hospitals (http://medicalsegmentation.com/covid19/). The PyMuPDF software was used to extract images from the manuscripts, in order to maintain high quality. Meta data were manually extracted and associated with each image: patient age, gender, location, medical history, scan time, severity of COVID-19, and medical report. A total of 349 images were collected, from 216 patients.
Regarding healthy and non-covid patients, the authors collected images from two other datasets (MedPix dataset, LUNA dataset), from the Radiopaedia website and from other articles and texts available at PubMed Central (PMC). A total of 463 images were collected from 55 patients.
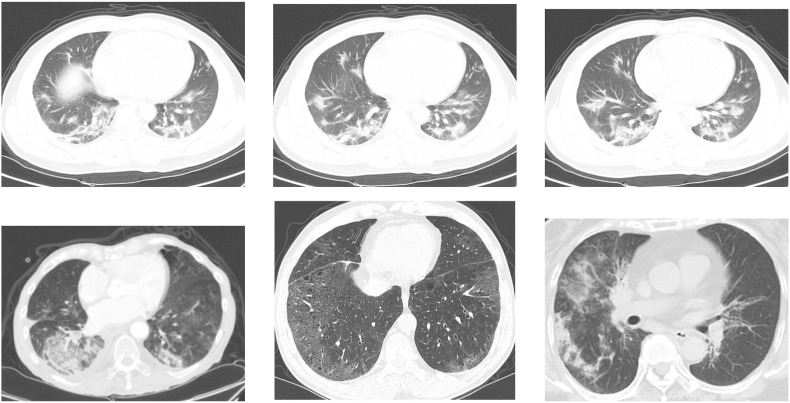
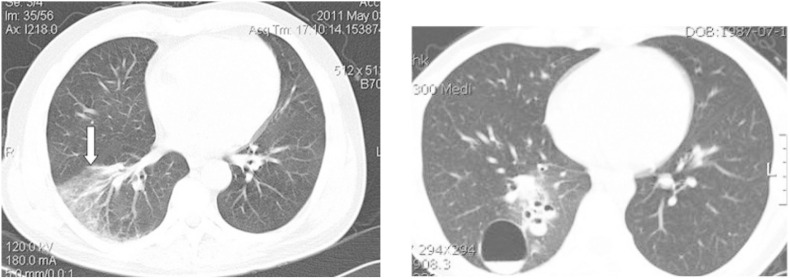
Analogous to the previous dataset, the COVID-CT dataset has defined standard for image size and contrast. Fig. 3 shows some examples. It is also important to highlight that some images contain textual information which may interfere with model prediction. See Fig. 4 .
Fig. 3.
Examples of CT images that are: positive for COVID-19 (top) and non-COVID-19 (bottom) from COVID-CT dataset.
Fig. 4.
Example of images with textual information.
A protocol is proposed for the creation of training, validation, and test sets. The COVID-19 images that were donated by hospitals and extracted directly from medical equipment (LUNA and Radiopaedia) were selected to compose the validation and test sets. The remaining - extracted from scientific articles and manuscripts - were reserved to compose the training set. The dataset is available at https://github.com/UCSD-AI4H/COVID-CT.
2.3. Final regards
Table 1 summarizes the datasets presented in this section. In the table is possible to observe the issues identified in the datasets, and the relation between the number of patients and the amount of images of each class (COVID and Non-COVID).
Table 1.
Datasets distribution.
3. Methodology
In this section, the proposed methodology for COVID-19 screening based on CT scans is presented. To this end, we extended an architecture of the EffiecientNet family and we trained the models with CT images from healthy and SaR-CoV-2 infected patients. The CT images come from the datasets described in the previous section and undergo the pre-processing procedure described below.
3.1. Pre-processing
Pre-processing is a very common process in computer vision applications. Pre-processing techniques can be useful for removing unwanted noise, emphasize aspects of the image that can help with the recognition task, or even help with the deep learning training phase.
In this work, a simple pixel intensity normalization in the range of is applied. This pre-processing is necessary for model convergence during the training phase.
For convolutional network models, the input images are often resized to maintain compatibility with the network architectures. Since EfficientNets have a low computational cost in terms of latency and memory, it makes it possible to exploit higher resolution input images. Thus, we also investigate the impact of varying the input resolution in the quality of the model. In this way, this pre-processing step becomes another parameter of the network.
