Abstract
Abnormal metabolism is common in cancer cells and often correlates with mutations in genes encoding for enzymes involved in small-molecule metabolism. Isocitrate dehydrogenase 1 (IDH1) is the most frequently mutated metabolic gene in cancer. Cancer-associated substitutions in IDH1 and IDH2 impair wild-type production of 2-oxoglutarate and reduced nicotinamide adenine dinucleotide phosphate (NADPH) from isocitrate and oxidised nicotinamide adenine dinucleotide phosphate (NADP+ ), and substantially promote the IDH variant catalysed conversion of 2-oxoglutarate to d-2-hydroxyglutarate (d-2HG). Elevated d-2HG is a biomarker for some cancers, and inhibition of IDH1 and IDH2 variants is being pursued as a medicinal chemistry target. We provide an overview of the types of cancer-associated IDH variants, discuss some of the proposed consequences of altered metabolism as a result of elevated d-2HG, summarise therapeutic efforts targeting IDH variants and identify areas for future research.
Keywords: 2-Hydroxyglutarate, Isocitrate dehydrogenase, Cancer metabolism, 2-oxoglutarate, Alpha-ketoglutarate, JmjC demethylase, Epigenetics, Hypoxia-inducible factor, Glioma, Acute myeloid leukaemia
Graphical abstract
Introduction to IDH mutations and cancer
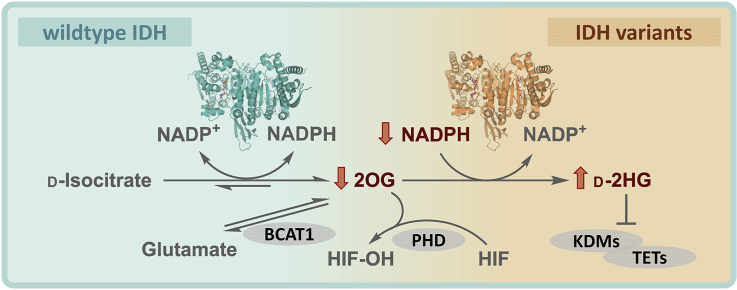
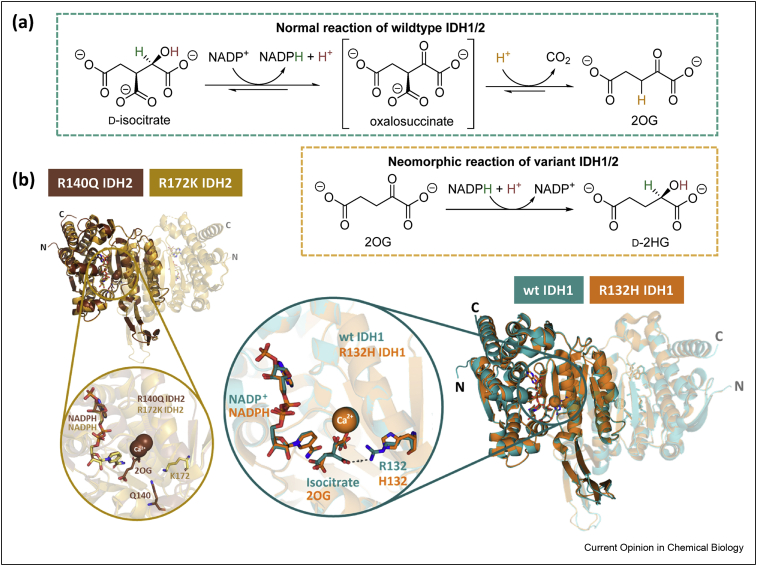
Abnormal metabolism of cancer cells often correlates with mutations in genes encoding for metabolic enzymes, including those involved in the tricarboxylic acid (TCA) cycle and related metabolism, such as succinate dehydrogenase and fumarate hydratase [1]. The isocitrate dehydrogenase 1 (IDH1) gene is the most frequently identified mutated metabolic gene in cancer; IDH1 and IDH2 mutations cause active site substitutions with consequent profound effects on IDH activity, cellular metabolism and cancer development [2, 3, 4, 5]. There are three human IDH isoforms, that is, the closely related homodimeric IDH1 and IDH2 (∼70% identity) and the more distantly related heterotetrameric (2α,1β,1γ) IDH3. IDH1 localises to the cytoplasm and peroxisomes; IDH2 and IDH3 localise to mitochondria. IDH1 and IDH2 undergo mutations correlating with >80% of low-grade glioma (LGG) [6] and ∼20% of acute myeloid leukaemia (AML) cases [7]. By contrast, no tumour-associated IDH3 mutations are reported [8]. IDH3 catalyses the NAD+-dependent oxidative decarboxylation of d-isocitrate giving 2-oxoglutarate (2OG) in the TCA cycle, a reaction reported to be irreversible under physiological conditions [9]. IDH1 and IDH2 catalyse the reversible oxidised nicotinamide adenine dinucleotide phosphate (NADP+)-dependent oxidative decarboxylation of d-isocitrate to 2OG [10], in a manner regulating isocitrate and 2OG levels and which provides reduced nicotinamide adenine dinucleotide phosphate (NADPH) [10]. Cancer-associated substitutions in IDH1 and IDH2 impair wild-type (wt) activity–producing 2OG by promoting a ‘neomorphic’ reaction that converts 2OG to d-2-hydroxyglutarate (d-2HG), using NADPH as a cosubstrate [11] (Figure 1a).
Figure 1.
Reactions catalysed by wild-type (wt) and variant isocitrate dehydrogenases. (a) Oxidative and reductive reactions catalysed by wt and variant IDH1/2, respectively. The reversible conversion of isocitrate to 2OG and CO2 by wt IDH1/2 proceeds via NADP+-mediated oxidation of isocitrate giving unstable oxalosuccinate, which undergoes β-keto decarboxylation giving 2OG. IDH1/2 variants catalyse reduction of 2OG to d-2HG using NADPH. IDH reactions require Mg2+/Mn2+ [11]. (b) Overall and expanded active site views from crystal structures of wt IDH1 (teal, PDB 1T0L) [12], R132H IDH1 (orange, PDB 3INM) [11], R140Q IDH2 (brown, PDB 5I95) [13] and R172K IDH2 (gold, PDB 5SVN) [14]. One monomer in the homodimer is differentiated by a different transparency level. Each active site is bound to a cofactor (NADP+ for wt IDH1; NADPH for R132H IDH1, R140Q IDH2 and R172K IDH2), a substrate (isocitrate for wt IDH1; 2OG for R132H IDH1 and R140Q IDH2) and an inhibitory Ca2+ (positioned to coordinate to the substrate). 2OG, 2-oxoglutarate; IDH, isocitrate dehydrogenase; wt, wild-type.
The nature of IDH substitutions varies with the cancer type; in many cancers IDH mutations are rare or not observed; the reasons for these differences are unclear [4,5]. In AML, for example, IDH substitutions are common, whereas with multiple myeloma, another blood cancer, they are rare. In LGG, the majority (>80%) of IDH mutations occur in the IDH1 gene, being dominated by R132H IDH1 [15]. Less frequently, substitutions occur at IDH2 R172 [6,16], which is located at a structurally analogous position to IDH1 R132 (Figure 1b). This contrasts with AML where IDH2 mutations occur at a similar or higher frequency compared with IDH1 mutations [15]. The most common IDH substitution in AML is IDH2 R140Q. The analogous IDH1 R100Q variant is rarer, being only found in grade II/III gliomas [17,18] Interestingly, IDH1 and IDH2 mutations appear to be mutually exclusive [19]. All the substituted arginine residues (IDH1 R132/R100 and IDH2 R172/R140) are likely directly or indirectly involved in binding isocitrate and 2OG at the IDH1/2 active sites [12] (Figure 1b). The precise details of how substitutions impact on the individual steps of the complex Mg2+-using IDH mechanisms are unclear.
