Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) seminal studies in mammalian cells1, 2, 3 have resulted in gene editing being broadly adopted in basic research. However, it has become apparent that the CRISPR/Cas9 system induces unintended off- and on-target genomic alterations4 and that there is a need for stricter clone screening methods before phenotypic characterisation is made, particularly before the technology is adopted for clinical purposes. Caution is also needed when working with cancer cell lines, as these often have underlying genomic instability and deficiencies in DNA repair or other safeguarding mechanisms which may permit large genomic deletions or rearrangements. Here, we report that CRISPR/Cas9 targeting of genes in close proximity to telomeres can result in chromosome arm truncations. We suggest assessing heterozygous single-nucleotide polymorphisms (SNPs) downstream of targeted genes to select clones without arm truncations. This screening approach could be applied alongside initial genotype assessment via sequencing at early stages of the experiment, prior to cell line expansion.
We generated CRISPR/Cas9-mediated ZNF516 knockout (KO) cell lines to characterise the role of ZNF516 in colorectal cancer. We used an HCT116 cell line harbouring doxycycline-inducible Cas9 (HCT116-Cas9) to restrict temporal expression of the endonuclease and minimise off-target effects. Cells were transfected with either a pool of four CRISPR RNAs (crRNAs) against ZNF516 (sites A–D in Figure 1A) or a pool of five nontargeting crRNAs, trans-activating crRNA, and treated with doxycycline for 5 days to induce Cas9 expression. After single-cell sorting, clones were expanded and screened for indel mutations using Sanger sequencing. ZNF516 protein levels were assessed by western blotting and messenger RNA expression levels assessed using quantitative PCR.
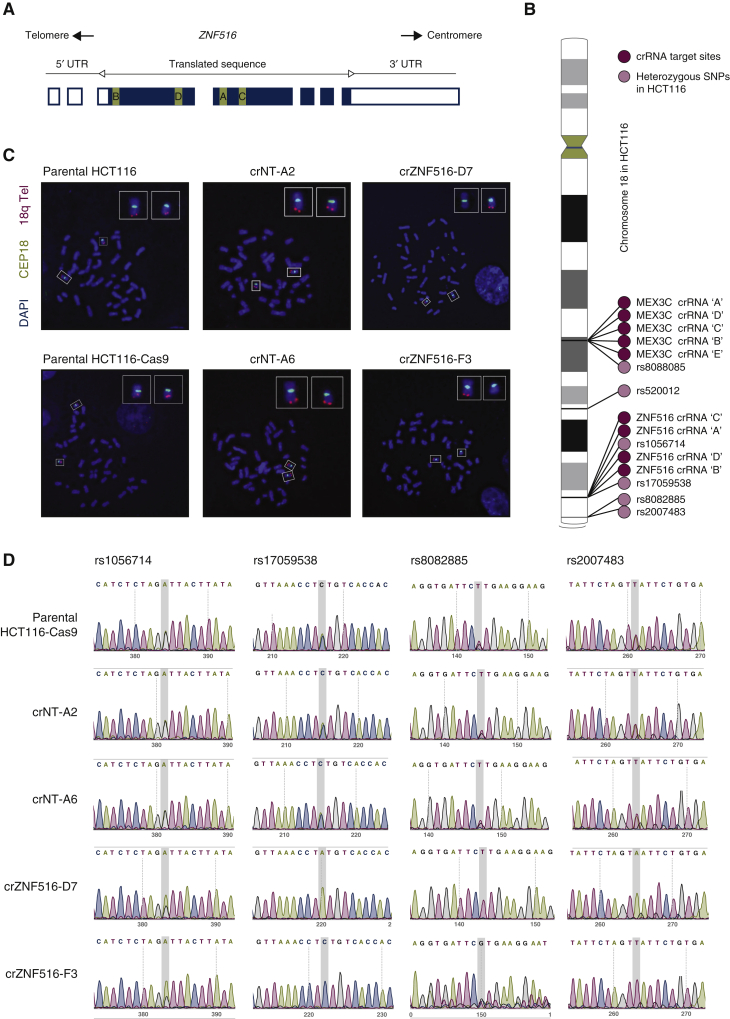
Figure 1.