3.2. EfficientCovidNet
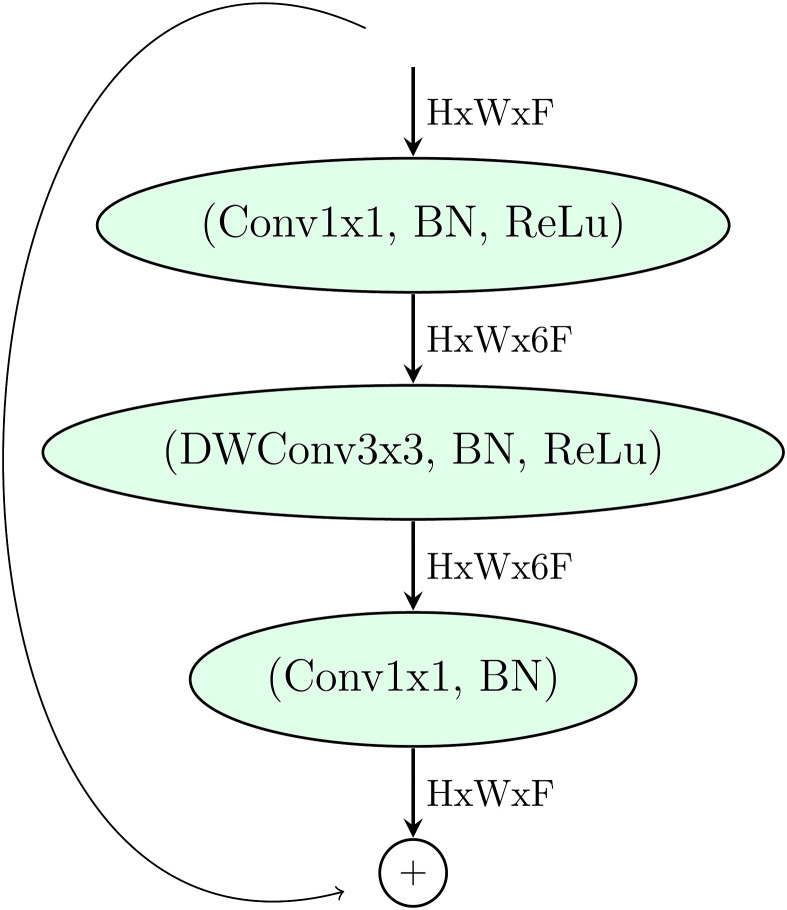
The EfficientNets are a family of artificial neural networks in which the basic building block is the Mobile Inverted Bottleneck Conv Block, MBconv [16], as depicted in Fig. 5 .
Fig. 5.
MBConv Block [16]. DWConv stands for depthwise conv, k3x3/k5x5 defines the kernel size, BN is batch normalization, represents the tensor shape (height, width, depth), and x1/2/3/4 is the multiplier for number of repeated layers.
Table 2 presents a typical EfficientNet architecture, particularly the B0 model. The main idea to achieve the EfficientNet architecture was to start from one high quality yet compact baseline model presented in Table 2 and progressively scale each of its dimensions, in a systematical manner, with a fixed set of scaling coefficients.
Table 2.
EfficientNet baseline network: B0 architecture.
| Stage | Operator | Resolution | #channels | #layers |
|---|---|---|---|---|
| 1 | Conv3x3 | 224x224 | 32 | 1 |
| 2 | MBConv1,k3x3 | 112x112 | 16 | 1 |
| 3 | MBConv6,k3x3 | 112x112 | 24 | 2 |
| 4 | MBConv6,k5x5 | 56x56 | 40 | 2 |
| 5 | MBConv6,k3x3 | 28x28 | 80 | 3 |
| 6 | MBConv6,k5x5 | 14x14 | 112 | 3 |
| 7 | MBConv6,k5x5 | 14x14 | 192 | 4 |
| 8 | MBConv6,k3x3 | 7x7 | 320 | 1 |
| 9 | Conv1x1/Pooling/FC | 7x7 | 1 |
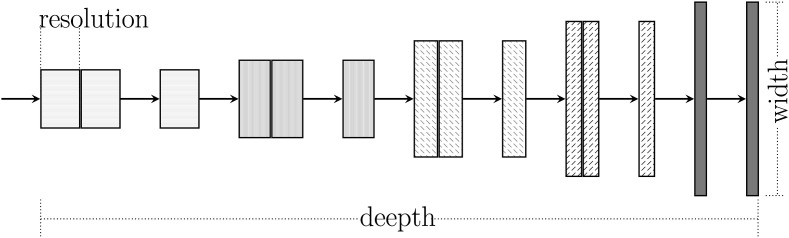
An EfficientNet can be defined by three dimensions: (i) depth; (ii) width; and (iii) resolution as illustrated in Fig. 6 .