The metabolic consequences of IDH mutations
Elevated d-2HG levels
Amongst the multifaceted cellular impacts of IDH mutations in malignancies (Figure 2), the substantially increased levels of d-2HG stand out, leading to its description as an ‘oncometabolite’ and the proposal that elevated d-2HG levels promote tumorigenesis [20]. Studies using metabolomics mass spectrometryanalyses demonstrated that the d-isomer of 2HG ((R)-2HG) accumulates in >100-fold excess relative to normal levels in cells/tissues of patients with LGG and AML, harbouring IDH1/2 mutations [11,21,22]. Most, but not all, studies report a less substantial 2OG reduction, with other TCA cycle intermediate levels being relatively unchanged [23]. Although variant IDHs consume 2OG, cellular 2OG stocks can be replenished from other sources, including glutamine [24]. On the other hand, whilst d-2HG produced in normal cells (where its roles are unclear) can be cleared by d-2HG dehydrogenase (D2HGDH) catalysed conversion to 2OG, it seems the normal clearance rate of D2HGDH is insufficient to suppress the high levels of d-2HG produced in IDH variant–bearing cells [11,25]. The mitochondrial localisation of D2HGDH might further contribute to its ineffectiveness in clearing cytosolic d-2HG produced by IDH1 variants [25].
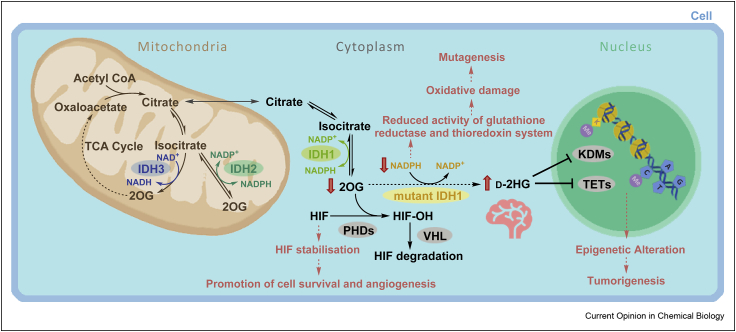
Figure 2.
Roles of wt IDH1/2/3 and some of the potential multiple effects of IDH mutation in cells (exemplified for IDH1). Different subcellular localisations (IDH1: cytoplasm; IDH2/3: mitochondria) and cosubstrate usage (IDH1/2: NADP+; IDH3: NAD+) distinguish the 3 human IDHs. The effects of IDH1 variants, including promotion of tumorigenesis, are proposed to manifest because of metabolic changes including d-2HG accumulation, depletion of NADPH and/or reduced 2OG. Changes in d-2HG/2OG levels are proposed to inhibit 2OG oxygenases involved in regulation of expression, for example, PHD, JmjC KDM, or TET enzymes. 2OG, 2-oxoglutarate; d-2HG, d-2-hydroxyglutarate; IDH, isocitrate dehydrogenase; HIF, hypoxia-inducible factor; HIF–OH, hydroxylated HIF; PHD, HIF prolyl hydroxylase domain enzyme; KDM, histone lysine demethylase; NADP+, oxidised nicotinamide adenine dinucleotide phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; TCA, tricarboxylic acid; TET, ten-eleven translocation oxygenase; VHL, Von Hippel-Lindau; wt, wild-type.
Elevated d-2HG levels serve as a robust biomarker for IDH1/2 mutations in gliomas [26]. d-2HG levels in plasma/serum can be analysed by liquid chromatography-mass spectrometry (LC-MS) and in the case of gliomas, d-2HG can be imaged by magnetic resonance spectroscopy [27,28]. Antibodies for IDH variants, in particular for IDH1 R132H, are also potentially useful for diagnosis. Aside from diagnosis and medicinal chemistry opportunities, the discovery of the elevated d-2HG levels was exciting from a cancer biochemistry perspective, because it opened opportunities to link readily quantifiable (in vitro and in vivo) changes in the levels of a specific metabolite (d-2HG), with cellular processes relevant to cancer, such as tumorigenesis and epigenetic regulation.
2-oxoglutarate–dependent oxygenases
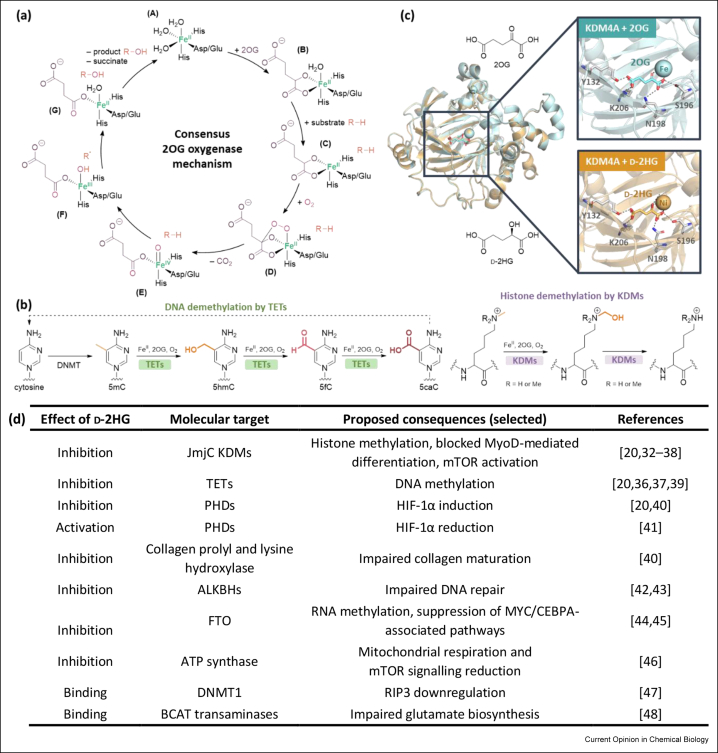
Before the work on IDH variant neomorphic activity, cancer-linked loss-of-function mutations to TCA cycle enzymes other than IDH, succinate dehydrogenase and fumarate hydratase, were identified [29]. The consequently elevated levels of succinate and/or fumarate are proposed to inhibit human 2OG and Fe(II) dependent oxygenases [30]. Given the structural similarity between d-2HG and 2OG, it was also proposed that elevated d-2HG levels competitively inhibit 2OG oxygenases in manner relevant to cancer (Figure 3c). 2OG oxygenases couple two electron substrate oxidations, typically hydroxylation or demethylation via hydroxylation, to the conversion of 2OG and dioxygen to succinate and carbon dioxide (Figure 3a). Human 2OG oxygenases have roles in collagen biosynthesis, lipid metabolism, DNA/RNA damage repair/modification, ribosomal/translation machinery modification, the hypoxic response, and epigenetics/chromatin biology [31].
Figure 3.