Detection of chromosome 18q arm truncation following CRISPR/Cas9 targeting of ZNF516.
(A) Schematic of 7 exons of ZNF516 on the reverse strand, with coding sequence shown as filled rectangles, and untranslated regions as empty rectangles. crRNA target sites are indicated in green in exon 3 and 4. (B) Schematic of heterozygous SNP location on chromosome 18 in HCT116. Schematic was made with Phenogram created by Ritchie Lab (2012). (C) FISH on metaphase spreads stained with DAPI (blue) and hybridised to a chromosome 18 centromere probe (green) and 18q subtelomeric region probe (red). (D) Sanger sequencing traces of four heterozygous SNPs in HCT116. rs1056714 is located in the ZNF516 intron between crRNA-targeted exons 3 and 4. The rest of the SNPs are located distal to ZNF516 and towards the telomere. crRNA, CRISPR (clustered regularly interspaced short palindromic repeats) RNA; DAPI, 4′,6-diamidino-2-phenylindole; SNP, single-nucleotide polymorphism; UTR, untranslated region.
To undertake biological characterisation of ZNF516 we performed RNA-seq in two ZNF516 KO clones (crZNF516-D7 and crZNF516-F3) and two nontargeting clones (crNT-A2 and crNT-A6), to characterise gene expression changes upon ZNF516 KO. Unexpectedly, we observed that a large proportion of the most significantly downregulated genes were located downstream of ZNF516 towards the telomere of 18q. This observation was suggestive of a large-scale deletion or chromosome arm truncation following Cas9-induced double-strand break (DSB) in ZNF516.
To investigate the potential chromosome 18q arm truncation, we assessed loss of heterozygosity (LOH) of established heterozygous HCT116 SNPs, the location of which was obtained from the Sanger COSMIC Cell Line Project's genotyping calls (https://cancer.sanger.ac.uk/cell_lines/download). We examined four heterozygous SNPs, rs1056714 located in the ZNF516 intron between targeted exons 3 and 4, and rs17059538, rs8082885 and rs2007483 distal to ZNF516 (Figure 1B). Sanger sequencing revealed that both ZNF516 KO clones had LOH of all SNPs downstream of ZNF516. Interestingly the ZNF516 KO clones had lost a different allele, suggesting that it was equally possible to lose either of the chromosome arms. Furthermore, rs1056714 remained heterozygous, suggesting that arm truncation was due to a Cas9-induced DSB in exon 3 of ZNF516.
To further confirm arm truncation, we carried out FISH on metaphase spreads from parental cell lines, ZNF516 KO and nontargeting clones, with chromosome 18 centromeric probes and 18q subtelomeric region probe. We detected subtelomeric probe signals on both copies of chromosome 18 in all cells (50 analysed) from parental cell lines and nontargeting clones, indicating the expected presence of 18q on both chromosomes 18 in HCT116 cells. By contrast, crZNF516 clones had detectable signal on only one chromosome 18 in all cells (50 analysed; Figure 1C). Since the homologous chromosomes 18 pairs have different morphologies in the HCT116 cell line, it was possible to confirm that different chromosomes were affected in the crZNF516-D7 and crZNF516-F3 clones (Figure 1C), confirming the result from Sanger sequencing.
To identify the prevalence of this phenomenon, we utilised next-generation sequencing and sequenced DNA fragments spanning ZNF516 crRNA target sites to assess the mutation rate, and fragments spanning heterozygous SNPs to assess chromosome 18q arm truncations in parallel. Of the 155 crZNF516 clones analysed (a sum of two independent experiments), 89 (57%) had indel mutations in at least one target site. Ten (6%) crZNF516 clones had LOH of all three SNPs below ZNF516, of which five also had LOH of the intronic SNP, suggesting arm truncation could occur due to DSB either in exon 3 or exon 4 (Table 1). Of the 107 crNT clones analysed (a sum of two independent experiments), none had indel mutations and one (1%) had LOH of all four SNPs analysed, suggesting that active Cas9 and/or transfection with nontargeting crRNAs could also induce large deletions.