Fig. 6.
Efficient net compound scaling on three parameters (Adapted from Ref. [17]).
Each dimension is scaled by the parameter φ according to Equation (1) where and are constants obtained experimentally by a grid search. Varying φ, one can find other derived networks. For instance, gives rise to the EfficientNet B1, gives rise to the EfficientNet B2, and so on.
According to Ref. [17], Eq. (1) provides a nice trade-off between computational cost and performance.
| (1) |
In [18], four new blocks are added to the baseline model to improve COVID-19 recognition on x-ray images. Here, we proposed modifications aimed at CT images, and six new blocks are added to an EfficientNet B0 architecture. These blocks were achieved by a grid search and can be seen in Table 3 and Table 4 . The considered search space was: Layers [1 to 6], Number of Neurons [100 to 1024], Activation Function [ReLu, Sigmoid, Swich], Dropout [0–80%], Batch Normalization [yes or no].
Table 3.
Smaller architecture - example of achieved blocks on the SARS-CoV-2 CT-scan dataset - EfficientNet-CT model. (NC = Number of Classes).
| Stage | Operator | Resolution | #channels | #layers |
|---|---|---|---|---|
| 1–9 | EfficientNet B3 | 300x300 | 32 | 1 |
| 10 | BN/Dropout | 7x7 | 1280 | 1 |
| 11 | FC/BN/Swich/Dropout | 1 | 512 | 1 |
| 12 | FC/BN/Swich | 1 | 128 | 1 |
| 13 | FC/Softmax | 1 | NC | 1 |
Table 4.
Deeper architecture - example of achieved blocks on the SARS-CoV-2 CT-scan dataset - EfficientNet-CT model. (NC = Number of Classes).
| Stage | Operator | Resolution | #channels | #layers |
|---|---|---|---|---|
| 1–9 | EfficientNet B0 | custom input | 32 | 1 |
| 10 | BN/Dropout | 7x7 | 1280 | 1 |
| 11 | FC/BN/Swich/Dropout | 1 | 2048 | 1 |
| 11 | FC/BN/Swich/Dropout | 1 | 1024 | 1 |
| 12 | FC/BN/Swich | 1 | 512 | 1 |
| 13 | FC/Softmax | 1 | NC | 1 |
Two searches are carried out. One aiming at a shallower architecture (number of layers clipped to 4 - Table 3) and the other deeper (Table 4).
On top of the model a new fully connected layer (FC) is added to adapt the classification task to a new domain. We highlight the following operations that compose the blocks: Batch normalization (BN), dropout, and swish activation functions.
The batch normalization operation constrains the output of the layer in a specific range, forcing zero mean and standard deviation one. That works as a regularization, increasing the stability of the neural network, and accelerating the training [19].
The Dropout [20] operation also act as a regularization, by inhibiting a few neurons and thus emulating a bagged ensemble of multiple neural networks, for each mini-batch on training. The dropout parameter defines the number of inhibited neurons (0–100 percent of the neurons of one layer).
Despite Rectified Linear Unit (ReLU) is considered the most popular activation function, here we explore the swish activation function [21]. ReLu can be formally defined as , while the swish function is defined by the equation:
| (2) |
The swish activation produces a smooth curve during the minimization loss process and contrary to that, the ReLu produces an abrupt change. Also, the swish function does not zero out small negative values. We believe those factors may be relevant for capturing patterns underlying the data [21].
3.3. Training
Due to its complexity, Deep learning models require a large number of instances to avoid overfitting. However, for the majority of real-life problems, data is not abundant. In fact, few are the situations where there is an abundance of data, such as the ImageNet [22] dataset. To overcome this issue, one could rely on two techniques: data augmentation and transfer learning. In this work, we made use of both techniques and we describe below.