2-hydroxyglutarate may inhibit 2-oxoglutarate oxygenases in a cancer-relevant manner. (a) Consensus mechanism for the 2OG oxygenases. The substrate is hydroxylated in the presence of Fe(II), 2OG and O2, giving succinate and CO2 as by-products. (b) Examples of reactions catalysed by 2OG oxygenases involved in chromatin regulation, as catalysed by TETs and JmjC KDM methyl-group modifying enzymes; DNA cytosine demethylation is catalysed by TETs and histone lysine demethylation catalysed by the JmjC KDMs. (c) Views from crystal structures of the JmjC KDM4A in complex with 2OG or d-2HG showing their analogous binding modes. Both 2OG (pale teal, PDB 2YBK [32]) and d-2HG (pale orange, PDB 2GP5, half maximal inhibitory concentration (IC50) against KDM4A = 24 μM [49]) occupy the same binding site and interact with KDM4A Y132, S196, N198 and K206. (d) Some of the multiple cellular targets and pathways potentially affected by d-2HG accumulation. There is mixed evidence on whether the mammalian target of rapamycin (mTOR) [35,46] and hypoxia-inducible factor-1α (HIF-1α) [20,40,41] are activated by d-2HG [42, 43, 44, 45∗∗,47]. ALKBH, 2OG-dependent AlkB homologue; 2OG, 2-oxoglutarate; BCAT, branched-chain amino acid transferase; CEBPA, CCAAT enhancer binding protein alpha; d-2HG, d-2-hydroxyglutarate; DNMT1, DNA methyltransferase 1; FTO, fat mass and obesity-associated protein; HIF, hypoxia-inducible factor; HIF–OH, hydroxylated HIF; IDH, isocitrate dehydrogenase; JmjC KDM, Jumonji C domain-containing histone lysine demethylase; KDM, histone lysine demethylase; mTOR, the mammalian target of rapamycin; PHD, HIF prolyl hydroxylase domain enzyme; RIP3, receptor-interacting protein 3; TET, ten-eleven translocation oxygenase [42, 43, 45∗∗,47,83, 84, 85, 86, 87, 88, 89].
It is proposed that elevated levels of d-2HG competitively inhibit 2OG oxygenases involved in epigenetic regulation, including the Jumonji C domain–containing histone lysine demethylases (JmjC KDMs) and the ten-eleven translocation (TET) oxygenases, which regulate expression by catalysing histone demethylation and oxidation of N-methylcytosine in DNA, respectively (Figure 3b,d) [20,32, 33, 34∗∗, 35, 36, 37, 38∗∗, 39]. Competitive inhibition of the JmjC KDMs and the TETs by d-2HG could contribute to the histone and DNA hypermethylation states manifested in IDH variant gliomas and AML [20,32, 33, 34∗∗, 35,39,50]. Such chromatin states are proposed to block differentiation of (potential) cancer cells, enabling them to grow and proliferate, thereby promoting tumorigenesis [51]. Figure 3d summarises the diverse cellular consequences proposed to result from elevated levels of d-2HG.
The combined studies indicate that the effects of d-2HG on chromatin are potentially complex, though in some contexts dysregulation of specific enzymes may be particularly important, leading to candidate medicinal chemistry targets. It should be noted that d-2HG is not a potent inhibitor of, at least many, 2OG-dependent oxygenases, even though its high concentrations in (specific regions of) tumour cells may compensate for its weak inhibition against isolated enzymes.
IDH variants and regulation of hypoxia-inducible factors
Hypoxia-inducible factor (HIF) is an α,β-heterodimeric transcription factor that is a central regulator of the chronic human hypoxic response. HIF is reported to be elevated in multiple types of cancer, in a manner often, but not always, associated with hypoxia and/or mutations to TCA cycle enzymes, including succinate dehydrogenase and IDHs [52]. Both the levels and transcriptional activity of HIFs (human HIF-1α, -2α and -3α) are directly regulated by 2OG oxygenases. Catalysis by the HIFα prolyl hydroxylase domain enzymes (human HIF prolyl hydroxylase domain enzyme [PHD] 1–3) signals for HIFα degradation in an oxygen availability–limited manner, a property enabling them to act as hypoxia sensors (Figure 2). The factor inhibiting HIF, a JmjC HIFα asparginyl hydroxylase, regulates HIF transcriptional activity by limiting its interactions with transcriptional coactivators [53].
There is mixed evidence on the effects of d-2HG on HIF activity in IDH variant gliomas. Some studies show no correlation between HIF levels and IDH variant gliomas [54,55]; others show either upregulation [40,56] or downregulation [41] of HIF-1α in cells expressing mutant IDH1, possibly reflecting variations in O2 availability in different studies. It is proposed that microvascular proliferation in glioblastoma multiforme is in part due to vascular endothelial growth factor upregulation due to increased HIF [57]. In a perhaps counterintuitive mechanism for HIF downregulation, PHD catalysis is proposed to be coupled to d-2HG oxidation, that is, d-2HG behaves as a PHD agonist rather than an inhibitor; however, the mechanism(s) underlying the cellular observations is unclear because other studies do not find d-2HG to be a PHD substrate [41,58]. Note that the reported half maximal inhibitory concentration (IC50) of d-2HG for PHD2 is high (∼7.3 mM [32]), and d-2HG concentration in IDH mutant glioma is 5–35 mM [11]. Thus, pathophysiologically relevant PHD inhibition by d-2HG is possible, but if this level of inhibition is biologically relevant one might expect multiple enzymes to be inhibited causing cytotoxicity. One possible reason for the cellular observations concerning the agonist activity of d-2HG is its nonenzymatic or enzymatic conversion to 2OG [58], although whether this can occur at sufficient levels is unclear. HIF is often viewed as an oncogene (or proto-oncogene) that is predicted to be activated in cancers and HIF-1α– and HIF-2α– suppressing drugs are being pursued [59]. However, some studies suggest that, at least in particular contexts, HIF-α has a tumour suppressor role, including in glioma [60] and leukaemic [61] cells. For example, inhibition of HIF-2α reduces angiogenesis but enhances tumour growth in glioma cells, partly because of a reduction in tumour cell apoptosis [60]. The expression levels of many HIF target genes are also decreased in IDH mutant gliomas [62]. Collectively, these findings raise the possibility that HIF activation via PHD inhibition, by direct or indirect means, may impair mutant IDH tumour growth [41].
IDH variants and alterations in the NADPH/NADP+ ratio
Mutations in IDH genes result in a reduction in the cellular NADPH/NADP+ ratio. IDH1 and IDH2 are important sources of NADPH in the cytoplasm and mitochondria, respectively [63]. The overall neomorphic conversion of isocitrate to d-2HG does not necessarily directly change the NADP+/NADPH ratio, whereas the normal forward IDH reactions produce NADPH for use in fatty acid biosynthesis and protection against oxidative damage; hence IDH mutant–bearing cells may be unusually sensitive to redox-related damage [40,64,65]. One study suggests that the R132H substitution in IDH1 results in ∼38% reduction of its NADPH production capacity in glioblastoma tissue samples [64]. Consumption of NADPH for d-2HG synthesis is reported to decrease NADPH-dependent fatty acid synthesis, thereby increasing pentose phosphate pathways to support the NADPH demands and sensitising IDH1 mutant cells to oxidative stress [66].
Depletion of NADPH also impairs regeneration of the thiol form of glutathione (GSH) (γ−glutamyl-cysteinyl-glycine) from its disulfide form; GSH is a thiol-containing reducing agent which protects cells from oxidative stress by neutralising reactive oxygen species (ROS). GSH is present at lower levels in both R132H IDH1 overexpressing glioma cells [65] and knock-in mice [40]. Reduction in NADPH and GSH levels can result in oxidative damage, which may contribute to mutations, ultimately promoting tumorigenesis [67]. The reduction in GSH may be exacerbated by depletion of glutamate (a GSH component) in IDH mutant–bearing cells [24], where glutamine is converted to glutamate and then to 2OG to replenish its role in the TCA cycle. In support of this, isotope-labelling experiments indicate that d-2HG produced by variant IDH1/2 is derived not only from glucose, but also from glutamine via glutamate [11,66].
IDH variants and branched-chain amino acid transferase-1
A related link between IDH1 mutant cells and amino acids, concerns branched-chain amino acid transferase-1 (BCAT1) which is a 2OG-dependent enzyme catalysing the transamination of branched-chain amino acids (valine, leucine and isoleucine) with 2OG giving glutamate and branched-chain α-ketoacids. In glioma, IDH1 mutations correlate with lower levels of BCAT1 [68]; d-2HG is also reported to directly inhibit BCAT1, although only weakly [48]. The impairment of BCAT1 catalysis, however, has an impact on cellular metabolism, in particular, an increase in branched-chain amino acid levels and a decrease in glutamate levels [48]. Levels of other amino acids and other metabolites, including lipids, are reported to be changed in IDH mutant–bearing cells, although results are sometimes conflicting and the disease relevance of these changes are unclear.