Table 1.
Prevalence of LOH of heterozygous SNPs following CRISPR/Cas9 targeting.
| Cell line | Targeted gene | crRNA | Clones analysed | Clones with indels | Number of clones with indels at |
Any SNP LOH | All dsa SNP LOH | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Target site ‘A’ | Target site ‘B’ | Target site ‘C’ | Target site ‘D’ | Target site ‘E’ | |||||||
| HCT116 | Nontargeting | Pool of 5 (Exp 1) | 66 | 0 | 0 | 0 | 0 | 0 | — | 2 (3%) | 0 |
| HCT116 | ZNF516 | Pool of 4 (Exp 1) | 96 | 65 (68%) | 0 | 57 (59%) | 21 (22%) | 22 (23%) | — | 35 (36%) | 6 (6%) |
| HCT116 | Nontargeting | Pool of 5 (Exp 2) | 41 | 0 | 0 | 0 | 0 | 0 | — | 1 (2%) | 1 (2%) |
| HCT116 | ZNF516 | Pool of 4 (Exp 2) | 59 | 24 (40%) | 20 (33%) | 6 (10%) | 1 (2%) | 2 (3%) | — | 6 (10%) | 4 (7%) |
| HCT116 | ZNF516 | crRNA ‘A’ | 58 | 33 (57%) | 33 (57%) | — | — | — | — | 5 (9%) | 4 (7%) |
| HCT116 | ZNF516 | crRNA ‘B’ | 48 | 17 (35%) | — | 17 (35%) | — | — | — | 2 (4%) | 1 (2%) |
| HCT116 | ZNF516 | crRNA ‘C’ | 47 | 4 (9%) | — | — | 4 (9%) | — | — | 3 (6%) | 1 (2%) |
| HCT116 | ZNF516 | crRNA ‘D’ | 53 | 13 (25%) | — | — | — | 13 (25%) | — | 3 (6%) | 2 (4%) |
| HCT116 | Nontargeting | Pool of 5 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (2%) | 0 |
| HCT116 | MEX3C | Pool of 5 | 52 | 15 (29%) | 0 | 5 (10%) | 2 (4%) | 10 (19%) | 1 (2%) | 0 | 0 |
crRNA, CRISPR (clustered regularly interspaced short palindromic repeats) RNA; LOH, loss of heterozygosity; SNP, single-nucleotide polymorphism.
ds, downstream from the targeted gene towards the telomere.
In parallel, we transfected HCT116-Cas9 cells with an individual crRNA (‘A’, ‘B’, ‘C’ or ‘D’; target sites shown in Figure 1A) to investigate whether this would have the same effect as the pool of four crRNAs. Depending on the crRNA used, mutation rates varied from 9% for crRNA ‘C’ to 57% for crRNA ‘A’ (Table 1). The proportion of samples affected by LOH of all heterozygous SNPs downstream of ZNF516 varied between 2% for crRNA ‘C’ and 7% for crRNA ‘A’, indicating that perhaps the efficiency of a particular crRNA to induce mutation could correlate with its efficiency of chromosome arm truncation. Results for all crRNA target sites are summarised in Table 1.
Finally, we targeted MEX3C located further away from the telomere than ZNF516, with a pool of five crRNAs (Figure 1B) to investigate whether proximity to the telomere influences arm truncation prevalence. Of the 52 crMEX3C clones analysed, 15 (29%) had indel mutations and none had LOH of the two SNPs analysed below MEX3C (rs8088085 and rs520012; Figure 1B; Table 1). Of the 58 crNT clones analysed, none had indel mutations and one (2%) had LOH of rs8088085, again suggesting that active Cas9 and/or transfection with nontargeting crRNAs could also induce large deletions.