3.3.1. Data augmentation
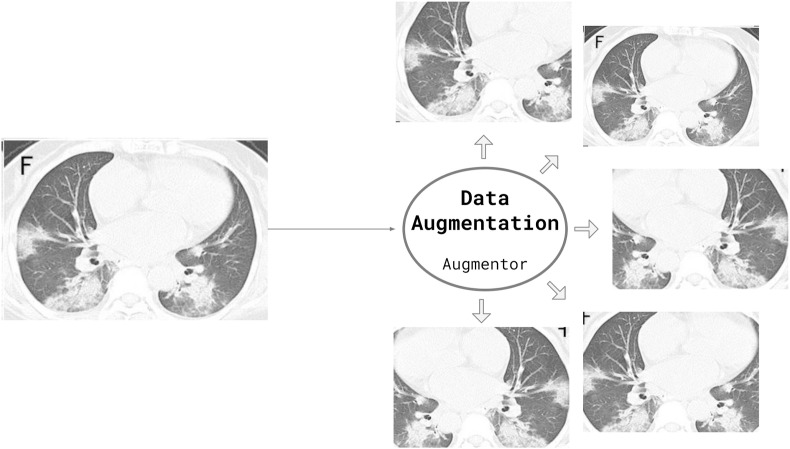
Data augmentation consists of increasing the training samples by transforming the images without losing semantic information. In this work, we applied three transformations to the training samples: rotation, horizontal flip, and scaling. Fig. 7 presents an example of the applied data augmentation. Such transformations preserve the images and would not prevent a physician from interpreting the images.
Fig. 7.
Data augmentation applied using the Augmentor python package. The transformations applied to the images are: rotation (0–15° clockwise or anticlockwise), 20% Zoom or horizontal flipping. All or none changes may be applied/combined according to a probability. (Figure created by the authors).
3.3.2. Transfer learning
Starting from a pre-trained neural network and re-training it to fit other datasets or other domains is called transfer learning [23]. Performing a fine-tune from a pre-trained network can enable the use of deep architectures when there is little training data, as the network has already learned filters in other domains/problems that can be reused [24]. In the present work, we have few images to carry out the training, especially of the COVID-19 class. Thus, transfer learning becomes imperative.
Our models inherit several layers from EfficientNet (See Table 2) and the new layers are randomly initialized with zero mean. EfficientNets were originally trained for the Imagenet dataset [22]. Thus, we follow the steps to transfer leanring from one domain to another:
-
1.
Copy the weights from one EfficientNet model to the new model;
-
2.
Modify the architecture of the new model, including new layers on top;
-
3.
Random initialize the new layers;
-
4.
Define which layers will pass through the learning process and which one will be frozen; and
-
5.
Perform the learning process, by updating the weights according to the loss function and optimization algorithm.
Here, the weights are updated with Adam Optimizer with a maximum learning rate of . We schedule the learning rate to decrease by a factor of 10 in the event of stagnation. The number of epochs is fixed at 10.
3.4. Evaluation metrics
Five metrics are used here to evaluate models: accuracy (), COVID-19 sensitivity (), COVID-19 positive prediction (), F1-score (), and Area Under the Receiver Operating Curve (), i.e.,
| (3) |
wherein , , , and stand for the COVID-19 samples correctly classified, non-COVID-19 samples correctly classified, the COVID-19 samples classified as non-COVID-19, the non-COVID-19 classified as COVID-19.
To comparison with the literature, we also report the result in terms of Area Under the Receiver Operating Curve (AUC). The Receiver Operating Curve is a plot of true positive rate (A.K.A. sensitivity = ) versus false positive rate (FPR). The FPR is define by Equation (4).
| (4) |
Higher the AUC, better the model is at distinguishing among image categories.
4. Experiments and discussion
Experiments were carried on an Intel(R) Core(TM) i7-5820K CPU 3.30 GHz, 64 GB Ram, one Titan X Pascal with 12 GB, and the TensorFlow/Keras framework for Python. The source code and pre-trained models are available in https://github.com/ufopcsilab/EfficientCovidNet. In the following subsections, we present the three experimental setups explored in this work. The training model start with a learning rate of 0.001 with the Adam Optimizer for 20 epochs using the categorical cross-entropy loss.
In a first setup, in Section 4.1, we investigate the discrepancy regarding the results reported by the methods considered state-of-the-art for the two studied datasets. The best approach for the COVID-CT dataset reports 86.0% of accuracy [9]. For the SARS-CoV-2 CT-scan dataset, the state-of-the-art method achieves 97.38% of accuracy [14]. However, the SARS-CoV-2 CT-scan dataset has significantly more images than the COVID-CT dataset and the same number of patients (individuals). To assess whether this difference is due to the evaluation protocol, we perform two experiments. We investigate the impact of selecting samples/images for training and test sets at random and in a second step, we evaluate the impact of performing the selection guided by individuals, that is, ensuring that there are no samples from the same individual simultaneously in the training and test sets.