Therapeutic advances with variant IDH1/2 inhibitors
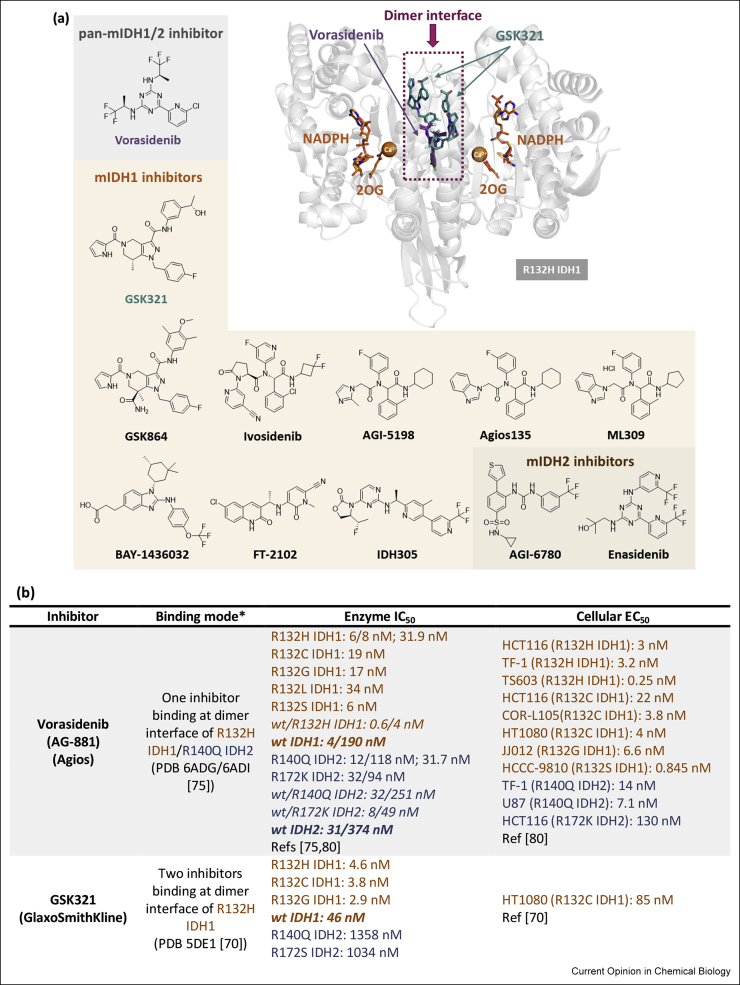
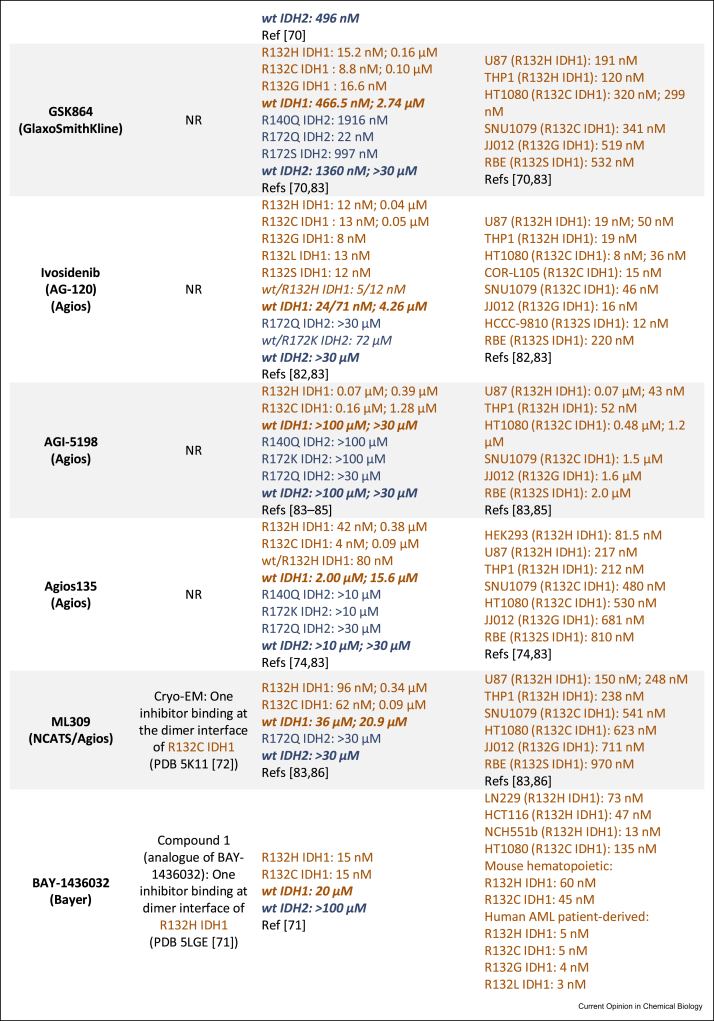
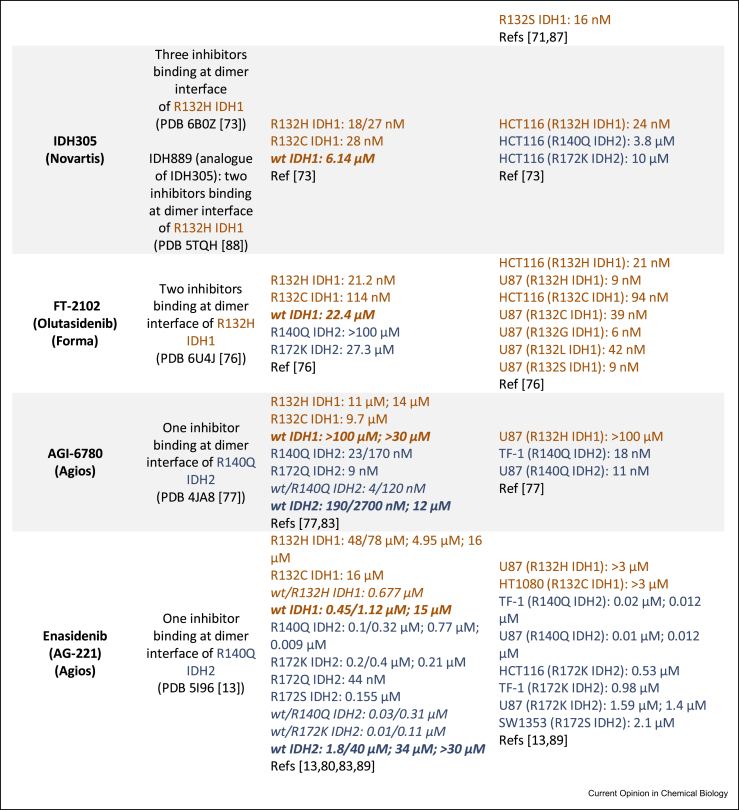
Following the identification of IDH mutations in gliomas and AML, multiple drug discovery campaigns targeting variant IDH1/2 were initiated. The inhibitors developed can successfully reduce d-2HG levels as shown by studies in cells and animals [69]. The majority of potent (IC50 ≤ 100 nM) R132H IDH1 inhibitors for which crystal structures are available inhibit via an allosteric mechanism, involving binding at the dimer interface, instead of the more typical active-site binding mode. This is interesting given the structural diversity in the allosteric inhibitors [14,70, 71, 72, 73] (Figure 4a). The allosteric inhibition is proposed to involve disruption of the binding of catalytically required metal ion (Mg2+ or Mn2+) at the active site [74]. Crystallographic data for ivosidenib and analogues (AGI-5198, Agios135, ML309) is lacking, although cryogenic electron microscopy (cryo-EM) data for ML309 [72] suggest it binds at the dimer interface like other allosteric inhibitors (vorasidenib [75], GSK321 [70], BAY-1436032 [71], FT-2102 [76], Novartis 305 [73], AGI-6780 [77], enasidenib [13]) (Figure 4b).