In this study we demonstrate that CRISPR/Cas9 targeting can induce inadvertent arm truncation. While the existence of a low-background LOH of 18q in parental HCT116-Cas9 cells is a possibility even without CRISPR/Cas9 intervention, it seems that targeting genes with close proximity to telomeres could elevate the extent of chromosomal arm deletions compared with a commonly used nontargeting control.
Two studies5,6 previously reported incidental chromosome arm truncation following CRISPR/Cas9 targeting telomere-proximal genes: POLE5 located 11 kb and UROS6 located 6 million bp away from chromosome arm end. ZNF516 is located 6 million bp from the telomere, whereas MEX3C, targeting which did not induce arm truncation, is located 31 million bp away from the arm end. This could suggest that targeting genes close to telomeres could result in arm truncations. Reports by Rayner et al.5 and Cullot et al.6 suggested detection of arm truncation with FISH analysis for targeted genes; however, this can only be achieved after cell colony expansion and involves several experimental steps. We suggest assessing heterozygous SNPs downstream of targeted genes in addition to initial mutation analysis and genotype confirmation via sequencing. This methodology could preselect clones with correct genotype and without arm truncations before further cell line expansion for future downstream characterisation.
ACKNOWLEDGEMENTS
We thank Dr Ming Jiang and Dr Michael Howell for provision of the HCT116-Cas9 cell line; Dr Tom Watkins for provision of resources for heterozygous SNPs selection; Jeff Li and Haoran Zhai for contribution to MiSeq library preparation; Dr Maria Greco and Laura Cubitt for processing samples before sequencing; Dr Panos Zalmas, Dr Eva Grönroos, Dr Su Kit Chew and Dr Harshil Patel for their continued advice.
Conflict of Interest
J.P, N.K and A.R. declare no conflict of interests. R.R. has stock options in and has consulted for Achilles Therapeutics. C.S. acknowledges grant support from AstraZeneca, BMS, Roche-Ventana, Boehringer-Ingelheim and Ono Pharmaceutical and Pfizer. C.S. has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, BMS, Celgene, AstraZeneca, Illumina, Genentech, Roche-Ventana, GRAIL, Medicxi, and the Sarah Cannon Research Institute. C.S. has stock option in Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options and is co-founder of Achilles Therapeutics.
Funding acknowledgment and disclosure
This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169).
J.P. PhD studentship was funded by the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844).
A.R. receives funding from the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169).
R.R. receives funding from the Royal Society Enhancement Award.
N.K. receives funding from Cancer Research UK.
C.S. is Royal Society Napier Research Professor. C.S. is funded by Cancer Research UK (TRACERx, PEACE and CRUK Cancer Immunotherapy Catalyst Network), Cancer Research UK Lung Cancer Centre of Excellence, the Rosetrees Trust, Butterfield and Stoneygate Trusts, NovoNordisk Foundation (ID16584), the NIHR BRC at University College London Hospitals, and the CRUK-UCL Centre, Experimental Cancer Medicine Centre, and the Breast Cancer Research Foundation (BCRF). This research is supported by a Stand Up To Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. C.S. receives funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844), European Commission ITN (FP7-PloidyNet 607722), an ERC Advanced Grant (PROTEUS) from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 835297), and Chromavision from the European Union’s Horizon 2020 research and innovation programme (grant agreement 665233).
References
- 1.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2013:1–9. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cong L., Ran F.A., Cox D. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P., Yang L., Esvelt K.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rayner E., Durin M.A., Thomas R. CRISPR-Cas9 causes chromosomal instability and rearrangements in cancer cell lines, detectable by cytogenetic methods. CRISPR J. 2019;2:406–416. doi: 10.1089/crispr.2019.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullot G., Boutin J., Toutain J. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]