In a second setup, in Section 4.2, we investigate a very important aspect, which is the generalization power of a model. A model is only useful if it can also generalize to data from other distributions or other datasets. In this regard, we evaluate how the model, trained with the SARS-CoV-2 CT-scan dataset, behaves when it is faced with images from another dataset, the COVID-CT Dataset. We follow the data-split protocol proposed in Ref. [15].
Finally, for the third setup, we explore our EfficientCovidNet model only with the COVID-CT Dataset, considering the protocol proposed in Refs. [15]. This setup aims to expand the comparison of the proposed approach with the literature since this dataset is the most popular to date. Here we also explore the impact of varying the size of the input images.
4.1. Setup 1 : 5-fold evaluation on a large dataset
To evaluate the performance of the proposed approach, we tested the protocol proposed by Soares et al. [14] and three different scenarios using a 5-fold cross-validation: (i) “Random”, (ii) “Slices”, and (iii) “Voting”. The “Random” evaluation divide the data into training and test sets randomly. The “Slice” evaluation consider all the CT images independent of each other but consider the patient division, that is, we prevent samples from one individual simultaneously in the training and test sets. In this manner, the model will always be evaluated with samples from unknown individuals. Finally, the “Voting” evaluation consider all images of an individual and a voting scheme to achieve a diagnosis per individual instead of by instance or image. Considering that several CT images are acquired in a single exam for a single individual, we believe that the disease patterns will not be present on all instances. Thus, an evaluation using a voting scheme, considering all possible instances of one individual, could increase the chances of success.
4.1.1. Results
Following the protocol proposed in Refs. [14] the data were divided into training, validation and test. The proposed approach in this work enhanced all metrics on test set as shown in Table 5 .
Table 5.
Classification protocol proposed in Ref. [14].
| Approach | (%) | (%) | (%) |
|---|---|---|---|
| Soares et al. [14] | 97.38 | 95.53 | 99.16 |
| Proposed Approach | 98.99 | 98.80 | 99.20 |
Despite the outstanding results presented in Table 5, we believe that such results are overestimated. Upon this fact, we introduce a 5-fold classification and some changes in the original protocol as described and with the results presented in Table 6.
The “Random” evaluation presents better results when compared to the two other approaches (“Slice” and “Voting”). One of the reasons is due to data from the same patient/individual in both training and test sets, which leads to an overestimated result. Upon this fact, our hypothesis is that an approach tends to learn the patterns related to the individuals instead the COVID patterns.
In the “Slice” evaluation, the samples are classified as an isolated instance, such as the “Random” one but ensuring that all samples of an individual are exclusively present only on one data partition: training or test set. A downgrade is observed which clearly shows an overestimation from the “Random” evaluation.
On the opposite to the “Slice” evaluation, the “Voting” one considers all images of an individual to decide whether the individual is infected or not. It is worth to emphasize that the same model is used in both approaches, that is, the model trained by image (only one “ slice ” of the lung).
Due to the nature of CT scans, we believe the disease patterns will not manifest in all slices (instance/images) of an individual CT exam, and results of “Slice” and “Voting” evaluation reflect that. We believe this can generate false positives/false negatives and therefore impact the figures of approaches (See Table 6 . Besides, this problem can be seen as a multiple instance learning (MIL) problem [25] and that a MIL-based approach can be a promising path for future work.
Table 6.
5-fold classification by slicing and with voting.
| Approach | (%) | (%) | (%) |
|---|---|---|---|
| Random | 98.5 0.4 | 98.6 0.6 | 98.4 0.6 |
| Slice | 86.6 10.1 | 94.8 4.5 | 79.7 20.9 |
| Voting | 89.6 5.1 | 92.0 10.0 | 77.5 23.3 |
Comparing the results of both Table 5, Table 6, we believe the presence of samples from the same individual in training and test tends to lead an overestimation of an approach. To circumvent this issue, it is necessary to ensure the division of the dataset considering the individual, and the use of a cross-dataset approach.
4.2. Setup 2: cross-dataset evaluation
For this experiment, we investigate the impact of learning a model in one data distribution and evaluate on another one. This scenario is closer to reality since it is almost impossible to train a model with images acquired from all available sensors, environments and individuals.