Figure 4.
Summary of selected variant IDH1 and IDH2 inhibitors. (a) Structures of variant IDH (mIDH) inhibitors and a crystallography-derived representation of how two allosteric inhibitors (superimposed) bind at the dimer interface of R132H IDH1. Vorasidenib (purple, PDB 6ADG [75]) and GSK321 (teal, PDB 5DE1 [70]) have binding stoichiometries of 1 and 2 inhibitors per R132H IDH1 dimer, respectively. The dimer interface where most reported variant IDH inhibitors bind is indicated by the dotted magenta region and is proximate to the active site (Ca2+ is inhibitory). (b) Binding modes, biochemical half maximal inhibitory concentration (IC50) and cellular half maximal effective concentration (EC50) values of variant IDH inhibitors. Note: Different IC50 values may be (partly) attributed to the different enzyme concentrations/assay conditions. “/” in IC50 values refers to measurements from different incubations times from the same report; “;” separates values from different reports. “wt/mutant” refers to a heterodimer, with other entries representing a homodimer. Data for IDH1 and IDH2 are shown in brown or navy respectively [83-89]. NR: not reported. ∗As observed by crystallography except where “cryo-EM” is stated. 2OG, 2-oxoglutarate; IDH, isocitrate dehydrogenase; wt, wild-type [42, 43, 45∗∗,47,83, 84, 85, 86, 87, 88, 89].
Ivosidenib [78] and enasidenib [79], which target variant IDH1 and IDH2, respectively, received FDA approval for AML treatment. Ivosidenib is in ongoing clinical trials for glioma treatment among other malignancies, in some cases as a combination therapy, for example, with vorasidenib (NCT03343197) and nivolumab (NCT04056910). Enasidenib is in clinical trials mostly for AML and haematological malignancies, including in combination therapy with azacitidine (NCT03683433). Vorasidenib is the only reported inhibitor that targets both variants of IDH1 and IDH2 [80]. Given its blood–brain barrier penetrating ability [80], vorasidenib is promising for glioma treatment, and it is currently in a phase 3 clinical trial for residual and recurrent grade 2 glioma (NCT04164901) and a phase 1 clinical trial for advanced solid tumours including gliomas (NCT02481154). BAY-1436032 has completed a phase 1 clinical trial for advanced AML (NCT03127735) and is currently in a phase 1 clinical trial for advanced solid tumours (NCT02746081). FT-2102 is in phase 1/2 clinical trials for advanced solid tumours and gliomas (NCT03684811), AML and myelodysplastic syndrome (MDS) (NCT02719574). Similarly, IDH305 is in a phase 1 clinical trial for advanced malignancies including gliomas, AML/MDS (NCT02381886); unfortunately, dose-limiting toxicities appear to have halted its clinical development [81]. One series developed by GlaxoSmithKline (e.g. GSK321 and the more bioavailable analogue GSK864) shows low selectivity between wt and variant IDH1 [70], potentially hindering its clinical development —although low wt/variant selectivity is also observed for vorasidenib and ivosidenib (Figure 4b). Preclinical compounds including AGI-5198 and AGI-6780 serve as useful tool compounds but lack clinical applications because of poor metabolic stability [82] and lack of an in vivo response, respectively.
Conclusions
The breakthrough discovery that cancer-linked mutations to IDHs cause major metabolic changes, notably increased d-2HG, has opened up exciting new therapeutic and diagnostic possibilities. It also provided an opportunity for research to connect in vitro and in vivo small-molecule biochemistry with the pathophysiology of cancer. From an IDH-variant drug development perspective, work has progressed rapidly with compounds approved for use in AML. Resistance to ivosidenib and enasidenib in the form of a second mutation at the IDH1/2 dimer interface has emerged [90], potentially compromising the long-term efficacy of similar IDH variant inhibitors. It is, however, important to state that the optimal patient populations for deployment of mutant IDH inhibitors have likely not yet been identified. There is also scope for developing new types of IDH inhibitor, including molecules that target the active site, which might manifest reduced resistance compared with the current allosteric type inhibitors.
At least in model systems, it is also of interest to explore inhibition of wt IDH, for which no (selective) inhibitors has been developed. In part, this is because nearly all reported cancer-linked IDH gene mutations are heterozygous [6,91], and both the variant homodimer and the wt/variant heterodimer of IDH1 can generate d-2HG. For heterozygous IDH1 mutant tumours, a significantly lower d-2HG level is found in gliomas that undergo loss of the wt IDH1 allele [91]. Thus, at least in some circumstances, inhibiting the remaining wt IDH1 allele to reduce local 2OG availability may alleviate protumour effects of IDH1 mutations. Wild-type IDH inhibitors are also of interest because there is evidence TCA cycle disruption holds promise for cancer treatment, for example, via inhibition of the 2OG dehydrogenase complex, which converts 2OG to succinyl CoA [92].
Despite the rapid progress in the IDH mutant field important basic questions remain. These include the definition of exactly how IDH mutations promote tumorigenesis. Studies to date have highlighted the potential of d-2HG to compete with 2OG in its role as a cosubstrate for enzymes involved in epigenetic/transcriptional regulation and metabolism. However, small molecules other than d-2HG may well be involved, and the role of altered metabolism in tumorigenesis and cancer progression is likely context dependent. Although it has not shown to be relevant in humans, the recent discovery that the lysine metabolite 2-oxoadipate can be converted into d-2HG via oxygenase catalysis in bacteria reveals the potential for discovery of new metabolic processes relating to 2OG/2HG and cancer metabolism [93].
One interesting observation is that glioma patients with IDH1/2 mutations are associated with an increase in overall survival compared with those with wt IDH genes [6,64,94]. Consistent with the observations for human gliomas, mouse models expressing R132H IDH1 manifest increased median survival [95]; in addition to increased d-2HG production, they manifest increased DNA cytosine methylation and reduced infiltration of immune cells [96]. By contrast, wt IDH1 gliomas correlate with high levels of chemokines and interleukins that stimulate infiltration of immune cells, consistent with poor prognosis [96,97]. Other studies attribute improved prognosis of IDH mutant gliomas to their higher sensitivity to chemotherapy [65] and radiotherapy [98]. This may be due to the R132H IDH1–induced depletion of NADPH and GSH and/or increased reactive oxygen species generation. These observations highlight the need for detailed context-dependent studies on the biochemistry of tumorigenesis and subsequent events in cancer progression.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
CJS thanks Cancer Research UK and the Wellcome Trust for funding. SL thanks the Agency for Science, Technology and Research (A∗STAR, Singapore) for a National Science Scholarship. The authors apologise for incomplete citations due to the focused nature of this short review.
This review comes from a themed issue on Chemical Genetics and Epigenetics
Edited by Akane Kawamura and Arasu Ganesan
References
- 1.Anderson N.M., Mucka P., Kern J.G., Feng H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell. 2018;9:216–237. doi: 10.1007/s13238-017-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oermann E.K., Wu J., Guan K.L., Xiong Y. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol. 2012;23:370–380. doi: 10.1016/j.semcdb.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietlein F., Weghorn D., Taylor-Weiner A., Richters A., Reardon B., Liu D., Lander E.S., Van Allen E.M., Sunyaev S.R. Identification of cancer driver genes based on nucleotide context. Nat Genet. 2020;52:208–218. doi: 10.1038/s41588-019-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tommasini-Ghelfi S., Murnan K., Kouri F.M., Mahajan A.S., May J.L., Stegh A.H. Cancer-associated mutation and beyond: the emerging biology of isocitrate dehydrogenases in human disease. Sci Adv. 2019;5:eaaw4543. doi: 10.1126/sciadv.aaw4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molenaar R.J., Maciejewski J.P., Wilmink J.W., van Noorden C.J.F. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene. 2018;37:1949–1960. doi: 10.1038/s41388-017-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medeiros B.C., Fathi A.T., DiNardo C.D., Pollyea D.A., Chan S.M., Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–281. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Khallaf H. Isocitrate dehydrogenases in physiology and cancer: biochemical and molecular insight. Cell Biosci. 2017;7:37. doi: 10.1186/s13578-017-0165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabriel J.L., Zervos P.R., Plaut G.W.E. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism. 1986;35:661–667. doi: 10.1016/0026-0495(86)90175-7. [DOI] [PubMed] [Google Scholar]
- 10.Reitman Z.J., Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Zhao J., Xu Z., Peng B., Huang Q., Arnold E., Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 13.Yen K., Travins J., Wang F., David M.D., Artin E., Straley K., Padyana A., Gross S., DeLaBarre B., Tobin E. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 2017;7:478–493. doi: 10.1158/2159-8290.CD-16-1034. [DOI] [PubMed] [Google Scholar]
- 14.Xie X., Baird D., Bowen K., Capka V., Chen J., Chenail G., Cho Y., Dooley J., Farsidjani A., Fortin P. Allosteric mutant IDH1 inhibitors reveal mechanisms for IDH1 mutant and isoform selectivity. Structure. 2017;25:506–513. doi: 10.1016/j.str.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Losman J.A., Kaelin W.G. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann C., Meyer J., Balss J., Capper D., Mueller W., Christians A., Felsberg J., Wolter M., Mawrin C., Wick W. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 17.Gupta R., Flanagan S., Li C.C., Lee M., Shivalingham B., Maleki S., Wheeler H.R., Buckland M.E. Expanding the spectrum of IDH1 mutations in gliomas. Mod Pathol. 2013;26:619–625. doi: 10.1038/modpathol.2012.210. [DOI] [PubMed] [Google Scholar]
- 18.Bhavya B., Anand C.R., Madhusoodanan U.K., Rajalakshmi P., Krishnakumar K., Easwer H.V., Deepti A.N., Gopala S. To be wild or mutant: role of isocitrate dehydrogenase 1 (IDH1) and 2-hydroxy glutarate (2-HG) in gliomagenesis and treatment outcome in glioma. Cell Mol Neurobiol. 2020;40:53–63. doi: 10.1007/s10571-019-00730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chotirat S., Thongnoppakhun W., Promsuwicha O., Boonthimat C., Auewarakul C.U. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.-H., Ito S., Yang C., Wang P., Xiao M.-T. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Canc Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward P.S., Patel J., Wise D.R., Abdel-Wahab O., Bennett B.D., Coller H.A., Cross J.R., Fantin V.R., Hedvat C.V., Perl A.E. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Canc Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross S., Cairns R.A., Minden M.D., Driggers E.M., Bittinger M.A., Jang H.G., Sasaki M., Jin S., Schenkein D.P., Su S.M. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S., Tominaga K., Sakashita E., Urabe M., Onuki Y., Gomi A., Yamaguchi T., Mieno M., Mizukami H., Kume A. Comprehensive metabolomic analysis of IDH1R132H clinical glioma samples reveals suppression of β-oxidation due to carnitine deficiency. Sci Rep. 2019;9:9787. doi: 10.1038/s41598-019-46217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; Work linking IDH mutations to impaired fatty acid β-oxidation due to reduced formation of carnitine / acyl-carnitine, possibly due to d-2HG inhibition of 2OG oxygenases involved in carnitine biosynthesis.
- 24.Ohka F., Ito M., Ranjit M., Senga T., Motomura A., Motomura K., Saito K., Kato K., Kato Y., Wakabayashi T. Quantitative metabolome analysis profiles activation of glutaminolysis in glioma with IDH1 mutation. Tumor Biol. 2014;35:5911–5920. doi: 10.1007/s13277-014-1784-5. [DOI] [PubMed] [Google Scholar]
- 25.Lin A.P., Abbas S., Kim S.W., Ortega M., Bouamar H., Escobedo Y., Varadarajan P., Qin Y., Sudderth J., Schulz E. D2HGDH regulates alpha-ketoglutarate levels and dioxygenase function by modulating IDH2. Nat Commun. 2015;6:7768. doi: 10.1038/ncomms8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalinina J., Carroll A., Wang L., Yu Q., Mancheno D.E., Wu S., Liu F., Ahn J., He M., Mao H. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med. 2012;90:1161–1171. doi: 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X., Voets N., Larkin S., de Pennington N., Plaha P., Stacey R., McCullagh J., Schofield C., Clare S., Jezzard P. A noninvasive comparison study between human gliomas with IDH1 and IDH2 mutations by MR spectroscopy. Metabolites. 2019;9:35. doi: 10.3390/metabo9020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emir U.E., Larkin S.J., de Pennington N., Voets N., Plaha P., Stacey R., Al-Qahtani K., Mccullagh J., Schofield C.J., Clare S. Noninvasive quantification of 2-hydroxyglutarate in human gliomas with IDH1 and IDH2 mutations. Cancer Res. 2016;76:43–49. doi: 10.1158/0008-5472.CAN-15-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardaci S., Ciriolo M.R. TCA cycle defects and cancer: when metabolism tunes redox state. Int J Cell Biol. 2012;2012:1–9. doi: 10.1155/2012/161837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schofield C.J., Ratcliffe P.J. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 31.Islam M.S., Leissing T.M., Chowdhury R., Hopkinson R.J., Schofield C.J. 2-Oxoglutarate-Dependent oxygenases. Annu Rev Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury R., Yeoh K.K., Tian Y.-M., Hillringhaus L., Bagg E.A., Rose N.R., Leung I.K.H., Li X.S., Woon E.C.Y., Yang M. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.M., Reuter V.P., Koche R.P., Thompson C.B. 2-hydroxyglutarate inhibits MyoD-mediated differentiation by preventing H3K9 demethylation. Proc Natl Acad Sci. 2019;116:12851–12856. doi: 10.1073/pnas.1817662116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Work showing IDH2 mutant bearing mesenchymal cells manifest increased H3K9 methylation resulting in impaired transcription factor mediated regulation of cell differentiation.
- 35.Carbonneau M., Gagné L M., Lalonde M.-E., Germain M.-A., Motorina A., Guiot M.-C., Secco B., Vincent E.E., Tumber A., Hulea L. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nat Commun. 2016;7:12700. doi: 10.1038/ncomms12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey B.W., Finley L.W.S., Cross J.R., Allis C.D., Thompson C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeung B.H., Huang J., An S.S., Solway J., Mitzner W., Tang W. Role of isocitrate dehydrogenase 2 on DNA hydroxymethylation in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2020 doi: 10.1165/rcmb.2019-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkowski P.L., Oeck S., Dow J., Economos N.G., Mirfakhraie L., Liu Y., Noronha K., Bao X., Li J., Shuch B.M. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature. 2020 doi: 10.1038/s41586-020-2363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed study linking elevated small-molecule oncometabolite (2HG, succinate, fumarate) levels with DNA damage repair via JmjC KDM inhibition.