On this setup, the SARS-CoV-2 CT-scan dataset [14] is used only for training/validation, and none image of this dataset is present on the test set. For the test set, we use the dataset presented in Ref. [15], the COVID-CT, since it is a dataset used by several authors in the literature. We follow the protocol proposed in Ref. [15] to split the COVID-CT in train and test sets, however, we highlight that for training the model only images from the SARS-CoV-2 CT-scan dataset is used. We also evaluated other test configurations, such as using the COVID-CT training partition as a test and also combining both partitions from the COVID-CT dataset as a larger test set (See Table 7 ). We also test the opposite scenario, in which we use all images from the COVID-CT dataset [15] for training and all images of SARS-CoV-2 CT-scan dataset [14] to test.
Table 7.
Cross-dataset results.
| Training dataset | Test dataset | (%) | (%) | (%) |
|---|---|---|---|---|
| SARS-CoV-2 CT-scan dataset [14] | COVID-CT [15] (Train) | 59.12 | 64.14 | 54.95 |
| SARS-CoV-2 CT-scan dataset [14] | COVID-CT [15] (Test) | 56.16 | 53.06 | 54.74 |
| SARS-CoV-2 CT-scan dataset [14] | COVID-CT [15] (Train + Test) | 58.31 | 61.03 | 54.90 |
| COVID-CT [15] (Train + Test) | SARS-CoV-2 CT-scan dataset [14] | 45.25 | 54.39 | 46.36 |
4.2.1. Results
As one can see, the model performance is drastically reduced when we compare cross-dataset evaluation against an intra dataset one. We believe that the reason for this behavior is due to data acquisition diversity. Images from different datasets can be acquired by different equipment, different image sensors, and thus, change relevant features on the images impairing recognition. The model could learn how to identify portions and patterns of one image that may indicate the presence (or absence) of COVID-19, although, those patterns may no appear in a different dataset.
Training on COVID-CT [15] and testing in SARS-CoV-2 CT-scan dataset [14] presents even worse results since COVID-CT training set is smaller.
We believe such test should be mandatory for all methods aiming at COVID-19 recognition with CT images, since it is the one that most resembles a real test.
4.3. Setup 3: impact of input resolution
In this setup, we evaluate the protocol presented in Ref. [15] only on COVID-CT dataset. Zhao et al. [15] proposes to divide the COVID-CT dataset into three sets: training, validation, and testing. We also applied data augmentation by rotating (max 0.15° for each side), randomly zooming (80% of the are) with 20% of chance and horizontal flipping with a probability of 50%. We stress that the data augmentation is applied only for training data. The final number of training images totalized 2968 images (1442 of COVID and 1408 of NonCOVID). Using the protocol in Refs. [15], the test set consists of 203 images (98 of COVID and 105 of NonCOVID).
4.3.1. Results
In Table 8 , we report the results of the proposed approach using the protocol described in Ref. [15]. One may observe that the experiments with the same approach used in Setups 1 and 2 (EfficientNet-B3) has a worse performance when compared with the ones available in the literature.
Table 8.
Custom input using the EfficientNet-B0 as the base network.
| Depth | Input size | (%) | (%) | (%) | (%) |
|---|---|---|---|---|---|
| EfficientNet-B3 | 300x300 | 77.34 | 69.39 | 80.95 | 74.72 |
| Architecture 1 | 224x224 | 79.31 | 70.41 | 84.15 | 76.70 |
| 300x300 | 76.85 | 69.39 | 80.00 | 74.32 | |
| 350x350 | 80.79 | 79.59 | 80.41 | 80.00 | |
| 400x400 | 83.25 | 80.61 | 84.04 | 82.29 | |
| 450x450 | 83.25 | 81.63 | 83.33 | 82.47 | |
| 500x500 | 83.74 | 83.67 | 82.83 | 83.25 | |
| Architecture 2 | 224x224 | 83.74 | 77.55 | 87.36 | 82.16 |
| 300x300 | 81.28 | 79.59 | 81.25 | 80.41 | |
| 350x350 | 86.21 | 81.63 | 88.89 | 85.10 | |
| 400x400 | 80.30 | 74.49 | 82.95 | 78.49 | |
| 450x450 | 77.34 | 75.51 | 77.08 | 76.29 | |
| 500x500 | 87.68 | 79.59 | 93.98 | 86.19 |
Aiming to reduce the incidence of overfitting during training of “Architecture 1”, we propose a deeper network. In most of the cases, when the deeper network is used (see “Architecture 2” in Table 8), rather than a Architecture 1 one (see “Architecture 1” in Table 8), a gain is observed on all reported figures.