- 39.Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Canc Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki M., Knobbe C.B., Itsumi M., Elia A.J., Harris I.S., Chio I.I.C., Cairns R.A., McCracken S., Wakeham A., Haight J. d-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koivunen P., Lee S., Duncan C.G., Lopez G., Lu G., Ramkissoon S., Losman J.A., Joensuu P., Bergmann U., Gross S. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., Wu J., Ma S., Zhang L., Yao J., Hoadley K.A., Wilkerson M.D., Perou C.M., Guan K.-L., Ye D. Oncometabolite d-2-hydroxyglutarate inhibits ALKBH DNA repair enzymes and sensitizes IDH mutant cells to alkylating agents. Cell Rep. 2015;13:2353–2361. doi: 10.1016/j.celrep.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F., Bian K., Tang Q., Fedeles B.I., Singh V., Humulock Z.T., Essigmann J.M., Li D. Oncometabolites d- and l-2-hydroxyglutarate inhibit the AlkB family DNA repair enzymes under physiological conditions. Chem Res Toxicol. 2017;30:1102–1110. doi: 10.1021/acs.chemrestox.7b00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elkashef S.M., Lin A.-P., Myers J., Sill H., Jiang D., Dahia P.L.M., Aguiar R.C.T. IDH mutation, competitive inhibition of FTO, and RNA methylation. Canc Cell. 2017;31:619–620. doi: 10.1016/j.ccell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]; Work showing d-2HG can exert anti-luekaemic acitvity in a manner linked to altered RNA N6-methyl ademosine status via inhibition of the fat mass ansd obesity protein (FTO) nicely illustrating context dependent effects of d-2HG.
- 46.Fu X., Chin R.M., Vergnes L., Hwang H., Deng G., Xing Y., Pai M.Y., Li S., Ta L., Fazlollahi F. 2-Hydroxyglutarate inhibits ATP synthase and mTOR signaling. Cell Metab. 2015;22:508–515. doi: 10.1016/j.cmet.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z., Jiang B., Wang Y., Ni H., Zhang J., Xia J., Shi M., Hung L.-M., Ruan J., Mak T.W. 2-HG inhibits necroptosis by stimulating DNMT1-dependent hypermethylation of the RIP3 promoter. Cell Rep. 2017;19:1846–1857. doi: 10.1016/j.celrep.2017.05.012. [DOI] [PubMed] [Google Scholar]
- McBrayer S.K., Mayers J.R., DiNatale G.J., Shi D.D., Khanal J., Chakraborty A.A., Sarosiek K.A., Briggs K.J., Robbins A.K., Sewastianik T. Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell. 2018;175:101–116.e25. doi: 10.1016/j.cell.2018.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; Work linking 2OG dependent transaminase inhibition and altered metabolism in IDH1 mutant bearing cells.
- 49.Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q., Pan C.-H., Dai S. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 50.Turcan S., Rohle D., Goenka A., Walsh L.A., Fang F., Yilmaz E., Campos C., Fabius A.W.M., Lu C., Ward P.S. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oberley L.W., Oberley T.D., Buettner G.R. Cell differentiation, aging and cancer: the possible roles of superoxide and superoxide dismutases. Med Hypotheses. 1980;6:249–268. doi: 10.1016/0306-9877(80)90123-1. [DOI] [PubMed] [Google Scholar]
- 52.Mills E., O'Neill L.A.J. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014;24:313–320. doi: 10.1016/j.tcb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Webb J.D., Coleman M.L., Pugh C.W. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci. 2009;66:3539–3554. doi: 10.1007/s00018-009-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metellus P., Colin C., Taieb D., Guedj E., Nanni-Metellus I., de Paula A.M., Colavolpe C., Fuentes S., Dufour H., Barrie M. IDH mutation status impact on in vivo hypoxia biomarkers expression: new insights from a clinical, nuclear imaging and immunohistochemical study in 33 glioma patients. J Neuro Oncol. 2011;105:591–600. doi: 10.1007/s11060-011-0625-2. [DOI] [PubMed] [Google Scholar]
- 55.Williams S.C., Karajannis M.A., Chiriboga L., Golfinos J.G., von Deimling A., Zagzag D. R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1α upregulation in adult glioma. Acta Neuropathol. 2011;121:279–281. doi: 10.1007/s00401-010-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science (80- ) 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Das S., Marsden P.A. Angiogenesis in glioblastoma. N Engl J Med. 2013;369:1561–1563. doi: 10.1056/NEJMcibr1309402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarhonskaya H., Rydzik A.M., Leung I.K.H., Loik N.D., Chan M.C., Kawamura A., McCullagh J.S.O., Claridge T.D.W., Flashman E., Schofield C.J. Non-enzymatic chemistry enables 2-hydroxyglutarate-mediated activation of 2-oxoglutarate oxygenases. Nat Commun. 2014;5:3423. doi: 10.1038/ncomms4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu T., Tang B., Sun X. Development of inhibitors targeting hypoxia-inducible factor 1 and 2 for cancer therapy. Yonsei Med J. 2017;58:489. doi: 10.3349/ymj.2017.58.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acker T., Diez-Juan A., Aragones J., Tjwa M., Brusselmans K., Moons L., Fukumura D., Moreno-Murciano M.P., Herbert J.-M., Burger A. Genetic evidence for a tumor suppressor role of HIF-2α. Canc Cell. 2005;8:131–141. doi: 10.1016/j.ccr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Song L.P., Zhang J., Wu S.-F., Huang Y., Zhao Q., Cao J.-P., Wu Y.-L., Wang L.-S., Chen G.-Q. Hypoxia-inducible factor-1α-induced differentiation of myeloid leukemic cells is its transcriptional activity independent. Oncogene. 2008;27:519–527. doi: 10.1038/sj.onc.1210670. [DOI] [PubMed] [Google Scholar]
- 62.Kickingereder P., Sahm F., Radbruch A., Wick W., Heiland S., Deimling A von, Bendszus M., Wiestler B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep. 2015;5:16238. doi: 10.1038/srep16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jo S.H., Son M.-K., Koh H.-J., Lee S.-M., Song I.-H., Kim Y.-O., Lee Y.-S., Jeong K.-S., Kim W.B., Park J.-W. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. [DOI] [PubMed] [Google Scholar]
- 64.Bleeker F.E., Atai N.A., Lamba S., Jonker A., Rijkeboer D., Bosch K.S., Tigchelaar W., Troost D., Vandertop W.P., Bardelli A. The prognostic IDH1 R132 mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol. 2010;119:487–494. doi: 10.1007/s00401-010-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi J., Sun B., Shi W., Zuo H., Cui D., Ni L., Chen J. Decreasing GSH and increasing ROS in chemosensitivity gliomas with IDH1 mutation. Tumor Biol. 2015;36:655–662. doi: 10.1007/s13277-014-2644-z. [DOI] [PubMed] [Google Scholar]
- 66.Gelman S.J., Naser F., Mahieu N.G., McKenzie L.D., Dunn G.P., Chheda M.G., Patti G.J. Consumption of NADPH for 2-HG synthesis increases pentose phosphate pathway flux and sensitizes cells to oxidative stress. Cell Rep. 2018;22:512–522. doi: 10.1016/j.celrep.2017.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hollinshead K.E.R., Munford H., Eales K.L., Bardella C., Li C., Escribano-Gonzalez C., Thakker A., Nonnenmacher Y., Kluckova K., Jeeves M. Oncogenic IDH1 mutations promote enhanced proline synthesis through PYCR1 to support the maintenance of mitochondrial redox homeostasis. Cell Rep. 2018;22:3107–3114. doi: 10.1016/j.celrep.2018.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayers J.R., Vander Heiden M.G. BCAT1 defines gliomas by IDH status. Nat Med. 2013;19:816–817. doi: 10.1038/nm.3263. [DOI] [PubMed] [Google Scholar]
- 69.Golub D., Iyengar N., Dogra S., Wong T., Bready D., Tang K., Modrek A.S., Placantonakis D.G. Mutant isocitrate dehydrogenase inhibitors as targeted cancer therapeutics. Front Oncol. 2019;9 doi: 10.3389/fonc.2019.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okoye-Okafor U.C., Bartholdy B., Cartier J., Gao E.N., Pietrak B., Rendina A.R., Rominger C., Quinn C., Smallwood A., Wiggall K.J. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878–886. doi: 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pusch S., Krausert S., Fischer V., Balss J., Ott M., Schrimpf D., Capper D., Sahm F., Eisel J., Beck A.-C. Pan-mutant IDH1 inhibitor BAY 1436032 for effective treatment of IDH1 mutant astrocytoma in vivo. Acta Neuropathol. 2017;133:629–644. doi: 10.1007/s00401-017-1677-y. [DOI] [PubMed] [Google Scholar]
- 72.Merk A., Bartesaghi A., Banerjee S., Falconieri V., Rao P., Davis M.I., Pragani R., Boxer M.B., Earl L.A., Milne J.L.S. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cho Y.S., Levell J.R., Liu G., Caferro T., Sutton J., Shafer C.M., Costales A., Manning J.R., Zhao Q., Sendzik M. Discovery and evaluation of clinical candidate IDH305, a brain penetrant mutant IDH1 inhibitor. ACS Med Chem Lett. 2017;8:1116–1121. doi: 10.1021/acsmedchemlett.7b00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deng G., Shen J., Yin M., McManus J., Mathieu M., Gee P., He T., Shi C., Bedel O., McLean L.R. Selective inhibition of mutant isocitrate dehydrogenase 1 (IDH1) via disruption of a metal binding network by an allosteric small molecule. J Biol Chem. 2015;290:762–774. doi: 10.1074/jbc.M114.608497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma R., Yun C.H.H. Crystal structures of pan-IDH inhibitor AG-881 in complex with mutant human IDH1 and IDH2. Biochem Biophys Res Commun. 2018;503:2912–2917. doi: 10.1016/j.bbrc.2018.08.068. [DOI] [PubMed] [Google Scholar]
- 76.Caravella J.A., Lin J., Diebold R.B., Campbell A.-M., Ericsson A., Gustafson G., Wang Z., Castro J., Clarke A., Gotur D. Structure-Based design and identification of FT-2102 (olutasidenib), a potent mutant-selective IDH1 inhibitor. J Med Chem. 2020;63:1612–1623. doi: 10.1021/acs.jmedchem.9b01423. [DOI] [PubMed] [Google Scholar]
- 77.Wang F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 78.FDA . 2019. FDA approves ivosidenib as first-line treatment for AML with IDH1 mutation. [Google Scholar]
- 79.FDA . 2017. FDA granted regular approval to enasidenib for the treatment of relapsed or refractory AML. [Google Scholar]
- Konteatis Z., Artin E., Nicolay B., Straley K., Padyana A.K., Jin L., Chen Y., Narayaraswamy R., Tong S., Wang F. Vorasidenib (AG-881): a first-in-class, brain-penetrant dual inhibitor of mutant IDH1 and 2 for treatment of glioma. ACS Med Chem Lett. 2020;11:101–107. doi: 10.1021/acsmedchemlett.9b00509. [DOI] [PMC free article] [PubMed] [Google Scholar]; Description of the first brain penetrant inhibitor of both IDH1/IDH2 variants suitable for evaluation in glioma treatment.
- 81.DiNardo C.D., Stein E.M. SOHO state of the art update and next questions: IDH therapeutic targeting in AML. Clin Lymphoma Myeloma Leuk. 2018;18:769–772. doi: 10.1016/j.clml.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 82.Popovici-Muller J., Lemieux R.M., Artin E., Saunders J.O., Salituro F.G., Travins J., Cianchetta G., Cai Z., Zhou D., Cui D. Discovery of AG-120 (Ivosidenib): a first-in-class mutant IDH1 inhibitor for the treatment of IDH1 mutant cancers. ACS Med Chem Lett. 2018;9:300–305. doi: 10.1021/acsmedchemlett.7b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Urban D.J., Martinez N.J., Davis M.I., Brimacombe K.R., Cheff D.M., Lee T.D., Henderson M.J., Titus S.A., Pragani R., Rohde J.M. Assessing inhibitors of mutant isocitrate dehydrogenase using a suite of pre-clinical discovery assays. Sci Rep. 2017;7:12758. doi: 10.1038/s41598-017-12630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rohle D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science (80- ) 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Popovici-Muller J., Saunders J.O., Salituro F.G., Travins J.M., Yan S., Zhao F., Gross S., Dang L., Yen K.E., Yang H. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett. 2012;3:850–855. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davis M.I., Gross S., Shen M., Straley K.S., Pragani R., Lea W.A., Popovici-Muller J., DeLaBarre B., Artin E., Thorne N. Biochemical, cellular, and biophysical characterization of a potent inhibitor of mutant isocitrate dehydrogenase IDH1. J Biol Chem. 2014;289:13717–13725. doi: 10.1074/jbc.M113.511030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chaturvedi A., Herbst L., Pusch S., Klett L., Goparaju R., Stichel D., Kaulfuss S., Panknin O., Zimmermann K., Toschi L. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia. 2017;31:2020–2028. doi: 10.1038/leu.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Levell J.R., Caferro T., Chenail G., Dix I., Dooley J., Firestone B., Fortin P.D., Giraldes J., Gould T., Growney J.D. Optimization of 3-Pyrimidin-4-yl-oxazolidin-2-ones as allosteric and mutant specific inhibitors of IDH1. ACS Med Chem Lett. 2017;8:151–156. doi: 10.1021/acsmedchemlett.6b00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.FDA Centre for drug evaluation and research . 2017. 209606Orig1s000 Multi-discipline review. [Google Scholar]
- Intlekofer A.M., Shih A.H., Wang B., Nazir A., Rustenburg A.S., Albanese S.K., Patel M., Famulare C., Correa F.M., Takemoto N. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature. 2018;559:125–129. doi: 10.1038/s41586-018-0251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Resistance to the 'allosteric' inhibitors, ivosidenib and enasidenib, is shown to be via mutations at the IDH1 and IDH2 variant dimer interface respectively.
- 91.Jin G., Reitman Z.J., Duncan C.G., Spasojevic I., Gooden D.M., Rasheed B.A., Yang R., Lopez G.Y., He Y., McLendon R.E. Disruption of wild-type IDH1 suppresses d-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson N.M., Li D., Peng H.L., Laroche F.J.F., Mansour M.R., Gjini E., Aioub M., Helman D.J., Roderick J.E., Cheng T. The TCA cycle transferase DLST is important for MYC-mediated leukemogenesis. Leukemia. 2016;30:1365–1374. doi: 10.1038/leu.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thompson M.G., Blake-Hedges J.M., Cruz-Morales P., Barajas J.F., Curran S.C., Eiben C.B., Harris N.C., Benites V.T., Gin J.W., Sharpless W.A. Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. MBio. 2019;10 doi: 10.1128/mBio.02577-18. e02577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P., Dong Q., Zhang C., Kuan P.-F., Liu Y., Jeck W.R., Andersen J.B., Jiang W., Savich G.L., Tan T.-X. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Núñez F.J., Mendez F.M., Kadiyala P., Alghamri M.S., Savelieff M.G., Garcia-Fabiani M.B., Haase S., Koschmann C., Calinescu A.-A., Kamran N. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaq1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Amankulor N.M., Kim Y., Arora S., Kargl J., Szulzewsky F., Hanke M., Margineantu D.H., Rao A., Bolouri H., Delrow J. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ceccarelli M., Barthel F.P., Malta T.M., Sabedot T.S., Salama S.R., Murray B.A., Morozova O., Newton Y., Radenbaugh A., Pagnotta S.M. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tran A.N., Lai A., Li S., Pope W.B., Teixeira S., Harris R.J., Woodworth D.C., Nghiemphu P.L., Cloughesy T.F., Ellingson B.M. Increased sensitivity to radiochemotherapy in IDH1 mutant glioblastoma as demonstrated by serial quantitative MR volumetry. Neuro Oncol. 2014;16:414–420. doi: 10.1093/neuonc/not198. [DOI] [PMC free article] [PubMed] [Google Scholar]