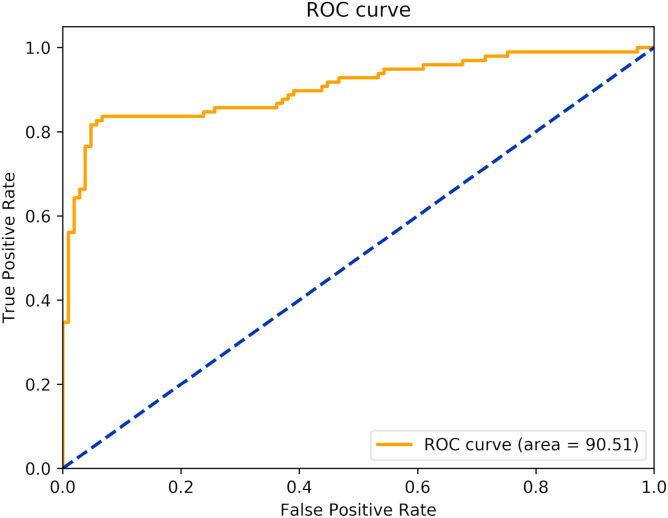
The best model is the one with the Architecture 2 with input size of 500x500 (source available at https://github.com/ufopcsilab/EfficientCovidNet). The ROC curve of the model is presented in Fig. 8 .
Fig. 8.
ROC curve of the proposed approach.
We present in Table 9 a comparison of the best proposed approach against the ones available in the literature. Despite the results presented by Amyar et al. [12] and Mobiny et al. [10], both evaluated their approach with only 105 images (47 COVID and 58 NonCovid) and, therefore, they cannot be directly compared to the present work. Thus, the best results previously obtained in this setup were presented in Refs. [9]. Although the work proposed here overcomes it in terms of accuracy and F1-score on COVID-CT dataset using a significantly smaller model (3 smaller). The base model proposed in Ref. [9] needs 14,149,480 parameters while the one proposed here only 4,779,038 parameters. The training loss curves for the best model can be seen in Fig. 9 .
Table 9.
Comparison with literature. # - Evaluated with a different test set: only 105 images (47 COVID and 58 NonCovid).
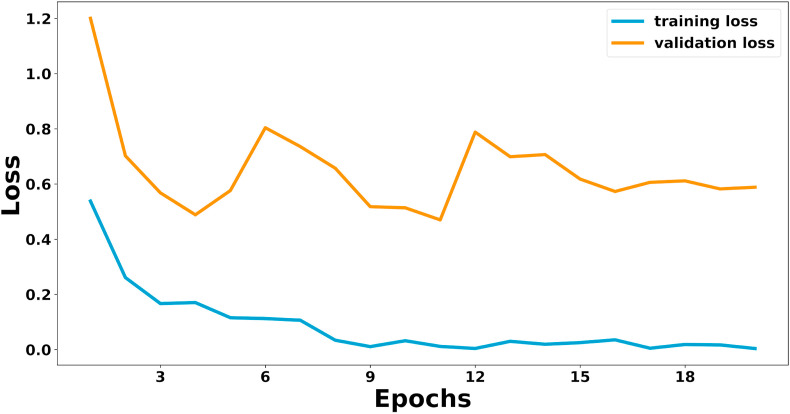
Fig. 9.
Training loss curve of Architecture 2 with input of 500x500.
5. Conclusion
In this work, a model for the detection of COVID-19 patterns in CT images, namely EffiecintCovidNet, is proposed along with a voting based approach and a cross-dataset analysis. The proposed model presents comparable results to the state-of-the-art methods and the highest accuracy to date on both datasets. Also, it is three times smaller (with 4.78 million parameters against 14.15 million of He et al. [9]) and has a latency of 0.010 s. This model could enable the use on devices with low computational power, such as smartphones and tablets or even facilitate integration with the Radiology PACS.
Our model was evaluated on three setups and with the two largest public datasets, including a cross-dataset analysis. To the best of our knowledge, this is the first work to carry out such analysis for the present task and we believe that this is a major contribution of our work. The cross-dataset approach is of paramount importance for the methods aiming to detect COVID-19 in CT images since the approach resembles a real scenario and unveils the limitations of the methods (for instance, the accuracy drops from 87.68% to 56.16% in this scenario for the COVID-CT test set). Our analysis suggests that the methods that aim COVID-19 detection in CT images have to improve significantly to be considered as a clinical option. The proposed voting-base approach favors the detection of false positives and false negatives and thus contributes to improving accuracy.
In this study, we show the potential of Deep Learning models for the task of COVID-19 detection on CT images. We also emphasize that larger and more diverse datasets are needed in order to evaluate the methods in a more realistic manner. As a future research path, we intend to build a very large CT image datasets from several Brazilian centers, in order to try to cover a larger spectrum of equipment (sensors), ethnic groups and acquisition processes and thus, properly validate our method.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank UFOP, UFPR, FAPEMIG, CAPES and CNPq (Grant #428333/2016–8 and #313423/2017–2) for the financial support. The authors also would like NVIDIA for the donation of one GPU Titan Black and two GPU Titan X.
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., Lui M.M.-s., Lo C.S.-Y., Leung B., Khong P.-L. Imaging profile of the covid-19 infection: radiologic findings and literature review. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images; p. 11055. arXiv preprint arXiv:2003. [Google Scholar]
- 4.Farooq M., Hafeez A. 2020. Covid-resnet: a deep learning framework for screening of covid19 from radiographs; p. 14395. arXiv preprint arXiv:2003. [Google Scholar]
- 5.Li T., Han Z., Wei B., Zheng Y., Hong Y., Cong J. 2020. Robust screening of covid-19 from chest x-ray via discriminative cost-sensitive learning; p. 12592. ArXiv abs/2004. [Google Scholar]
- 6.Abbas A., Abdelsamea M., Gaber M. medRxiv; 2020. Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Wong A. 2020. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luz E., Silva P.L., Silva R., Silva L., Moreira G., Menotti D. 2020. Towards an effective and efficient deep learning model for covid-19 patterns detection in x-ray images. arXiv:2004.05717. [Google Scholar]
- 9.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., Xie P. medRxiv; 2020. Sample-efficient deep learning for covid-19 diagnosis based on ct scans. [Google Scholar]
- 10.Mobiny A., Cicalese P.A., Zare S., Yuan P., Abavisani M., Wu C.C., Ahuja J., de Groot P.M., Van Nguyen H. 2020. Radiologist-level covid-19 detection using ct scans with detail-oriented capsule networks. arXiv preprint arXiv:2004.07407. [Google Scholar]
- 11.Polsinelli M., Cinque L., Placidi G. 2020. A light cnn for detecting covid-19 from ct scans of the chest; p. 12837. arXiv preprint arXiv:2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amyar A., Modzelewski R., Ruan S. medRxiv; 2020. Multi-task deep learning based ct imaging analysis for covid-19: classification and segmentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., Xu B. medRxiv; 2020. A deep learning algorithm using ct images to screen for corona virus disease (covid-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. medRxiv; 2020. Sars-cov-2 ct-scan dataset: a large dataset of real patients ct scans for sars-cov-2 identification. [Google Scholar]
- 15.Zhao J., Zhang Y., He X., Xie P. 2020. Covid-ct-dataset: a ct scan dataset about covid-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 16.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L.-C. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018. Mobilenetv2: Inverted residuals and linear bottlenecks; pp. 4510–4520. [Google Scholar]
- 17.Tan M., Le Q.V. 2019. Efficientnet: rethinking model scaling for convolutional neural networks. arXiv preprint arXiv:1905.11946. [Google Scholar]
- 18.Luz E., Moreira G., Junior L.A.Z., Menotti D. Deep periocular representation aiming video surveillance. Pattern Recogn Lett. 2018;114:2–12. [Google Scholar]
- 19.Ioffe S., Szegedy C. 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167. [Google Scholar]
- 20.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929–1958. [Google Scholar]
- 21.Ramachandran P., Zoph B., Le Q.V. 2017. Searching for activation functions. arXiv preprint arXiv:1710.05941. [Google Scholar]
- 22.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. [Google Scholar]
- 23.Goodfellow I., Bengio Y., Courville A. MIT press; 2016. Deep learning. [Google Scholar]
- 24.Oquab M., Bottou L., Laptev I., Sivic J. Proceedings of the IEEE conference on computer vision and pattern recognition. 2014. Learning and transferring mid-level image representations using convolutional neural networks; pp. 1717–1724. [Google Scholar]
- 25.Sudharshan P., Petitjean C., Spanhol F., Oliveira L.E., Heutte L., Honeine P. Multiple instance learning for histopathological breast cancer image classification. Expert Syst Appl. 2019;117:103–111. [Google Scholar]